Abstract
Recent study proves that the combination of loop mediated isothermal nucleic acid amplification (LAMP) with one-step strand displacement (OSD) is of great help to improve the sequence specificity during genetic detection. However, because OSD is incapable of signal amplification, the signal-to-noise ratio or the observable signal change may be usually not significant enough to satisfy practical usage. With the purpose to overcome this challenge, herein a more advanced and practical sensing principle is developed with the OSD replaced by an amplifiable nucleic acid circuit, hybridization chain reaction (HCR). The very contagious norovirus (NoV) was employed as the model target. Compared with LAMP-OSD, the LAMP-HCR can detect as few as 30 copies of NoV gene in 2% fecal samples with significantly enlarged signal change and signal-to-background ratio. Therefore, more reliable detection is achieved. Moreover, due to the high compatibility of HCR, the final LAMP-HCR products can be flexibly transduced into different types of readouts, including fluorescence, flow cytometer (FCM) and even a personal glucose meter (PGM). This further enlarges the operating environments for the detection from hospital labs, bedsides, to potential off-the-shelf devices in local pharmacies. Especially when using FCM or PGM, with the assistance of magnetic beads (MBs), the detection shows even higher tolerance capability to complicated biological matrices.
Keywords: Norovirus detection, Loop-mediated isothermal amplification, Hybridization chain reaction, Flow cytometer, Personal glucometer
Graphical abstract
A versatile molecular diagnostic method is developed for sensitive norovirus (NoV) gene detection by coupling the advantages of loop-mediated isothermal amplification (LAMP) with enzyme-free signal amplification hybridization chain reaction (HCR) for robust signal readout via flow cytometer (FCM) and personal glucometer (PGM).
Highlights
-
•
Highly sensitive and selective detection method for norovirus gene.
-
•
Sensitivity down to 30 gene copies in 2% fecal samples.
-
•
Significantly enlarged signal change and signal-to-background ratio.
-
•
Higher tolerance capability to complicated biological matrices.
-
•
Exhibiting great potential to be used in the diagnosis of gastroenteritis in hospital or at home.
1. Introduction
Molecular diagnostic is a series of techniques which are used for sensitively analysing biomarkers in the genome and the proteome-genetic code of individuals in vitro [1]. In the past several decades, it was rapidly developed to guide patient care management in the field of infectious diseases, cancers and so on. The increasing demand for the information of genetic and genomic leads to the rapid advances of molecular techniques [2]. Among most molecular diagnostic methods for nucleic acid biomarkers detection, polymerase chain reaction (PCR) has been the most widely used method for vastly increasing the number of nucleic acid molecules [[3], [4], [5]]. To make the molecular diagnostic test simpler and more portable, a suite of isothermal amplification techniques have been invented, such as rolling circle amplification (RCA) [6], helicase-dependent amplification (HDA) [7], recombinase polymerase amplification (RPA) [8], and loop-mediated isothermal amplification (LAMP) [9,10]. These methods can realize the exponential amplification at constant temperature which could facilitate the point-of-care (POC) gene detection [[11], [12], [13], [14], [15], [16], [17]]. However, these assays are generally still impractical for real applications, because of the resource poor settings as well as the high frequency to generate false positive results [[18], [19], [20]]. With the purpose of improving the detection specificity and achieving more simple and reliable readout, we recently developed an innovation in which one-step nucleic strand displacement reaction (usually shortened as OSD) was adapted, instead of traditional fluorescent intercalating dyes, to probe the products of isothermal amplifications (e.g., LAMP). Due to the high sequence specificity, the possibility of false positive signals has been reduced at the maximum degree. Even though, unlike the intercalating dyes that can provide signal amplification via interacting with all the base pairs of the LAMP amplicons [[21], [22], [23]], one OSD probe can be activated by a single LAMP amplicon [[24], [25], [26]]. Therefore, the signal-to-noise ratio or the observable signal change may be usually not significant enough to satisfy practical usage.
In order to overcome the above potential shortage and further enable the detection practical in more possible operation environments and settings, herein we import another innovation through updating the OSD probe with an enzyme-free nucleic acid circuit, the hybridized chain reaction (HCR). As an integration of multiple OSDs, the HCR reaction was firstly invented by Pierce group [27] and immediately became a starring signal amplifier due to high amplification efficiency and compatibility to different readouts [[28], [29], [30], [31], [32]]. In assistance of HCR, a variety of proof-of-concept targets, including cancer cells, microRNA, metal ions, and many aptamer ligands have been detected with improved sensitivity [[33], [34], [35], [36]]. In this paper, through designing partial of LAMP amplicon sequence as the input sequence to trigger HCR reaction, we for the first time couple the LAMP and HCR together to achieve both ultra-sensitivity and significant signal change for general gene targets. And different transduction strategies were developed to make sure the products of LAMP-HCR could be flexibly monitored via many kinds of available settings (readouts), e.g. gel electrophoresis, flow cytometer (FCM) and even a commercial personal glucometer (PGM). The practicability of the method has thus been largely improved.
Recently, the very contagious noroviruses (NoVs) have become the leading cause of sporadic gastroenteritis across all age groups, especially pediatric populations. It was reported that 26% of 4000 fecal samples from Chinese pediatric out patients were NoV-positive with more than 98% belonging to GII [37,38]. As a response to the urgent demand for more accurate and reliable detection, here the gene segments of NoV GII was specifically employed as the model target [39]. In spite of their high sensitivity, earlier reports like electron microscopy [40,41], immunology-based detection (radioimmunoassay (RIA) [42,43], enzyme-linked immunosorbent assays (ELISA) [44,45]) and nucleic acids biomarkers detection based on gene amplification techniques (PCR [46,47], nucleic acid sequence-based amplification (NASBA) [48,49], LAMP [50,51]) always lack of specificity and false positive results can easily arise. Herein, the powerful exponential amplification of LAMP results in ultrasensitive NoV gene detection with a detection limit of 30 copies, while the HCR gives rise to very good specificity and reliable signal changes in all kinds of readouts. And all of the three readouts can be compatible with fecal samples. Compared with fluorescence (gel electrophoresis and fluorescent reader), the FCM and PGM signals were more stable and interference-resistant because of the importance of magnetic beads (MBs) separation.
