Abstract
The amyloid precursor protein (APP), that plays a critical role in the development of senile plaques in Alzheimer disease (AD), and the gp41 envelope protein of the human immunodeficiency virus (HIV), the causative agent of the acquired immunodeficiency syndrome (AIDS), are single-spanning type-1 transmembrane (TM) glycoproteins with the ability to form homo-oligomers. In this review we describe similarities, both in structural terms and sequence determinants of their TM and juxtamembrane regions. The TM domains are essential not only for anchoring the proteins in membranes but also have functional roles. Both TM segments contain GxxxG motifs that drive TM associations within the lipid bilayer. They also each possess similar sequence motifs, positioned at the membrane interface preceding their TM domains. These domains are known as cholesterol recognition/interaction amino acid consensus (CRAC) motif in gp41 and CRAC-like motif in APP. Moreover, in the cytoplasmic domain of both proteins other α-helical membranotropic regions with functional implications have been identified. Recent drug developments targeting both diseases are reviewed and the potential use of TM interaction modulators as therapeutic targets is discussed.
Keywords: gp41, Amyloid precursor protein, Transmembrane segment, Membrane, HIV, Alzheimer
1. Introduction
Biological membranes are complex mixtures composed primarily of lipids and proteins. Although membrane proteins represent approximately one third of all proteins encoded in the human genome, and are involved in almost every aspect of cell biology and physiology, there is still little knowledge about how these proteins act and interact in biological membranes [1]. This is despite that more than half of currently marketed pharmaceuticals are targeting membrane proteins [2].
The vast majority of membrane proteins are anchored to cellular membranes through transmembrane (TM) domains that predominantly adopt an α-helical secondary structure [3]. Membrane-spanning α-helices, rather than serving merely as featureless hydrophobic stretches required for anchorage and facilitating insertion of proteins in membranes, have recognized functions well beyond these classical roles (for recent overviews see [4], [5]). The organization and number of TM segments varies between membrane proteins, but it is generally believed that van der Waals interactions play an important role in the packing of TM domains. These interactions compensate for the lack of the hydrophobic effect that drives the folding of water-soluble proteins. Modulation of TM helix–helix interactions provides new exciting means to regulate the functions of membrane proteins. It is well established that homo- or heterodimerization, trimerization and other types of TM associations play important roles in different biological processes [5]. In the present review, we will discuss recent results on the structure, packing determinants and assembly of the TM domains of HIV gp41 and APP. These are both membrane proteins implicated in human diseases of paramount importance. The development of exogenous agents that recognize TM domains can be used for rational drug design [1], [6], and by interfering with TM interactions new targeted therapeutics should be expected in the near future.
2. HIV envelope (Env) glycoproteins
Human immunodeficiency virus type-1 (HIV-1) is an enveloped virus that gains entry into target cells by mediating the fusion of viral and cellular membranes. Entry into cells is directed by the envelope (Env) glycoproteins, which are present on the surface of HIV-1 virions as trimers [7]. HIV-1 Env complex is synthesized as a type-1 TM gp160 precursor, which undergoes oligomerization, disulfide bond formation and extensive glycosylation, and is then post-translationally cleaved into the surface receptor binding subunit gp120 and the TM fusion protein gp41 [8], which remain non-covalently associated [9].
