Abstract
Dengue virus (DV) infection depends on a step of membrane fusion, which occurs in the acidic environment of the endosome. This process is mediated by virus surface envelope glycoprotein, in which the loop between residues D98–G112 is considered to be crucial, acting as a fusion peptide. Here, we have characterized functionally and structurally the interaction between the DV fusion peptide and different model membranes by fluorescence and NMR. Its interaction was strongest in dodecylphosphocholine (DPC) micelles and anionic phosphatidylcholine/phosphatidylglycerol vesicles, the only vesicle that was fused by DV fusion peptide. The three-dimensional structure of DV fusion peptide bound to DPC micelles was solved by solution homonuclear NMR with an r.m.s.d. of 0.98 Å. The most striking result obtained from the solution structure was the hydrophobic triad formed by residues W101, L107, and F108, pointing toward the same direction, keeping the segment between G102 and G106 in a loop conformation. The interaction of DV fusion peptide with phosphatidylcholine/phosphatidylglycerol vesicles was also mapped by transfer-nuclear Overhauser enhancement (NOE) experiments, in which the majority of the NOE cross-peaks were from the hydrophobic triad, corroborating the DPC-bound structure. Substitution of the residue W101 by an alanine residue completely abolished membrane binding and, thus, fusion by the peptide and its NOE cross-peaks. In conclusion, the 15-residue DV fusion peptide has intrinsic ability to promote membrane fusion, most likely due to the hydrophobic interaction among the residues W101, L107, and F108, which maintains its loop in the correct spatial conformation.
Abbreviations used: DV, dengue virus; E, envelope glycoprotein; HIV, human immunodeficiency virus; DPC, dodecylphosphocholine; PC, phosphatidylcholine; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; PS, phosphatidylserine; 3D, three-dimensional; Rh-PE, N-(lissamine Rhodamine B sulfonyl) phosphatidylethanolamine; NBD-PE, N-(7-nitro-2,1,3-benzoxadiazol-4-yl) phosphatidylethanolamine; LUV, large unilamellar vesicle; TOCSY, total correlation spectroscopy; NOE, nuclear Overhauser enhancement; NOESY, nuclear Overhauser effect spectroscopy
Keywords: dengue virus, fusion peptide, membrane fusion, NMR, transfer NOE
Introduction
Despite a century of scientific advances, infectious diseases are still among the main causes of death worldwide.1, 2 Among mosquito-transmitted diseases, those caused by the dengue virus (DV) pose the most serious public health hazard. The classic form of the disease caused by DV is dengue fever, a fever of moderate severity, frequently accompanied by intense pains in muscles, bones, and joints. The more severe forms, dengue hemorrhagic fever and dengue shock syndrome, can be lethal, and include hemorrhage, thrombocytopenia, and hemoconcentration,3, 4 whose molecular mechanisms are far from being fully understood.
DV is a member of the Flaviviridae family, a group of enveloped viruses containing a positive-sense single-stranded RNA genome, which is translated into a polyprotein that is processed in three viral structural proteins [capsid (C), membrane (M), and envelope (E)] and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5).5 As for other enveloped viruses, DV infection depends on a step of membrane fusion, which is mediated by the virus surface E glycoprotein. DV is internalized by receptor-mediated endocytosis, and fusion occurs between the viral and the endosome membranes in the acidic environment of this compartment. Insights into this protein-mediated process have been achieved by the determination of the pre- and post-fusion structures of the flavivirus E glycoproteins.6, 7, 8, 9, 10 Exposure to low pH triggers the dissociation of E dimers that lie flat on the virion's surface and promotes a change in their conformation, causing E-protein trimers to form and protrude from the envelope.11 The segment between residues 98 and 112 forms a loop in domain II of E glycoprotein8 and has been considered to be the fusion peptide of the flaviviruses because (i) it presents almost 100% homology among all the members of the Flaviviridae family (it is identical in all of them, except for a single residue in the tick-borne encephalitis virus); (ii) site-directed mutagenesis in that region prevents the virus fusion;11 and (iii) it is located at the tip of the trimeric fusogenic structure of the protein.8 It has been suggested that the rearrangement caused by exposure of E protein to low pH starts the fusion process by causing the fusion peptide to become exposed and bind to the target membrane.8, 11, 12
Without a doubt, X-ray crystallography has been of fundamental importance in solving the three-dimensional (3D) structures of viral proteins in pre- and post-fusion states, providing substantial information concerning the mechanism of interaction between viral glycoproteins and cellular membranes during the fusion process. This has revealed new potential target molecules for rational drug design,13 as demonstrated in the development of a peptide inhibitor of human immunodeficiency virus type 1 (HIV-1) fusion,12, 14 a new class of anti-HIV drugs.15, 16 However, the mechanism of membrane interaction is indirectly inferred, since the structure of viral proteins is not determined the moment they are bound to membranes. To fill this gap, several groups have used solution NMR to solve the 3D structure of micelle-bound fusion peptides, such as those from influenza hemaglutinin17 and HIV gp41,18 which are class I viral fusion glycoproteins. However, no solution structure is available yet for class II fusion peptides, which include flavivirus and alphavirus glycoproteins.
In a previous study, we have characterized by steady-state and time-resolved fluorescence spectroscopy the interaction between a DV E-glycoprotein fragment of 36 amino acids containing the fusion loop and phospholipid bilayers.19 We showed that, despite being only a small fragment of the 394-amino-acid-long E glycoprotein, the peptide was fusogenic by itself. In addition, we found evidence for oligomerization of this fragment in the membrane, which might reflect the in vivo trimerization of the E glycoprotein. In the present work, we have extended this study with a fragment that exactly comprises the conserved fusion peptide, aiming to characterize both structurally and functionally its interaction with membrane models as a means to better understand the role of the fusion loop in the process of the DV infection. It is worth mentioning that, despite the DV E-glycoprotein structure having been solved in pre- and post-fusion states by X-ray crystallography, none of the structures involved interaction with phospholipids or mimetic membranes. Therefore, using NMR techniques, we determined the structure of the peptide interacting with dodecylphosphocholine (DPC) micelles and mapped the residues that interact with phospholipid vesicles in fusogenic conditions, revealing a crucial role for W101.
