Abstract
Severe acute respiratory syndrome coronavirus (SARS‐CoV) encodes a highly basic nucleocapsid (N) protein of 422 amino acids. Similar to other coronavirus N proteins, SARS‐CoV N protein is predicted to be phosphorylated and may contain nuclear localization signals, serine/arginine‐rich motif, RNA binding domain and regions responsible for self‐association and homo‐oligomerization. In this study, we demonstrate that the protein is posttranslationally modified by covalent attachment to the small ubiquitin‐like modifier. The major sumoylation site was mapped to the 62lysine residue of the N protein. Further expression and characterization of wild type N protein and K62A mutant reveal that sumoylation of the N protein drastically promotes its homo‐oligomerization, and plays certain roles in the N protein‐mediated interference of host cell division. This is the first report showing that a coronavirus N protein undergoes posttranslational modification by sumoylation, and the functional implication of this modification in the formation of coronavirus ribouncleoprotein complex, virion assembly and virus–host interactions.
Keywords: SARS-CoV, Nucleocapsid protein, Sumoylation, Oligomerization, Multinucleated cells, Cytokinesis
1. Introduction
A novel coronavirus was identified as the etiological agent of severe acute respiratory syndrome (SARS) [6, 13, 17, 21]. SARS coronavirus (SARS‐CoV) is an enveloped virus with a single strand, positive‐sense RNA genome of 29.7 kb in length. In SARS‐CoV‐infected cells, a 3′‐coterminal nested set of nine mRNAs species, including the genome‐length mRNA (mRNA1) and eight subgenomic mRNA species (mRNA2–9), is expressed. The genome‐length mRNA1 encodes two overlapping replicase proteins in the form of polyproteins 1a and 1a/b, which are processed by virus‐encoded proteinases into at least 16 putative nonstructural proteins (NSP1–NSP16) [24, 29]. The four structural proteins, spike (S), envelope (E), membrane (M) and nucleocapsid (N), are encoded by subgenomic mRNA 2, 4, 5, and 9, respectively. In addition, eight putative nonstructural proteins, 3a, 3b, 6, 7a, 7b, 8a, 8b, 9b, are encoded by subgenomic mRNA3, 6, 7, 8, and 9 [24, 29].
All coronaviruses encode an extensively phosphorylated, highly basic protein. It varies from 377 to 455 amino acids in length and has high serine content (7–11%), which are potential targets for phosphorylation. Although the primary sequence conservation of the N proteins within the genus is low, three structural domains can be identified based on sequence comparisons [14]. Of which the middle domain is a potential RNA‐binding domain, capable of binding both coronavirus‐ and non‐coronavirus‐derived RNA sequences in vitro [26]. The functions of domains I and III remain unknown. However, putative motifs for ribosome binding and nucleolar localization signals (NuLs) have been identified in domain III region [31]. In cells expressing the N protein, it localizes either to the cytoplasm alone or to the cytoplasm and nucleolus [9]. This nucleolar localization has been shown to be a common feature of the coronavirus family [31]. Multiple functions have been postulated for the coronavirus N protein throughout the virus life cycle. Primarily, the protein can be associated with the genomic RNA to form a ribonucleoprotein complex (RNP) and viral core [5, 7, 19, 20]. It plays an important role in the replication of the genomic RNA [3], and in the transcription and translation of subgenomic RNAs (sgRNA) [1, 26, 28]. In addition, N protein might inhibit host cell proliferation or delay cell growth, possibly by disrupting cytokinesis [4, 31]. The protein is generally phosphorylated at multiple positions and is one of the most abundant structural proteins [14]. It can also stimulate strong humoral and cellular immune response, making it a potential vaccine candidate [12].
Similar to other coronaviruses, SARS‐CoV N protein is a highly basic protein (Fig. 1 A). Among its 422 amino acids, there are several basic amino acid‐rich regions, that may function as NuLs and RNA‐binding motifs, and a serine/arginine (S/R) rich motif (Fig. 1A). Sequence comparison and available evidence showed that the N‐terminal one third region (from amino acid 49 to 178) may contain the RNA binding domain and the C‐terminal half (amino acid 213 to 422) may be responsible for self‐association and homo‐oligomerization of the protein (Fig. 1A) [11, 27].
Figure 1.

(A) Schematic representation of the SARS‐CoV N protein, showing the SUMO‐1 conjugation site (K62), three putative nucleolar localization signals, the S/R‐rich motif, the RNA‐binding domain and self‐association domain. (B) Analysis of the expression of N protein in bacterial and mammalian cells. Plasmids pGEX‐5X‐1 and pGEX‐N were transformed into E. coli BL21, and expressed by induction with IPTG. The GST (lane 1) and GST‐N (lane 2) proteins were affinity‐purified and separated on an SDS–10% polyacrylamide gel. The protein was visualized by staining the gel with Coomassie blue. Total cell lysates prepared from HeLa cells transfected with pKT0‐Flag empty plasmid (lane 3) and Flag‐tagged N protein (lane 4) were separated by SDS–PAGE and analyzed by Western blotting with anti‐Flag antibody. Numbers on the left and right indicate molecular masses in kilodaltons.
In this study, the SARS‐CoV N protein was cloned and expressed in bacterial and mammalian cells. In Escherichia coli BL21 cells, the protein was expressed as a single protein species. However, multiple protein bands with a wide range of molecular masses were detected when the N protein was expressed in mammalian cells, indicating that it may have undergone posttranslational modification in addition to phosphorylation and may form homo‐oligomers. Biochemical characterization and mutagenesis studies demonstrated that the protein was posttranslationally modified by covalent attachment to the small ubiquitin‐like modifier (SUMO). The sumoylation site was mapped to the 62lysine residue of the N protein. Further expression and characterization of wild type N protein and K62A mutant revealed that sumoylation drastically promotes homo‐oligomerization of the protein. Sumoylation of the N protein may also play certain roles in its interference of host cell division. As self‐association and homo‐oligomerization of the N protein are essential for the formation of viral RNP and nucleocapsid assembly, sumoylation of this protein may play an important regulatory role in the SARS‐CoV replication cycles.