2. Experimental section
2.1. Materials and instruments
All of the chemicals used in this work are of analytical grade unless otherwise indicated. Betanie was purchased from Sigma-Aldrich (St. Louis, MO, U.S.A.). Bst 2.0 DNA polymerase, dNTP, MgSO4, 10 × isothermal buffer (10 × iso buffer) were purchased from New England Biolabs (Ipswich, MA, U.S.A.). All the oligonucleotides used in this work were ordered from Sangon Biotech (Shanghai, China), and the sequences were listed in Table S1 (in Supporting Information (SI)). All modified DNA sequences were purified by high performance liquid chromatography (HPLC). All unmodified DNA sequences were purified with polyacrylamide gel electrophoresis. All oligonucleotides were stored in 1 × TE (pH 7.5) at −20 °C. Buffers used here were: 1 × iso buffer (20 mM Tris-HCl, 10 mM (NH4)2SO4, 50 mM KCl, 4 mM MgSO4, 0.1% Tween 20, pH 8.8). GelRed was purchased from Biotium (Hayward, CA, U.S.A.). BioMag® Plus streptavidin particles (MBs, 1.0 μm in average diameter) was purchased from Polysciences (Warrington, PA, U.S.A.). SuperBlock™ (TBS) Blocking Buffer was purchased from Thermo Scientific (Wilmington, DE, U.S.A.). Bayer Contour Next Blood Glucose Test Strips and Bayer Contour Next Blood Glucose Monitoring System were used for the glucose tests in this work. The concentrations of the DNA suspensions were measured by Nanodrop OneC (Thermo Fisher Scientific, Inc., Wilmington, DE, U.S.A.). Fluorescence signals were recorded with the LightCycler 96 System (Roche, Inc., Basel, Switzerland) and COYOTE PCR system (Coyote Bioscience, Inc., Beijing, China). A flow cytometer Accuri C6 (BD, Inc., Ann Arbor, MI, U.S.A.) was used to analyze the fluorescence. Gels were detected using iBright FL1000 imaging system (Thermo Fisher Scientific, Inc., Wilmington, DE, U.S.A.).
2.2. Standard LAMP reaction and electrophoretic characterizations
A 25 μL LAMP reaction mixtures containing different copies of templates, 1.4 μM each BIP and FIP primers, 0.35 μM each B3 and F3 primers, 1 M betaine, and 0.6 mM dNTPs in a total volume of 24 μL 1 × iso buffer were incubated at 95 °C for 5 min, followed by chilling on ice for 2 min. And then 1 μL of 8000 units/mL Bst 2.0 DNA polymerase were added to initiate the LAMP reaction solution, followed by a constant 65 °C reaction for 1 h. Then, the samples were analysed by electrophoresis on a 1% agarose gel. Each well was loaded with 4 μL of sample and an additional 1 μL of 6 × Orange Loading Dye (40% glycerol, 0.25% Orange G). The electrophoresis was developed at 100 V for 25 min, then agarose gels were stained with GelRed.
2.3. Real-time LAMP reaction by one-step strand displacement (OSD)
The standard procedures of LAMP reaction were optimized by real-time fluorescence with the OSD reaction. Annealed OSD reporter (at a final concentration of 60 nM Reporter-F, 120 nM Reporter-Q) was added to the LAMP reagents with different ratios of primers and dNTPs, or with different concentrations of MgSO4. 1 μL of 8000 units/mL Bst 2.0 DNA Polymerase were added to initiate the LAMP reaction. And then, 20 μL of the LAMP-OSD solutions were transferred into a 96-well plate, which was maintained at different constant temperatures (50, 55, 60 or 65 °C). Fluorescence signals were recorded with a LightCycler 96. LAMP reactions in different concentration of fecal samples were performed using the similar protocol to the Real-Time LAMP reaction. Here, the human fecal was diluted to a concentration of 10 wt% with 1 × PBS, and then centrifuged 15 min at 13,000 rpm. The supernatant was collected and filtered with 0.45 μm Millipore filter, and then stored at −20 °C until further use.
2.4. HCR reactions
For the standard HCR reaction, the fluorescein modified H1 and H2 (i.e., FAM-H1 and FAM-H2) were annealed in 1 × iso buffer, respectively. Mixtures in 1 × iso buffer in a total volume of 15 μL containing 500 nM FAM-H1, 500 nM FAM-H2, 0.5 M NaCl and certain amount of DNA mimic (i.e., Mimic-T) were incubated 12 h at 25 °C. To analyze the HCR product, 15 μL of the HCR products were mixed with 3 μL 6 × Orange Loading Dye and loaded into the 8% native PAGE gel. After the electrophoresis, the gel was then stained with GelRed.
2.5. LAMP to HCR reactions
LAMP products were heated to 95 °C for 3 min, followed by chilling on ice for 2 min. The HCR reaction mixture containing 5 μL aliquot of the 25 μL LAMP products, 500 nM annealed FAM-H1 (or H1), 500 nM annealed FAM-H2 (or P15–H2, a well-designed DNA sequence with 15-base extension at the 5’ end of H2), 0.5 M NaCl in a total volume of 15 μL 1 × iso buffer were incubated at 25 °C for 12 h. The reactions were evaluated by electrophoresis in a 1% agarose gel. Each well was loaded with 4 μL of sample and an additional 1 μL of 6 × Orange Loading Dye. The electrophoresis was developed at 100 V for 25 min, and then agarose gels was stained with GelRed.
2.6. HCR-FCM measurements
Firstly, 4 μL of 5 mg/mL MBs were washed 3 times with 1 × iso buffer. The MBs were isolated by using an external magnetic rack and re-suspended in 10 μL 1 × iso buffer. Then, 6 μL of 0.75 μM annealed biotinylated H1 (Bio-H1) was incubated with the MBs on a rotator for 1 h. The unbound Bio-H1 was removed by washing the MBs/Bio-H1 in 1 × iso buffer at least 6 times. Then, 10 μL certain amount of Mimic-T, 0.5 M NaCl, 500 nM annealed FAM-H1 and 500 nM annealed FAM-H2 were added into the MBs/Bio-H1, rotating at 25 °C for 12 h to conduct the HCR reaction. After at least 6 times of washing with 100 μL 1 × iso buffer, each sample was diluted to 300 μL buffer before subjected to the flow cytometer. For analysis, 20,000 events were collected for each sample. Fluorescence signals of the HCR products-anchored MBs were detected by FL1 channel with 488 nm laser excitation. The mean fluorescence intensities (MFI) of the fluorescent histograms were used for the quantitative analysis of Mimic-T. This protocol was also suitable to LAMP products, except that Mimic-T was replaced by 5 μL aliquot of the 25 μL LAMP products.