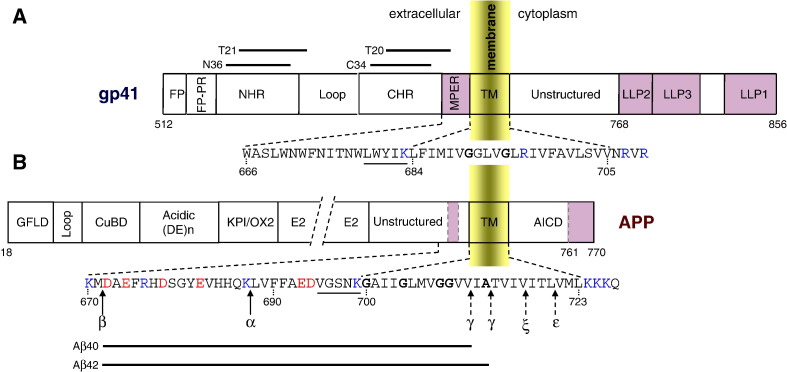
The full-length monomeric gp41 TM glycoprotein consists of three domains (Fig. 1A): an ectodomain (ECD), a TM domain and a large cytoplasmic domain (CTD). Several regions in the ECD are important for membrane fusion activity (see refs [10], [11] for recent reviews): a highly conserved (glycine-rich) hydrophobic fusion peptide (FP), located at the extreme N-terminus; N- and C-terminal heptad repeat (HR) regions (NHR and CHR), connected by a glycosylated 30–40 residue disulfide-bonded loop; and a tryptophan-rich membrane-proximal ectodomain region (MPER). Binding of gp120 to the CD4 cellular receptor on the surface of target cells triggers a series of conformational changes in gp120 subunit that facilitate gp120 binding to a co-receptor, CXCR4 or CCR5, and the exposure of the hydrophobic gp41 fusion peptide. The dynamics of gp41 conformational changes triggering membrane fusion have been reviewed extensively [11], [12], [13]. Briefly, three gp41 NHR regions can adopt a parallel triple-stranded coiled-coil configuration that enables penetration of the gp41 fusion peptide into the membrane of the target cell. Subsequent refolding of gp41 heptad repeat (HR) regions into a six-helix bundle structure (trimer-of-hairpins) forces the juxtaposition of the viral and cell membranes, promoting their fusion [14]. Recently, the C-terminal boundary of this six-helix bundle fusion conformation in an ongoing dynamic fusion process has been demonstrated [15]. At present, it is thought that the structural rearrangements in the gp41 TM glycoprotein are crucial for the membrane fusion process and viral entry [16].
Fig. 1.
Schematic representation of HIV-1 gp41 (A) and APP (B). TM domain and membranotropic sequences in each protein are depicted darker. TM and juxtamembrane regions are enlarged and for APP the sequence involved in processing is shown with the major sites of cleavage by α-, β-, and γ-secretases highlighted. Dashed arrows indicate APP intramembrane cleavage sites. The CRAC motifs are underlined. TM glycines and alanine residues involved in GxxxG/A motifs are shown in bold. Locations of gp41 inhibitor peptides and Aβ 40/42 peptides are depicted with dark lines. Numbering refers to HIV gp160 precursor, BH10 isolate (A) and human APP770 isoform (B). See text for details.
3. gp41 membranotropic sequences
Although gp41 six-helix bundle formation is the main driving force for the fusion process, other gp41 regions in the ECD may regulate fusion activity in numerous ways. The role of the N-terminal fusion peptide region and its implication in membrane destabilization and fusogenic activity has been analyzed in recent reviews [10], [11]. The membrane conformation of the fusion peptide (α-helical/β-strand/disordered) deduced from chemically synthesized FPs in model membranes is controversial [17], [18], [19], probably due to an effect of microenvironment composition dictating the adopted conformation [20]. In this context, gp41 FP, which is unstructured in solution, adopts an α-helical structure in micelles [21], [22], inserting its N-terminal residues in an α-helical conformation and presenting a flexible hinge reminiscent of the kinked structure proposed for several N-terminal fusion peptides [23], [24], [25], [26]. Structural plasticity of gp41 FP has been also observed depending on peptide concentration. When bound to lipid bilayers at low concentration gp41 FP is largely α-helical, however, at higher protein/lipid ratios the domain is partially converted to form β-structures [19]. A 13C FTIR study have demonstrated that this peptide adopts an intermolecular parallel β-sheet structure in membranes when stabilized by the adjacent N-terminal heptad repeat [27]. Recent 2D correlation spectra and distance measurements from solid-state NMR-spectroscopy in cholesterol-containing host-cell-like membranes indicated that the fusion region had predominantly a β-strand conformation [28], with 50–60% population of antiparallel strand orientation [29], that allows close proximity of the A525–G527 region with the lipid headgroups, which would likely perturb the cell membrane [30]. Perhaps both α-helical and β-sheet structures are relevant at different stages of HIV fusion process. Given its α-helix to β-sheet interconversion, elevated alanine and glycine levels, fusogenicity and plaque formation, gp41 FP has been proposed as an ‘amyloid homolog’ (or ‘amylog’) [31], [32], and more recent results suggest that bound to membranes, FP may contribute to cytopathicity of HIV through an amyloid-type mechanism [33].