2. Materials and methods
2.1. Cells and cell culture
HeLa cells were cultured in complete Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% newborn calf serum (Sterile) and 1% penicillin/streptomycin (Invitrogen) and maintained at 37 °C in humidified 5% CO2.
2.2. Transient expression of viral protein in mammalian cells
Constructs containing plasmid DNA under the control of a T7 promoter were transiently expressed in mammalian cells using a vaccinia virus‐T7 system. Briefly, semiconfluent monolayers of HeLa cells were infected with 10 plaque forming units/cells of recombinant vaccinia virus (vTF7‐3), which expresses the T7 RNA polymerase gene, for 2 h at 37 °C prior to transfection. The plasmid DNA was transfected into vTF7‐3‐infected cells using Effectene transfection reagent according to the manufacturer's instructions (Qiagen).
2.3. Western blot analysis
Total protein from transfected HeLa cells was lysed with 2× SDS loading buffer and subjected to 10% SDS–PAGE. Protein was transferred to PVDF membrane (Bio‐Rad) by using a semi‐dry transfer cell (Bio‐Rad, Trans‐blot SD), and blocked overnight at room temperature with 10% nonfat milk in PBS–T. The membrane was probed with anti‐SARS N, anti‐Flag, or anti‐Myc antibodies followed by anti‐mouse or anti‐rabbit antibodies conjugated with horseradish peroxidase (Sigma). Membrane‐bound antibodies were detected with the SuperSignal west pico chemiluminescence substrate kit (Pierce).
2.4. Immunoprecipitation
Transiently transfected HeLa cells in 100‐mm dishes were lysed in 1 ml of lysis buffer (150 mM NaCl, 1% NP‐40, and 50 mM Tris–HCl, pH 8.0) with 0.5% protease inhibitor cocktail (Sigma). The lysates were centrifuged at 12 000 rpm for 20 min at 4 °C. The supernatants were added with anti‐SUMO‐1 (Zymed), anti‐Flag M2 (Stratagene), or anti‐Myc (Biomed Diagnostics) antibodies at 4 °C for 2 h. Protein‐A agarose beads (40 μl) (KPL) were added to the lysates and incubated with shaking for 1 h at 4 °C. The beads were collected by centrifugation and washed for three times with RIPA buffer (150 mM NaCl, 1% NP‐40, 0.5% sodium deoxycholate, 0.05% SDS, and 50 mM Tris–HCl, pH 8.0). Proteins binding to the beads were eluted by adding 2× SDS loading buffer and analyzed by Western blotting with anti‐Flag antibody.
2.5. Expression of GST fusion protein
The SARS‐CoV N protein was cloned into pGEX‐5X‐1 (Amersham Pharmacia Biotech) and expressed as GST‐N fusion protein in E. coli BL21 cells. Both GST‐N and GST alone were purified by affinity chromatography using glutathione–Sepharose 4B (Amersham Pharmacia Biotech).
2.6. Determination of cell division
HeLa cells grown on coverslips were transfected with appropriate constructs expressing the N protein or SUMO‐1, and fixed with 4% paraformaldehyde. The number of cells undergoing cell division was determined by counting a total of 300 cells that expressing the green fluorescence protein (GFP). Statistical analysis was performed using Student's t test, and P values less than 0.05 were considered to be statistically significant.
2.7. Indirect immunofluorescence
SARS‐CoV N protein was transiently expressed in HeLa cells grown on 4‐well chamber slides (IWAKI). After rinsing with phosphate‐buffered saline (PBS), cells were fixed with 4% paraformaldehyde for 15 min at room temperature and permeabilized with 0.2% Triton X‐100, followed by incubation with specific antibodies diluted in fluorescence dilution buffer (PBS with 5% newborn calf serum) at room temperature for 2 h. Cells were then washed with PBS and incubated with FITC‐conjugated anti‐rabbit or anti‐mouse secondary antibodies (Dako) in fluorescence dilution buffer at 4 °C for 1 h before mounting.
2.8. Construction of plasmids
Plasmid pcDNA3‐N, which covers the SARS‐CoV N sequence, was constructed by cloning an EcoRI/NotI digested PCR fragment into EcoRI/NotI digested pcDNA3.1(+). The PCR fragment was generated using primers (5′‐CGGAATTCCGATGTCTGATAATG GACCC‐3′) and (5′‐AATAAATAGCGGCCGCTGCCTGAGTTG AATC‐3′). pFlag‐N was created by cloning a PstI/EcoRI digested PCR fragment into PstI/EcoRI digested pKT0‐Flag. The PCR fragment was generated using primers (5′‐AACTGCAGCATGTCTG ATAATGGACCCC‐3′) and (5′‐CGGAATTCCGTTATGCCTGAGTTGAATCAGC‐3′). Plasmid pEGFP‐N was generated by cloning an EcoRI‐ and BamHI‐digested PCR fragment into EcoRI/BamHI digested pEGFP‐N1 (Clontech). The two primers used to generate the PCR fragments are (5′‐CGGAATTCCGATGTCTGATAATGGACCC‐3′) and (5′‐CGGGATCCCGTGCCTGAGTTGA ATCAGC‐3′). Plasmid pGEX‐N was made by cloning a BamHI‐ and EcoRI‐digested PCR fragment into BamHI/EcoRI digested pGEX‐5X‐1 (Pharmacia). The two primers used are (5′‐CGGGATCCCGATGTCTGATAATGGACCC‐3′) and (5′‐CGGAATTCTGCCTGAGTTGAA TCAGC‐3′). The K62A mutant was introduced by two rounds of PCR as described in Liu et al. [16]. All constructs were confirmed by automated nucleotide sequencing.
