Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that negatively regulate gene expression at post-transcriptional level. miRNA dysregulation plays a causal role in cancer progression. In this study, miR-208-3p was highly expressed and directly repressed ARID2 expression. As a result, ARID2 expression in hepatocellular carcinoma (HCC) was decreased. In vitro, miR-208-3p down-regulation and ARID2 over-expression elicited similar inhibitory effects on HCC cell proliferation and invasion. In vivo test results revealed that miR-208-3p down-regulation inhibited HCC tumorigenesis in Hep3B cells. Moreover, ARID2 was possibly a downstream element of transforming growth factor beta1 (TGFβ1)/miR-208-3p/ARID2 regulatory pathway. These findings suggested that miR-208-3p up-regulation is associated with HCC cell progression and may provide a new target for liver cancer treatment.
Keywords: Hepatocellular carcinoma, miR-208-3p, ARID2, Proliferation, Invasion
Highlights
-
•
miR-208-3p was highly expressed and directly repressed the expression of ARID2 in HCC.
-
•
miR-208-3p contributed to HCC cell progression both in vitro and in vivo.
-
•
Over-expression of ARID2 inhibited the HCC cell proliferation and invasion.
-
•
Restoration of ARID2 partly reversed the the effect of miR-208-3p down-regulation on HCC cells.
-
•
Newly regulatory pathway: miR-208-3p mediated the repression of ARID2 by TGFβ1 in HCC cells.
1. Introduction
With a high death rate attributed to high mortality and unsatisfactory treatment options, hepatocellular carcinoma (HCC) is one of the most prevalent malignant cancers worldwide [1], [2], [3]. So effective therapeutic tools, such as gene therapy, should be developed for HCC treatment. For instance, microRNAs (miRNAs) are a class of small non-coding RNAs that regulate gene expression at a post-transcriptional level by binding to complementary sequences in mRNAs [4], [5]. The dysregulation of miRNAs play critical roles in tumor initiation and development. One of the well-known miRNAs is miR-21, which is highly expressed and plays important role in various types of cancers, including colon cancer, prostate cancer, colorectal cancer, and HCC [6], [7], [8], [9]. In contrast, miR-34a, miR-15-/16 cluster, and let-7 miRNA are usually expressed at low levels in different malignant cancers [10], [11], [12], [13]. However, few studies have focused on cancer regulation of miR-208, which promotes cell proliferation of human esophageal squamous cell carcinoma and contributes to cell metastasis and invasion by inducing epithelial-to-mesenchymal transition (EMT) in pancreatic cancer cells [14], [15].
ARID2 is located in chromosome 12q and is composed of 21 exons; this domain is a subunit of a polybromo-associated BRG1-associated factor (PBAF) chromatin-remodeling complex, a switch/sucrose non-fermenting (SWI/SNF) chromatin-remodeling complex involved in ligand-dependent transcriptional activation of nuclear receptors [16]. Human ARID superfamily includes 15 members, which are subunits of SWI/SNF complexes. Human ARID2 protein contains a conservative N-terminal ARID, an RFX-type winged helix, a proline- and glutamine-rich region, and two conservative C-terminal C2H2 Zn-finger motifs [17]. ARID-containing proteins are involved in various biological processes, including embryonic development, cell lineage gene regulation, and cell cycle control. ARID2 is also implicated in tumor progression, as described in previous studies. For example, ARID2 have been found to be mutated and functions as a tumor suppressor gene in HCC [18], [19]. In non-small cell lung cancer and colorectal cancer, loss-of-function mutations of ARID2 have also frequently occurred [20], [21]. Although the crucial roles of ARID2 in cancer have been extensively investigated, the correlation between miR-208-3p and ARID2 in HCC remains unclear.
In this study, miR-208-3p, which is regulated by TGFβ1 at transcriptional level, was highly expressed and inversely correlated with ARID2 expression in HCC. In addition, miR-208-3p down-regulation inhibited HCC cell proliferation and invasion. This down-regulation of miR-208-3p also suppressed tumor growth in nude mice. Morever, ARID2 was confirmed to be regulated by miR-208-3p directly, and the depletion of ARID2 could reverse the effect of miR-208-3p down-regulation in HCC cells. These results indicated the important role of miR-208-3p/ARID2 in HCC and may also provide a basis to develop new and potent therapeutic agents for HCC treatment.
2. Materials and methods
2.1. Cell culture and clinical specimens
HepG2 and Hep3B HCC cell lines were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). HepG2 was maintained in Dulbecco's modified eagle's medium (GIBCO, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS) at 37 °C in 5% CO2. Hep3B was grown in minimum essential medium (GIBCO, Grand Island, NY, USA) containing 10% FBS at 37 °C in 5% CO2. Human HCC specimens (n=20) and paired adjacent control tissues (n=20), with documented informed consent from each case, were obtained from the Southwest Hospital, Third Military Medical University. The information of speciments were listed in Table S1.
2.2. Small interfering RNA (siRNA), mimics, and DNA construction
miRNA inhibitor was purchased from Genewiz (Suzhou, China). miRNA mimics and siRNA used in this study were purchased from GenePharma Co., Ltd. (Shanghai, China). The DNA fragments of full-length ARID2 (5508 bp) were synthesized and cloned in pcDNA3.1 vector by using NotI and XbaI to construct miR-208-3p and ARID2 expression plasmids, respectively. Wild-type 3′-untranslated region (UTR; 156 bp) of ARID2 and mutated 3′-UTR (156 bp) of ARID2 were also synthesized by Genewiz (Suzhou, China) and then separately inserted in pmirGLO dual-luciferase reporter vector (Promega, Madison, WI, USA) by using NheI and SalI, respectively.
2.3. Luciferase reporter assays
Hep3B cells were seeded in a 96-well plate at a final concentration of 1×103 cells per well and maintained at 37 °C in 5% CO2. After 24 h, luciferase reporter plasmids were co-transfected with miRNA expression plasmid or its control vector. The cells were harvested 48 h after transfection; firefly and Renilla luciferase activities were determined using a Dual-Glo® luciferase assay system (Promega, Madison, WI, USA). Firefly luciferase was normalized to Renilla luciferase activity. Transfections were performed thrice.
2.4. RNA extraction and quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNAs were isolated from tissues, and HCC cells were isolated using Trizol (Sigma, St. Louis, MO, USA) according to the manufacturer's instructions. RNAs were used as samples in reverse transcription (RT) by using oligo(dT) or miR-208-3p-specific RT primer. cDNAs were then used to examine ARID2 and miR-208-3p expressions. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 small nuclear RNA (snRNA) were used as controls. Expression levels were quantified using the 2−ΔΔCt method. Each experiment was performed in triplicate. All of the primers used in qRT-PCR are listed in Table 1.