The gp41 ECD region preceding the TM domain is designated as MPER (membrane-proximal ECD region), or pre-transmembrane, preTM region (reviewed in [34]). This region, which is predicted to form an α-helical conformation on membrane contact (Fig. 2A), contains highly conserved hydrophobic residues and is unusually rich in tryptophan residues (see Fig. 1A). The basis for the invariant nature of the tryptophans appears to be at the level of glycoprotein incorporation into virions, since mutants in this region reduce glycoprotein incorporation and drop the efficiency of virus entry while having no significant effect on syncytium formation [35]. The MPER region actively participates in the clustering of gp41 within the HIV-1 envelope, and in destabilization of the bilayer architecture at the loci of fusion. Interestingly, the MPER carboxy-terminus has a LWYIK sequence immediately preceding the TM segment (see Fig. 1A) that can be identified as a ‘cholesterol recognition/interaction amino acid consensus’ (CRAC) motif [36], [37], [38], [39]. A number of studies have shown that cholesterol-enriched microdomains (lipid rafts) play important roles in both early and late phases of the HIV lifecycle (reviewed in [40], [41]). MPER–cholesterol complex might form at the interface of the external viral membrane monolayer, with the potential of inducing membrane perturbations upon self-assembly [35], [42]. Monolayer intrinsic curvatures could hypothetically change their sign from positive to negative curvature upon MPER desorption, thus facilitating fusion pore opening [34]. Recent mutational studies suggest that both structural (for gp41 stability and incorporation) and functional (membrane disruption) constraints may contribute to the highly conserved nature of the membrane-proximal ECD region [43]. More recently, it has been shown that the highly conserved LWYIK motif acts as a structural determinant in modulating membrane fusion and post-fusion events [44], and that intact plasma membrane cholesterol and lipid raft microdomains are essential for HIV entry in macrophages, a critical target cell type for HIV-1 [45].
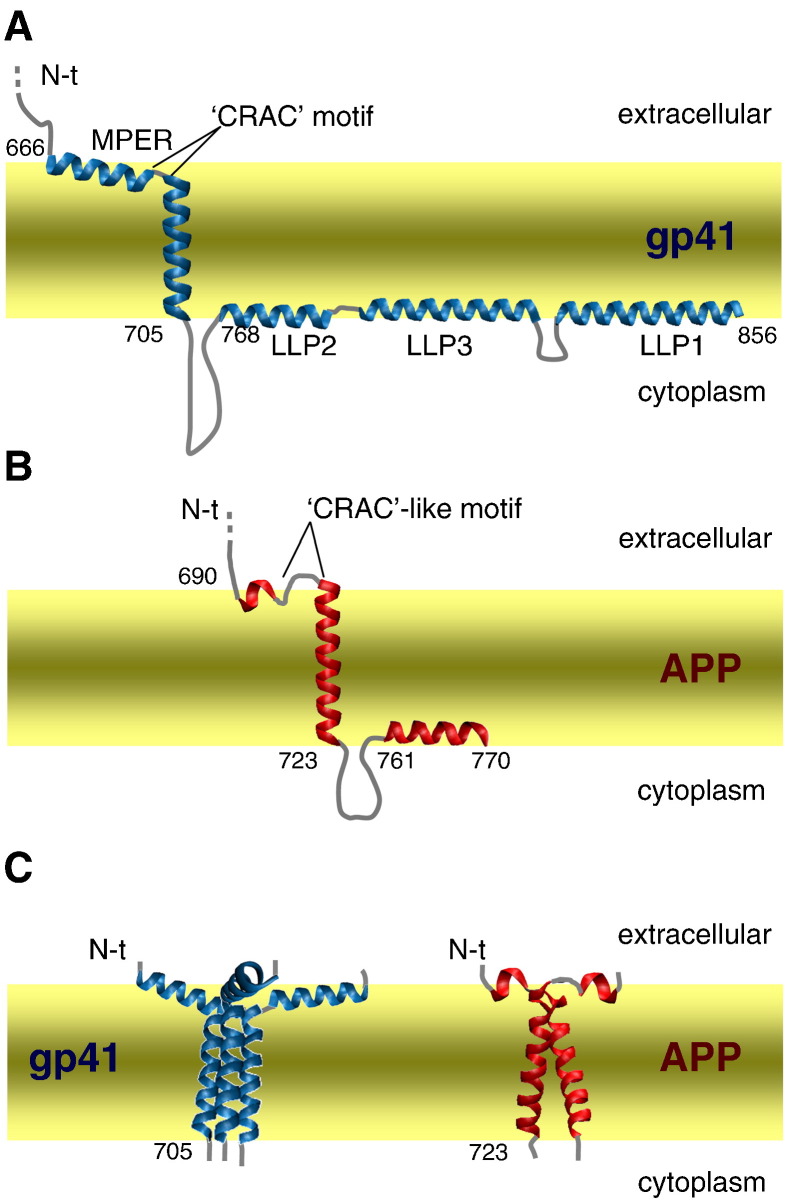
Fig. 2.