SUMO‐1 was amplified from human cDNA derived from HeLa cells by PCR with primers (5′‐TATCGGATCCCATGTCTGACCAGGCAAAACC‐3′) and (5′‐CGGATC CTCGAGCTAAACTGTTGAATGACCCCCCGT‐3′). The purified PCR product was digested with BamHI and XhoI and cloned into BamHI/XhoI digested pcDNA3.1(+) to generate pcDNA3‐SUMO‐1. The construct was confirmed by automated nucleotide sequencing.
3. Results
3.1. Expression of SARS‐CoV N protein in bacterial and mammalian cells
To study its biochemical properties and functions in viral replication, virion assembly and virus–host interaction, we cloned and expressed the SARS‐CoV N protein in bacterial and mammalian cells. As seen in Fig. 1B, expression of the protein in E. coli BL21 cells as a GST fusion protein showed the detection of a single band of approximately 70 kDa, representing the GST‐N fusion protein (Fig. 1B, lane 2). The protein could be purified by using the GST resin (Fig. 1B, lane 2). Expression of the N protein tagged with the nine amino acid Flag tag at its N‐terminus in mammalian cells showed the detection of multiple bands instead of a single protein band (Fig. 1B, lane 4). Similar to the recently reported detection of three major isoforms [15], a major protein species of approximately 48 kDa and two slightly less abundant species, which migrate more rapidly than the 48‐kDa band, were detected (Fig. 1B, lane 4). These three isoforms may represent the full‐length and posttranslationally modified forms of the N protein. In addition, three other species with approximately molecular masses of 175, 85, and 82 kDa, which may represent oligomerization of the N protein, were sometimes detectable under reducing conditions (Fig. 1B, lane 4).
3.2. Posttranslational modification of SARS‐CoV N protein by sumoylation
The detection of multiple protein species with a wide range of molecular masses when the N protein was expressed in mammalian cells indicates that the protein may undergo other posttranslational modification, in addition to the known phosphorylation. As N protein does not contain any cysteine residue, the detection of potential oligomers suggests that it may form higher‐order structures through other interactions. Sumoylation is one of the posttranslational modifications that affect the migration of the protein. To detect if the N protein is modified by sumoylation, cells transfected with pFlag‐N were lysed either with a lysis buffer containing two isopeptidase inhibitors, iodoacetamide (IAA, 10 mM) and N‐ethylmaleimide (NEM, 20 mM), or with the Laemmli protein gel loading buffer preheated to 80 °C, and subjected to Western blot analysis with anti‐Flag antibody. As can be seen in Fig. 2 A, in addition to the three major isoforms of N protein that were detected under all conditions, a protein species of approximately 65 kDa was detected in cell lysates prepared with lysis buffer containing IAA and NEM (Fig. 2A, lanes 2, 3, and 5). Occasionally, a band of approximately 55 kDa was also detected in cell lysates prepared with buffer containing IAA and NEM (Fig. 2A, lane 3). Co‐expression of N protein with SUMO‐1 led to the detection of significant more 65 kDa species (Fig. 2A, lanes 2 and 3). The 65‐kDa band was also detected when cells were lysed directly with preheated SDS loading buffer (Fig. 2A, lane 6). During the course of this study, we noted that the 65‐kDa band could also be efficiently detected under nonreducing conditions (Fig. 2A, lanes 5 and 6). Under both reducing and nonreducing conditions, the three major isoforms and the 65‐kDa band are migrating at the same positions. This would allow detection of N proteins in the subsequent co‐immunoprecipitation experiments using SDS–PAGE under non‐reducing conditions. The reason for choosing the non‐reducing conditions in these assays is that the IgG heavy chain would mask the detection of the three major N isoforms under reducing conditions.
Figure 2.
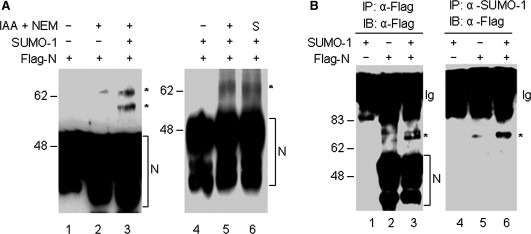
Modification of SARS‐CoV N protein by SUMO‐1. (A) Analysis of sumoylation of N protein by Western blotting. Cell lysates prepared from HeLa cells overexpressing either the Flag‐tagged N protein alone (lanes 1 and 2) or together with SUMO‐1 (lanes 3, 4, 5, and 6) were prepared either in the presence (lanes 2, 3 and 5) or absence (lanes 1 and 4) of the isopeptidase inhibitors IAA and NEM or by direct lysis in preheated SDS loading buffer (lane 6). The polypeptides were separated by SDS–PAGE under either reducing (lanes 1–3) or non‐reducing (lanes 4–6) conditions and analyzed by Western blotting with anti‐Flag antibody. The three major isoforms of N protein are indicated by brackets and the major SUMO‐1 modified form of N protein is indicated by asterisks. Numbers on the left indicate molecular masses in kilodaltons. (B) Analysis of sumoylation of N protein by immunoprecipitation and Western blotting. Total cell lysates were prepared, in the presence of the isopeptidase inhibitors IAA and NEM, from HeLa cells expressing SUMO‐1 (lanes 1 and 4), Flag‐N (lanes 2 and 5), and Flag‐N + SUMO‐1 (lanes 3 and 6), and immunoprecipitated with either anti‐Flag (lanes 1, 2 and 3) or anti‐SUMO‐1 antibody (lanes 4, 5, and 6). The immunoprecipitated proteins were separated by SDS–PAGE under nonreducing conditions and analyzed by Western blotting with anti‐Flag antibody. The major SUMO‐1 modified forms of N protein are indicated by asterisks, and the immunoglobulin is indicated by Ig.