Results and Discussion
Kinetics of vesicle fusion induced by wild-type DV fusion peptide
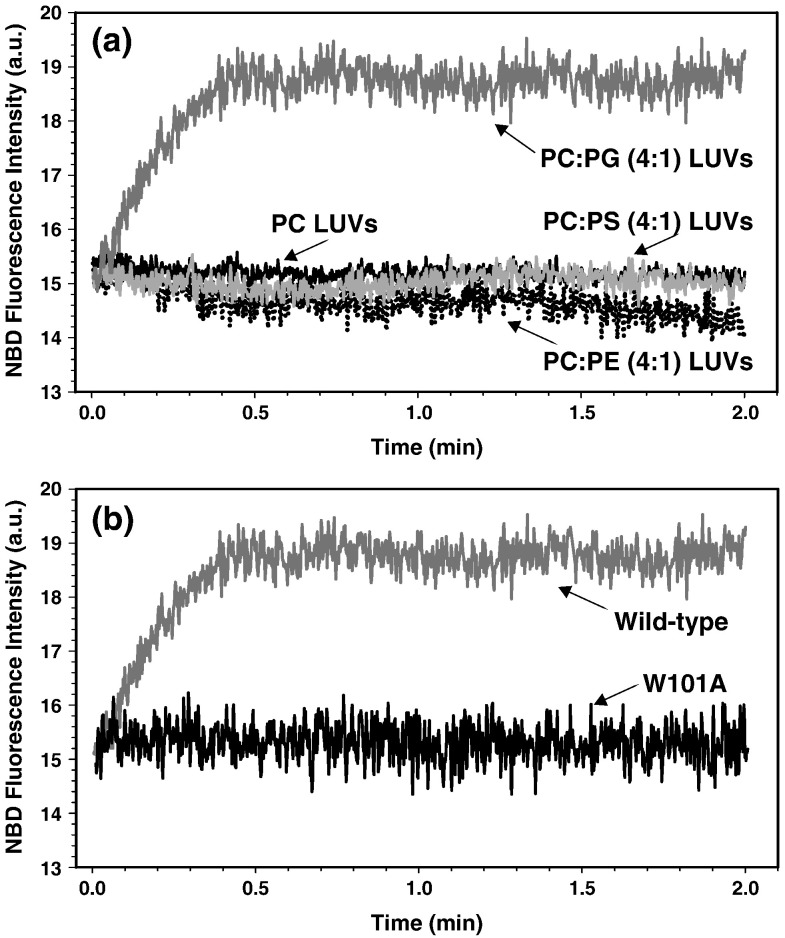
Before solving the 3D structure of the DV fusion peptide, we assessed the fusogenic ability of this very short peptide using a fluorescence resonance energy transfer-based assay for lipid mixing (Fig. 1 a). The result clearly demonstrated that the DV fusion peptide was able to induce vesicle fusion when the large unilamellar vesicles (LUVs) were composed of the anionic phospholipid phosphatidylglycerol (PG), but not phosphatidylcholine (PC), phosphatidylethanolamine (PE), or phosphatidylserine (PS). The requirement of PG for fusogenic activity may be related to specific features of this phospholipid such as the head group itself rather than a charge effect, since no fusion was observed for vesicles containing PS, another negatively charged phospholipid.
Fig. 1.
Kinetics of membrane fusion induced by DV fusion peptide. (a) Comparison of peptide-induced fusion of vesicles of different composition: PC/PG (4:1) (dark gray curve), PC (black curve), PC/PE (4:1) (dotted black curve), or PC/PS (4:1) (gray curve) LUVs. (b) Comparison of the membrane fusion kinetics between the wild-type (dark gray curve) and W101A mutant (black curve) DV fusion peptides in PC/PG (4:1) LUVs.
Although the fusogenic capability of the E glycoprotein depends on an intact fusion loop,11 it persists even after stripping the protein down to the loop itself, suggesting that the fusion-active structure is present in that small segment. This hypothesis has been reinforced by the recent solution of the structure of the prM–E complex, which showed that the M protein binds exactly on the fusion peptide region, blocking premature fusion.20
Fluorescence analysis of the fusion peptide–lipid interaction
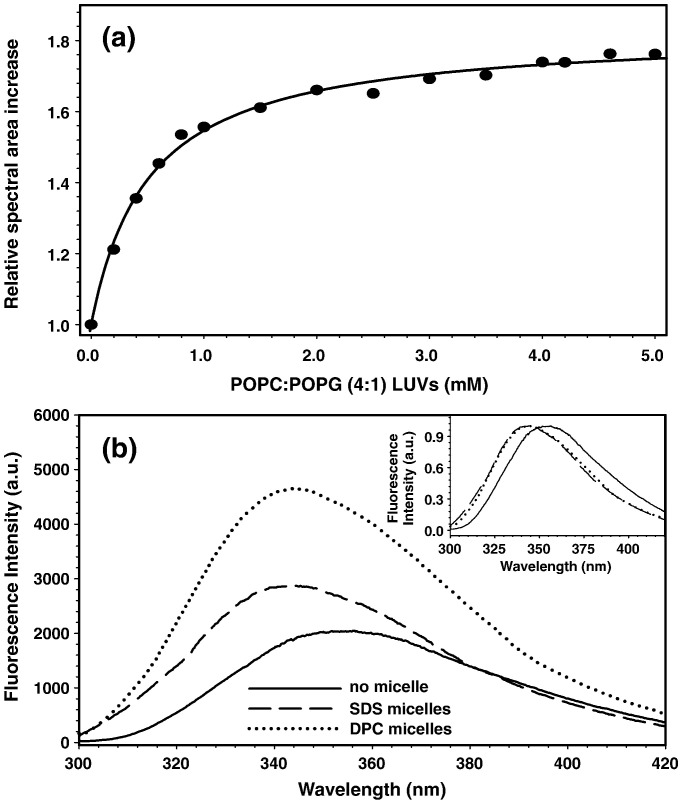
The flavivirus E glycoproteins have a tryptophan residue in the fusion peptide sequence that is fully conserved among the members of this family. Tryptophan is known to be an excellent intrinsic fluorescence probe because its fluorescence spectrum shifts drastically according to the degree of partition between the solution milieu and the hydrophobic environment of a membrane, as well as its ensemble-averaged fluorescence quantum yield.21, 22 The spectroscopic properties of W101 present in the DV fusion peptide were used to monitor its interaction with LUVs. Fluorescence intensity increased upon titration of the peptide with PC/PG (4:1) LUVs (Fig. 2 a). A K p of (2.6 ± 0.2) × 103, indicative of a relatively strong binding, was obtained after fitting by Eq. (1). Knowing that the fusion peptide is in fast equilibrium exchange between its free state in solution and being bound to the LUVs [see below, with transfer-nuclear Overhauser enhancement (NOE) experiments], it is plausible to conclude that PG favors a strong interaction with the fusion peptide, thus leading to the membrane fusion observed in Fig. 1a.
Fig. 2.
Interaction of DV fusion peptide with membrane models monitored by fluorescence spectroscopy. (a) Partition of DV fusion peptide toward LUVs composed of 1-palmitoyl-2-oleyl-sn-glycero-3-phosphocholine/1-palmitoyl-2-oleyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPC/POPG) (4:1) in 20 mM MES, 30 mM Tris buffer (pH 5.5), monitored as the relative increase in Trp fluorescence emission spectra area. The line represents a fit according to Eq. (1) developed elsewhere.22 (b) Trp fluorescence emission spectra of the DV fusion peptide in buffer (continuous trace) and in the presence of SDS (dashed trace) or DPC (dotted trace) micelles. Inset, normalized spectra.
