Abstract
Coronavirus nucleocapsid proteins are basic proteins that encapsulate viral genomic RNA to form part of the virus structure. The nucleocapsid protein of SARS-CoV is highly antigenic and associated with several host-cell interactions. Our previous studies using nuclear magnetic resonance revealed the domain organization of the SARS-CoV nucleocapsid protein. RNA has been shown to bind to the N-terminal domain (NTD), although recently the C-terminal half of the protein has also been implicated in RNA binding. Here, we report that the C-terminal domain (CTD), spanning residues 248–365 (NP248-365), had stronger nucleic acid-binding activity than the NTD. To determine the molecular basis of this activity, we have also solved the crystal structure of the NP248-365 region. Residues 248–280 form a positively charged groove similar to that found in the infectious bronchitis virus (IBV) nucleocapsid protein. Furthermore, the positively charged surface area is larger in the SARS-CoV construct than in the IBV. Interactions between residues 248–280 and the rest of the molecule also stabilize the formation of an octamer in the asymmetric unit. Packing of the octamers in the crystal forms two parallel, basic helical grooves, which may be oligonucleotide attachment sites, and suggests a mechanism for helical RNA packaging in the virus.
Abbreviations: NTD, N-terminal domain containing residues 45–181; CTD, C-terminal domain containing residues 248–365; SARS-CoV, severe acute respiratory syndrome coronavirus; IBV, infectious bronchitis virus; NP270-370, fragment of SARS-CoV nucleocapsid protein containing residues 270–370; 15N-HSQC, 15N-edited heteronuclear single-quantum coherence; SeMet, selenomethionine; MAD, multiwavelengh anomalous dispersion
Keywords: severe acute respiratory syndrome coronavirus, nucleocapsid, RNA-binding domain, oligomerization, viral packaging
Introduction
To protect the genome and to ensure its timely replication and reliable transmission, viruses package their genomic material with specific structural proteins to form a ribonucleoprotein complex known as the nucleocapsid (or capsid). Nucleocapsids contain a large number of copies of the structural protein(s), which often polymerize through a self-assembly mechanism. Some viruses form helical capsids. For some viruses, such as the tobacco mosaic virus, the mechanism of this helical packaging is relatively well understood.1., 2., 3. For others, including the influenza virus and severe acute respiratory syndrome-associated coronavirus (SARS-CoV), the molecular mechanism by which the helical packaging is achieved remains unclear. The interaction between nucleic acid binding and protein oligomerization is central to this problem. High-resolution structures of capsid proteins provide a starting point for elucidation of the packaging mechanism of these clinically important viruses.
Severe acute respiratory syndrome (SARS) is the first infectious disease to emerge in the 21st century, has a fatality rate of about 8% and is caused by a novel SARS-associated coronavirus (SARS-CoV).4., 5. One of the key processes in the assembly of SARS-CoV and other coronaviruses is the packaging of viral RNA. The nucleocapsid (N) protein of SARS-CoV enters the host cell together with the viral RNA and interferes with several cellular processes.6., 7., 8. Some of these processes involve interactions between SARS-CoV N protein and host-cell proteins.9 It has also been demonstrated that the SARS-CoV N protein can bind to DNA in vitro.10 These interactions might have a role in the pathology of SARS. The nucleocapsid protein of SARS-CoV packages the viral RNA to form a helical capsid and is essential for viability. Previous nuclear magnetic resonance (NMR) studies have shown that the SARS-CoV N protein contains two structural domains flanked by disordered segments, as shown in Figure 1(a).11 The two structural domains have characteristics common to all coronavirus N proteins, such as order–disorder profiles and predicted secondary structure. Structural studies of the N-terminal domain (NTD, residues 45–181) of the SARS-CoV N protein have shown that it acts as a putative RNA-binding domain, whereas the C-terminal domain (CTD, residues 248–365) acts as a dimerization domain.12., 13. The recently determined structure of the C-terminal domain fragment containing residues 270–370 (NP270-370) shows a core stabilized by multiple hydrophobic interactions.14 Similar structures to those of the SARS-CoV N protein have also been reported for the NTD and CTD of avian infectious bronchitis virus (IBV) N protein (NTD: residues 19–162 in IBV, analogous to residues 45–181 in SARS-CoV; CTD: residues 219–349 in IBV, analogous to residues 248–365 in SARS-CoV), indicating that these structural arrangements are common among coronaviruses.15., 16.
Figure 1.
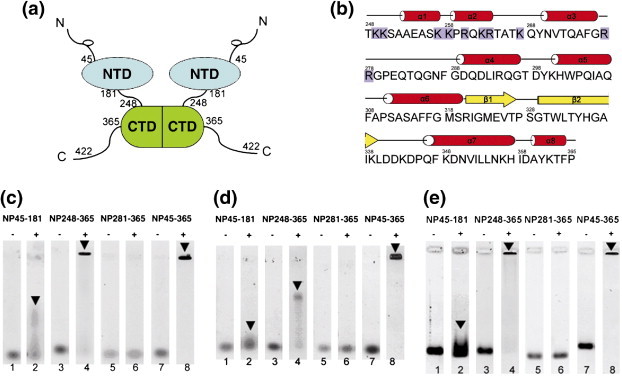
Nucleic acid-binding assay of various SARS-CoV N protein fragments. (a) Schematic of the domain architecture of SARS-CoV NP. NTD: N-terminal domain comprising residues 45–181. CTD: C-terminal dimerization domain comprising residues 248–365. (b) Sequence of the SARS-CoV CTD. The secondary structure elements are shown above the sequence and indicated by red cylinders for α-helices and yellow arrows for β-strands. The positively charged residues within the region 248–280 are shaded in blue. (c) Gel-mobility-shift assay of the 32-mer ssRNA. (+) Lanes have a 16-fold molar excess of protein compared with control (–). Arrows denote shifted bands. (d) Gel-mobility-shift assay of 32-mer ssDNA. Notations are the same as in (c). (e) Gel-mobility-shift assay of 32-mer dsDNA. Notations are the same as in (c). 2 μM of ssDNA or ssRNA in phosphate buffer (10 mM sodium phosphate, 50 mM NaCl, 1 mM EDTA, 0.01% NaN3, pH 7.4) was heated to 95 °C and immediately put on ice to destroy its secondary structure. The oligonucleotides were then mixed with a 16-fold molar excess of various proteins (indicated on the top) and separated on 1% agarose gels.