Table 1.
Primers used in this study.
| Primer name | Primer sequence |
|---|---|
| ARID2 siRNA | 5′-AACACUCGAGGUGCUAUACAU-3′ |
| Ctrl siRNA | 5′-GUAUAUAAGCA AGCAUUACUU-3′ |
| miR-208-3p mimics | 5′-AUAAGACGAGCAAAAAGCUUGU-3′ |
| Ctrl mimics | 5′-UUCUCCGAACGUGUCACGUTT-3′ |
| miR-208-3p inhibitor | 5′-ACAAGCTTTTTGCTCGTCTTAT-3′ |
| NC | 5′-ACATACTCCTTTCTCAGAGTCCA-3′ |
| miR-208-3p RT primer | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACACAAGCT-3′ |
| miR-208-3p forward primer | 5′-TGCGGATAAGACGAGCAAAAAG-3′ |
| miR-132 RT primer | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACCGACCAT-3′ |
| miR-132 forward primer | 5′-TGCGGTAACAGTCTACAGCCATG-3′ |
| miR-155 RT primer | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACACCCCTA-3′ |
| miR-155 forward primer | 5′-TGCGGTTAATGCTAATCGTGAT-3′ |
| miR-203 RT primer | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACCTAGTGG-3′ |
| miR-203 forward primer | 5′-TGCGGGTGAAATGTTTAGGAC-3′ |
| U6 snRNA RT primer | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAATATGGAAC-3′ |
| U6 snRNA forward primer | 5′-TGCGGGTGCTCGCTTCGGCAGC-3′ |
| Reverse primer | 5′-CCAGTGCAGGGTCCGAGGT-3′ |
2.5. Cell proliferation assay
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and colony formation assays were performed to determine HCC cell proliferation. For MTT assay, cells were seeded in a 96-well plate at final concentrations of 2×103 cells per well. At 24, 48, and 72 h after transfection, the cells were treated with MTT (0.5 mg/mL) for 4 h at 37 °C. Absorbance was determined at 490 nm. For colony formation assay, cells were seeded in a six-well plate at final concentrations 2×103 cells per well. At 7 d (for Hep3B cells) or 10 d (for HepG2 cells) after transfection, the clones were fixed using methanol and stained with 2% Giemsa solution (Merck, Germany).
2.6. 5-Bromo-2-deoxyuridine (BrdU) labeling and flow cytometry analysis
Cells were seeded in a six-well plate at a final concentration of 2×105 cells per well. Transfected cells were incubated with BrdU at a final concentration of 10 µM for 40 min, stained with Brdu-fluorescein isothiocyanate antibodies and 7-aminoactinomycin D by using BrdU flow kits from BD Pharmingen (San Diego, CA), and analyzed by flow cytometry.
2.7. Cell invasion assay
Transfected cells were seeded in Matrigel-coated transwell cell-culture inserts (Invitrogen Carlsbad, CA, USA) with DMEM/MEM containing 2% FBS. The bottom chamber was filled with DMEM containing 10% FBS. The cells were allowed to shift for 8 h. Afterward, the inserts were washed gently and stained with crystal violet. The passed cells were photographed under a microscope.
2.8. Western blot analysis
Total proteins (20 µg) were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore, MA, USA). The membranes were blocked and incubated with a rabbit polyclonal antibody for ARID2 (Abcam, Cambridge, UK), TGFβ1 (Abcam, Cambridge, UK), and GAPDH (Abcam, Cambridge, UK). The membranes were then incubated with goat horseradish peroxidase-labeled anti-rabbit IgG secondary antibody (CST Inc., Danvers, MA, USA). Proteins were detected by chemiluminescence with enhanced chemiluminescent and Western blot detection reagents (Amersham Biosciences UK, Ltd., Little Chalfont, Buckinghamshire, UK); the detected proteins were then exposed to a chemiluminescent film.
2.9. Tumorigenesis assay in nude mice
Tumorigenic potentials of the cells were examined using five-week-old to six-week-old BALB/c nude mice purchased from the Animal Center of the Chinese Academy of Science (Shanghai, China). The mice were randomly assigned to one of the four groups (8 mice per group) and maintained under pathogen-free conditions. Cells (1×107 cells in 100 μL PBS) were injected subcutaneously to the nude mice. The nude mice were monitored at an interval of 3 d for the appearance of tumors. The mice were sacrificed 28 d after injection, and tumor sizes were measured. Tumor size was calculated using the following equation: tumor size=width2×length×0.5.
2.10. Statistical analysis
Statistical analyses were performed using SPSS® (SPSS Inc., Chicago, IL, USA). All data were expressed as mean±standard deviation. Student's t-test or one-way ANOVA test was performed to determine significant differences. A P-value<0.05 was considered statistically significant.