Albeit LUVs mimic more realistically the cellular environment encountered by DV during infection, their large correlation time hampers their use in the structure determination of proteins/peptides at high atomic resolution by NMR spectroscopy, since the proton signals of the bound peptide are broadened out. To overcome the peak broadening, surfactants are used instead of vesicles, since they can form micelles of sizes suitable for the NMR technique. The ability of the DV fusion peptide to interact with surfactants was clear from the tryptophan fluorescence change upon addition of either DPC or SDS micelles (Fig. 2b). The spectral blue shift indicated the incorporation of the tryptophan indole side chain into a less polar environment. Nonetheless, although both surfactants promote a blue shift in the W101 fluorescence spectra (Fig. 2b, inset), the increase of its fluorescence intensity in DPC was twice that in SDS, which indicates a more extensive partition to DPC and/or a more hydrophobic local microenvironment for the tryptophan residue. It is worth mentioning that micellar systems mimic biological membranes quite well, in the sense that micelles provide a noncontinuous environment with a water–aliphatic chain interface. However, the combination of the fast dynamic and high curvature of micellar systems makes the aliphatic hydrophobic chains much more available to interact with peptides, meaning that the micelles stabilize the bound conformational state, which is favorable for NMR structural determination. On the other hand, the interaction of the peptide with LUVs is much more sensitive to the chemical characteristics of the phospholipids and, thus, much more informative about the peptide specificity toward different membranes.23
Interactions of DV fusion peptide with detergent micelles monitored by 1D 1H NMR
NMR spectroscopy is an excellent technique to map interactions between macromolecules because the nuclei chemical shifts, especially 1H nuclei, are very sensitive to any change around their environment. More importantly, this technique can identify which nuclei are involved in such interactions. Differences in the 1H chemical shifts of the peptide upon addition of micelles were an indication of interaction and also showed that the signal from the micelle-bound peptide protons could be measured, a necessary condition for the determination of the interacting peptide structure. In order to determine the best [detergent]/[peptide] ratio to be used for solving the 3D structure of the DV fusion peptide, titrations of 2 mM peptide with increasing concentrations of d25-SDS or d38-DPC were carried out. In the titration with d25-SDS, there were significant 1H chemical-shift changes when 15 mM of this detergent was added, but no further change was observed above this concentration. Conversely, in the titration with d38-DPC, large changes in the 1H chemical shifts of the DV fusion peptide were detected as the detergent concentration was increased from 0 to 64 mM. We also observed greater chemical-shift dispersion with d38-DPC, suggestive of gain of structure. Increasing the concentration of this detergent to 100 mM did not change chemical-shift dispersion, but led to sharp lines, indicating a decrease in conformational exchange that usually is interpreted as the stabilization of the bound state of the peptide. No further significant changes were observed above this detergent concentration. Based not only on these titrations experiments but also on the fluorescence experiment, we concluded that the interaction between the DV fusion peptide and d38-DPC would mimic more realistically its interaction with vesicles and, thus, with cellular membranes, which led us to choose DPC micelles to perform the structure determination. To guarantee that all peptide molecules would be bound to the micelles, 100 mM d38-DPC was chosen in order to obtain a [detergent]/[peptide] ratio of 50:1.
Structure determination of DV fusion peptide in d38-DPC micelles by NMR
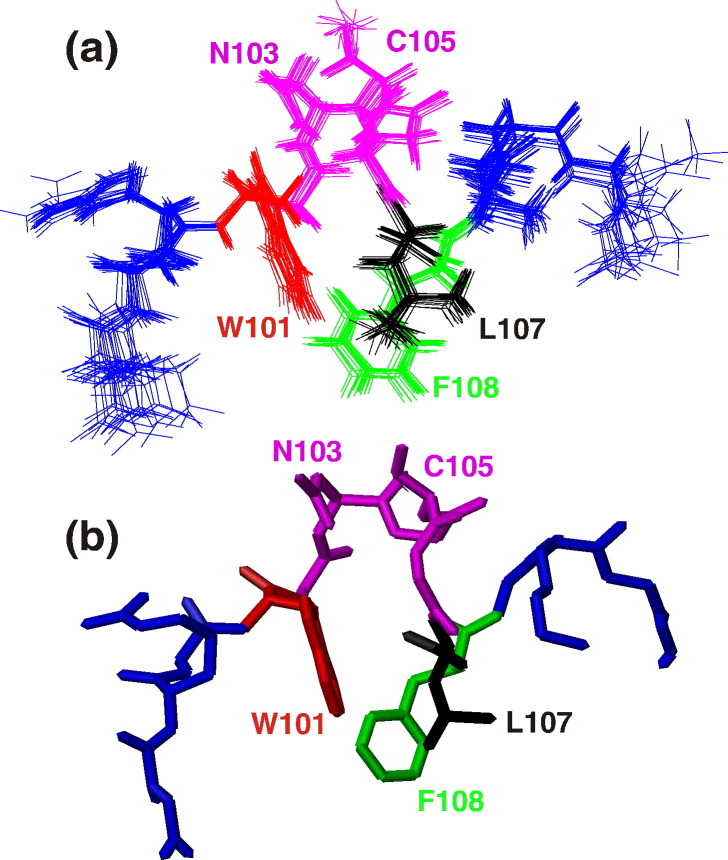
The structure of the DV fusion peptide in d38-DPC micelles was solved by 2D homonuclear NMR spectroscopy. There is a good overlap of the backbone of the 20 lowest-energy structures after simulated annealing, despite some divergence of the last two residues from the N and C termini, due to lack of restraints involving these residues (Fig. 3 a and b). From a total of 221 assigned NOE cross-peaks, 114 nonredundant upper-limit constraints were obtained for DV fusion peptide under the fusogenic condition (Table 1 ). The average structure of the 20 lowest-energy conformers in DPC micelles at pH 5.5 obtained from the initial 200 calculated structures is shown in Fig. 3b. They did not violate NOEs or dihedral angle constraints. The overall energy of the selected structures was 42.21 ± 0.87 kcal/mol. The r.m.s.d. among all peptide residues was 1.16 Å for the backbone, decreasing to 0.19 Å when residues 2–13 were taken into account. Moreover, the Ramachandran plot analysis showed that 96.5% of the residues were in favored or allowed regions and no residue is in disallowed region (Table 1).
Fig. 3.
Solution structure of the DV fusion peptide bound to d38-DPC micelles at fusogenic pH. The 20 lowest-energy structures showing all atoms are represented in (a), and the average structure showing only the heavy atoms is shown in (b). The structures are colored as follows: D98–G100 and G109–G112 in blue, W101 in red, L107 in black, F108 in green, and the loop formed by residues G102–G106 in magenta. The NMR spectra were acquired at 5 °C.
Table 1.
Summary of structural statistics of the DV fusion peptide at pH 5.5 in DPC micelles
| Total NOE distance constraints | 224 |
| NOE inter-residue constraints | 114 |
| Energies (kcal/mol) | |
| Overall | 42.21 ± 0.87 |
| Bond | 3.22 ± 0.05 |
| Angle | 18.51 ± 0.12 |
| Improper | 1.87 ± 0.05 |
| VDW (LJ) | 8.24 ± 0.96 |
| NOE | 6.80 ± 0.13 |
| Backbone pairwise r.m.s.d. (Å)a | |
| Residues 1–15 | 0.98 |
| Residues 2–13 | 0.11 |
| Heavy pairwise r.m.s.d. (Å)a | |
| Residues 1–15 | 1.16 |
| Residues 2–13 | 0.19 |
| % of residues in: | |
| Most favored regionsb | 33.6 |
| Additional allowed regionsb | 62.9 |
| Generously allowed regionsb | 3.6 |
| Disallowed regionsb | 0 |
r.m.s.d. values are from MOLMOL.
Data from PROCHECK-NMR.