We have previously shown that SARS-CoV N protein fragments containing the dimerization domain (residues 236–384) could also bind to an RNA packaging signal.17 This suggests that this domain may also have a role in the packaging of SARS-CoV viral RNA. The basic region between residues 248–280 is one of the most positively charged regions of the N protein, and thus represents a likely site for RNA binding, as shown in Figure 1(b). We have shown previously that the 15N-HSQC NMR spectra of the C-terminal domain containing residues 248–365 (CTD) and a shorter fragment containing residues 281–365 (NP281-365) are different, indicating that residues 248–280 form part of the complete dimerization domain structure, although residues 281–365 are sufficient for dimerization.13 Here, we report that the CTD region, which contains both the dimerization core (residues 281–365) and the charge-rich region of the dimerization domain (residues 248–280), is capable of binding to single-stranded RNA (ssRNA), single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) with greater affinities than the NTD. This binding capacity can be abolished by deletion of residues 248–280. To determine the molecular basis of the binding activity, we have solved the X-ray crystal structure of CTD to a resolution of 2.5 Å. The structure shows that residues 248–280 form a positively charged patch, similar to that observed in IBV.15 Unlike the other crystal structures of coronavirus dimerization domains, residues 248–280 also participate in inter- and intramolecular interactions within the NP248-365 crystal, resulting in formation of an octameric asymmetric unit. Molecular packing displays the formation of a helical multimeric core often observed in other virus capsids, which suggests a possible mechanism for the helical packaging of viral RNA by SARS-CoV N protein.18., 19.
Results
Residues 248–280 are necessary for the nucleic acid-binding activity of the CTD
We have shown previously that the C-terminal half of the SARS-CoV N protein can bind a putative packaging signal within the viral RNA.17 However, the precise location of the RNA-binding site within the C-terminal portion has not been identified. To assess the nucleic acid-binding affinity of the C-terminal portion, we conducted gel-shift assays in the presence of a 32-mer stem-loop II motif (s2m) single-stranded RNA (ssRNA) (Figure 1(c)) and its 32-mer ssDNA mimic (Figure 1(d)), using the NTD of SARS-CoV as a positive control.12 s2m ssRNA is a highly conserved sequence among coronaviruses and has been used to map the putative RNA-binding domain of SARS-CoV N protein.12., 20. Significant band shifts were observed for s2m in the presence of both NTD (lane 2 of Figure 1(c)) and CTD (lane 4 of Figure 1(c)). However, s2m bound to the CTD shows a clear shift, whereas binding to NTD shows only a smeared band, indicating that the CTD binds to s2m with higher affinity than NTD. Similar results were observed when NTD and CTD were added to ssDNA (Figure 1(d)), although the shifts are markedly smaller compared with those of s2m. The longer construct NP45-365 shows even higher affinities to both s2m (lane 8 of Figure 1(c)) and ssDNA (lane 8 of Figure 1(d)). NP45-365 includes the NTD, the interdomain linker and the CTD. The stronger affinity observed with this construct indicates that NTD and CTD bind to s2m and ssDNA with increased apparent affinity. Similar results were also observed for binding of these three constructs to dsDNA (Figure 1(e)).
CTD contains ten positively charged residues in the region 248–280; thus, the N terminus of the CTD is highly basic and could be a nucleic acid-binding site. To test this hypothesis, a deletion mutant, NP281-365, was subjected to the same studies as the CTD. This segment is highly structured and retains dimerization activity, indicating that the dimerization core is intact.13 When this fragment was added to ssRNA (lane 6 of Figure 1(c)), ssDNA (lane 6 of Figure 1(d)) or dsDNA (lane 6 of Figure 1(e)), we observed no retardation of the oligonucleotide band. This indicates that all the oligonucleotides bind to the same region of the CTD, residues 248–280. The strong electrostatic character of residues 248–280 and the fact that both single-stranded and double-stranded oligonucleotides bind to CTD strongly indicates that oligonucleotide binding is based on non-specific charge interactions between the positively charged protein and the negatively charged nucleic acid backbone.
Organization of the SARS-CoV CTD octamer in the crystal
The crystal structure of the CTD of SARS-CoV nucleocapsid protein was determined by the multiple-wavelength anomalous diffraction (MAD) method using phasing applied to selenomethionine (SeMet) and refined to 2.5 Å resolution. The diffraction parameters and refinement statistics are shown in Table 1 . Each asymmetric unit consists of an octamer formed by four homo-dimers, denoted I–IV, related by two pseudo 2-fold symmetry (Figure 2 ). The structure of the monomeric subunit consists of eight α-helices and two β-strands (Figure 1(b)), and is in general agreement with previous NMR studies, except for three short helices at the termini (residues 252–257, 259–263 and 360–364) that could not be observed by NMR.13 The root mean square (r.m.s.) deviation of Cα atoms between any two of the eight monomers in the asymmetric unit ranged from 0.52 to 0.80 Å, indicating that the structures of each monomer within an asymmetric unit are similar. Superimposition of individual subunits showed variations in the structure occurring primarily at both termini and the β-hairpin loops. These regions also have higher B-factors.
Table 1.
Data collection and refinement statistics
| Data collection | |||
|---|---|---|---|
| Space group | C2 | ||
| a, b, c (Å), β (°) | 159.42, 84.21, 105.19, 131.17 | ||
| Peak | Inflection | Remote | |
| Wavelength (Å) | 0.9798 | 0.9800 | 0.9645 |
| Resolution (Å) | 2.5 | 2.5 | 2.5 |
| Rmerge (%)a | 8.4 (26.4)b | 11.0 (55.5) | 8.8 (38.7) |
| <I/σ (I)> | 14.18 (3.73) | 10.99 (2.19) | 13.70 (3.36) |
| Completeness | 98.9 (98.2) | 99.6 (98.8) | 99.7 (99.9) |
| Redundancy | 3.2 (3.1) | 3.5 (3.1) | 3.5 (3.4) |
| Refinement statistics | |||
| Resolution (Å) | 30–2.5 | ||
| No. reflections | 31,405 | ||
| Rwork/Rfree (%) | 24.3/25.7 | ||
| No. atoms | |||
| Protein | 7171 | ||
| Water | 858 | ||
| B-factor | |||
| Protein | 28.56 | ||
| Water | 28.86 | ||
| r.m.s deviations | |||
| Bond lenths (Å) | 0.023 | ||
| Bond angles (°) | 2.2 | ||
Rsym = ∑h∑i|Ii–I|∑h∑II, where I is the mean intensity of the i observations of reflection h.