The molecular mass of 65 kDa and its biochemical properties suggest that this species may represent the sumoylation of the N protein. To confirm this possibility further, cell lysates were subjected to immunoprecipitation with either anti‐Flag or anti‐SUMO‐1 antibody, and then analyzed by Western blotting with anti‐Flag antibody. The results showed that anti‐Flag antibody specifically precipitated the three major isoforms of N protein from cells transfected with pFlag‐N (Fig. 2B, lanes 2 and 3). In addition, the 65‐kDa species was also detected (Fig. 2B, lanes 2 and 3). Analysis of the anti‐SUMO‐1 precipitates by Western blotting with anti‐Flag antibody showed that only the 65‐kDa band was detected (Fig. 2B, lanes 5 and 6). Once again, co‐expression of N protein with SUMO‐1 greatly increased the detection of the 65 kDa species (Fig. 2B, lanes 2, 3, 5, and 6). No N protein bands were detected from cells transfected with SUMO‐1 alone (Fig. 2B, lanes 1 and 4). These results confirmed that the 65 kDa band represents the sumoylated N protein.
3.3. Mapping of the sumoylation site on SARS‐CoV N protein
The consensus motif for sumoylation has been defined as a tetrapeptide ΨKXE (where Ψ is usually a hydrophobic residue with exceptions, and X is any amino acid) that surrounds the acceptor lysine in target proteins [23]. Analysis of the N protein sequence showed that it contains 27 lysine residues. One lysine residue at amino acid position 62, K62, lies roughly within the consensus SUMO‐1 modification sequence (GKEE) (Fig. 1A). To determine whether this lysine was responsible for the modification of N protein by sumoylation, it was mutated to an Ala by site‐directed mutagenesis. Proteins extracted from cells transfected with wild‐type N and K62A mutant were immunoblotted with anti‐Flag antibody. As shown in Fig. 3 , similar amounts of the three isoforms of N protein were detected from cells transfected with either wild type or mutant N constructs (Fig. 3, lanes 1 and 2). The 65 kDa sumoylated band was detected from cells transfected with wild‐type N protein only (Fig. 3, lane 1); no 65 kDa sumoylated form was detected from cells expressing the K62A mutant (Fig. 3, lane 2). These results demonstrate that the K62 residue is the major sumoylation site of N protein.
Figure 3.

Mapping of the major sumoylation site on SARS‐CoV N protein. Cell lysates from HeLa cells overexpressing wild type (lane 1) and K62A mutant N protein (lane 2) together with SUMO‐1 were prepared in the presence of IAA and NEM. The polypeptides were separated by SDS–PAGE under nonreducing conditions and analyzed by Western blotting with anti‐Flag antibody. The three major isoforms of N protein are indicated by brackets and the major SUMO‐1 modified form of N protein is indicated by asterisks. Numbers on the left indicate molecular masses in kilodaltons.
3.4. Promotion of homo‐oligomerization of SARS‐CoV N protein by sumoylation
It has been well documented that the ability of viral nucleocapsid protein to interact with itself to form homo‐oligomers is fundamental to the process of viral particle assembly. Recent studies showed that SARS‐CoV N protein exhibits intrinsic properties of self‐interaction [8, 27]. Multimerization of N protein was observed both in vitro and in vivo [8, 27]. To study the effects of sumoylation on the homo‐oligomerization of N protein, cells expressing N protein alone or together with SUMO‐1 were analyzed. As shown in Fig. 4 , Western blot analysis of cells expressing wild type N protein showed the detection of the three major isoforms of N protein and the 65 kDa sumoylated bands (Fig. 4A and B, lanes 1 and 2). In addition, two bands of approximately 85 and 175 kDa were detected (Fig. 4A and B, lanes 1 and 2). Based on their apparent molecular masses, they may represent dimers and tetramers, respectively, of the N protein.
Figure 4.
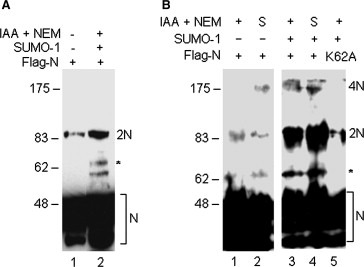
Analysis of the homo‐oligomerization of SARS‐CoV N protein. (A) HeLa cells overexpressing wild type N protein only (lane 1) or together with SUMO‐1 (lane 2) were lysed in the absence (lane 1) or presence of IAA and NEM (lane 2). The polypeptides were separated by SDS–PAGE under reducing conditions and analyzed by Western blotting with anti‐Flag antibody. The three major isoforms of N protein are indicated by brackets and the major SUMO‐1 modified form of N protein is indicated by asterisks. The dimers are also indicated. Numbers on the left indicate molecular masses in kilodaltons. (B) HeLa cells overexpressing wild type N protein only (lanes 1 and 2), wild type + SUMO‐1 (lanes 3 and 4) or the K62A mutant N protein + SUMO‐1 (lane 5) were lysed in the presence of IAA and NEM (lanes 1, 3 and 5) or with preheated SDS loading buffer (lanes 2 and 4). The polypeptides were separated by SDS–PAGE under nonreducing conditions and analyzed by Western blotting with anti‐Flag antibody. The three major isoforms of N protein are indicated by brackets and the major SUMO‐1 modified form of N protein is indicated by asterisks. The dimers and tetramers are also indicated. Numbers on the left indicate molecular masses in kilodaltons.