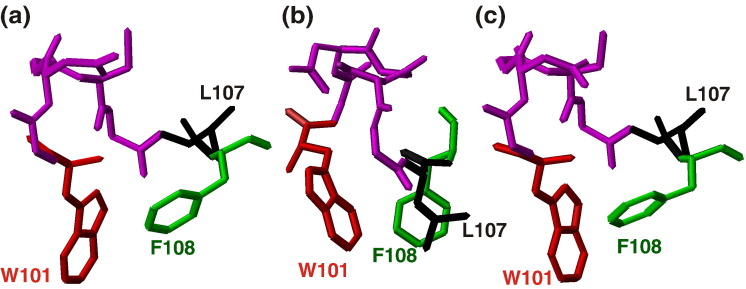
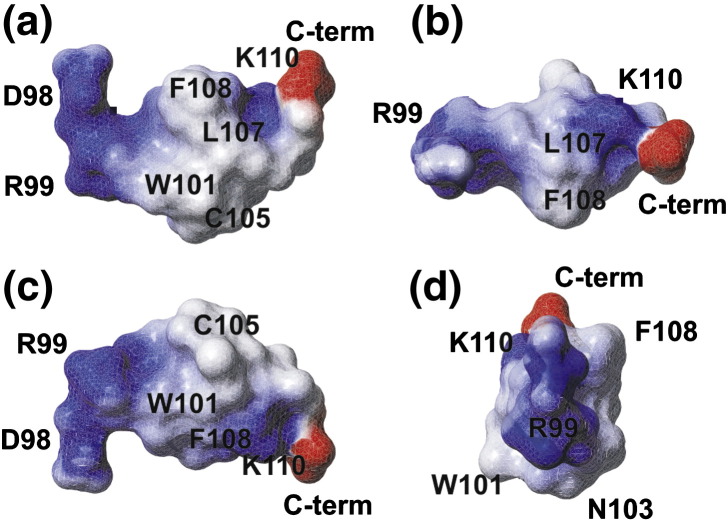
The most striking result obtained from the solution structure of DV fusion peptide is the formation of a hydrophobic triad among the residues W101 (red), L107 (black), and F108 (green), all pointing to the same direction (Fig. 5). Moreover, the central region (segment W101–F108) of the DPC-bound DV fusion peptide correlates very well to the structure of this region when it is attached to the full-length E-glycoprotein fusion loop, as solved by X-ray crystallography in either pre- or post-fusion state. The r.m.s.d. between the NMR and crystallographic structure is 3.41 Å; the r.m.s.d. between the pre- and post-fusion states of this segment in the crystal structures is 0.24 Å.7, 8 The loop conformation formed by the segment 101–108 (Fig. 3, magenta residues) was maintained, as well as the spatial confluence of the W101 and F108 residues. The interactions among these hydrophobic residues are based on long-range cross-peak NOEs between W101 and L107 protons, namely, HN–HN, Hδ–Hɛ, and Hδ–Hζ NOEs. A long-range interaction was also observed between Hζ of W101 and Hα of G106. Additionally, the protons of the residues L107 and F108 make several i, i + 1 contacts such as Hα–Hα, HN–HN, Hα–Hβ, and Hα–Hδ. However, no cross-peak NOE between W101 and F107 side chains was assigned, although they are very close to each other according to the solution structure (Table 2 ). One explanation for this result may be the large number of overlapping peaks in the NOE spectroscopy (NOESY) spectrum and/or the intrinsic flexibility of the side chain. Furthermore, the segments W101–G102 and G106–L107 establish the majority of the longer-range NOE interactions, in agreement with the hydrogen bond formed between the Trp101 Hɛ1 and the Gly106 carbonyl observed in the X-ray structure; it was also observed that the loop in the 103–105 residues, present in both NMR and X-ray structures, is formed due to the interactions between flanking residues 101–102 and 106–107, despite not being involved in many long-range interactions itself. According to the electrostatic surface potential of the DV fusion peptide bound to DPC micelles in the fusogenic condition (Fig. 4 a–d), there is a hydrophobic patch formed by the loop residues W101–F108, which might be responsible for the E-glycoprotein penetration into the cellular membrane during the fusion process, as already hypothesized by Modis and coworkers.8 Furthermore, the hydrophobic patch is surrounded by positively charged residues, characteristic of membrane-interacting peptides.23
Fig. 5.
3D conformation of the DV fusion peptide segment W101–F108 of the full-length E protein at different fusogenic conditions. In (a) and (c) are represented the DV fusion peptide in pre- and post-fusogenic states, respectively, solved by X-ray crystallography,7, 8 and in (b) when it is bound to DPC micelles in the fusogenic state solved by NMR. The residue colors are the same as in Fig. 3. The distances between the hydrophobic residues are presented in Table 2.
Table 2.
Distances between the side-chain protons of hydrophobic triad residues W101, L107, and F108 of the dengue fusion peptide in pre-fusion, post-fusion, and micelle-bound states
| Pre-fusion state (Å) | Post-fusion state (Å) | Micelle-bound state (Å) | |
|---|---|---|---|
| Hζ3 (W101)–Hδ21 (L107) | 11.9 | 11.2 | 2.4 |
| Hζ3 (W101)–Hɛ2 (F108) | 5.7 | 4.0 | 1.8 |
| Hδ21 (L107)–Hɛ2 (F108) | 7.9 | 8.6 | 3.4 |
Fig. 4.
Electrostatic surface potential of the DV fusion peptide bound to d38-DPC micelles at fusogenic pH. Four views of the surface potential of the average structure shown in Fig. 3b: (a) front view; (b) rotated 90° along the x-axis; (c) rotated 180° along the x-axis; and (d) rotated 90° along the z-axis. Surfaces in red, blue, and white represent respectively negatively charged, positively charged, and neutral residues.
In spite of these similarities, there are major differences when the structure of the fusion loop segment (W101–F108) of the pre- and post-fusion conformations of the full-length E glycoprotein,7, 8 which are very similar to each other (Fig. 5b), is compared to the structure of the same region bound to DPC micelles (Fig. 5 a–c). The major prominent difference is the L107 orientation relative to the other hydrophobic residues: its side chain is facing the opposite direction in relation to the aromatic ring of W101 in the pre- and post-fusion states; the ring lies 11.9 and 11.2 Å away from the L107 side chain in the two crystallographic structures, but only about 2.4 Å away in the DPC-bound state, as shown in Fig. 4b and in Table 2. This difference in the orientation of the residue L107 among the three structures might be a consequence of the milieu surrounding the fusion peptide, since it is surrounded by water molecules in the conditions used to crystallize the pre- and post-fusion states of the full-length glycoprotein, whereas it is surrounded by long hydrophobic chains of the DPC detergent molecules in the structure determination in solution by NMR. It may be that the detergent side chains drive thermodynamically the clustering of the hydrophobic triad W101, L107, and F108 present in the DV fusion peptide. We speculate that this driving force might exist in vivo as well, since the 15-residue peptide maintained its fusogenic activity, as shown in Fig. 1.