3. Results
3.1. High miR-208-3p expression directly decreased ARID2 expression in HCC
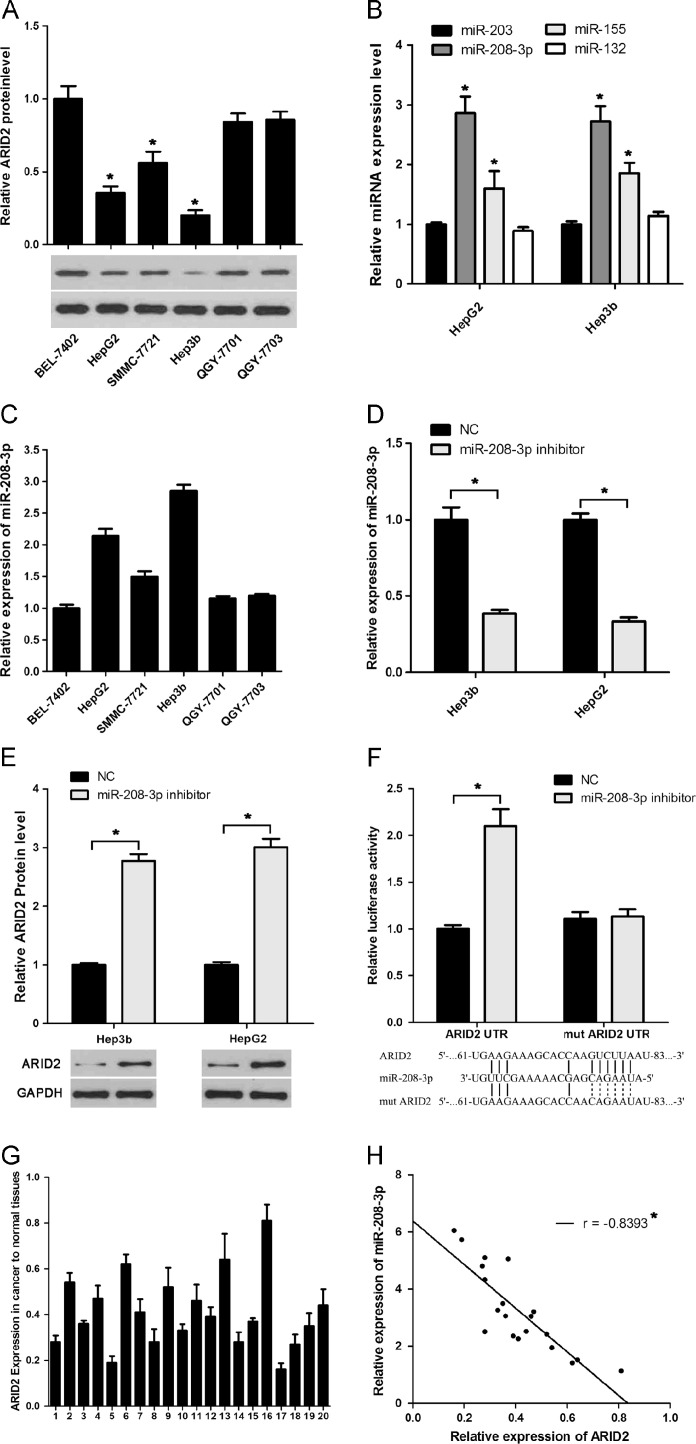
To determine the miRNA that regulates ARID2 expression, we selected four miRNAs, miR-203, miR-208-3p, miR-132, and miR-155, which contain ARID2 binding sites. Western blot analysis results in HCC cells showed that ARID2 protein level was lower in HepG2 and Hep3B than that in the three other cell lines ( Fig. 1A). qRT-PCR results showed that two cell lines, HepG2 and Hep3b, with high expression level of miR-208-3p. In addition, miR-208-3p expression was higher than those of the three other miRNAs in HepG2 and Hep3B cell lines (Fig. 1B and C). To further confirm the correlation between miR-208-3p and ARID2, we transfected miR-208-3p inhibitor in HepG2 and Hep3B cell lines. qRT-PCR and Western blot results showed that ARID2 protein levels were up-regulated after miR-208-3p was inhibited (Fig. 1D and E). Luciferase reporter assay was then performed to confirm that miR-208-3p directly bound to the 3′-UTR of ARID2 (Fig. 1F). Furthermore, we found that ARID2 mRNA levels were lower in HCC speciments and inversely correlated miR-208-3p expression (Fig. 1G and H). These observations demonstrated that miR-208-3p could directly bind to the 3′-UTR of ARID2 and repress its expression.
Fig. 1.
Highly expressed miR-208-3p is inversely correlated with ARID2 in HCC. (A) ARID2 expression levels in six HCC cell lines were analyzed by Western blot. GAPDH was used as a loading control. (B) Expression of miRNAs predicted to bind to ARID2 was detected by miRNA-specific qRT-PCR of HepG2 and Hep3B cell lines. U6 snRNA was used for normalization. (C) Expression of miR-208-3p was analyzed in 6 HCC cell lines by miRNA-specific qRT-PCR. U6 snRNA was used for normalization. (D, E) miR-208-3p inhibitor was transfected in Hep3B and HepG2 cells; then the expression of miR-208-3p and ARID2 were detected by qRT-PCR (D) and Western blot (E). (F) ARID2 3′-UTR luciferase reporter vector was co-transfected with miR-208-3p inhibitor or NC in Hep3B and HepG2 cells for 48 h, then luciferase activity was analyzed. (G) The expression of ARID2 mRNA in HCC speciments and control speciments were analyzed by qRT-PCR, GAPDH was used for normalization. (H) Relationship of ARID2 and miR-208-3p was analyzed by Pearson’s correlation. The experiments were independently repeated thrice. *P<0.05.
3.2. miR-208-3p down-regulation inhibited HCC cell proliferation and invasion
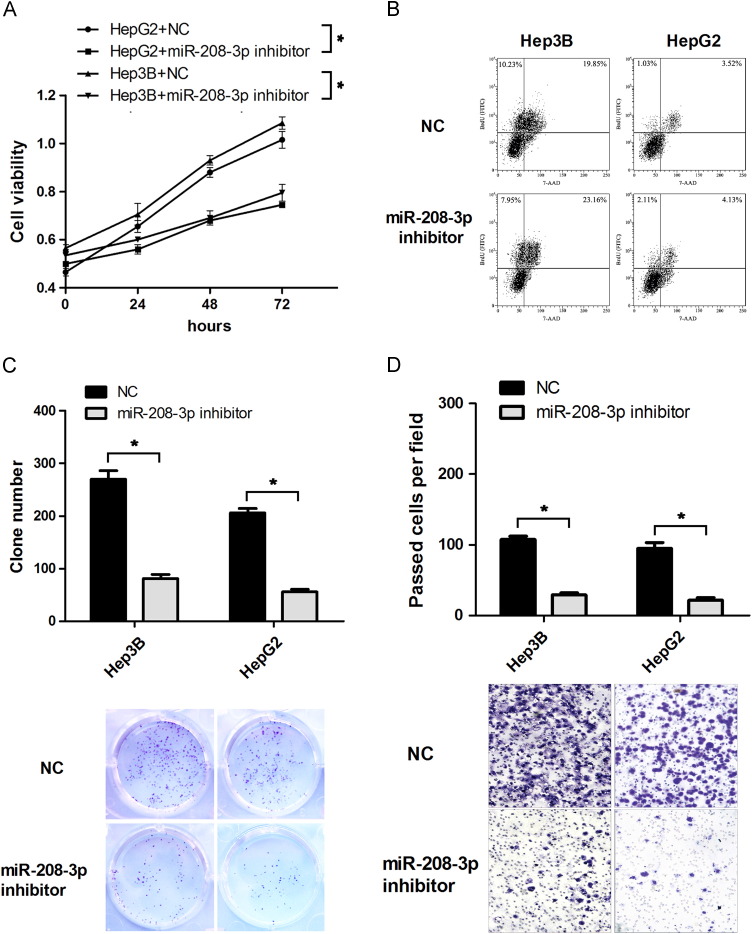
To investigate the function of miR-208-3p dysregulation on HCC, we transfected miR-208-3p inhibitor in HepG2 and Hep3B cell lines. Then MTT, colony formation, transwell, and flow cytometry assays were conducted to analyze the proliferation and the invasion ability of HCC cells. MTT, colony formation assay and flow cytometry analysis results suggested that miR-208-3p down-regulation could repress HCC cell proliferation ( Fig. 2A–C). Transwell analysis results showed that miR-208-3p inhibitor inhibited HCC cell invasion (Fig. 1D).