Simplified membrane topology models for TM and membranotropic regions. (A) Monomeric HIV-1 gp41. (B) Monomeric APP. (C) TM segment oligomeric schemes: gp41 trimer (left) and APP dimer (right). The predicted location of gp41 TM segment, α-helical MPER region and LLP helices are shown. The two surface-membrane-associated helices of APP are depicted according to [107].
The MPER may act in conjunction with other regions in the gp41 ECD to maintain the native and fusion states. The FP and MPER sequences can assemble restricting FP-mediated fusion [46], [47]. Extensive studies with synthetic peptides have recently pointed out that the MPER W666–N677 segment interacts with a fusion peptide proximal region S528–Q540 (FP-PR, Fig. 1A), and that this interaction contributes to stabilize gp41 six-helix bundle formation [48]. FP-PR and MPER can act synergistically in forming a fusion-competent gp120/gp41 complex and in stabilizing the membrane-interactive end of the trimer-of-hairpins [49]. In fact, hydrophobicity-at-interface analysis of gp41 MPER mutants [34] predicts the existence of a gp41 region anchored to the viral membrane through an interfacial sequence, amphipathic at its N-terminus, which is followed by the hydrophobic TM domain (Fig. 2A).
Other membranotropic regions have been located in the gp41 cytoplasmic domain (Fig. 2A). Thus, the lentiviral lytic peptide (LLP) sequences [50], [51], [52], three amphipathic helical segments, might function in modulating fusogenicity during envelope processing and viral membrane fusion or budding [11], [53]. Also, progressive truncations and point mutations in the gp41 cytoplasmic tail have demonstrated that the C-terminal end plays a key role in coupling HIV-1 fusion competence to virion maturation [54]. Furthermore, photoinduced chemical reaction studies with a membrane-embedded probe provide a further demonstration that portions of gp41 cytoplasmic tail are partially inserted into the viral membrane [55].
4. gp41 transmembrane domain
The gp41 TM domain is one of the most conserved regions of the gp41 sequence among independent isolates of HIV-1, and is primarily responsible for anchoring the envelope glycoproteins into the viral membrane (Fig. 2A). In contrast to the membrane-spanning region of most viral envelope proteins that consist of stretches of hydrophobic amino acids, the TM region of lentivirus envelope glycoproteins is generally interrupted by charged residues [56]. In this regard, HIV-1 not only has one lysine (K683) and two arginines (residues 707 and 709) flanking the membrane-spanning region, but also contains one basic residue (R696) that is predicted to be located within the hydrophobic TM region (Fig. 1A). In the absence of high-resolution structural information on the HIV-1 TM domain, the exact length of this region remains unresolved. At present, two models have been proposed for the membrane spanning of HIV-1 gp41 protein. In the first model 25 amino acid residues were suggested to cross the membrane as an α-helix, with a length similar to the thickness of the lipid bilayer [57]. In this model R696 is placed in the hydrophobic core of the bilayer, without any known mechanism to neutralize the basic groups. In this context, C-terminal truncation studies of simian immunodeficiency virus (SIV) [58] and HIV-1 [59] indicate that the entire 25-amino acid region is not required for the biological function of Env glycoproteins. Therefore, a second model was proposed [58] based on the capacity of polar residues to ‘snorkel’, i.e. they may bury their aliphatic part in the hydrophobic region of the lipid bilayer, while positioning the charged group in the more polar interface. In this model, twelve amino acid residues will form the hydrophobic helical core buried within the membrane [59], with K683 and R696 (BH10 isolate numbering) residues flanking the hydrophobic core region. Thus, membrane-embedded charged residues can be neutralized by side-chain interactions with negatively charged lipid polar head groups. This latter membrane disposition precludes any polar residues from being placed at the hydrophobic lipid core since the polypeptide membrane-spanning region is significantly reduced, probably accompanied by elastic distortion of the surrounding lipid chains [60]. Interestingly, influenza virus hemagglutinin (HA) studies have indicated similar TM requirements. The presence of a single arginine residue allowed a shortened HA TM domain to span the bilayer, most likely by an interaction between the guanidinium group with phosphate head groups of the viral membrane [61].