The effects of sumoylation on the formation of these oligomers were then analyzed by expression of wild type and K62A mutant constructs in cells in the presence of SUMO‐1. The results showed that co‐expression of wild type N protein with SUMO‐1 dramatically increased the detection of the 65‐kDa sumoylated band and the 85‐kDa/175‐kDa oligomers (Fig. 4B, lanes 3 and 4). Co‐expression of the K62A mutant with SUMO‐1, once again, showed no detection of the 65‐kDa sumoylated N protein (Fig. 4B, lane 5). Interestingly, only a trace amount of the 85‐ and 175‐kDa species was detected (Fig. 4B, lane 5). These results suggest that abolishment of the sumoylation of N protein by mutating the K62 sumoylation site significantly decrease homo‐oliogomerization of the protein.
3.5. Further characterization of sumoylation‐mediated homo‐oligomerization of SARS‐CoV N protein
The effect of sumoylation on homo‐oligomerization of N protein was further characterized by two independent co‐immunoprecipitation experiments. First, cells expressing wild type and K62A mutant constructs were lysed with buffer containing IAA and NEM, immunoprecipitated with anti‐Flag antibody. The immunoprecipitated proteins were separated in SDS–PAGE and analyzed by Western blotting with anti‐Flag antibody. As shown in Fig. 5 A, similar amounts of the three isoforms of N protein were detected from cells expressing the wild type and mutant constructs (Fig. 5A, lanes 2 and 3). However, significantly more 85‐kDa dimers were detected in cells expressing the wild type construct than did from cells expressing the mutant (Fig. 5A, lanes 2 and 3).
Figure 5.

Further analysis of the homo‐oligomerization of SARS‐C‐V N protein. (A) HeLa cells overexpressing the empty pKT0‐Flag (lane 1), wild type N protein (lane 2), or the K62A mutant N protein (lane 3) were lysed in the presence of IAA and NEM. The lysates were immunoprecipitated with anti‐Flag antibody. The precipitated polypeptides were separated by SDS–PAGE under nonreducing conditions and analyzed by Western blotting with anti‐Flag antibody. The three major isoforms of N protein, the N protein dimer, and the immunoglobulin are indicated. Numbers on the left indicate molecular masses in kilodaltons. (B) HeLa cells overexpressing the Flag‐tagged wild type N protein alone (lane 1), the c‐Myc‐tagged wild type N protein alone (lane 2), the Flag‐tagged wild type and c‐Myc‐tagged wild type N protein (lane 3), and the Flag‐tagged K62A mutant and c‐Myc‐tagged wild type N protein (lane 4) were lysed in the presence of IAA and NEM. Polypeptides were immunoprecipitated with anti‐Myc antibody, separated by SDS–PAGE under nonreducing conditions, and analyzed by Western blotting with anti‐Flag antibody. The N protein dimer and the immunoglobulin are indicated. A band migrating at approximately 50 kDa, which may represent the antibody heavy chain, is also indicated. (C) HeLa cells overexpressing the Flag‐tagged wild type N protein alone (lanes 1 and 5), the c‐Myc‐tagged wild type N protein alone (lanes 2 and 6), the Flag‐tagged wild type and c‐Myc‐tagged wild type N protein (lanes 3 and 7), and the Flag‐tagged K62A mutant and c‐Myc‐tagged wild type N protein (lanes 4 and 8) were lysed in the presence of IAA and NEM. Polypeptides were separated by SDS–PAGE under nonreducing conditions, and analyzed by Western blotting with either anti‐Flag antibody (lanes 1–4), or anti‐Myc antibody (lanes 5–8). The three major isoforms of N protein and the dimer are indicated.
Second, the Flag‐tagged wild type and mutant N constructs were co‐expressed with a c‐Myc‐tagged wild type N construct. Cells were then lysed with buffer containing IAA and NEM, immunoprecipitated with anti‐Myc antibody. The immunoprecipitated proteins were separated in SDS–PAGE and analyzed by Western blotting with anti‐Flag antibody. As shown in Fig. 5B, the 85‐kDa dimers were readily detected in cells co‐expressing the Flag‐tagged wild type and Myc‐tagged wild type N protein (Fig. 5B, lane 3). Only a trace amount of the 85‐kDa dimers was detected in cells co‐expressing the Flag‐tagged K62A mutant and Myc‐tagged wild type N protein (Fig. 5B, lane 4). In addition, a band migrating at approximately 50 kDa position was consistently detected (Fig. 5B). It might represent the heavy chain of the anti‐Myc antibody used.
To make sure that similar levels of N protein were expressed in the transfected cells, total cell lysates were analyzed by Western blotting with anti‐Flag (Fig. 5C, lanes 1–4) and anti‐Myc (Fig. 5C, lanes 5–8) antibodies. The results showed that the expression levels of flag‐tagged wild type and mutant N protein and the Myc‐tagged N protein are approximately the same (Fig. 5C, lanes 1–4 and 5–8). Interestingly, Western blot analysis of cells co‐expressing Flag‐ and Myc‐tagged wild type N protein with anti‐Flag antibody showed readily detection of the 85‐ and 175‐kDa oligomers (Fig. 5C, lane 3). Analysis of the same cell lysates with anti‐Myc antibody, however, led to much less detection of the two forms (Fig. 5C, lane 7). The two bands were only detectable after prolonged exposure of the gel (data not shown).
3.6. Effects of sumoylation on the subcellular localization of SARS‐CoV N protein
Subcellular distribution of N protein was first studied by cloning and expressing wild type and K62A mutant N protein as a fusion protein with the enhanced green fluorescent protein (EGFP). The plasmid was transfected into HeLa cells and incubated at 37 °C for 36 h. The majority of the SARS‐CoV N‐EGFP fusion protein was observed to be distributed throughout the cytoplasm (Fig. 6 B). A certain proportion of the fusion protein was also observed to be localized to the nucleolus (Fig. 6B). Similar cytoplasmic localization was observed when K62A‐EGFP fusion protein was expressed in HeLa cells (Fig. 6C). However, much less, if any, nucleolar localization was seen in cells expressing this mutant construct (Fig. 6C).