Numbers in parentheses refer to the highest resolution shell.
Figure 2.
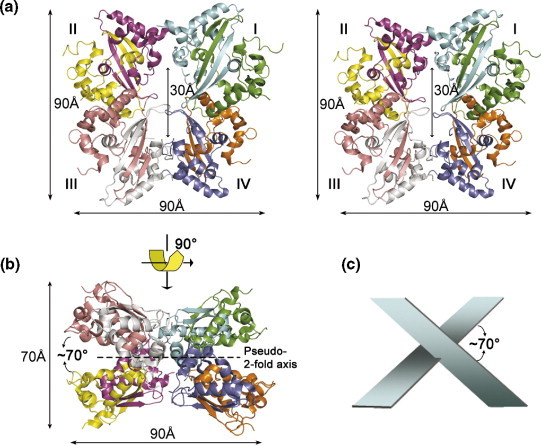
Structural overview of the SARS-CoV CTD octamer. (a) Top view of a stereo-pair of the octamer. There are eight molecules in an asymmetric unit. Each subunit of the octamer is colored differently: A, green; B, cyan; C, magenta; D, yellow; E, pink; F, silver; G, blue; and H, orange. The eight monomers form four tetramers, I–IV, as shown. (b) Side view of the octamer. The pseudo-2-fold axis is indicated by the broken line. (c) Schematic representation of the arrangement between the two tetramers in the octamer shown in (b).
Figure 2(a) shows the stereo-pair of the top view of the octamer in an asymmetric unit and Figure 2(b) shows its side view. The top view of the octamer shows a cylinder-like structure with an outer diameter of ∼90.0 Å and an inner-cavity diameter of ∼30.0 Å. The upper part of the octamer consists of dimers I and II, which contact at an apex to form a butterfly-shaped tetramer. The bottom half of the octamer is also a butterfly-shaped tetramer, formed by dimers III and IV. Viewing from the side, the octamer has the shape of a tilted cross with dimensions of 90 Å × 70 Å (Figure 2(b)). In this orientation, the butterfly-shaped tetramer assumes a rectangular shape and stacks at the bottom of the III–IV tetramer at an angle of ∼70°, as shown schematically in Figure 2(c). The octamer is held together through hydrophobic interactions and hydrophilic contacts among the four dimers. The contact surface areas between pairs of dimers are: ∼1135 Å2 for dimers I and II or III and IV (Figure 3 ); ∼414 Å2 for dimers II and III or I and IV; and ∼120 Å2 for dimers I and III and II and IV. Networks of inter-dimer hydrogen bonds further help stabilize the octamer (Figure 3). The backbone carbonyl of Lys267 forms a hydrogen bond with the side-chain of Arg277, which in turn forms a hydrogen bond with Gln273. An additional inter-dimer hydrogen bond is formed between the side-chains of Gln290 and Arg294. Although the interactions between dimers seem weak when examined individually, the multitude of interactions compensate for the weakness and provide the basis for octamer formation in the crystal.
Figure 3.
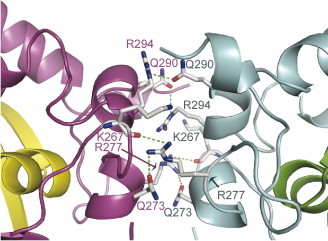
Residues involved in dimer–dimer interactions of the tetramer. Residues belonging to different protomers are labeled with letters color-coded the same as their respective ribbon colors.
The dimer is the building block
The dimer has the shape of a rectangular slab with dimensions of 45 Å × 35 Å × 28 Å and in which the four-stranded β-sheet forms one 45 Å × 35 Å face of the slab and the α-helices form the opposite face (Figure 4 ). The two C termini are located at the diagonal apices on the β-sheet face and the two N termini are located at the center of two opposing 45 Å edges of the slab (Figure 4(a)). The dimerization interface of the CTD is composed of four β-strands and six α-helices, also in general agreement with results from solution NMR analyses.13 Each protomer contributes one β-hairpin and helices α5, α6 and α7 to form the interface. The two β-hairpins form a four-stranded intermolecular β-sheet that is stabilized through extensive hydrogen bonding. The other part of the dimerization interface is composed of helices α5 and α6, where strong hydrophobic interactions involving Trp302, Ile305, Pro310, Phe315 and Phe316 were observed (Figure 4(b)). The dimer is further stabilized by hydrophobic interactions between the longest helix, α7, and the intermolecular β-sheet. Helix α7 is amphipathic and its hydrophobic residues, including Phe347, Val351, Leu354 and Ile358, interact with the hydrophobic side-chains of Ile321, Met323, Thr330 and Leu332 from β1 and β2 of the opposite protomer (Figure 4(c)). In addition, Arg320, which is located in β1, forms a strong intermolecular hydrogen bond with Gln284 and has an important role in dimer formation. Residues 248–270 also have a role in stabilizing the dimer structure through the formation of intra-monomer and intra-dimer hydrogen bonds with the rest of the molecule (Figure 4(d)). The combination of hydrogen bonds and hydrophobic interactions results in a very stable dimer with a buried surface area of ∼5280 Å2. Thus, the dimer structure seems to be the most stable structure in solution, in agreement with previous results.11., 13., 21.
Figure 4.