Fig. 2.
miR-208-3p down-regulation inhibits HCC cell proliferation and invasion. Hep3B and HepG2 cells were transfected with miR-208-3p inhibitor or NC. (A) Cell viability was analyzed by MTT assay at different time points. (B, C) Cell proliferation was analyzed by flow cytometry (B) and colony formation assays(C). (D) The effect of miR-208-3p on HCC cells was analyzed by transwell assay. The experiments were independently repeated thrice. *P<0.05.
3.3. ARID2 over-expression exhibited similar effect to that of miR-208-3p down-regulation in HCC cells
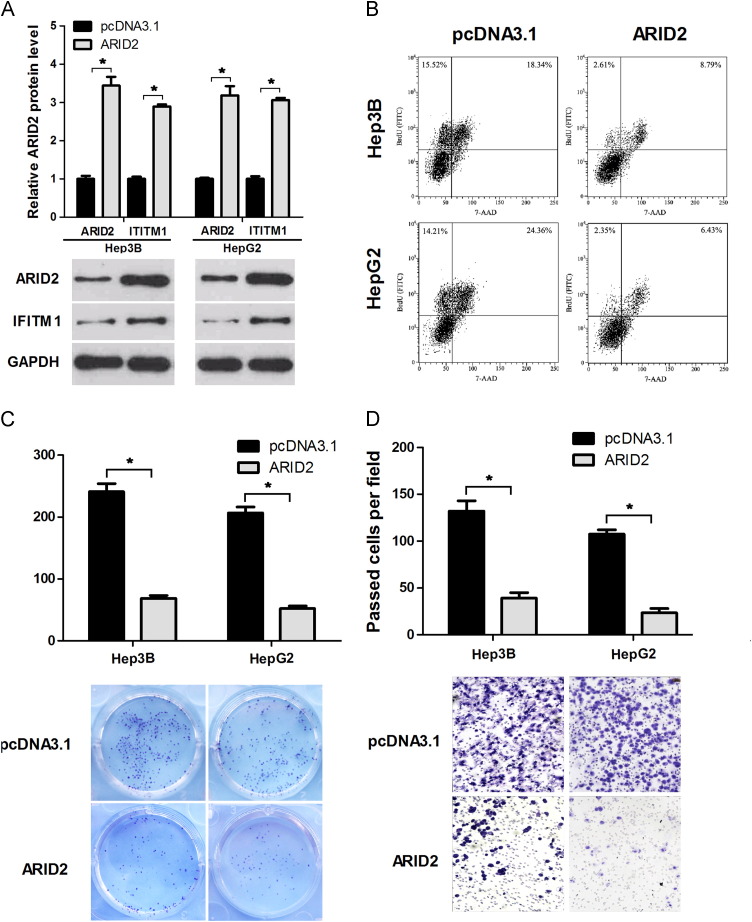
The above results indicated that miR-208-3p directly decreased ARID2 expression and promoted HCC cell progression. Considering that miRNAs are regulatory genes, we speculated that this miR-208-3p effect on HCC cells was possibly exhibited by ARID2. to verify this hypothesis, we transfected ARID2 expression plasmid in HepG2 and Hep3B cells. Results of western blot analysis showed that ARID2 and IFITM1 were up-regulated after transfection of ARID2 expression plasmid ( Fig. 3A). Colony formation and flow cytometry analysis results showed that ARID2 over-expression inhibited HCC cell proliferation (Fig. 3B and C). HCC cell Invasive ability was also inhibited in over-expressed ARID2 groups compared with those in the control group (Fig. 3D).
Fig. 3.
ARID2 ectopic expression inhibits HCC cell proliferation and invasion. ARID2 expression plasmid or pcDNA3.1 empty vector was transfected in Hep3B and HepG2 cells. At 48 h after transfection, the cells were harvested and analyzed. (A) ARID2 and IFITM1 protein level was detected by Western blot; GAPDH was used as a loading control. (B, C) Cell proliferation was analyzed by flow cytometry (B) and colony formation assays(C). (D) Transwell assay was performed to analyze the invasion ability of HCC cells. The experiments were independently repeated thrice. *P<0.05.
3.4. ARID2 restoration partly reversed the effect of miR-208 down-regulation on HCC cells
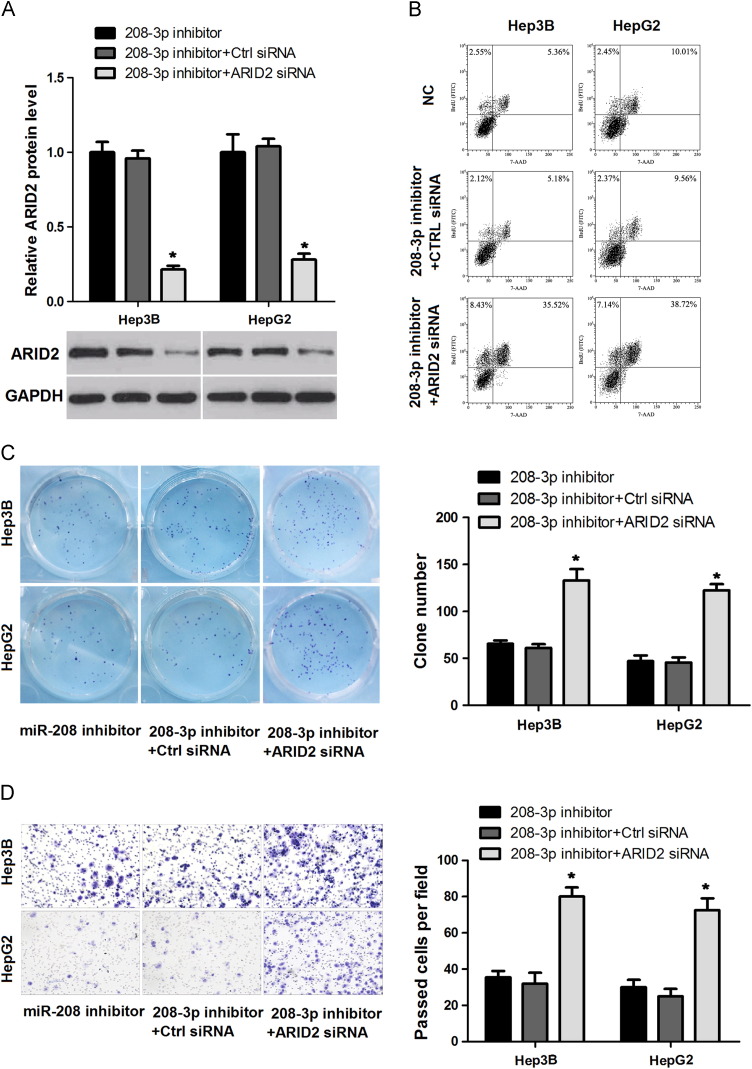
To further confirm the vital role of ARID2 in miR-208-3p regulation network, we negatively regulated ARID2 expression in miR-208-3p inhibitor pretreated HepG2 and Hep3B cells by using siRNA. The transfection of ARID2-specific siRNA decreased ARID2 protein levels compared with that of the control group in miR-208-pretreated HCC cells ( Fig. 4A). Furthermore, we evaluated the effects of ARID2 restoration on cell proliferation and invasion. The results showed that ARID2 knock-down could rescue cell proliferation (Fig. 4B and C) and invasion ability (Fig. 4D) of HCC cells that inhibited by miR-208-3p inhibitor. These data suggested that the effects of miR-208-3p on HCC cells were achieved by targeting ARID2 to a certain degree.