Figure 6.

Subcellular localization of SARS‐CoV N protein. HeLa cells expressing EGFP (A), N‐EGFP (B) and K62A‐EGFP fusion protein (C) were detected directly under the fluorescence microscope at 36 h posttransfection. Indirect immunofluorescent staining of HeLa cells transfected with empty plasmid (D), wild type N protein (E) and K62A mutant (F), was carried out at 18 h posttransfection with rabbit anti‐SARS N antisera and FITC‐labeled goat anti‐rabbit antibodies. The multinucleated cells are indicated by arrows.
The wild type and K62A mutant N protein was then cloned into pcDNA3.1(+) and expressed in HeLa cells. After incubation at 37 °C for 18 h, cells were analyzed by indirect immunofluorescence using rabbit anti‐SARS‐CoV N antiserum, followed by FITC‐labeled goat anti‐rabbit antibody. Similar cytoplasmic localization pattern to the N‐EGFP fusion protein was observed in cells expressing wild type and mutant N protein (Fig. 6E and F). In some cells, wild type N protein exhibited typical nucleolar localization (Fig. 6E). Once again, less obvious nucleolar staining was observed in cells expressing the K62A mutant (Fig. 6F). These results suggest that sumoylation of the N protein might affect its nucleolar localization.
3.7. Nucleolar localization of SARS‐CoV N protein
The nucleolar localization pattern of wild type N protein observed above was confirmed by its colocalization with fibrillarin in the nucleolus. As shown in Fig. 7 , similar cytoplasmic and nuclear localization pattern was observed in cells expressing the Flag‐tagged N protein (A). Immunofluorescent staining with anti‐fibrillarin antibodies showed typical nucleolar staining of both transfected and untransfected cells (Fig. 7B). Interestingly, strong cytoplasmic staining was also observed in cells expressing the N protein using the same antibodies (Fig. 7B). The staining patterns co‐aligned well with the patterns observed with anti‐Flag antibodies (Fig. 7C). These results confirm the nucleolar localization of the N protein, and further demonstrate that N protein may physically interact with fibrillarin, resulting in the retention of fibrillarin in the cytoplasm of cells expressing the N protein.
Figure 7.

Nucleolar localization of SARS‐CoV N protein. Indirect immunofluorescent staining of HeLa cells expressing the Flag‐tagged N protein was carried out at 18 h posttransfection with mouse anti‐Flag (A) and rabbit anti‐fibrillarin (B) antisera. The N protein was then detected by TRITC‐conjugated anti‐mouse secondary antibodies and fibrillarin was detected by FITC‐labeled anti‐rabbit antibodies. (C) represents the merged images. All images were taken using a Zeiss LSM510 META laser scanning confocal microscope.
3.8. Effects of sumoylation on SARS‐CoV N protein‐mediated disruption of host cell division
Similar to other coronavirus N protein, overexpression of SARS‐CoV N protein in mammalian cells disrupts the cell division, as certain proportion of cells expressing the protein at various time points posttransfection were apparently undergoing cell division (Fig. 6). Furthermore, much less multinucleated cells were observed in cells expressing the K62A mutant (Fig. 6). To quantitate the multinucleated cells, HeLa cells transfected with pEGFP, pEGFP‐N, pEGFP‐N + SUMO‐1, and pEGFP‐N(K62A) + SUMO‐1 were examined under the fluorescence microscope at 24, 36, 48 and 60 h posttransfection, respectively, and the multinucleated cells were counted among 300 cells expressing GFP. As shown in Fig. 8 , over 25% of HeLa cells expressing pEGFP‐N were observed to be undergoing cell division at all time points. The percentages of cells undergoing cell division increased to 31–36% among GFP‐positive cells when pEGFP‐N were co‐expressed with SUMO‐1. However, the percentages of cells undergoing cell division were markedly reduced to 11–15% in cells overexpressing the K62A mutant and SUMO‐1 (Fig. 8), significantly less than (P < 0.05) the multinucleated cells observed when the wild type construct was expressed. In a control experiment, the percentages of multinucleated cells that overexpress GFP only were between 2% and 3%. These results indicate that sumoylation of the N protein may play certain roles in its interference and disruption of host cell division.
Figure 8.

Effects of sumoylation of SARS‐CoV N protein on its interference of host cell division. Percentages of multinucleated cells among HeLa cells expressing EGFP, N‐EGFP, N‐EGFP + SUMO‐1, and K62A‐EGFP + SUMO‐1 were calculated by counting the multinucleated cells among 300 green cells under the fluorescence microscope. The percentages and S.D. are results of three repeated experiments.
4. Discussion
In this study, we showed that, in addition to phosphorylation, the SARS‐CoV N protein was modified by covalent attachment of SUMO to its 62lysine residue. Evidence provided demonstrated that sumoylation may promote homo‐oligomerization of the protein. It may also play certain roles in the N protein‐mediated interference of host cell division.
Sumoylation is a highly regulated process in all eukaryotes, involving in diverse regulatory events, such as nuclear transport, transcriptional regulation, chromosome segregation and cell‐cycle control [18, 23, 30]. Conjugation of SUMO to a protein involves the formation of an isopeptide bond between the C‐terminal glycine of SUMO and the ε‐amino group of lysine residue in the target protein. Although SUMO‐1 has an estimated molecular mass of approximately 12 kDa, earlier studies demonstrated that many SUMO‐conjugated proteins usually have a size increase of approximately 20 kDa after conjugation by one SUMO‐1 molecule. SUMO conjugated proteins are typically unstable and can be rapidly hydrolyzed by SUMO‐1 hydrolase during purification. This modification usually alters or regulates the main function of the target proteins.