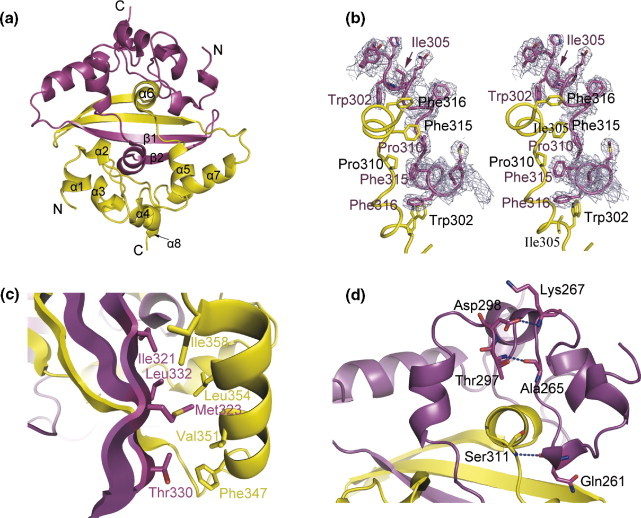
Structural features of the SARS-CoV NP248-365 dimer. (a) Ribbon diagram of the dimer structure of the SARS-CoV CTD. The two monomers are colored in yellow and magenta, respectively. (b) Stereo view of the 2Fo–Fc electron density showing the hydrophobic dimerization interaction between helix α5 and α6. The map is contoured at 1.0σ. (c) Residues involved in hydrophobic interactions between β1, β2 of one protomer, and α7 of the adjacent protomer in a dimer. (d) Ribbon diagram showing the intra-monomer and intra-dimer interactions between residues 248 and 270 and other regions of the dimer. Ala265 and Lys267 form intra-monomer hydrogen bonds with Thr297 and Asp298, whereas Gln261 forms an intra-dimer hydrogen bond with Ser311. These interactions may have a role in stabilizing the secondary structures of residues 248–270.
Structural basis of RNA binding to SARS-CoV NP248-365
We have defined a putative RNA-binding site between residues 248–280 of NP248-365 (shown in Figure 1), which contains a large number of basic amino acids. Electrostatic analysis of the CTD dimer structure reveals a region with significant clustering of positive charges (Figure 5(a)). This clustering of charges is due to the eight positively charged lysine and arginine residues (shown in Figure 1(b)), which are absent from the NP270-370 construct reported by Yu et al.14 (Figure 5(b)). The electrostatic surface is similar to that found in the C-terminal domain of the IBV N protein (Figure 5(c)), but the electrostatic area of the SARS-CoV N protein is markedly larger. This is partly due to the presence of additional negatively charged residues in the N protein of IBV and partly due to the absence of residues 215–218 from the IBV construct; this region contains two lysine residues and can be aligned to residues 248–251 of our SARS construct. Another difference is in the position of Asp298 of the SARS-CoV N protein. In SARS-CoV, this negatively charged residue forms isolated electronegative islets flanking the putative RNA-binding site. The corresponding IBV residue, Asp264, is located in the same region but its negative charge is partially modulated by the presence of a flanking Lys263. By contrast, Asp298 of SARS-CoV N protein is relatively isolated from the other positively charged residues and the two Asp298 residues in the dimer structure of SARS-CoV are ∼30 Å apart, which is comparable to the dimension of dsDNA (23–25 Å in diameter).22 The two Asp298 residues could act as molecular guides to position oligonucleotides in the binding groove in a preferred orientation.
Figure 5.
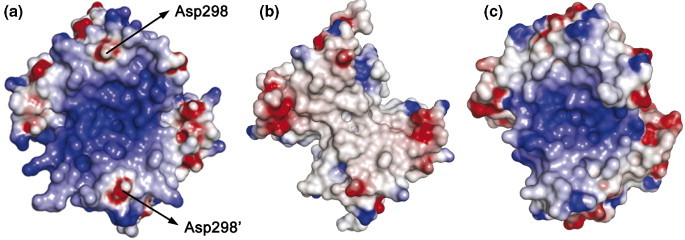
Electrostatic surface potential of the SARS-CoV CTD dimer structure compared with previously published coronavirus dimerization domain structures. Surfaces are colored according to the local electrostatic potential, from −10 kBT−1 (red) to +10 kBT−1 (blue). The orientations are the similar to that shown in Figure 4(a). (a) SARS-CoV CTD. (b) SARS-CoV NP270-370 (PDB ID: 2G1B). Note the absence of the electropositive patch compared with (a). (c) IBV dimerization domain (PDB ID: 2CA1). The relative electropositive region has a smaller area than that of the SARS-CoV CTD shown in (a).
NP248-365 packs in the crystal as a helix
Unlike the N protein of IBV, where multiple packing modes were observed under different crystallization conditions, we can obtain only one crystal form with a single packing mode.15 The crystal packing of SARS-CoV CTD resembles a twin helix formed by translation stacking of octamers (as shown in Figure 2(b)) in the vertical direction (along the b axis of Figure 6(a)). Each octamer is formed by two tetramers, colored yellow and magenta, respectively, wound around each other, as shown schematically in Figure 6(b). The separation between adjacent helices is ∼70 Å. This is a novel architecture that has not previously been reported for coronavirus N protein structures. Surface-potential calculations of the helical supercomplex show two positively charged grooves wound around the helical core (Figure 6(c)). The grooves are mainly formed by the N-terminal residues of NP248-365 and provide continuous potential RNA-binding sites. Each helix has an outer diameter of ∼90 Å and an inner diameter of ∼45 Å, with a pitch of 140 Å, giving the groove a depth of ∼22.5 Å. It also contains an oblong central pore with a long axis of ∼30 Å, as shown in Figure 2(a). The N terminus of one protomer of the dimer is located at the inner base of the groove, whereas the N terminus of the other protomer is located on the outside of the groove. The C termini of the octamer are located in the interfacial regions between adjacent dimers half way in the groove. Coronavirus nucleocapsids have been reported to have a diameter of 9–16 nm with 3–4 nm diameter hollow cores.23 Thus, although the biological significance of this packing mode is still unclear, the dimensions of the helical octamer core reported here are in good agreement with those observed previously. The diameter of the full SARS-CoV nucleocapsid, including the N-terminal RNA-binding domain and disordered regions that are likely to cover the helical superstructure, would also give a total diameter consistent with the recently reported 15 nm diameter of the SARS-CoV ribonucleoprotein complex.24
Figure 6.