Altogether, these results show that, despite being a 15-amino-acid fragment, DV fusion peptide interacting with micelles (or bilayers; see next section) adopts a structure with several features similar to those of the fusion loop in the full-length E glycoprotein. Moreover, this structural similarity indicates that the local conformation adopted by the segment W101–F108 upon membrane binding is dependent only on interactions established by the fusion loop itself; the remaining domains of the E glycoprotein, namely, the adjacent domain II, play only an ancillary role in defining the structure of the loop. This might explain the loop's fusogenic activity being independent of the presence of those domains, and makes the self-assembling region W101–F108 a putative structural motif responsible for membrane fusion.
Interaction of DV fusion peptide with LUVs monitored by transfer-NOE experiments
As mentioned previously, the large rotational correlation time of the vesicle-bound DV peptide hampers the use of NMR to solve its structure at high resolution. Alternatively, exchange-transfer NOESY is a convenient method to obtain structural information on small molecules in association with high molecular weight receptors such as proteins or biomolecular assemblies like phospholipid vesicles.24 This technique permits the detailed structural analysis of high molecular mass complexes that are not feasible by common NMR methods due to fast relaxation processes that ultimately result in broad lines. The method is particularly useful for structural analysis of proton-rich, flexible ligands that usually have rotational correlation time (τc) near the value for the NOE null condition (ωο × τc = 1.12, where ω is the Larmor frequency) or shorter, which corresponds to a molecular mass of ∼ 5 kDa.24 Exchange-transfer NOESY can be applied to systems in fast exchange on the chemical-shift time scale, that is, in which the dissociation rate, k off, is larger than the chemical-shift difference between the free and the bound forms of the ligand,24 so that ligand protons show a single resonance peak averaged over free and bound states. The buildup of the transferred NOEs requires fast exchange between the bound and free states of the ligand, corresponding to a K D > 10− 6 M for the complex. Therefore, this strategy is particularly useful for low-affinity complexes (micromolar to millimolar range).23, 25, 26, 27
In this context, 1D 1H spectra of 400 μM DV fusion peptide were recorded upon addition of increasing concentrations of either PC or PC/PG LUVs, so that peptide spectra were acquired in buffer solution, in 6 and 15 mM PC, and in 6 and 15 mM PC/PG (4:1) LUVs (data not shown). Clearly, changes and broadening in the proton chemical shifts of the DV fusion peptide were more pronounced when it was incubated with 15 mM PC/PG LUVs; that is, it interacts preferentially with this type of vesicle although some binding also seemed to occur with PC LUVs. This result is in agreement with the observed vesicle fusion behavior shown in Fig. 1a, the moderately high value of K p (Fig. 2a), and the extent of partition reported for the longer DV fusion peptide K88–K123 toward these membranes.19
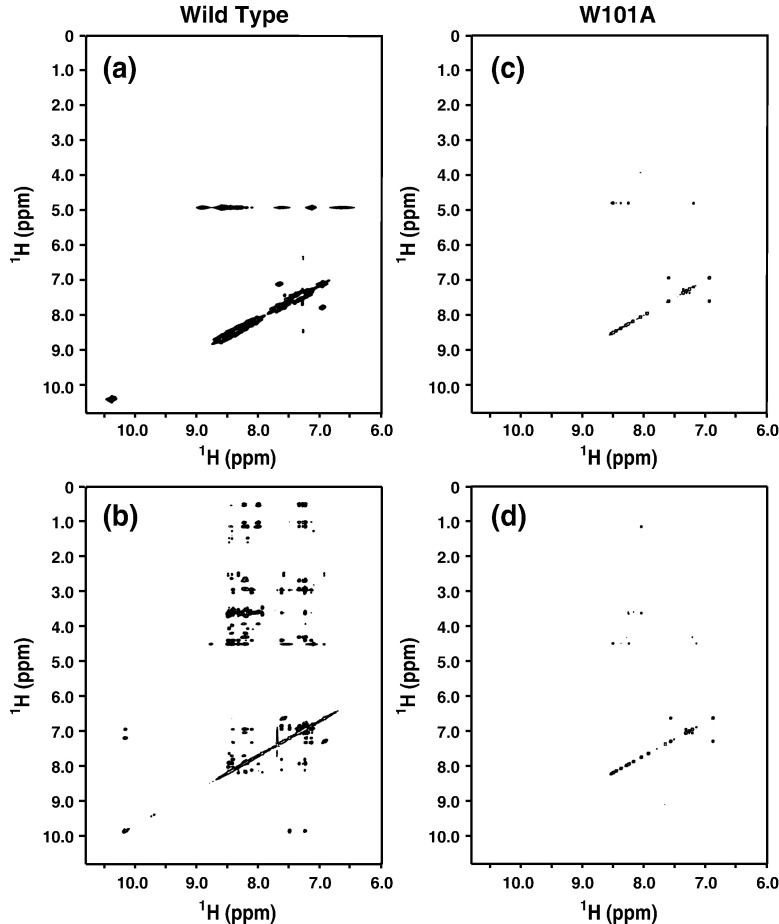
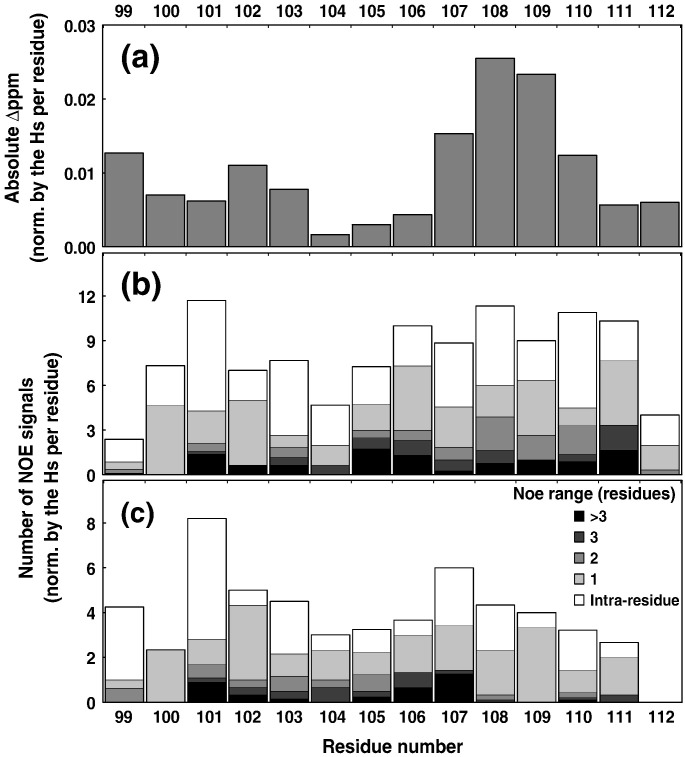
Several exchange-transfer NOEs were observed in the 2D 1H NOESY spectra of the DV fusion peptide in the presence of 15 mM PC/PG (4:1) LUVs (Fig. 6 b), but not in buffer solution (Fig. 6a), clearly indicating a transient interaction with this vesicle as well as the adoption of an ordered structure upon binding. Although we have not yet calculated a structure from the NOE data of the fusion peptide in the lipid vesicles yet, structural information could be obtained from both the total correlation spectroscopy (TOCSY) and NOESY spectra. The changes in the DV fusion peptide chemical shifts observed by 1H TOCSY upon its binding to the vesicles were used to map the regions of the peptide that interact most with the bilayer (Fig. 7 a). This result agrees with the number of inter-residue NOEs per proton per residue, since the segments W101–G102 and L107–F108, which gain the most structure upon binding to DPC micelles, are also those that undergo the largest chemical-shift changes upon vesicle binding (Fig. 7b and c). There is excellent agreement between the profiles of the number of NOEs per residue for LUV and micelle interactions. On the other hand, the segment G103–G106 showed the smallest chemical-shift changes, reinforcing the idea that this part of the fusion peptide makes fewer contacts with the rest of the peptide, as already observed in the post-fusion state of the E glycoprotein.8 While a structure was not calculated for the fusion peptide interacting with LUVs, the NOE data validate the use of DPC micelles to mimic the peptide–bilayer interaction, since the interaction pattern is the same in both cases.