2.7. Non-specific adsorption measurements
4 μL of 5 mg/mL MBs were washed 3 times with 1 × iso buffer. Then 10 μL of different samples in 1 × iso buffer were added into the MBs, respectively, for 12 h incubation and rotation at 25 °C. After at least 6 washes, the FCM detections were performed.
2.8. HCR-PGM measurements
MBs/Bio-H1 were prepared almost in the same manner as HCR-FCM measurement. Then 50 μL SuperBlock™ (TBS) Blocking Buffer was added to MBs/Bio-H1 for 1 h rotation on a rotator at 25 °C. The magnetically separated MBs/Bio-H1 was then incubated with 10 μL 0.5 M NaCl, 500 nM annealed H1, 500 nM annealed P15–H2 and certain amount of Mimic-T in 1 × iso buffer, rotating at 25 °C for 12 h. The unbound DNA were removed by washing for at least 6 times, and then the MBs were suspended in 9 μL buffer and incubated with 1 μL of the 5 mg/mL P15-Inv (thermostable invertase-labelled P15-SH) conjugates for 1.5 h on a rotator at 25 °C. After at least 6 washes to remove the excess P15-Inv, the MBs were re-suspended in 20 μL buffer and transferred into an equal volume of 500 mM sucrose. The mixture was incubated for 60 min at 55 °C to allow invertase-mediated catalytic conversion of sucrose to glucose. Subsequently, 1.5 μL of the reaction mixture was transferred to a glucometer strip and the amount of glucose was measured by using personal glucometer. The above protocols for detecting Mimic-T were also suitable for LAMP products, except that Mimic-T was replaced by 5 μL aliquot of the 25 μL LAMP products.
2.9. OSD-FCM measurements
4 μL of 5 mg/mL MBs were washed 3 times and re-suspended in 10 μL buffer. Then, 6 μL mixture of 0.75 μM annealed Bio-Reporter-F and Reporter-Q was incubated with MBs on a rotator for 1 h, and unbound DNA were removed by washing MBs with buffer for at least 6 times. After that, the MBs were re-suspended in 5 μL buffer and 5 μL LAMP products to a total volume of 10 μL, performing FCM detection.
3. Results and discussion
3.1. Sensing principle
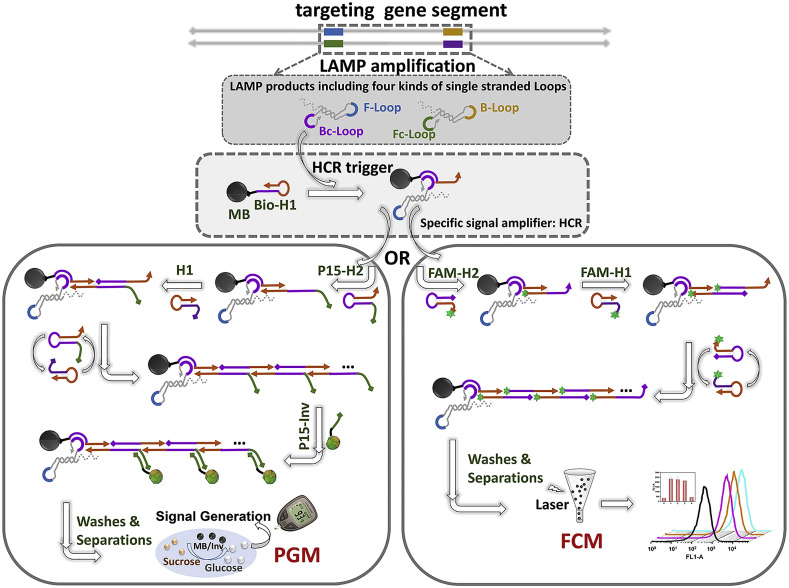
Our strategy was built as the following steps (Scheme 1 ): The standard LAMP reaction was carried out for producing flower-like amplicons which contain four different single strand loops (i.e., F-loop, Fc-loop; B-loop; Bc-loop). A standard HCR reaction uses one single stranded oligonucleotide as trigger, and two hairpin oligonucleotides, H1 and H2, as substrates. The trigger successively opens H1 and H2 via two OSD reactions, and then starts an elongation cascade reaction continuously to form long, nicked duplex products. Herein the sequence of H1 and H2 were designed by using the Bc-Loop as the trigger. H1, H2, and the LAMP amplicons were mixed with Bio-H1-labelled MBs to trigger the HCR at the same time.
Scheme. 1.
Schematic illustration on coupling LAMP with HCR for molecular diagnostic via flow cytometer (FCM) and personal glucometer (PGM).
In detail, the hairpin structure of Bio-H1 could be opened by the hybridization with Bc-loop of the amplicons, leading to the subsequent cascade of hybridization reaction between H1 and H2. The nicked double stranded DNA (dsDNA) could be finally formed on the surface of MBs. On one hand, if the probe H1 and H2 were modified with fluorochrome, the HCR triggered by LAMP could result in the accumulation of large amount of fluorophores on the MBs. By reading the FCM signals of the fluorescent MBs, the NoV gene can be directly analysed. On the other hand, the well-designed P15–H2 serves as a tail probe and the 15 bases extension could not disturb the LAMP to trigger HCR. In this case, after the HCR, the numerous tailed sequences, which were extended from the nicked dsDNA on the MBs’ surface, could hybridize with its complementary DNA labelled with invertase (i.e., P15-Inv). Followed by the washing and separation steps, the re-suspended MBs with invertase could hydrolyze sucrose into glucose which can be detected by using the commercial PGM.