Fig. 4.
Repression of ARID2 expression partly counteracts the effect of miR-208-3p inhibitor on HCC cells. Hep3B and HepG2 cells were transfected with miR-208-3p inhibitor or co-transfected with ARID2 siRNA or control siRNA; ARID2 expression and cell progression were analyzed after 48 h. (A) ARID2 protein levels in HCC cells were detected by Western blot; GAPDH was used for normalization. (B, C) The effects of ARID2 restoration on cell proliferation were evaluated by flow cytometry (B) and colony formation assays (C). (D) HCC cell invasion ability was analyzed by transwell assay. The experiments were independently repeated thrice. *P<0.05.
3.5. miR-208 inhibited HCC tumorigenesis in vivo
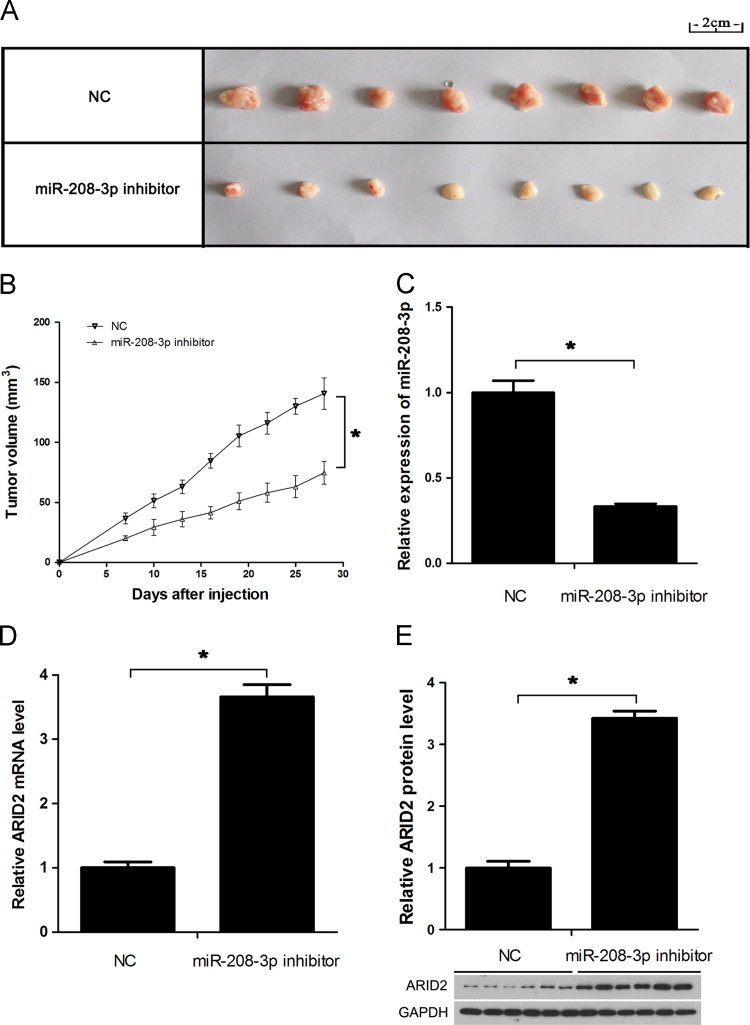
To investigate the role of miR-208-3p in vivo, we injected control or miR-208-3p-inhibitor transfected HepG2 and Hep3B cells subcutaneously into BALB/c nude mice. No solid tumors were detected in HepG2-injected mice. After 28 d, Hep3B-injected mice were sacrificed, and the tumors were removed and photographed. Tumor volumes in mice with low miR-208-3p levels were smaller than those of control group ( Fig. 5A and B). RT-PCR and western blot were performed to analyze the expression of miR-208-3p and ARID2 in 6 pair solid tumors. The results indicated that miR-208-3p expressions were lower in miR-208-3p inhibitor-transfected groups (Fig. 5C), in which ARID2 was over-expressed (Fig. 5D and E). These results suggested that down-regulation of miR-208-3p could suppress HCC tumorigenesis in vivo.
Fig. 5.
miR-208-3p down-regulation inhibits HCC tumorigenesis in nude mice. miR-208-3p inhibitor-transfected Hep3B and HepG2 cells were injected subcutaneously in nude mice at a concentration of 7×106 in 100 µL of PBS. Tumor volume was monitored at an interval of 3 d, and the mice were sacrificed 28 d after injection. There were no obvious tumor in HepG2 cells injection groups. (A) Images of dissected tumors from nude mice were photographed. (B) Tumor growth curve was analyzed in all the mice that injected with Hep3B cells. (C) miR-208-3p expression levels in 6 pair solid tumors were analyzed by qRT-PCR. (D, E) ARID2 mRNA and protein levels in 6 pair solid tumors were analyzed by real-time qRT-PCR (D) and Western blot (E). U6 snRNA or GAPDH was used as control. *P<0.05.