Fig. 6.
Transfer-NOE experiments of wild-type and W101A DV fusion peptide. Amidic and aromatic proton regions of the NOESY spectra of 1 mM wild-type (spectra a and b) and W101A (spectra c and d) DV fusion peptides in phosphate buffer (spectra a and c) or in 15 mM PC/PG (4:1) LUVs (spectra b and d).
Fig. 7.
(a) Chemical-shift deviation of DV fusion peptide upon membrane binding. Absolute chemical-shift change from 1H TOCSY spectra (not shown) of wild-type DV fusion peptide upon PC/PG (4:1) vesicle addition, averaged per residue proton. (b and c) Average number of NOE signals per residue proton, differentiated according to NOE range, upon interaction with PC/PG (4:1) LUVs (b) or DPC micelles (c).
Mutation of W101 in the DV fusion peptide abrogates membrane fusion
The NOE information used to calculate the DV fusion structure by NMR shows that W101 makes important contacts with several residues in the peptide, mainly L107 and F108. The interaction/proximity of this triad of hydrophobic amino acids keeps the residues between G102 and G106 in the fusion loop conformation.8 The essential role of the interaction among the three hydrophobic amino acids in peptide fusogenic activity was confirmed by the complete inhibition of the peptide-induced fusion when W101 was mutated to alanine (Fig. 1b). Moreover, no 1H peak broadening was observed when the W101A peptide was incubated with LUVs of either PC only or PC/PG (4:1), strongly suggesting that the replacement of W101 by alanine disrupts the peptide binding to the membrane. This result was further confirmed by the absence of exchange-transfer NOEs when 1 mM mutant peptide was incubated with 15 mM PC/PG (4:1) LUVs (Fig. 6b).
Concluding remarks on the structure and function of the DV fusion peptide
The activity observed for the DV fusion peptide is remarkable, considering its size both in absolute terms (15 residues long) and relative to the whole protein (∼ 3.8%). The fact that it does display less activity than the longer peptide K88–K12319 is not surprising; our work shows that there are membrane-interacting residues almost up to the termini (Fig. 3). The removal of the 10 to 11 residues flanking the fusion loop in the longer fusion peptide will perturb these interacting residues, both because of the observed increase in flexibility (Fig. 3a) and because of the introduction of the terminal NH3 + and COO− charges in a region usually occupied by apolar residues in the full-length E glycoproteins. Nonetheless, our results enable us to propose a putative self-assembling fusion motif in flavivirus fusion peptide, making its interaction with membrane models a very attractive target in the development of antiviral inhibitors.
Although several authors have shown that single-point mutations in the fusion peptide sequence (98–112) of flavivirus E glycoproteins can abolish virus entry,11 to our knowledge, the importance of W101 residue to the membrane binding and thus to the fusion process has never been directly demonstrated as it is here. Among the 15 residues of the peptide, 12 are polar and 3 are hydrophobic (W, F, L). Three residues have membrane-anchoring properties (W, R, K) due to their ability to locate at membrane interfaces.28 However, tryptophan is the residue with the greatest tendency to locate at the membrane interface,29 near the carbonyl groups.30 The combined effect of size, rigidity, and aromaticity is the main reason for this positioning.28 These properties of tryptophan residues make them appropriate for stabilizing the interaction of integral membrane proteins with lipid bilayers;31 in some cases, an “aromatic belt” is present.28 They may also make crucial contributions to the conformational stability of peptides and proteins,32 making them key players in protein/peptide–membrane interactions. Tryptophan residues serve as anchors at interfaces and have a significant effect on conformation, which, in turn, is crucial for the adaptation and stabilization of peptides and transmembrane protein segments present in lipid bilayers.
Some membrane-active sequences of viral proteins are Trp-rich.33, 34 Here we show that tryptophan mutation leads to the abrogation of fusion either by suppression of peptide–lipid interaction or destabilization of the adsorbed or intercalated conformations. The idea that conformational freedom of membrane-active sequences plays a role in fusion has been largely overlooked but is supported by data from other viruses, such as SARS-CoV.35
Experimental Procedures
Chemicals
Phosphatidylcholine, phosphatidylglycerol, phosphatidylethanolamine, and phosphatidylserine from egg yolk were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Synthetic 1-palmitoyl-2-oleyl-sn-glycero-3-phosphocholine and 1-palmitoyl-2-oleyl-sn-glycero-3-[phospho-rac-(1-glycerol)] were purchased from Avanti Polar-Lipids (Alabaster, AL, USA), used in the extent of partition in LUVs. N-(Lissamine Rhodamine B sulfonyl) phosphatidylethanolamine (Rh-PE) and N-(7- nitro-2,1,3-benzoxadiazol-4-yl) phosphatidylethanolamine (NBD-PE) were purchased from Molecular Probes, Inc. (Eugene, OR, USA). d25-SDS and d38-DPC were purchased from CIL (Boston, MA, USA). A 15-amino-acid fragment corresponding to the conserved region of the E protein consisting of residues 98 to 112 (DV fusion peptide, sequence DRGWGNGCGLFGKGG) and a mutant of tryptophan 101 (W101A) were used in this study. The peptides were purchased from Genemed Synthesis, Inc. (South San Francisco, CA, USA).
Vesicle preparation and fusion assays
Large unilamellar vesicles of about 100-nm diameter were prepared by rapid extrusion of phospholipid suspensions previously subjected to freeze/thaw cycles, as described elsewhere.36 Extrusions were carried out through a 0.1 μm pore diameter Whatman Nuclepore polycarbonate Track-Etch membrane (Newton, MA, USA) using an Avanti Mini-Extruder (Alabaster, AL, USA). Vesicles were prepared in 20 mM MES, 30 mM Tris–HCl buffer (pH 5.5) for the fusion assays or in 20 mM phosphate buffer (pH 5.5) for the NMR measurements. For fusion assays, equal amounts of unlabeled vesicles and vesicles labeled with both 20 μM Rh-PE and 20 μM NBD-PE (final concentration of labeled vesicles, 1 mol% lipid), adjusted to a final phospholipid concentration of 100 μM, were mixed and the reaction was initiated by addition of the peptide to a final concentration of 18 μM. Fusion was monitored by the resonance energy transfer assay as described elsewhere.37 Fluorescence intensity was monitored using λexc = 470 nm (NBD absorption) and λem = 530 nm (Rh emission). Control measurements in the absence of peptide, for determination of spontaneous background fusion, or with 0.2% Triton X-100 (Sigma Chemical), for determination of total fusion, were carried out in all cases.