3.2. Verification of LAMP process
The steps were initially verified separately. Here the four primers set of LAMP for NoV GII gene was self-designed, using 30 copies to 3 × 106 copies as the template. The standard procedures of LAMP reaction were optimized by real-time fluorescence with the OSD reaction (Fig. S1, SI). The OSD reporter was designed for Bc-Loop sequence (Fig. 1 A). After the optimization, 1.4 μM each BIP and FIP primers, 0.35 μM each B3 and F3 primers, 1 M betaine, 0.6 mM dNTPs, 2 mM MgSO4, and 1 μL of 8000 units/mL Bst 2.0 DNA Polymerase in 1 × iso buffer were employed as the LAMP reagents, followed by a constant 65 °C reaction. Under the optimal conditions, as few as 30 copies nucleic acid templates of NoV could be detected within 1 h in 0.1% fecal samples (Fig. 1B and C). It should be noted actually the LAMP-OSD is already a ready-to-use method for efficient NoV detection. However, the concern is even with plenty of optimizations, the signal magnitude of the fluorescence signal (NoV positive) over background (NoV negative) is still not satisfied. Sometimes the signals may even be too small to be distinguished from the non-specific fluorescence increase generated by unknown interferences, especially when fecal sample concentrations are increased (Fig. 1D and E) or non-specific amplifications (Fig. S2, in SI) are induced. Actually, this problem may commonly exist in the detection for many other gene targets. That is the reason why amplifiable circuit (HCR) has to be imported as an enhancement to the detection reliability.
Fig. 1.
Sensitive detection of NoV DNA templates. (A) Schematic illustration on coupling LAMP with OSD for diagnostic. (B) The function of time with varying amounts of the NoV templates in 0.1% fecal samples by LAMP-OSD assay. (C) The same samples from (B) are separated on a 1% agarose gel and stained with gel red. Lanes: (M) DNA marker; (1) 0, (2) 30, (3) 3 × 102, (4) 3 × 104 and (5) 3 × 106 copies of NoV LAMP products. (D) The function of time with 3 × 106 copies of NoV templates in the different concentrations of fecal samples by LAMP-OSD assay. (E) The same samples from (D) are separated on a 1% agarose gel and stained with gel red. Lanes: (M) DNA marker; (1) 0 copies of NoV virus LAMP products; (2) 3 × 106 copies of LAMP reactions in 0% fecal samples; (3) 3 × 106 copies of LAMP reactions in 0.1% fecal samples; (4) 3 × 106 copies of LAMP reactions in 0.5% fecal samples; (5) 3 × 106 copies of LAMP reactions in 1% fecal samples; (6) 3 × 106 copies of LAMP reactions in 2% fecal samples. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Verification of HCR process
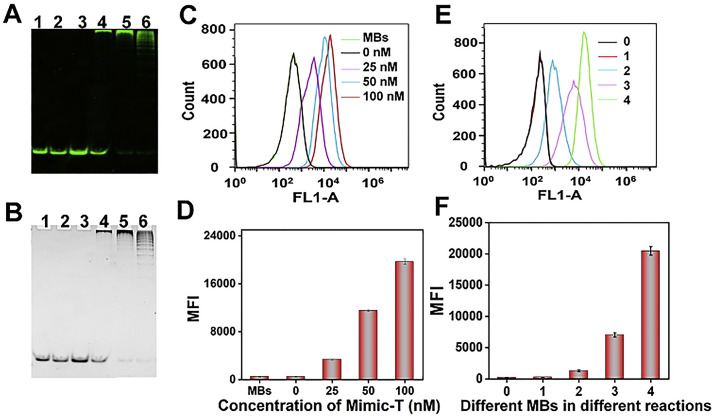
The efficiency of HCR was assessed by using Mimic-T with the same sequence as Bc-Loop (Fig. 2 ). The 8% native PAGE gel (without or with GelRed stain) results show that HCR reaction took place with the Mimic-T introduced (Fig. 2A and B, lane 4–6). However, the H1 and H2 could exist steadily without Mimic-T (Fig. 2A and B, lane 1–3). FCM analysis also shows the same results. In Fig. 2C and D, the fluorescence increased significantly with the increased concentration of Mimic-T. While there is no obvious signal increase in the absence of the Mimic-T compared to the bare MBs.
Fig. 2.
Verification of HCR process. (A–B) 8% native PAGE gel results without and with GelRed stained, lanes: (1) 500 nM FAM-H1; (2) 500 nM FAM-H2; (3) 500 nM FAM-H1 + 500 nM FAM-H2; (4) 25 nM Mimic-T + 500 nM FAM-H1 + 500 nM FAM-H2; (5) 50 nM Mimic-T + 500 nM FAM-H1 + 500 nM FAM-H2; (6) 100 nM Mimic-T + 500 nM FAM-H1 + 500 nM FAM-H2. (C–D) FCM histograms of fluorescence response of MBs for different concentrations of Mimic-T. (E–F) FCM histograms of fluorescence response of MBs for different conditions: (0) MBs/Bio-H1; (1) MBs/Bio-H1 + 500 nM FAM-H1 + 100 nM Mimic-T; (2) MBs/Bio-H1-Mimic-T + 100 nM FAM-H2; (3) MBs/Bio-H1 + 500 nM FAM-H2 + 100 nM Mimic-T; (4) MBs/Bio-H1 + 500 nM FAM-H1 + 500 nM FAM-H2 + 100 nM Mimic-T.
To testify the cascade HCR amplification on MBs, some control experiments were carried out. Fig. 2E and F show about 16 orders of magnitude the MFI increase than that of one-step reaction where the pre-hybridized Bio-H1 and Mimic-T were immobilized on the MBs, with 100 nM FAM-H2 serving as the target to form a 1: 1 binding. And there is about 3 orders of magnitude signal increase than that of the assay only using FAM-H2 where no additional FAM-H1 was introduced during the HCR process together with 100 nM Mimic-T. When FAM-H1 and 100 nM Mimic-T were mixed with Bio-H1-labelling MBs, no signal increase was observed compared with bare MBs. These results clearly demonstrate the signal amplification in HCR approach. Note that, the non-specific adsorption of DNAs on MBs (Fig. S3, in SI) and the optimization of the concentration of HCR reagents (Fig. S4, in SI) were also well investigated.