3.6. miR-208-3p mediated the repression of ARID2 by TGFβ1 in HCC cells
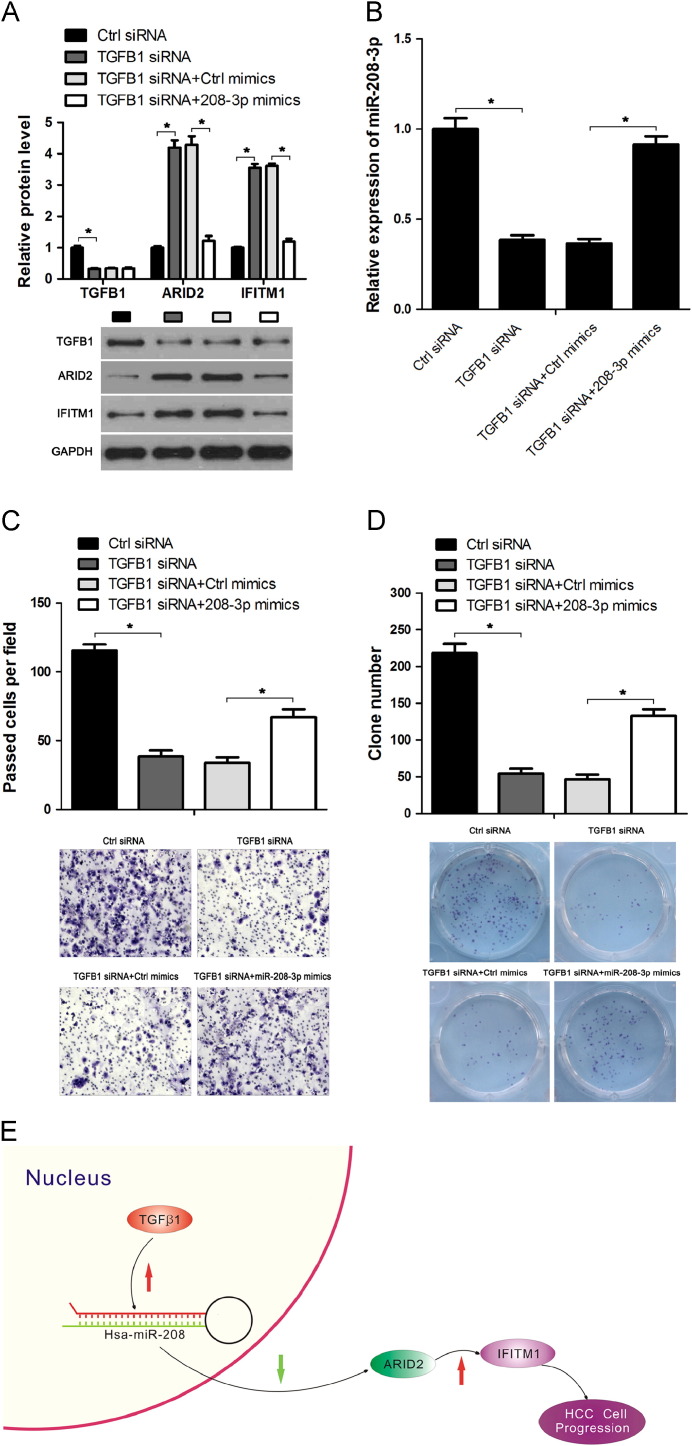
TGFβ1-specific siRNA was transfected into Hep3B cells separately or co-transfected with miR-208-3p mimics or control mimics to confirm whether ARID2 is a downstream response element of TGFβ1/miR-208-3p regulatory network. Western blot analysis was performed to analyze TGFβ1, ARID2 and IFITM1 protein levels, and qRT-PCR was conducted to detect miR-208-3p expression. The transfection of TGFβ1-specific siRNA could repress TGFβ1 expression, repress miR-208-3p expression, and upregulate the protein level of ARID2 and IFITM1, This effect of TGFβ1 siRNA on miR-208-3p, ARID2 and IFITM1 could be reversed by miR-208-3p mimics ( Fig. 6A and B). The results of transwell and colony formation assay indicated that depeted TGFβ1 inhibited HCC cell invasion and colony formation ability, but the over-expression of miR-208-3p could reverse the effect of TGFβ1 down-regulation on HCC cells (Fig. 6C and D). These data suggested a new regulatory pathway, in which ARID2 is regulated by TGFβ1 via miR-208-3p (Fig. 6E).
Fig. 6.
ARID2 is a downstream response element of TGFβ1/miR-208-3p/ARID2 regulatory network. Hep3B cells were transfected with TGFβ1 siRNA or co-transfected with miR-208-3p mimics or control mimics. 48 h after transfection, cells were harvested and analyzed. (A) Western blot were performed to detect TGFβ1, ARID2 and and IFITM1 protein levels, GAPDH was used as loading control. (B) miR-208-3p expression levels were analyzed by qRT-PCR, U6 snRNA were used as loading control. (C and D) HCC cell invasion and proliferation ability were evaluated by transwell (C) and colony formation assays (D). (E) A new miR-208-3p-mediated TGFβ1/miR-208-3p/ARID2 regulatory pathway in HCC. In this regulatory network, miR-208-3p was firstly activated by TGFβ1, then miR-208-3p directly targeting ARID2 and repressed its expression. In HCC cells, both TGFβ1 and miR-208-3p were expressed at a high level, which leaded to the ARID2 down-regulation. The dysregulation of ARID2 finally resulted in the promotion of HCC cell progression via IFITM1. *P<0.05.
4. Discussion
In the recent years, more and more researchers focused on the selection of potent therapeutic miRNAs for cancer treatment. miR-122a is minimally expressed and is inversely correlated with cyclin G1 expression in HCC [22]. In addition, miR-124 and miR-203 are down-regulated and cell growth is inhibited by regulating multiple targets during hepatocarcinogenesis [23]. By contrast, miR-181b, a TGFβ-activated miRNA is up-regulated and can promote HCC cell invasion and proliferation by targeting TIMP3 [24]. Moreover, miR-21 and miR-17-92 exhibit increased expressions in HCC, and cell proliferation is significantly reduced when these two miRNAs were knocked down [9]. Previous studies showed the role of miR-208 in myocardial injury, EMT, and cancer biological regulation [14], [15], [25]. However, limited information is available regarding the role of miR-208-3p in HCC. In this study, high levels of miR-208-3p were detected in HCC speciments compared to that in paired normal hepatic tissues. In HepG2 and Hep3B cells, down-regulation of miR-208-3p inhibited HCC cell proliferation and invasion. In addition, the decreased miR-208-3p inhibited HCC tumorigenesis in nude mice. Furthermore, miR-208-3p expression was inversely correlated with ARID2 expression level in HCC specimens.