Fluorescence measurements
Intrinsic fluorescence measurements were recorded using Hitachi F-4500 or Varian Cary Eclipse fluorescence spectrophotometers. Intrinsic fluorescence was measured by exciting samples at 280 nm and scanning emission between 300 and 420 nm using a bandwidth of 8 nm for the Hitachi F-4500 or 10 nm for the Varian Cary Eclipse. After addition of the peptide, the system was equilibrated for ∼ 5 min before fluorescence was measured. Partition data were extracted from peptide titrations with LUVs according to Eq. (1), based on a Nernst partition formalism22
| (1) |
where [L] is the total phospholipid concentration available for the peptide to interact with, K p is the Nernst partition constant, and γ L is the phospholipids molar volume. I is the global integrated fluorescence intensity of the system, and I L and I W are the integrated fluorescence intensities the mixture would display if all the peptide was in the lipidic or aqueous phase, respectively. I W can be readily obtained from the measurement of I in the absence of lipid; I L, however, is a limit value of fluorescence intensity as [L] → ∞ and is determined as a system parameter together with K p.
NMR spectroscopy
NMR measurements were performed on a Bruker (Rheinstetten, Germany) Avance DRX600 or on a Varian (Palo Alto, CA, USA) Inova AS600, both operating at 600 MHz. For the studies in micelles, the sample temperature was maintained at 5 °C. The samples were prepared with 2 mM DV fusion peptide and 100 mM D38-DPC (CIL, Cambridge, MA, USA) detergent in 20 mM sodium phosphate buffer (pH 5.5), 10 mM deuterated DTT, 10% D2O, and 0.01% NaN3. 1H 2D TOCSY spectra (spin-lock time of 100 ms) were acquired using the MLEV-17 pulse sequence.38 1H 2D NOESY spectra were acquired using a 150-ms mixing time.39, 40 Water suppression was achieved using the WATERGATE technique;40 the spectra were collected with 4096 and 512 data points in the direct and indirect dimensions, respectively, with 16 transients. For the studies in LUVs, the sample temperature was maintained at 25 °C. 1H 2D TOCSY spectra (spin-lock time of 100 ms) were acquired using the MLEV-17 pulse sequence. 1H 2D NOESY spectra were acquired using a mixing time of 140 ms. The NMR data were processed with NMRPipe.41 Resolution enhancement was achieved by apodization of the free induction decays with cosine bell multiplication and zero filling. The chemical-shift assignments were carried out using the NMRView program version 5.0,42 and the DV fusion peptide structure bound to DPC micelles was calculated by Crystallography & NMR System (CNSsolve) program release 1.0.43 The solution structure quality was evaluated using the PROCHECK program (NMR version).44 The figures were prepared using the MOLMOL program.45
Accession numbers
The atomic coordinates for the solution structure of the DV fusion peptide bound to d38-DPC micelles are available in the Biological Magnetic Resonance Databank under BMRB 20047.
Acknowledgements
We thank Drs. Ana Carolina Zeri and Mauricio Sforça of the National Laboratory of Synchrotron Light (LNLS) for help with the acquisitions on the Varian spectrometer. We also thank Dr. Jerson Lima Silva of the Medical Biochemistry Institute (Universidade Federal do Rio de Janeiro) and Dr. Roberto De Guzman of the University of Kansas for helpful insights in this project, and Dr. Martha Sorenson (Medical Biochemistry Institute, Universidade Federal do Rio de Janeiro) for proofreading the manuscript. This work was supported by the following funding agencies: WHO/TDR, CNPq, FAPERJ, TWAS, IMBEBB, PRONEX-FAPERJ, and FINEP (GENOPROT Dengue), the National Institutes of Science and Technology in Dengue (INCT-Dengue) and in Structural Biology and Bioimaging (INCT-INBEB). The authors acknowledge a grant from FCT (Portugal) to M.N.M. and funding from CAPES/GRICES (Brazil, Portugal) and FCT project PTDC/QUI/69937/2006. F.J.R.S. holds a Ph.D. fellowship from CNPq (Brazil).
Edited by A. G. Palmer III
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2009.07.035
Appendix A.
1H 1D NMR spectra of 2 mM peptide upon titration with increasing concentrations of either d25-SDS (left column) or d38-DPC (right column) micelles. The experiment was carried out at 25 °C.
Schematic representation of the unambiguously assigned inter-residue NOEs. The thicker bars correspond to stronger NOEs.
1H NOESY spectrum regions showing some of the major long-range NOE cross-peaks between (a) W101 HN and L107 HN, (b) W101 Hζ2 and L107 Hδ22, and (c) G105 HN and K110 Hδ1, as well as G105 HN and K110 Hβ#.
1H NMR spectra of 400 μM wild-type (spectra a–e) and W101A (spectra f–j) DV fusion peptides upon titration with either PC or PC/PG (4:1) LUVs. Spectra (a) and (f) were obtained in phosphate buffer without LUVs; (b) and (g) in 6 mM PC LUVs; (c) and (h) in 15 mM PC LUVs; (d) and (i) in 6 mM PC/PG (4:1) LUVs; and (e) and (j) in 15 mM PC/PG (4:1) LUVs. The experiments were carried out at 25 °C.