3.4. LAMP-HCR FCM assay
Following these steps, we integrated LAMP and HCR for NoV templates detection. The agarose gel electrophoretic characterization of the amplification products reveals the success of this LAMP to HCR signal transduction (Fig. S5, in SI). In the FCM assay, the LAMP amplicons (5 μL), 500 nM each of FAM-H1 and FAM-H2 were mixed with Bio-H1-modified MBs and incubated for 12 h prior to magnetic washing and separation. The completely washed MBs with HCR products were used for FCM measurement to the definitive detection of the synthetic template of NoV. Fig. 3 A shows the fluorescence signal of the MBs detected in FL1 channel. The MFI of the tested MBs of ∼338 represents the negative controls (1 × iso buffer with no template or noncognate Rotavirus LAMP products) (Fig. 3B). While as low as 30 copies of the NoV template could be detected (MFI is 2330) without the dose-discrimination, because the primers have been consumed in the given LAMP reaction time (1 h). To detect the NoV templates in fecal samples, rather than in buffer, the NoV templates were spiked into different percentages of fecal samples (from 0.1% to 2%). The agarose gel electrophoretic result shows that it makes no difference of LAMP reactions in fecal samples compared with the LAMP reaction in buffer (Fig. 1E).
Fig. 3.
Sensitive and specific detection of NoV DNA plasmid using LAMP-HCR FCM assay. (A–B) FCM histograms of fluorescence response of MBs for negative control (no template, 0), different copies of NoV templates ((1), 30 copies; (2), 3 × 103 copies; (3), 3 × 105 copies) and (4), 3 × 105 copies of rota virus LAMP products. (C–D) FCM histograms of mean fluorescence intensity of MBs in 2% fecal sample with different copies of NoV template. (E–F) Reproducibility of FCM assay with 2 × 105 copies of NoV DNA (positive signal) and buffer controls on different days. On each day, three parallel assays were carried out with both NoV DNA and buffer controls. A total of nine sets of assays were performed for both NoV DNA and buffer. Assay conditions were the same as the normal procedure.
Following the optimization of the reaction, the NoV template spiked into 2% human fecal also gave consistent FCM results without any loss in signal intensity and detection sensitivity (Fig. 3C and D). These results exhibited the MBs based LAMP-HCR strategy realized robust NoV detection and was more resistant to interference than the real-time fluorescence assay, where the fluorescence signal could be decreased with the increased concentrations of fecal samples (Fig. 1D and Fig. S6, in SI). The reproducibility test performed in 2% human fecal consists of three parallel assays, detecting both NoV positive samples (2 × 105 copies of NoV DNA) and buffer negative controls on three days, respectively (Fig. 3E and F). As expected, only small standard deviations were yielded by all nine sets of measurements (i.e., 94.0 MFI and 278 MFI for negative response (448 MFI) and positive response (2031 MFI), respectively). Besides sensitivity, selectivity and reproducibility, the re-usability of the Bio-H1-labelled MBs after detection is easy to regenerate by heating the MBs at 80 °C for 15 min, immediately followed by a quick separation of the Bio-H1-labelled MBs from other reagents (i.e., FAM-H1, FAM-H2 and LAMP products), leading to the regenerated modified MBs. The successful regeneration could be verified by the subsequent detection with the same result as that of the first one (Fig. S7, in SI). Note that, the Bio-H1-labelled MBs could be reused at least but may be not limited to twice. The aggregation of MBs could occur under too much repeated regeneration, resulting in unreliable detection results. Encouragingly, the cost of per analysis could be controlled within 1 dollar, which makes it suitable for POC gene biomarker analysis.
3.5. LAMP-HCR PGM assay
As we know, the most commonly used commercially available POC device is the PGM because of its portable size, low-cost and easy operation. Lu group has firstly reported the aptasenors by using PGM to detect a series of targets [52]. Previously, we developed the sweet spot for molecular diagnostics by coupling LAMP with OSD, which could directly transduce Middle East respiratory syndrome coronavirus and Zaire Ebola virus templates into glucose signals [53]. However, it shows that the signal amplitude (ΔGlucose meter signal, ΔGMS = positive signal – negative signal) is not high (∼60 mg/dl).
In our LAMP-HCR to PGM assay, the HCR reaction on MBs could be triggered by LAMP amplicons in the presence of H1 and P15–H2. Here, P15–H2 with a tail sequence can hybridize with its complementary DNA labelled with invertase (P15-Inv). After washing and separation steps, the invertase attached on MBs hydrolyzed sucrose into glucose, which could be detected by PGM. Firstly, Mimic-T was chosen for investigating the HCR process (Fig. 4 A). With the increasing concentration of Mimic-T, the signal amplitude (ΔGMS) of PGM gradually increased. Then, instead of Mimic-T, LAMP amplicons could also trigger the HCR on the surface of MBs, following the hybridization of P15-Inv with the HCR products on the MBs. Fig. 4B shows a positive signal compared with negative control (non-cognate Rotavirus LAMP products). Remarkably, as few as 30 copies of NoV in 2% human fecal sample can produce an obvious amplitude, reaching up to 156 mg/dl. The agarose gel electrophoretic result also shows that HCR reaction occurred with NoV LAMP amplicons introduced (Fig. 4C). The reproducibility test of this PGM assay was also carried out under the same experimental condition as the FCM assay. The ΔGMS values for detecting 2 × 105 copies of NoV in 2% human fecal on three days were comparable (Fig. 4D), showing a very low standard deviation of 17 mg/dl.
Fig. 4.