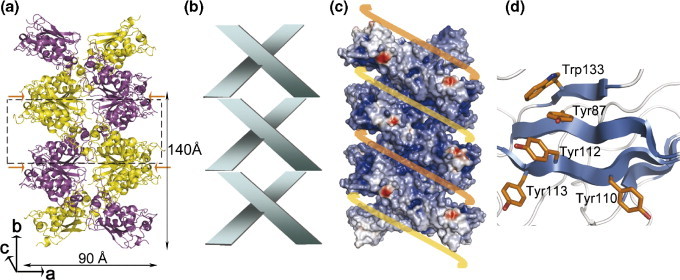
Crystal packing of the SARS-CoV NP248-365 forms a helical supercomplex structure. (a) Ribbon diagram. The asymmetric unit is denoted by the broken box with the arrows pointing at the crystal packing interfaces. (b) Schematic representation of the tetramers in the helical supercomplex shown in (a). (c) Proposed RNA-binding mode. The yellow and orange lines represent two viral RNA strands wrapped around the helical supercomplex nucleocapsid protein. The pitch of the helix corresponds to two octamers with a total height of 140 Å. The electrostatic regions of the helical supercomplex are colored, with positive charges colored blue and negative charges colored red. (d) Putative binding surface of the NTD of SARS-CoV N protein (residues 45–181). Aromatic side-chains are shown in a stick model.
Discussion
SARS-CoV N protein interacts with RNA at multiple sites
Packaging of nucleocapsid involves both specific (sequence-dependent) and non-specific (sequence-independent) binding of the nucleocapsid protein with RNA. Relatively little is known about the specific binding. The non-specific binding is likely to involve the interaction of positively charged residues of the nucleocapsid protein (NP) with RNA. There are three highly positively charged regions in SARS-CoV NP: the SR-rich region (residues 176–204, +6 charges), the N-terminal region of the CTD (residues 248–267, +7 charges) and the C-terminal disordered region (residues 370–389, +7 charges). The SR-rich region is located in the flexible linker region between the two structured domains and no data have reported binding of the SR-rich region to RNA. We have shown here that the CTD of SARS-CoV N protein has strong RNA-binding affinity (Figure 1). The C-terminal disordered region between residues 363 and 382 has also been shown to bind to RNA.25 Interestingly, in the crystal structure the C terminus of the CTD monomer protrudes out of the octamer near the putative RNA-binding groove, placing residues 363–382 in the vicinity of the putative RNA-binding groove and in a favorable position for interaction with the RNA genome. Although the biological significance of the helical packaging reported here is still unclear, the spatial proximity between residues 370–389 and 248–267 indicates that the RNA-binding site may be composed of both regions and that these two regions bind to RNA with increased apparent affinity. The electrostatic nature of the CTD, and probably also residues 370–382, indicates a non-specific binding mode, which could be involved in the packaging of the viral RNA genome.25 The NTD has also been shown to bind to RNA.12 This is confirmed here, and we further showed that NTD and CTD bind to nucleic acid with increased apparent affinity, indicating that more than one region of the nucleocapsid protein is involved in packaging of the RNA genome.
Oligomerization of SARS-CoV N protein
An important property of the coronavirus N protein is its ability to form oligomers. The oligomerization sequences have previously been mapped to residues 168–208 or residues 340–402.25., 26. Here, we observed the formation of an octamer in the asymmetric unit of the CTD crystal, which did not contain these oligomerization sequences. Instead, the stabilization is achieved mostly through the network of interactions involving the N-terminal residues of the CTD. Our previous NMR study at millimolar concentrations also showed that the CTD exists predominantly in the dimeric form.11., 13. However, we also found that the NMR resonances have T2 relaxation times shorter than would be expected for the dimer of 28 kDa, and deuterated CTD was needed to obtain quality spectra from the standard triple-resonance experiments for resonance assignments.13 The CTD is relatively compact, so the rapid transverse relaxation may be due to the rapid dynamic equilibrium between the dimeric form and the small fraction of higher-order oligomers, which cannot be observed due to rapid signal decay. Moreover, the concentration used for crystallization is radically higher than that used in the NMR studies, and the high viscosity of the mother liquor also slows the dynamic fluctuations observed in aqueous solutions. These conditions are conducive to the formation of higher-order structural entities, as observed here. It is interesting to note that the dimer–dimer and tetramer–tetramer interfaces are relatively small, ∼1000 Å2, indicating that the octamer is not a stable form of the CTD, even in the crystal. We should also highlight that the helical packaging of the CTD involves other regions of the N protein in inter-dimeric interactions. This is because the N and C termini of CTD in the crystal are solvent accessible, thereby allowing the extended sequence to interact with the adjacent molecules and stabilize higher-order oligomers. Furthermore, the presence of polynucleotides could induce the formation of oligomers by increasing the local concentration of the protein in solution, thus mimicking a high-concentration environment if the protein can bind to the polynucleotide. In Figure 7(a), we show that CTD does form higher-order oligomeric species when cross-linked with glutaraldehyde in the presence of poly(dT). However, there was little difference in higher-order oligomer formation when the putative RNA-binding region of residues 248–280 was deleted (Figure 7(b)). This indicates that, in the presence of polynucleotides, oligomeric species of CTD can also exist in solution.
Figure 7.
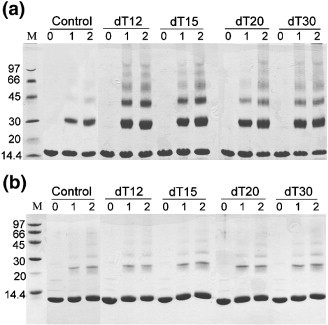
(a) Cross-linking of SARS-CoV CTD with oligonucleotides of different lengths as visualized by SDS-PAGE. Control, without oligonucleotides; dT12, 12-mer poly(dT); dT15: 15-mer poly(dT); dT20, 20-mer poly(dT); dT30, 30-mer poly(dT). Lane 0 contains no cross-linking reagent. Lanes 1 and 2 represent protein cross-linked with 0.01% glutaraldehyde and 0.02% glutaraldehyde, respectively. The sizes of the molecular mass markers (lane M) are indicated on the right in kDa. (b) Cross-linking of SARS-CoV NP281-365 with oligonucleotides of different lengths. The conditions and notations are the same as in (a).