Coronavirus N protein is a multi‐functional protein [14, 31]. Among them, the most prominent function of the protein is to wrap up the RNA genome to form RNP and assemble into the nucleocapsid core, due to its RNA‐binding activities and self‐association properties. Studies with other coronavirus N protein have mapped the RNA binding domain to the N‐terminal one third region of the protein [14]. A basic amino acid stretch between amino acids 238 and 293 may be responsible for the RNA binding activities of the coronavirus infectious bronchitis virus N protein [34]. Interactions between N protein and RNA are generally required for encapsidation of viral genomic RNA. It is possible that RNA could promote N–N interactions by neutralizing charge repulsions between the two stretches of basic amino acids. As the K62 residue is located in the putative RNA‐binding domain of the SARS‐CoV N protein, is the reduced homo‐oligomerization of the K62A mutant reported in this study due to its loss of the RNA binding activity? Although we do not know if this mutation could affect the RNA‐binding activities of the protein, co‐expression of the Flag‐tagged K62A mutant with the Myc‐tagged wild type N protein resulted in the detection of remarkably less 85‐kDa dimers than in cells co‐expressing the Flag‐ and Myc‐tagged wild type N protein. As two different antibodies were used in the immunoprecipitation and Western blot analyses, it virtually rules out the possibility that the detection of less 85‐ and 175‐kDa oligomers in Western blot studies involving a single antibody is due to a weaker RNA binding activity of the mutant construct.
Self‐association and homo‐oligomerization are another essential property of the coronavirus N protein. Two recent studies have shown that the S/R‐rich motif and the C‐terminal 209 amino acids are essential for self‐association and multimerization of the SARS‐CoV N protein [8, 27]. The findings in this report indicate that the N protein is capable of forming dimers and higher order multimers. Similar to the equivalent protein of equine arteritis virus and simian hemorrhagic fever virus, SARS N protein does not contain any cysteine in its 422 amino acid residues. This excludes the possibility that disulfide bonds are the primary force that mediates the initial N–N interactions. In fact, disulfide bonds do not appear to be relevant until the virus enters the secretory pathway (ER and Golgi) and/or egresses from the cell, both events occurred following the core particle assembly. Data reported in this study demonstrate that sumoylation of the SARS‐CoV N protein dramatically enhances the homo‐oligomerization of the protein. Promotion of oligomerization of protein by sumoylation has been speculated for a pathogenic protein, Huntingtin [25]. Since self‐association and homo‐oligomerization of N protein are essential for the assembly of nucleocapsid core, it suggests that sumoylation would play an important role in the SARS‐CoV replication cycles. Systematic testing this possibility would rely on the availability of an infectious cloning system, as developed by Yount et al. [33].
The apparent molecular weight of the dimer of the N protein detected in this report is 85 kDa, suggesting that it does not contain the SUMO conjugate. The failure to detect the sumoylated dimer in this study is unexpected, considering that sumoylation was shown to promote dimerization of the N protein. Two possibilities have been considered. First, sumoylation is a highly reversible process. The current data showed that only a small proportion of the N protein was dimerized compared to the monomers, and a certain proportion of the sumoylated dimer may be reversed during sample preparation and detection. The combination of these two factors would hamper the detection of the sumoylated dimers by the approaches used. The second possibility is that the sumoylated N protein may be not directly involved in the formation of dimers and other oligomers. Instead, it may target the N protein to different cellular compartments and facilitate the oligomerization of the N protein. Further studies are required to address these possibilities.
Site‐directed mutagenesis studies mapped the 62lysine residue as a major site for covalent attachment of SUMO to the protein, as the 65‐kDa sumoylated band cannot be detected when the mutant construct was expressed in cells. We do not know if other minor sumoylation sites may exist in the SARS‐CoV N protein. Potential sumoylation at these minor positions would compensate the effect of K62A mutation, and complicate the interpretation of the data generating from functional studies, such as the partial interference of cell division by the K62A mutant N protein. Sumoylation of protein at multiple sites was recently reported for several viral and host proteins. For example, the Epstein–Barr virus Rta protein was shown to be sumoylated at three alternating sites [2]. As the SARS‐CoV N protein contains a total of 27 lysine residues and no any other lysine residue is located in a consensus sequence context for sumoylation, it would be difficult to further define these sites, if any, by a conventional mutagenesis approach.
SARS‐CoV N protein has been found to be translocated to the nucleolus of host cells, resembling other coronavirus N proteins, such as IBV [10], MHV and TGEV [31]. This might be a common strategy for coronaviruses to control both host and viral RNA translation [10]. However, the functional significance and the mechanism by which the N protein translocates to the nucleolus are yet to be determined. Systematic investigation of NuLS of the SARS‐CoV N protein that might target it to the nucleolus will be carried out based on sequence comparison with other coronaviruses. An alternative possibility that may account for the nucleolar localization of a viral protein is its interaction with certain nucleolar antigens such as fibrillarin and nucleolin. This was demonstrated on IBV and MHV N proteins [4]. Sumoylation of certain host proteins can regulate their nucleo‐cytoplasmic shuttling [22, 32]. The K62A mutant exhibits much less nucleolus localization, indicating that sumoylation may regulate the subcellular localization of the SARS‐C‐V N protein.
Acknowledgments
This work was supported by the Agency for Science, Technology and Research, Singapore, and a grant from the Biomedical Research Council (BMRC 03/1/22/17/220), Agency for Science, Technology and Research, Singapore.