Sensitive and specific detection of NoV using LAMP-HCR PGM assay. (A) Detection of Mimic-T using the HCR-to-glucose transduction. (B) The histograms of PGM response for different copies of NoV template and 3 × 105 copies of rota virus LAMP products in 2% fecal sample. (C) Agarose gel electrophoretic characterization of HCR process in the presence of LAMP amplicons. Lanes: (M) marker; (1) LAMP amplicons; (2) LAMP amplicons + 500 mM NaCl; (3) LAMP amplicons + 500 mM NaCl + 500 nM H1; (4) LAMP amplicons + 500 mM NaCl + 500 nM P15–H2; (5) LAMP amplicons + 500 mM NaCl + 500 nM H1+ 500 nM P15–H2; (6) 500 mM NaCl + 500 nM H1+ 500 nM P15–H2. (D) Reproducibility of PGM assay with 2 × 105 copies of NoV DNA in 2% human fecal sample on different days. On each day, three parallel experiments were carried out with both NoV DNA and buffer controls.
3.6. LAMP-OSD FCM assay
The amplification effect of the HCR function was further confirmed using a LAMP to OSD detection by FCM. As shown in Fig. 5 A, an OSD duplex (hybridized between sequence Bio-Reporter-F and sequence Reporter-Q) was immobilized on MBs via the biotin labelled on Bio-Reporter-F. And for signal reporting, fluorophore FAM and its Quencher was, in respective, tagged onto Bio-Reporter-F and Reporter-Q. In presence of LAMP amplicons, the Bc-loop could initiate a strand displacement reaction and kick the Reporter-Q away from Bio-Reporter-F, finally leaving Bio-Reporter-F/Bc-loop (in amplicons) duplex on the MBs. Therefore, the MBs were lightened up with higher fluorescence emission. When such a process was transferred onto FCM assay, the sample with NoVs LAMP amplicons showed a tiny right-migration in the FL1 channel, indicating the success of the LAMP-OSD detection. However, the absolute (Fig. 5B) and relative MFI (Fig. 5C) values were all much smaller than those obtained from the LAMP-HCR detection.
Fig. 5.
Verification for the HCR amplification function. (A) Schematic illustration on coupling LAMP with OSD for molecular diagnostic via FCM. (B–C) FCM histograms of fluorescence response of MBs for LAMP to HCR and LAMP to OSD. Rectangle: Positive signal - Negative signal; Dot: Positive signal/Negative signal.
4. Conclusions
In summary, we have successfully developed a highly sensitive and selective detection method for NoVs both in buffers and fecal samples via the combination of LAMP with either OSD or HCR. In general, compared with various NoVs gene detection methods (Table S2, in SI), the present work proved much more reliable and reproducible. The LAMP reactions and transducers (i.e., OSD or HCR) we have developed are comparably sensitive but also sequence-specific to ensure low false positives. The sensitivity down to 30 gene copies is mainly contributed by the LAMP reaction. When the OSD signal suffers unsatisfied signal change, HCR reaction can further provide the ultra-selectivity and higher signal-to-background ratio which has usually and seriously affected the accuracy for LAMP and other enzymatic amplifications. The detection is very practical, low-cost and almost enough to be ready-to-use. And the readouts should be very flexible, exhibiting great potential to be used in the diagnosis of gastroenteritis in the hospital or even at home with the read out by the portable PGM.
Conflict of interest
The authors declare no conflict of interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was financially supported by the National Natural Science Foundation of China (21505129, 21605138 and 21874129), the Natural Science Foundation of Jilin Province (20160101296JC), the International Cooperation Project of Jilin Scientific and Technological Development Program (20190701059GH), and K. C. Wong Education Foundation. The authors declare no competing financial interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aca.2019.06.048.
Contributor Information
Dan Li, Email: lidan@ciac.ac.cn.
Bingling Li, Email: binglingli@ciac.ac.cn.
Yan Du, Email: duyan@ciac.ac.cn, duyan.bessie@gmail.com.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Kurkela S., Brown D.W. Molecular diagnostic techniques. Medicine. 2009;37:535–540. doi: 10.1016/j.mpmed.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar I., Misztal I., Johnson D.L., Legarra A., Tsuruta S., Lawlor T.J. A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of holstein final score. J. Dairy Sci. 2010;93:743–752. doi: 10.3168/jds.2009-2730. [DOI] [PubMed] [Google Scholar]
- 3.Saiki R.K., Bugawan T.L., Horn G.T., Mullis K.B., Erlich H.A. Analysis of enzymatically amplified globin and HLA-DQ DNA with allele-specific oligonucleotide probes. Nature. 1986;324:163–166. doi: 10.1038/324163a0. [DOI] [PubMed] [Google Scholar]
- 4.Gentsch J.R., Glass R.I., Woods P., Gouvea V., Gorziglia M., Flores J., Das B.K., Bhan M.K. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudecova I. Digital PCR analysis of circulating nucleic acids. Clin. Biochem. 2015;48:948–956. doi: 10.1016/j.clinbiochem.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Zhao W., Ali M.M., Brook M.A., Li Y. Rolling circle amplification: applications in nanotechnology and biodetection with functional nucleic acids. Angew. Chem. Int. Ed. 2008;47:6330–6337. doi: 10.1002/anie.200705982. [DOI] [PubMed] [Google Scholar]
- 7.Kivlehan F., Mavre F., Talini L., Limoges B., Marchal D. Real-time electrochemical monitoring of isothermal helicase-dependent amplification of nucleic acids. Analyst. 2011;136:3635–3642. doi: 10.1039/c1an15289k. [DOI] [PubMed] [Google Scholar]
- 8.Rosser A., Rollinson D., Forrest M., Webster B.L. Isothermal recombinase polymerase amplification (RPA) of schistosoma haematobium DNA and oligochromatographic lateral flow detection. Parasites Vectors. 2015;8:446. doi: 10.1186/s13071-015-1055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai S., Jung C., Bhadra S., Ellington A.D. Phosphorothioated primers lead to loop-mediated isothermal amplification at low temperatures. Anal. Chem. 2018;90:8290–8294. doi: 10.1021/acs.analchem.8b02062. [DOI] [PubMed] [Google Scholar]