References
- 1.Thomas S.J., Strickman D., Vaughn D.W. Dengue epidemiology: virus epidemiology, ecology, and emergence. Adv. Virus Res. 2003;61:235–289. doi: 10.1016/s0065-3527(03)61006-7. [DOI] [PubMed] [Google Scholar]
- 2.Mackenzie J.S., Gubler D.J., Petersen L.R. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- 3.Lei H.Y., Yeh T.M., Liu H.S., Lin Y.S., Chen S.H., Liu C.C. Immunopathogenesis of dengue virus infection. J. Biomed. Sci. 2001;8:377–388. doi: 10.1007/BF02255946. [DOI] [PubMed] [Google Scholar]
- 4.Pang T., Cardosa M.J., Guzman M.G. Of cascades and perfect storms: the immunopathogenesis of dengue haemorrhagic fever–dengue shock syndrome (DHF/DSS) Immunol. Cell Biol. 2007;85:43–45. doi: 10.1038/sj.icb.7100008. [DOI] [PubMed] [Google Scholar]
- 5.Lindenbach B.D., Thiel H.-J.R.C.M. Flaviviridae: the viruses and their replication. In: Fields B.N., Knipe D.M., Howley P.M., editors. chapter 33. Lippincott; Williams & Wilkins, Philadelphia, PA: 2007. pp. 1101–1152. (Virology). [Google Scholar]
- 6.Rey F.A., Heinz F.X., Mandl C., Kunz C., Harrison S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 7.Modis Y., Ogata S., Clements D., Harrison S.C. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl Acad. Sci. USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Modis Y., Ogata S., Clements D., Harrison S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 9.Bressanelli S., Stiasny K., Allison S.L., Stura E.A., Duquerroy S., Lescar J. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanai R., Kar K., Anthony K., Gould L.H., Ledizet M., Fikrig E. Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 2006;80:11000–11008. doi: 10.1128/JVI.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allison S.L., Schalich J., Stiasny K., Mandl C.W., Heinz F.X. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 2001;75:4268–4275. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perera R., Khaliq M., Kuhn R.J. Closing the door on flaviviruses: entry as a target for antiviral drug design. Antivir. Res. 2008;80:11–22. doi: 10.1016/j.antiviral.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z., Khaliq M., Zhou Z., Post C.B., Kuhn R.J., Cushman M. Design, synthesis, and biological evaluation of antiviral agents targeting flavivirus envelope proteins. J. Med. Chem. 2008;51:4660–4671. doi: 10.1021/jm800412d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michael N.L., Moore J.P. HIV-1 entry inhibitors: evading the issue. Nat. Med. 1999;5:740–742. doi: 10.1038/10462. [DOI] [PubMed] [Google Scholar]
- 15.Chan D.C., Kim P.S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 16.Kilby J.M., Hopkins S., Venetta T.M., DiMassimo B., Cloud G.A., Lee J.Y. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 17.Han X., Bushweller J.H., Cafiso D.S., Tamm L.K. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 18.Li Y., Tamm L.K. Structure and plasticity of the human immunodeficiency virus gp41 fusion domain in lipid micelles and bilayers. Biophys. J. 2007;93:876–885. doi: 10.1529/biophysj.106.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stauffer F., Melo M.N., Carneiro F.A., Sousa F.J., Juliano M.A., Juliano L. Interaction between dengue virus fusion peptide and lipid bilayers depends on peptide clustering. Mol. Membr. Biol. 2008;25:128–138. doi: 10.1080/09687680701633091. [DOI] [PubMed] [Google Scholar]
- 20.Li L., Lok S.M., Yu I.M., Zhang Y., Kuhn R.J., Chen J., Rossmann M.G. The flavivirus precursor membrane–envelope protein complex: structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 21.Lakowicz J.R. Principles of Fluorescence Spectroscopy. Plenum Press; New York NY: 1999. [Google Scholar]
- 22.Santos N.C., Prieto M., Castanho M.A. Quantifying molecular partition into model systems of biomembranes: an emphasis on optical spectroscopic methods. Biochim. Biophys. Acta. 2003;1612:123–135. doi: 10.1016/s0005-2736(03)00112-3. [DOI] [PubMed] [Google Scholar]
- 23.Cruzeiro-Silva C., Gomes-Neto F., Tinoco L.W., Cilli E.M., Barros P.V., Lapido-Loureiro P.A. Structural biology of membrane-acting peptides: conformational plasticity of anticoccidial peptide PW2 probed by solution NMR. Biochim. Biophys. Acta. 2007;1768:3182–3192. doi: 10.1016/j.bbamem.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 24.Post C.B. Exchange-transferred NOE spectroscopy and bound ligand structure determination. Curr. Opin. Struct. Biol. 2003;13:581–588. doi: 10.1016/j.sbi.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Vaynberg J., Qin J. Weak protein–protein interactions as probed by NMR spectroscopy. Trends Biotechnol. 2006;24:22–27. doi: 10.1016/j.tibtech.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Da Poian, A. T., Almeida, F. C. L., Valente, A. P., Mohana-Borges, R. & Neto, F. G. (2009). NMR to access the transient interactions between viral fusion peptides and their target membranes. In Structure and Function of Membrane-Active Peptides (Castanho, M., ed), chapter 19, IUL Publishers, La Jolla, CA, USA, in press.
- 27.Valente A.P., Miyamoto C.A., Almeida F.C. Implications of protein conformational diversity for binding and development of new biological active compounds. Curr. Med. Chem. 2006;13:3697–3703. doi: 10.2174/092986706779026147. [DOI] [PubMed] [Google Scholar]
- 28.Killian J.A., von Heijne G. How proteins adapt to a membrane–water interface. Trends Biochem. Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 29.Wimley W.C., White S.H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 30.de Planque M.R., Bonev B.B., Demmers J.A., Greathouse D.V., Koeppe R.E., Separovic F. Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide–lipid interactions. Biochemistry. 2003;42:5341–5348. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- 31.Liu W., Caffrey M. Interactions of tryptophan, tryptophan peptides, and tryptophan alkyl esters at curved membrane interfaces. Biochemistry. 2006;45:11713–11726. doi: 10.1021/bi0608414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemec K.N., Pande A.H., Qin S., Bieber Urbauer R.J., Tan S., Moe D., Tatulian S.A. Structural and functional effects of tryptophans inserted into the membrane-binding and substrate-binding sites of human group IIA phospholipase A2. Biochemistry. 2006;45:12448–12460. doi: 10.1021/bi061440r. [DOI] [PubMed] [Google Scholar]
- 33.Giannecchini S., Bonci F., Pistello M., Matteucci D., Sichi O., Rovero P., Bendinelli M. The membrane-proximal tryptophan-rich region in the transmembrane glycoprotein ectodomain of feline immunodeficiency virus is important for cell entry. Virology. 2004;320:156–166. doi: 10.1016/j.virol.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Lu Y., Neo T.L., Liu D.X., Tam J.P. Importance of SARS-CoV spike protein Trp-rich region in viral infectivity. Biochem. Biophys. Res. Commun. 2008;371:356–360. doi: 10.1016/j.bbrc.2008.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guillen J., Perez-Berna A.J., Moreno M.R., Villalain J. A second SARS-CoV S2 glycoprotein internal membrane-active peptide. Biophysical characterization and membrane interaction. Biochemistry. 2008;47:8214–8224. doi: 10.1021/bi800814q. [DOI] [PubMed] [Google Scholar]
- 36.Mayer L.D., Hope M.J., Cullis P.R. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim. Biophys. Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 37.Struck D.K., Hoekstra D., Pagano R.E. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 38.Bax A., Davis D.G. MLEV-17 based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 39.Piotto M., Saudek V., Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 40.Sklenar V., Piotto M., Leppik R., Saudek V. Gradient-tailored water suppression for 1H–15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. 1993;102:241–245. [Google Scholar]
- 41.Delaglio F., Grzesiek S., Vuister G.W., Guang Z., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 42.Johnson B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 43.Brunger A.T., Adams P.D., Clore G.M., De lano W.L., Gros P., Grosse-Kurnstleve R.W. Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 45.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1H 1D NMR spectra of 2 mM peptide upon titration with increasing concentrations of either d25-SDS (left column) or d38-DPC (right column) micelles. The experiment was carried out at 25 °C.
Schematic representation of the unambiguously assigned inter-residue NOEs. The thicker bars correspond to stronger NOEs.
1H NOESY spectrum regions showing some of the major long-range NOE cross-peaks between (a) W101 HN and L107 HN, (b) W101 Hζ2 and L107 Hδ22, and (c) G105 HN and K110 Hδ1, as well as G105 HN and K110 Hβ#.
1H NMR spectra of 400 μM wild-type (spectra a–e) and W101A (spectra f–j) DV fusion peptides upon titration with either PC or PC/PG (4:1) LUVs. Spectra (a) and (f) were obtained in phosphate buffer without LUVs; (b) and (g) in 6 mM PC LUVs; (c) and (h) in 15 mM PC LUVs; (d) and (i) in 6 mM PC/PG (4:1) LUVs; and (e) and (j) in 15 mM PC/PG (4:1) LUVs. The experiments were carried out at 25 °C.