Li Frank Qisheng,Xiao Han,Tam James P. and Liu D.X.(2005), Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus, FEBS Letters, 579, doi: 10.1016/j.febslet.2005.03.039
References
- 1. Baric R.S., Nelson G.W., Fleming J.O., Deans R.J., Keck J.G., Casteel N., Stohlman S.A., Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol., 62, (1988), 4280– 4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang L.K., Lee Y.H., Cheng T.S., Hong Y.R., Lu P.J., Wang J.J., Wang W.H., Kuo C.W., Li S.S., Liu S.T., Posttranslational modification of Rta of Epstein–Barr virus by SUMO-1. J. Biol. Chem., 279, (2004), 38803– 38812. [DOI] [PubMed] [Google Scholar]
- 3. Chang R.Y., Brian D.A., cis requirement for N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J. Virol., 70, (1996), 2210– 2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen H., Wurm T., Britton P., Brooks G., Hiscox J.A., Interaction of the coronavirus nucleocapsid with nucleolar antigens and the host cell. J. Virol., 76, (2002), 5233– 5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davies H.A., Dourmashkin R.R., MacNaughton R., Ribonucleoprotein of avian infectious bronchitis virus. J. Gen. Virol., 53, (1981), 67– 74. [DOI] [PubMed] [Google Scholar]
- 6. Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W., Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med., 348, (2003), 1967– 1976. [DOI] [PubMed] [Google Scholar]
- 7. Escors D., Ortego J., Laude H., Enjuanes L., The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol., 75, (2001), 1312– 1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N., Cutts T., Andonov A., Cao J., Booth T.F., Plummer F.A., Tyler S., Baker L., Li X., Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun., 316, (2004), 476– 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hiscox J.A., Wurm T., Wilson L., Cavanagh D., Britton P., Brooks G., The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol., 75, (2001), 506– 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hiscox J.A., The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res., 95, (2003), 13– 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T., Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry, 43, (2004), 6059– 6063. [DOI] [PubMed] [Google Scholar]
- 12. Kim T.W., Lee J.H., Hung C.F., Peng S., Roden R., Wang M.C., Viscidi R., Tsai Y.C., He L., Chen P.P., Boyd D.A.K., Wu T.C., Generation and Characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol., 78, (2004), 4638– 4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med., 348, (2003), 1953– 1966. [DOI] [PubMed] [Google Scholar]
- 14. Lai M.M., Cavanagh D., The molecular biology of coronaviruses. Adv. Virus Res., 48, (1997), 1– 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leung D.T.M., Tam F.C.H., Ma C.H., Chan P.K.S., Cheung J.L.K., Niu H., Tam J.S.L., Lim P.L., Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis., 190, (2004), 379– 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu D.X., Xu H.Y., Brown T.D.K., Proteolytic processing of the coronavirus infectious bronchitis virus 1a Polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J. Virol., 71, (1997), 1814– 1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L., The genome sequence of the SARS-associated coronavirus. Science, 300, (2003), 1399– 1404. [DOI] [PubMed] [Google Scholar]
- 18. Muller S., Hoege C., Pyrowolakis G., Jentsch S., SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol., 2, (2001), 202– 210. [DOI] [PubMed] [Google Scholar]
- 19. Narayana K., Maeda A., Maeda J., Makina S., Characterization of the coronavirus M protein and nuclocapsid interaction in infected cells. J. Virol., 74, (2000), 8127– 8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Riso C., Anton I.M., Enjuanes L., Carrascosa J.L., The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol., 70, (1996), 4773– 4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J., Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science, 300, (2003), 1394– 1399. [DOI] [PubMed] [Google Scholar]
- 22. Salinas S., Briancon-Marjollet A., Bossis G., Lopez M.A., Piechaczyk M., Jariel-Encontre I., Debant A., Hipskind R.A., SUMOylation regulates nucleo-cytoplasmic shuttling of Elk-1. J. Cell Biol., 165, (2004), 767– 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seeler J.S., Dejean A., Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol., 4, (2003), 690– 699. [DOI] [PubMed] [Google Scholar]
- 24. Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E., Unique and Conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol., 331, (2003), 991– 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steffan J.S., Agrawal N., Pallos J., Rockabrand E., Trotman L.C., Slepko N., Illes K., Lukacsovich T., Zhu Y.Z., Cattaneo E., Pandolfi P.P., Thompson L.M., Marsh J.L., SUMO modification of Huntingtin and Huntington's disease pathology. Science, 304, (2004), 100– 104. [DOI] [PubMed] [Google Scholar]
- 26. Stohlman S.A., Baric R.S., Nelson G.W., Soe L.H., Welter L.M., Deans R.J., Specific interaction between coronavirus leader RNA and nucleocapsid protein. J. Virol., 62, (1988), 4288– 4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Surjit M., Liu B., Kumar P., Chow V.T.K., La S.K., The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun., 317, (2004), 1030– 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tahara S.M., Dietlin T.A., Bergmann C.C., Nelson G.W., Kyuwa S., Anthony R.P., Stohlman S.A., Coronavirus translational regulation: leader affects mRNA efficiency. Virology, 202, (1994), 621– 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J., Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol., 84, (2003), 2305– 2315. [DOI] [PubMed] [Google Scholar]
- 30. Wilson V.G., Rangasamy D., Viral interaction with the host cell sumoylation system. Virus Res., 81, (2001), 17– 27. [DOI] [PubMed] [Google Scholar]
- 31. Wurm T., Chen H., Britton P., Brooks G., Hiscox J.A., Localization to the nucleolus is a common feature of coronavirus nucleoproteins and the protein may disrupt host cell division. J. Virol., 75, (2001), 9345– 9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiao Z., Watson N., Rodriguez C., Lodish H.F., Nucleocytoplasmic shuttling of smad1 conferred by its nuclear localization and nuclear export signals. J. Biol. Chem., 276, (2001), 39404– 39410. [DOI] [PubMed] [Google Scholar]
- 33. Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S., Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA, 100, (2003), 12995– 13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou M., Williams A.K., Chung S.I., Wang L., Collisson E.W., The infectious bronchitis virus nucleocapsid protein binds RNA sequences in the 3′ terminus of the genome. Virology, 217, (1996), 191– 199. [DOI] [PubMed] [Google Scholar]