- 10.Kitamura M., Kubo S., Tanaka J., Adachi T. Rapid screening method for male DNA by using the loop-mediated isothermal amplification assay. Int. J. Leg. Med. 2018;132:975–981. doi: 10.1007/s00414-017-1661-z. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y., Xianyu Y., Wu J., Dong M., Zheng W., Sun J., Jiang X. Double-enzymes-mediated bioluminescent sensor for quantitative and ultrasensitive point-of-care testing. Anal. Chem. 2017;89:5422–5427. doi: 10.1021/acs.analchem.7b00239. [DOI] [PubMed] [Google Scholar]
- 12.Du Y., Pothukuchy A., Gollihar J.D., Nourani A., Li B., Ellington A.D. Coupling sensitive nucleic acid amplification with commercial pregnancy test strips. Angew Chem. Int. Ed. Engl. 2017;56:992–996. doi: 10.1002/anie.201609108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung H.J., Castro C.M., Im H., Lee H., Weissleder R. A magneto-DNA nanoparticle system for rapid detection and phenotyping of bacteria. Nat. Nanotechnol. 2013;8:369–375. doi: 10.1038/nnano.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X., Zhang C., Liu K., Wang H., Lu C., Li H., Hua K., Zhu J., Hui W., Cui Y., Zhang X. Multiple SNPs detection based on lateral flow assay for phenylketonuria diagnostic. Anal. Chem. 2018;90:3430–3436. doi: 10.1021/acs.analchem.7b05113. [DOI] [PubMed] [Google Scholar]
- 15.Shin D.J., Trick A.Y., Hsieh Y.H., Thomas D.L., Wang T.H. Sample-to-answer droplet magnetofluidic platform for point-of-care hepatitis C Viral Load Quantitation. Sci. Rep. 2018;8:9793. doi: 10.1038/s41598-018-28124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin H.Y., Huang C.H., Park J., Pathania D., Castro C.M., Fasano A., Weissleder R., Lee H. Integrated magneto-chemical sensor for on-site food allergen detection. ACS Nano. 2017;11:10062–10069. doi: 10.1021/acsnano.7b04318. [DOI] [PubMed] [Google Scholar]
- 17.Min J., Nothing M., Coble B., Zheng H., Park J., Im H., Weber G.F., Castro C.M., Swirski F.K., Weissleder R., Lee H. Integrated biosensor for rapid and point-of-care sepsis diagnosis. ACS Nano. 2018;12:3378–3384. doi: 10.1021/acsnano.7b08965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda S., Takao S., Kuwayama M., Shimazu Y., Miyazaki K. Rapid detection of norovirus from fecal specimens by real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 2006;44:1376–1381. doi: 10.1128/JCM.44.4.1376-1381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parida M., Sannarangaiah S., Dash P.K., Rao P.L., Morita K. Loop-mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008;18:407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pham H.M., Nakajima C., Ohashi K., Onuma M. Loop-mediated isothermal amplification for rapid detection of newcastle disease virus. J. Clin. Microbiol. 2005;43:1646–1650. doi: 10.1128/JCM.43.4.1646-1650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian W., Meng Y., Lu Y., Wu C., Wang R., Wang L., Ying Y. Rapid, sensitive, and carryover contamination-free loop-mediated isothermal amplification coupled visual detection method for ‘candidatus liberibacter asiaticus’. J. Agric. Food Chem. 2017;65:8302–8310. doi: 10.1021/acs.jafc.7b03490. [DOI] [PubMed] [Google Scholar]
- 22.Kubota R., Alvarez A.M., Su W.W., Jenkins D.M. FRET-based assimilating probe for sequence-specific real-time monitoring of loop-mediated isothermal amplification (LAMP) Biol. Eng. Trans. 2011;4:81–100. [Google Scholar]
- 23.Oscorbin I.P., Belousova E.A., Zakabunin A.I., Boyarskikh U.A., L Filipenko M. Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification (qLAMP) Biotechniques. 2016;61:20–25. doi: 10.2144/000114432. [DOI] [PubMed] [Google Scholar]
- 24.Tang Y., Zhu Z., Lu B., Li B. Spatial organization based reciprocal switching of enzyme-free nucleic acid circuits. Chem. Commun. 2016;52:13043–13046. doi: 10.1039/c6cc07153h. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y.S., Bhadra S., Li B., Wu Y.R., Milligan J.N., Ellington A.D. Robust strand exchange reactions for the sequence-specific, real-time detection of nucleic acid amplicons. Anal. Chem. 2015;87:3314–3320. doi: 10.1021/ac504387c. [DOI] [PubMed] [Google Scholar]
- 26.Tang Y., Lu B., Zhu Z., Li B. Establishment of a universal and rational gene detection strategy through three-way junction-based remote transduction. Chem. Sci. 2018;9:760–769. doi: 10.1039/c7sc03190d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dirks R.M., Pierce N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J., Wu Y., Chen Y., Zhu Z., Yang X., Yang C.J., Wang K., Tan W. Pyrene-excimer probes based on the hybridization chain reaction for the detection of nucleic acids in complex Biological Fluids. Angew. Chem. Int. Ed. 2011;50:401–404. doi: 10.1002/anie.201005375. [DOI] [PubMed] [Google Scholar]
- 29.Li B., Jiang Y., Chen X., Ellington A.D. Probing spatial organization of dna strands using enzyme-free hairpin assembly circuits. J. Am. Chem. Soc. 2012;134:13918–13921. doi: 10.1021/ja300984b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu S., Tang Y., Yan M., Aguilar Z.P., Lai W., Xu H.A. Luorescent cascade amplification method for sensitive detection of salmonella based on magnetic Fe3O4 nanoparticles and hybridization chain reaction. Sensor. Actuator. B Chem. 2019;279:31–37. [Google Scholar]
- 31.Ren W., Liu H., Yang W., Fan Y., Yang L., Wang Y., Li Z.A. Cytometric bead assay for sensitive DNA detection based on enzyme-free signal amplification of hybridization chain reaction. Biosens. Bioelectron. 2013;49:380–386. doi: 10.1016/j.bios.2013.05.055. [DOI] [PubMed] [Google Scholar]