To investigate the potential association of miR-208-3p and ARID2, we performed luciferase reporter and Western blot assays. The results showed that miR-208-3p repressed ARID2 expression and directly targeted ARID2 by binding to its 3′-UTR. ARID2 undergoes mutation and functions as a tumor suppressor gene in small cell lung carcinoma and HCC [19], [20]. In this study, ARID2 expression was down-regulated, and ARID2 over-expression inhibited HCC cell proliferation and invasion. In addition, ARID2 expression could partly reverse the effect of miR-208-3p down-regulation on HCC cell progression. As a subunit of PBAF, ARID2 was also found to regulate the expression of interferon induced transmembrane protein 1 (IFITM1) [16]. IFITM1 is an IFN-induced antiviral protein which prevent multiple virus entry, including SARS coronavirus (SARS-CoV), Ebola virus (EBOV), Dengue virus (DNV), human immunodeficiency virus type 1 (HIV-1) and hepatitis C virus (HCV) [26], [27], [28], [29], [30]. IFITM1 was also found to regulate cancer cell proliferation and invasion. For example, in Head and neck squamous cell carcinoma (HNSCC) cells, IFITM1 overexpression promoted and IFITM1 knockdown suppressed the invasion of HNSCC cells in vitro [31]. While, IFITM1 could enhance the transcriptional activity of p53 and negatively regulated HCC cell cell proliferation and tumorigenesis [32]. miR-208 was shown to be regulated by TGFβ1 in rat cardiac myoblasts [33]. Transforming growth factor β (TGFβ) signaling pathway is a double-edged sword in tumor development. As a member of TGFβ family, dual-role of TGFβ1 has also been found in cancer progression. In metastatic prostate cancer cells, TGFβ1 was found to suppressed the growth and motility by inducing SMADs [34]. While in HCC, over-expression of TGF-β1 could promote cell progression and Hepatocarcinogenesis [35], [36]. Here we found that depletion of TGFβ1 by siRNA repressed miR-208-3p, upregulated ARID2 and IFITM1 expression. This up-regulation of ARID2 and IFITM1 were down-regulated by transfection of miR-208-3p mimics in TGFβ1 siRNA and miR-208-3p mimics co-transfection group. Furthermore, the inhibition of HCC cell invasion and colony formation ability caused by TGFβ1 siRNA were partly reversed by miR-208-3p mimics. These results may provide insights into ARID2 regulatory network in cancer.
In summary, this study demonstrated the importance of miR-208-3p and ARID2 dysregulation in HCC. We found the important role of TGFβ1-activated miR-208-3p in HCC cell progression by targeting ARID2 in vitro and in vivo. The newly identified TGFβ1-miR-208-3p-ARID2 regulatory pathway in HCC may be used to understand HCC; this pathway may also provide a new and potential therapeutic target for cancer treatment.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.yexcr.2015.07.008.
Appendix A. Supplementary material
Supplementary material
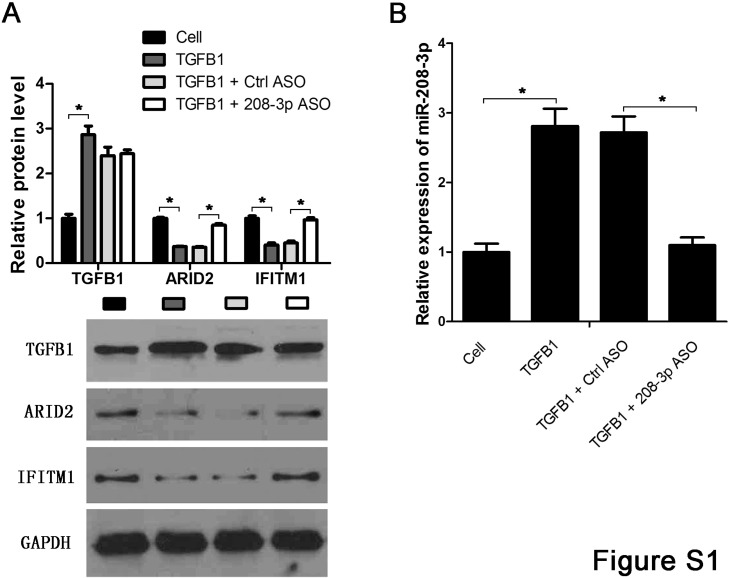
Fig. S1.
References
- 1.Worns M.A., Galle P.R. HCC therapies – lessons learned. Nat. Rev. Gastroenterol. Hepatol. 2014;11:447–452. doi: 10.1038/nrgastro.2014.10. [DOI] [PubMed] [Google Scholar]
- 2.Quetglas I.M., Moeini A., Pinyol R., Llovet J.M. Integration of genomic information in the clinical management of HCC, Best practice & research. Clin. Gastroenterol. 2014;28:831–842. doi: 10.1016/j.bpg.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathew S., Ali A., Abdel-Hafiz H., Fatima K., Suhail M., Archunan G., Begum N., Jahangir S., Ilyas M., Chaudhary A.G., Al Qahtani M., Mohamad Bazarah S., Qadri I. Biomarkers for virus-induced hepatocellular carcinoma (HCC) Infect. Genet. Evol. 2014;26:327–339. doi: 10.1016/j.meegid.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Lai E.C., Wiel C., Rubin G.M. Complementary miRNA pairs suggest a regulatory role for miRNA:miRNA duplexes. RNA. 2004;10:171–175. doi: 10.1261/rna.5191904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llave C., Xie Z., Kasschau K.D., Carrington J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 6.Kjaer-Frifeldt S., Hansen T.F., Nielsen B.S., Joergensen S., Lindebjerg J., Soerensen F.B., dePont Christensen R., Jakobsen A. The prognostic importance of miR-21 in stage II colon cancer: a population-based study. Br. J. Cancer. 2012;107:1169–1174. doi: 10.1038/bjc.2012.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribas J., Lupold S.E. The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle. 2010;9:923–929. doi: 10.4161/cc.9.5.10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong B., Cheng Y., Ma L., Zhang C. MiR-21 regulates biological behavior through the PTEN/PI-3K/Akt signaling pathway in human colorectal cancer cells. Int. J. Oncol. 2013;42:219–228. doi: 10.3892/ijo.2012.1707. [DOI] [PubMed] [Google Scholar]
- 9.Connolly E., Melegari M., Landgraf P., Tchaikovskaya T., Tennant B.C., Slagle B.L., Rogler L.E., Zavolan M., Tuschl T., Rogler C.E. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am. J. Pathol. 2008;173:856–864. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C., Kelnar K., Liu B., Chen X., Calhoun-Davis T., Li H., Patrawala L., Yan H., Jeter C., Honorio S., Wiggins J.F., Bader A.G., Fagin R., Brown D., Tang D.G. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bandi N., Vassella E. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol. Cancer. 2011;10:55. doi: 10.1186/1476-4598-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akao Y., Nakagawa Y., Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 13.Liu C., Kelnar K., Vlassov A.V., Brown D., Wang J., Tang D.G. Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res. 2012;72:3393–3404. doi: 10.1158/0008-5472.CAN-11-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Zheng D., Zhang B., Liu L., Ou J., Chen W., Xiong S., Gu Y., Yang J. Mir-208 promotes cell proliferation by repressing SOX6 expression in human esophageal squamous cell carcinoma. J. Transl. Med. 2014;12:196. doi: 10.1186/1479-5876-12-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu A., Shao C., Jin G., Liu R., Hao J., Song B., Ouyang L., Hu X. miR-208-induced epithelial to mesenchymal transition of pancreatic cancer cells promotes cell metastasis and invasion. Cell Biochem. Biophys. 2014;69:341–346. doi: 10.1007/s12013-013-9805-3. [DOI] [PubMed] [Google Scholar]