Comparison with other nucleocapsid protein structures
Two coronavirus CTD crystal structures have been published this year.14., 15. In addition to the differences in charge distribution, as discussed in Results, the crystal packing of these two previous structures differs from that observed in our structure. In the crystal structure reported by Yu et al. of a shorter construct spanning residues 270–370 of SARS-CoV N protein (NP270-370), dimers were only observed in the asymmetric unit (PDB ID: 2G1B).14 Comparisons between residues 270–365 of the two structures revealed a r.m.s. deviation of 0.61 Å for all Cα atoms, thus the two monomer structures are practically identical. The difference between these two constructs is the presence of an additional 22 residues at the N terminus and the absence of five residues from the C terminus of our construct. Inspection of the two structures showed that residues 248–269 contain additional structural elements that are crucial for multimerization; these residues are absent from the shorter construct but present in ours. These missing residues could account for the absence of higher-order oligomers from the crystal structure reported by Yu et al. In particular, there are several additional intra-monomer and intra-dimer interactions in the structure of NP248-365 (Figure 4(d)). The backbones of Ala265 and Thr297 are within hydrogen-bonding distance in the same monomer, and another intra-monomer hydrogen bond is formed between the backbone of Lys267 and the side-chain of Asp298. We also observe intra-dimer hydrogen bonds between the backbones of Gln261 and Ser311. Upon oligomerization, these interactions could have a role in stabilizing the secondary structure of residues 248–270, which was not observed in the previous NMR study, and could position these residues to form the inter-dimer contacts. However, although these secondary-structure elements are also present in the crystal structure of IBV N protein C-terminal domain, different ways of association were observed in the asymmetric unit, and none of them formed an octameric arrangement.15 The packing of SARS-CoV N protein CTD forms a contiguous electropositive surface, whereas the positive surface charges in the IBV N protein CTD packing are less clustered and do not form such a contiguous surface. The sequence differences between the SARS-CoV and IBV constructs are most likely to be responsible for this interspecies difference. For example, the side-chain of Arg277 in SARS-CoV N protein has an important role in the formation of inter-dimeric hydrogen bonds. However, the structurally equivalent position in IBV is Pro244, excluding the possibility of hydrogen-bond formation through its side-chain. Another example is the inter-dimeric hydrogen bond between the side-chains of Gln290 and Arg294 in the SARS-CoV N protein. The equivalent residues in IBV are Asp256 and Glu260, respectively. Electrostatic repulsion would deter the formation of any interaction between Asp256 and Glu260 in the IBV N protein. Loss of these inter-dimeric contacts could be the main reason that no higher-order oligomers were observed in the IBV studies.
The structural domains of coronavirus N proteins are well conserved at the sequence level and also at the structural level.11., 14., 15. Residues 248–280 of the SARS-CoV N protein also share marked similarity with other coronavirus N proteins (Figure 8 ). These similar sequences are always located at the N termini of the CTD, and all contain a large number of positively charged residues. The common location and electrostatic profile strongly suggest that these similar sequences are also capable of binding to nucleic acids. The recently reported structure of the C-terminal domain of IBV N protein, which can bind to RNA, supports this hypothesis because a positively charged region consisting of the N terminus of the IBV C-terminal domain is positioned on one side of the dimer.15., 16.
Figure 8.

Sequence alignment of residues 248–280 of SARS-CoV N protein and other coronavirus N proteins. From top to bottom: SARS- CoV (SwissProt: P59595), murine hepatitis virus (MHV) strain 1 (SwissProt: P18446), human coronavirus strain OC43 (HCoV OC43) (SwissProt: P33469), bovine coronavirus strain Quebec (SwissProt: P59712), porcine transmissible gastroenteritis virus (TGEV) strain FS772/70 (SwissProt: P05991) and avian infectious bronchitis virus (IBV) strain Gray (SwissProt: P32923). Positive residues are colored red and negative residues are colored blue.
Interestingly, the architecture of the SARS-CoV N protein CTD resembles that of the N protein of the porcine reproductive and respiratory syndrome virus (PRRSV). PRRSV N protein consists of 123 amino acid residues, is similar in length to the SARS-CoV CTD (118 residues), and also has a capsid-forming C-terminal half and a highly flexible N-terminal half, which presumably binds to RNA.27., 28. The C-terminal half forms an intertwined fold similar to the dimerization core of SARS-CoV N protein, whereas the N-terminal half contains several positively charged residues. The structure of the full-length PRRSV N protein has not yet been determined; however, the structure of the C-terminal capsid-forming region closely resembles that of the dimerization core of CTD. The architectural concept of an RNA-binding region followed by a dimerization core seems to be a common theme between the SARS-CoV N protein CTD and the PRRSV N protein, and by extension between coronavirus and arterivirus N proteins. Coronaviridae and Arteriviridae are both members of order Nidovirales and share common evolutionary roots. Although the full-length N proteins of the two families vary in length and protein sequence, it is possible that certain functional zones have been structurally conserved in both families, such as those of SARS-CoV N protein CTD and the PRRSV N protein. Therefore, the coronavirus N protein could be viewed as an extension of the arterivirus N protein, with additional modules (domains) attached to perform other functions.
Implication for helical capsid formation in coronaviruses
Coronaviruses form helical capsids that are resistant to RNase owing to the binding of the N protein with viral RNA. Within the crystal, the SARS-CoV N protein CTD forms a helical arrangement with a continuous binding surface that could potentially allow the RNA to bind to it through electrostatic interactions, as schematically shown in Figure 6(c). In this model the RNA molecule would wind around the outside of the helical core with the phosphate backbone lying deep inside the groove and the bases exposed to the solvent. One problem with this possibility is the susceptibility of the RNA to hydrolysis, because the RNA would now be wound around the outside of the helical core and the bases would be exposed. Examination of the sequence of the NTD and the unique domain architecture of the SARS-CoV NP suggests how the virus could overcome such a problem. The NTD contains an unusually high proportion of aromatic groups, such as Tyr87, Tyr88, Trp109, Tyr110, Phe111, Tyr112, Tyr113 and Trp133. Many of these aromatic residues are conserved in coronaviruses and it has been proposed that these aromatic residues may stabilize the RNA bases through stacking interactions.29 Inspection of the structure of the NTD (PDB ID: 1SSK) found that most of the conserved aromatic groups are located on the same exposed protein surface and arranged in such a way as to favor intercalation with a sequence of four consecutive bases (Figure 6(d)). Stacking of these aromatic rings with the bases has also being suggested for IBV.16 The long, flexible linker region between the two structured domains may function as a swing arm and allow the protruding NTD to wrap back and bind the RNA through stacking interactions between the aromatic groups and the RNA bases. Indeed, the area containing the conserved aromatic groups in the SARS-CoV N protein NTD has been identified as the RNA-binding site by Huang et al., and this is in agreement with the proposed role in stabilizing the RNA bases.12 As shown in Figure 1, the longer NP two-domain fragment containing both the NTD and CTD had the greatest nucleotide-binding affinity, indicating that the two domains bind with increased apparent affinity to the oligonucleotides, possibly by interacting with different parts of the nucleic acid, which would be expected if NTD interacted with the bases.