- 32.Li P., Zhang H., Wang D., Tao Y., Zhang L., Zhang W., Wang X. An efficient nonlinear hybridization chain reaction-based sensitive fluorescent assay for in situ estimation of calcium channel protein expression on bone marrow cells. Anal. Chim. Acta. 2018;1041:25–32. doi: 10.1016/j.aca.2018.08.031. [DOI] [PubMed] [Google Scholar]
- 33.Zhou G., Lin M., Song P., Chen X., Chao J., Wang L., Zuo X. Multivalent capture and detection of cancer cells with DNA nanostructured biosensors and multibranched hybridization chain reaction amplification. Anal. Chem. 2014;86:7843–7848. doi: 10.1021/ac502276w. [DOI] [PubMed] [Google Scholar]
- 34.Hou T., Li W., Liu X., Li F. Label-free and enzyme-free homogeneous electrochemical biosensing strategy based on hybridization chain reaction: a facile, sensitive, and highly specific microrna assay. Anal. Chem. 2015;87:11368–11374. doi: 10.1021/acs.analchem.5b02790. [DOI] [PubMed] [Google Scholar]
- 35.Ma C., Wang W., Mulchandani A., Shi C. A simple colorimetric DNA detection by target-induced hybridization chain reaction for isothermal signal amplification. Anal. Biochem. 2014;457:19–23. doi: 10.1016/j.ab.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y.M., Wu Z., Liu S.J., Chu X. Structure switching aptamer triggering hybridization chain reaction on the cell surface for activatable theranostics. Anal. Chem. 2015;87:6470–6474. doi: 10.1021/acs.analchem.5b01634. [DOI] [PubMed] [Google Scholar]
- 37.Luo J., Xu Z., Nie K., Ding X., Guan L., Wang J., Xian Y., Wu X., Ma X. Visual detection of norovirus genogroup II by reverse transcription loop-mediated isothermal amplification with hydroxynaphthol blue dye. Food Environ. Virol. 2014;6:196–201. doi: 10.1007/s12560-014-9142-8. [DOI] [PubMed] [Google Scholar]
- 38.Huang J., Xu X., Weng Q., Hong H., Guo Z., He S., Niu J. Pyrene-excimer probes based on the hybridization chain reaction for the detection of nucleic acids in complex biological fluids. Angew. Chem. Int. Ed. 2011;50:401–404. doi: 10.1002/anie.201005375. [DOI] [PubMed] [Google Scholar]
- 39.Chan M.C., Sung J.J., Lam R.K., Chan P.K., Lee N.L., Lai R.W., Leung W.K. Fecal viral load and norovirus associated gastroenteritis. Emerg. Infect. Dis. 2006;12:1278. doi: 10.3201/eid1208.060081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma S.H., Zheng L.J., Liu J.J., Wang W.H., Ma j., Cheng X.H., Ge L.L., Wang M.C., Huo Y.Q., Shen S. Chimeric GII. 3/GII. 6 norovirus capsid (VP1) proteins: characterization by electron microscopy, trypsin sensitivity and binding to histo-blood group antigens. Arch. Virol. 2014;163:3265–3273. doi: 10.1007/s00705-018-4002-8. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y.L., Chang P.J., Huang C.T. Small P particles formed by the Taiwan-native norovirus P domain overexpressed in Komagataella pastoris. Appl. Microbiol. Biotechnol. 2018;102:9707–9718. doi: 10.1007/s00253-018-9331-8. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi S., Fujiwara N., Takeda N., Minagawa H. Seroepidemiological study of norovirus infection in Aichi Prefecture, Japan. Microbiol. Immunol. 2009;53:356–359. doi: 10.1111/j.1348-0421.2009.00132.x. [DOI] [PubMed] [Google Scholar]
- 43.Rackoff L.A., Bok K., Green K.Y., Kapikian A.Z. Epidemiology and evolution of rotaviruses and noroviruses from an archival WHO Global Study in Children (1976–79) with implications for vaccine design. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng L., Wang W., Liu J., Chen X., Li S., Wang Q., Wang M. Characterization of a Norovirus-specific monoclonal antibody that exhibits wide spectrum binding activities. J. Med. Virol. 2018;90:671–676. doi: 10.1002/jmv.25001. [DOI] [PubMed] [Google Scholar]
- 45.Tin C.M., Yuan L., Dexter R.J., Parra G.I., Bui T., Green K.Y., Sosnovtsev S.V. A Luciferase Immunoprecipitation System (LIPS) assay for profiling human norovirus antibodies. J. Virol. Methods. 2017;248:116–129. doi: 10.1016/j.jviromet.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao X., Wang Z., Wang Y., Liu Z., Guan X., Ma Y., Xu Y. Surveillance of norovirus contamination in commercial fresh/frozen berries from Heilongjiang Province, China, using a TaqMan real-time RT-PCR assay. Food Microbiol. 2019;82:119–126. doi: 10.1016/j.fm.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 47.Loisy F., Atmar R.L., Guillon P., Cann P.Le, Pommepuy M., Guyader F.S. Real-time RT-PCR for norovirus screening in shellfish. J. Virol. Methods. 2005;123:1–7. doi: 10.1016/j.jviromet.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 48.Chung S.H., Baek C., Cong V.T., Min J. The microfluidic chip module for the detection of murine norovirus in oysters using charge switchable micro-bead beating. Biosens. Bioelectron. 2015;67:625–633. doi: 10.1016/j.bios.2014.09.083. [DOI] [PubMed] [Google Scholar]
- 49.Kou X., Wu Q., Zhang J., Fan H. Rapid detection of noroviruses in fecal samples and shellfish by nucleic acid sequence-based amplification. J. Microbiol. 2006;44:403–408. [PubMed] [Google Scholar]
- 50.Hanaki K.I., Ike F., Kajita A., Yasuno W., Yanagiba M., Goto M., Kyuwa S. Detection of murine norovirus by reverse transcription loop-mediated isothermal amplification. J. Virol. Methods. 2014;204:17–24. doi: 10.1016/j.jviromet.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeon S.B., Seo D.J., Oh H., Kingsley D.H., Choi C. Development of one-step reverse transcription loop-mediated isothermal amplification for norovirus detection in oysters. Food Control. 2017;73:1002–1009. [Google Scholar]
- 52.Xiang Y., Lu Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du Y., Hughes R.A., Bhadra S., Jiang Y.S., Ellington A.D., Li B. A sweet spot for molecular diagnostics: coupling isothermal amplification and strand exchange circuits to glucometers. Sci. Rep. 2015;5:11039. doi: 10.1038/srep11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.