- 16.Yan Z., Cui K., Murray D.M., Ling C., Xue Y., Gerstein A., Parsons R., Zhao K., Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohrmann L., Langenberg K., Krijgsveld J., Kal A.J., Heck A.J., Verrijzer C.P. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 2004;24:3077–3088. doi: 10.1128/MCB.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M., Zhao H., Zhang X., Wood L.D., Anders R.A., Choti M.A., Pawlik T.M., Daniel H.D., Kannangai R., Offerhaus G.J., Velculescu V.E., Wang L., Zhou S., Vogelstein B., Hruban R.H., Papadopoulos N., Cai J., Torbenson M.S., Kinzler K.W. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H., Wang J., Han Y., Huang Z., Ying J., Bi X., Zhao J., Fang Y., Zhou H., Zhou J., Li Z., Zhang Y., Yang X., Yan T., Wang L., Torbenson M.S., Cai J. ARID2: a new tumor suppressor gene in hepatocellular carcinoma. Oncotarget. 2011;2:886–891. doi: 10.18632/oncotarget.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manceau G., Letouze E., Guichard C., Didelot A., Cazes A., Corte H., Fabre E., Pallier K., Imbeaud S., Le Pimpec-Barthes F., Zucman-Rossi J., Laurent-Puig P., Blons H. Recurrent inactivating mutations of ARID2 in non-small cell lung carcinoma, Int. J. Cancer. 2013;132:2217–2221. doi: 10.1002/ijc.27900. [DOI] [PubMed] [Google Scholar]
- 21.Cajuso T., Hanninen U.A., Kondelin J., Gylfe A.E., Tanskanen T., Katainen R., Pitkanen E., Ristolainen H., Kaasinen E., Taipale M., Taipale J., Bohm J., Renkonen-Sinisalo L., Mecklin J.P., Jarvinen H., Tuupanen S., Kilpivaara O., Vahteristo P. Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. Int. J. Cancer. 2014;135:611–623. doi: 10.1002/ijc.28705. [DOI] [PubMed] [Google Scholar]
- 22.Gramantieri L., Ferracin M., Fornari F., Veronese A., Sabbioni S., Liu C.G., Calin G.A., Giovannini C., Ferrazzi E., Grazi G.L., Croce C.M., Bolondi L., Negrini M. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 23.Furuta M., Kozaki K.I., Tanaka S., Arii S., Imoto I., Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 24.Wang B., Hsu S.H., Majumder S., Kutay H., Huang W., Jacob S.T., Ghoshal K. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29:1787–1797. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montgomery R.L., Hullinger T.G., Semus H.M., Dickinson B.A., Seto A.G., Lynch J.M., Stack C., Latimer P.A., Olson E.N., van Rooij E. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., Longobardi L.E., Boltz D., Kuhn J.H., Elledge S.J., Bavari S., Denison M.R., Choe H., Farzan M. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhl A., Pohlmann S. How Ebola virus counters the interferon system. Zoonoses Public Health. 2012;59(Suppl. 2):S116–S131. doi: 10.1111/j.1863-2378.2012.01454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E., Adams D.J., Xavier R.J., Farzan M., Elledge S.J. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Compton A.A., Bruel T., Porrot F., Mallet A., Sachse M., Euvrard M., Liang C., Casartelli N., Schwartz O. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe. 2014;16:736–747. doi: 10.1016/j.chom.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raychoudhuri A., Shrivastava S., Steele R., Kim H., Ray R., Ray R.B. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J. Virol. 2011;85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatano H., Kudo Y., Ogawa I., Tsunematsu T., Kikuchi A., Abiko Y., Takata T. IFN-induced transmembrane protein 1 promotes invasion at early stage of head and neck cancer progression, Clinical cancer research: an official journal of the American Association for. Cancer Res. 2008;14:6097–6105. doi: 10.1158/1078-0432.CCR-07-4761. [DOI] [PubMed] [Google Scholar]
- 32.Yang G., Xu Y., Chen X., Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2007;26:594–603. doi: 10.1038/sj.onc.1209807. [DOI] [PubMed] [Google Scholar]
- 33.Shyu K.G., Wang B.W., Wu G.J., Lin C.M., Chang H. Mechanical stretch via transforming growth factor-beta1 activates microRNA208a to regulate endoglin expression in cultured rat cardiac myoblasts. Eur. J. Heart Fail. 2013;15:36–45. doi: 10.1093/eurjhf/hfs143. [DOI] [PubMed] [Google Scholar]
- 34.Miles F.L., Tung N.S., Aguiar A.A., Kurtoglu S., Sikes R.A. Increased TGF-beta1-mediated suppression of growth and motility in castrate-resistant prostate cancer cells is consistent with Smad2/3 signaling. Prostate. 2012;72:1339–1350. doi: 10.1002/pros.22482. [DOI] [PubMed] [Google Scholar]
- 35.Factor V.M., Kao C.Y., Santoni-Rugiu E., Woitach J.T., Jensen M.R., Thorgeirsson S.S. Constitutive expression of mature transforming growth factor beta1 in the liver accelerates hepatocarcinogenesis in transgenic mice. Cancer Res. 1997;57:2089–2095. [PubMed] [Google Scholar]
- 36.Benetti A., Berenzi A., Gambarotti M., Garrafa E., Gelati M., Dessy E., Portolani N., Piardi T., Giulini S.M., Caruso A., Invernici G., Parati E.A., Nicosia R., Alessandri G. Transforming growth factor-beta1 and CD105 promote the migration of hepatocellular carcinoma-derived endothelium. Cancer Res. 2008;68:8626–8634. doi: 10.1158/0008-5472.CAN-08-1218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material