In conclusion, we have identified an additional RNA-binding site in the C-terminal domain of SARS-CoV N protein. We found that residues 248–280 have a key role in the RNA binding and oligomerization of the protein, thus linking these two activities within a single structural domain. A model of RNA wrapping around a left-handed twin-helix nucleocapsid protein core is proposed based on the crystal structure of the CTD. Although the structure reported here contains only part of the sequence and the crystal packing may not reflect the true packaging of the structure, it shows features that are consistent with current data and is a good starting point for future studies. Further structure determination of the ribonucleoprotein complex will be required to gain a full understanding of the suprastructure, assembly and packaging of SARS-CoV.
Experimental Procedures
Protein expression and purification
SARS-CoV NP45-181, NP248-365, NP281-365 and NP45-365 were cloned into the pET6H vector as described.13 All clones contained a His-tag (MHHHHHHAMG) at the N terminus. The numbers denote the start and end amino acid number relative to the wild-type protein, excluding the His-tag. The fragments were expressed in Escherichia coli BL21(DE3) cells overnight at 37 °C in Luria-Broth media without inducing agents. Seleno-methionine (SeMet) substituted NP248-365 used for diffraction studies were expressed in E.coli B834(DE3) and grown in modified M9 media containing all amino acids except Met at concentrations of 50 μg/ml, 0.4% (w/v) glucose, 1 mM MgSO4, 4.2 μg/ml Fe2SO4, 1 μg/ml vitamin B mixture (B1, B2, B3, B6, B12), and 50 μg/ml SeMet. Protein purification was performed as reported.13
Gel-shift assay
32-mer s2m ssRNA (5′-CGAGGCCACGCGGAGUACGAUCGAGGGUACAG-3′) was purchased from Dharmacon (Lafayette, CO). Complementary 32-mer ssDNAs (5′-CGAGGCCACGCGGAGTACGATCGAGGGTACAG-3′ and 5′-CTGTACCCTCGATCGTAC TCCGCGTGGCCTCG-3′) were purchased from MDBio (Taipei, Taiwan). 2 μM ssDNA or ssRNA in phosphate buffer (10 mM sodium phosphate, 50 mM NaCl, 1 mM EDTA, 0.01% NaN3, pH 7.4) was heated to 95 °C and immediately put on ice to destroy its secondary structure. The oligonucleotides were then mixed with a 16-fold molar excess of protein and separated on 1% (w/v) agarose gels. Double-stranded DNA was prepared by mixing the two complementary ssDNA at equimolar concentrations, denaturing at 95 °C and renaturing at room temperature. The gels were stained with SYBR Green II dye (Cambrex, ME) in the case of single-stranded oligonucleotides and ethidium bromide for double-stranded oligonucleotides. Visualization was carried out using a UVP BioDoc IT Imaging System (Upland, CA).
Crystallization and data collection
Crystals of SeMet-substituted SARS-CoV NP248-365 were grown at 293 K using the hanging-drop vapor-diffusion method. Crystallization was performed with a 1 μl protein solution (50 mg/ml in 50 mM sodium phosphate (pH 7.4), 150 mM NaCl) mixed with 1 μl reservoir solution containing 30% (w/v) polyethylene glycol 4000, 0.2 M MgSO4, and 0.1 M Tris-HCl (pH 8.0). Plate-like crystals of diffraction quality appeared after four to ten days.
The crystals belonged to space group C2, with cell dimensions a = 159.4 Å, b = 80.2 Å, c = 105.2 Å, and β = 131.2°, and diffracted to 2.5 Å resolution. The structure of the SARS-CoV NP248-365 was determined by MAD phasing applied to the SeMet analogue. The MAD experiments for SeMet-NP248-365 were conducted at the Spring8 BL12B2 Taiwan beamline (Harima, Japan). A single crystal with approximate dimensions of 0.1 mm × 0.3 mm × 0.4 mm was flash-frozen at 110 K. The MAD data were collected at three wavelengths of 0.9798 Å (peak), 0.9800 Å (inflection), and 0.9646 Å (remote). The diffraction data were collected using Quantum 4R CCD (Area Detector System Corporation). All data sets were indexed and processed with the HKL2000 package.30
Structure determination and refinement
SHELX31 was used to locate the selenium sites and generate the initial MAD phase at 3.5 Å. Of a total of 16 selenium sites in one asymmetric unit of SARS-CoV NP248-365, ten sites were located by SHELX. The remaining six selenium sites were found through density modifications and phase extensions with RESOLVE, and a heavy-atom search with CNS.32., 33. To improve the quality of the initial phase, further density modification was performed with XtalView and the final model was built manually using XtalView/Xfit.34 Positional and temperature-factor crystallographic refinements were performed with CNS and REFMAC5.35 The structure was manually rebuilt after each round of refinement. Water molecules were added during the final stages of refinement with CNS. Processing and refinement statistics are summarized in Table 1. The structure contains 858 water molecules with an R-factor of 24.3% for all reflections above 2σ between 30.0 and 2.5 Å resolution, and an R free of 25.7% using 5% randomly distributed reflections. The final structure has good stereochemistry as assessed by PROCHECK.36 There are eight molecules forming an octamer in an asymmetric unit. The octamer structure includes all residues except for the first three to eight residues of the N termini of different subunits and the last three residues of subunits F and G, the electron densities of which could not be observed. All Figures were created with PyMOL (DeLano Scientific) and Swiss-PDB Viewer was used for structural superimpositions.37 The surface potential of SARS-CoV NP248-365 was calculated with GRASP.38
Cross-linking studies
SARS-CoV NP248-365 and NP281-365 were incubated with oligonucleotides of different lengths (12-mer, 15-mer, 20-mer, and 30-mer poly-deoxythymine with 4% oligonucleotide/protein ratio) for 2 h. The final protein concentration was 4.2 mg/ml in 50 mM sodium phosphate (pH 7.4), 150 mM NaCl. The protein/oligonucleotide mixtures were then cross-linked with 0.01% and 0.02% glutaraldehyde at room temperature for 5 min, and the control reactions were cross-linked under the same conditions. The reactions were quenched with 10 mM Tris-HCl and analyzed on 12.5% SDS-PAGE gels.
Multiple sequence alignment
The sequences of coronaviral nucleocapsid proteins were obtained from the SwissProt server. These sequences were aligned with ClustalW v1.83 as described.11
Protein Data Bank accession code
Atomic coordinates have been deposited with the Protein Data Bank, accession code 2cjr.
Acknowledgements
This work was supported in part by the Academia Sinica and by a grant NSC-92-2113-M-001-056 (T.-H.H.), NSC 95-2311-B-001 -066 -MY3 (T.-H.H) and NSC 94-2311-B-001-072 (C.-D.H.) from the National Science Council of the Republic of China. We thank the support staff for assistance in X-ray data collection at the Taiwan beamline BL12B2 (in SPring-8) of the National Synchrotron Radiation Center (NSRRC). The NMR spectra were obtained at the High-field Biomacromolecular NMR Core Facility, National Research Program for Genomic Medicine (NRPGM), Taiwan, Republic of China.
Edited by J. Doudna
References
- 1.Okada Y. Mechanism of assembly of tobacco mosaic virus in vitro. Advan. Biophys. 1975;7:1–41. [PubMed] [Google Scholar]
- 2.Caspar D.L., Namba K. Switching in the self-assembly of tobacco mosaic virus. Advan. Biophys. 1990;26:157–185. doi: 10.1016/0065-227x(90)90011-h. [DOI] [PubMed] [Google Scholar]
- 3.Klug A. The tobacco mosaic virus particle: structure and assembly. Phil. Trans. Roy. Soc. Lond. ser. B. 1999;354:531–535. doi: 10.1098/rstb.1999.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 6.Ng M.L., Tan S.H., See E.E., Ooi E.E., Ling A.E. Early events of SARS coronavirus infection in Vero cells. J. Med. Virol. 2003;71:323–331. doi: 10.1002/jmv.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He R., Leeson A., Andonov A., Li Y., Bastien N., Cao J. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2003;311:870–876. doi: 10.1016/j.bbrc.2003.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surjit M., Liu B., Jameel S., Chow V.T., Lal S.K. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 2004;383:13–18. doi: 10.1042/BJ20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surjit M., Liu B., Chow V.T., Lal S.K. The nucleocapsid protein of sars-coronavirus inhibits the activity of cyclin-CDK complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang T.K., Wu M.P., Chen S.T., Hou M.H., Hong M.H., Pan F.M. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics. 2005;5:925–937. doi: 10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang C.K., Sue S.C., Yu T.H., Hsieh C.M., Tsai C.K., Chiang Y.C. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 13.Chang C.K., Sue S.C., Yu T.H., Hsieh C.M., Tsai C. K., Chiang Y.C. The dimer interface of the SARS coronavirus nucleocapsid protein adapts a porcine respiratory and reproductive syndrome virus-like structure. FEBS Letters. 2005;579:5663–5668. doi: 10.1016/j.febslet.2005.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu I.M., Oldham M.L., Zhang J., Chen J. Crystal structure of the SARS coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between Corona- and Arteri- viridae. J. Biol. Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W., Lescar J., Prasad B.V. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X., Lescar J. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. 2005;13:1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudolph M.G., Kraus I., Dickmanns A., Eickmann M., Garten W., Ficner R. Crystal structure of the borna disease virus nucleoprotein. Structure. 2003;11:1219–1226. doi: 10.1016/j.str.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Martin-Benito J., Area E., Ortega J., Llorca O., Valpuesta J.M., Carrascosa J.L., Ortin J. Three-dimensional reconstruction of a recombinant influenza virus ribonucleoprotein particle. EMBO Rep. 2001;2:313–317. doi: 10.1093/embo-reports/kve063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson M.P., Igel H., Baertsch R., Haussler D., Ares M., Jr, Scott W.G. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 2005;3:e5. doi: 10.1371/journal.pbio.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu I.M., Gustafson C.L.T., Diao J., Burgner J.W., II, Li Z., Zhang J., Chen J. Recombinant severe acute respiratory syndrome (SARS) Coronavirus nucleocapsid protein forms a dimer through its C-terminal domain. J. Biol. Chem. 2005;280:23280–23286. doi: 10.1074/jbc.M501015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickerson R.E., Drew H.R., Conner B.N., Kopka M.L., Pjura P.E. Helix geometry and hydration in A-DNA, B-DNA, and Z-DNA. Cold Spring Harbor Symp. Quant. Biol. 1983;47:13–24. doi: 10.1101/sqb.1983.047.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Laude H., Masters P.S. The coronavirus nucleocapsid protein. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 141–163. [Google Scholar]
- 24.Neuman B.W., Adair B.D., Yoshioka C., Quispe J.D., Orca G., Kuhn P. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo H., Chen J., Chen K., Shen X., Jiang H. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization. Biochemistry. 2006;45:11827–11835. doi: 10.1021/bi0609319. [DOI] [PubMed] [Google Scholar]
- 26.He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;316:476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doan D.N., Dokland T. Structure of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus. Structure. 2003;11:1445–1451. doi: 10.1016/j.str.2003.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doan D.N., Dokland T. Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of the structural domain of the nucleocapsid N protein from porcine reproductive and respiratory syndrome virus (PRRSV) Acta Crystallog. sect. D. 2003;59:1504–1506. doi: 10.1107/s0907444903012721. [DOI] [PubMed] [Google Scholar]
- 29.Ollis D.L., White S.W. Structural basis of protein-nucleic acid interactions. Chem. Rev. 1987;87:981–995. [Google Scholar]
- 30.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Schneider T.R., Sheldrick G.M. Substructure solution with SHELXD. Acta Crystallog. sect. D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 32.Terwilliger T.C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 33.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallog. sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.McRee D.E. XtalView/Xfit–a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 35.Winn M.D., Isupov M.N., Murshudov G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallog. sect. D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallog. 1993;26:283–291. [Google Scholar]
- 37.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 38.Nicholls A., Sharp K.A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins: Struct. Funct. Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]