The process of membrane fusion initiated by gp120-CD4 binding seems to be dependent on the structural integrity of the TM domain [62], and it was found that a minimum length of the TM segment was crucial for the membrane fusion function of the protein [63]. Thus, in addition to membrane anchorage, gp41 TM domain may be directly or indirectly involved in membrane fusion events. In fact, the replacement of the HIV-1 TM segment with a glycophospholipid anchor abrogated both cell–cell fusion and virus–cell fusion, though the chimeric protein could be normally expressed, processed, and incorporated into virions [64]. Also, the complete substitution of gp41 TM segment by that of cellular glycophorin A (GpA), vesicular stomatitis virus G protein (VSV-G), or HA glycoprotein was found to severely impair the fusion activity of the chimeric molecules [65], [66].
According to the consensus sequence of HIV-1 TM segment [59], the twelve amino acid residues in the core region (L684–L695, Fig. 1A) are more conserved than those in the flanking residues. Within these hydrophobic residues, recent studies have focused on the GxxxG motif (G, glycine; x, other amino acid residues), since these glycine residues are the most conserved among all HIV-1 TM sequences [59], [65], [67]. This motif is often observed in TM α-helices of both cellular and viral proteins (Fig. 3 ), and has been proven to stabilize helix–helix interactions of membrane proteins [3]. In the case of GpA, this motif is critical for homodimerization [68], [69], although surrounding residues might ‘fine-tune’ the affinity of helix–helix interaction [70], [71], [72]. In fact, the length of the hydrophobic region has also been found to be critical for in vitro dimerization of the native GpA sequence [73], as well as in a polyleucine scaffold where the dimerization motif was grafted [74]. Similarly, the GxxxG motif in the E1 glycoprotein of hepatitis C virus (HCVE1) is important for the heterodimerization of its E1 and E2 envelope glycoproteins [75]. On the other hand, structural and functional studies of a gp41 construct comprising the complete C-terminal heptad repeat region, the connecting region, and the TM segment have shown that the TM domain of gp41 is sufficient to drive trimerization in vitro [57].
Fig. 3.
Amino acid sequences of membrane-spanning domains containing GxxxG and/or GxxxG-like motifs, for GpA and other membrane proteins from viral and neuronal origin. The amino acid residues of the putative TM domains are shown as upper case letters and flanking sequences are shown as lower case. Glycine and alanine residues in GxxxG/A motifs are in bold. HIV-1 consensus TM sequence (according to Shang et al. [59]); ScoV-S: SARS coronavirus spike protein; APP and ErbB4 are γ-secretase substrates; presenilin-1 (PS-1) and anterior pharynx defective protein-1 (APH-1) are components of the γ-secretase complex; p75: neurotrophin receptor protein [149]. For other abbreviations see text.
While the TM domain of HIV-1 gp41 is highly resistant to amino acid exchanges, mutations in the GxxxG motif (G690–G694) affected viral fusion events [65], [66], [67]. In particular, HIV-1 gp41 TM segment has a glycine residue at position 691 that forms a GGxxG sequence. Replacement of G691 with alanine, phenylalanine or leucine, decreased the efficiency of membrane fusion, with the major effect occurring with the leucine substitution [67]. Substitution with leucine residue also decreased the incorporation of gp41 protein into virions, suggesting that the steric nature of the side chain of the residue at position 691 is important for gp41 function. In the light of these results, a model for the association among three TM helices of gp41 has been proposed in which the GxxxG motif is facing inward of the trimeric structure and G691 locates itself near the helix–helix interface [67]. This model places the highly conserved arginine residue (R696) toward the lipid environment, which is in principle a thermodynamically unfavorable arrangement. However, arginine residues may be accommodated into a lipid bilayer more easily than expected [76], [77], [78]. In any case, since the atomic structure of the gp41 TM segment in lipid environment has not been yet solved, other putative arrangements of the trimeric gp41 TMs are also possible.
Recently, the hydrophobic core region of gp41 TM segment was replaced with 12 leucine residues (L12 mutant) and recovery-of-function mutants then constructed in which specific amino acid residues (including a GGxxG motif) were reintroduced [59]. Mutant L12 was defective in mediating virus infectivity and cell–cell fusion. The GGxxG motif was found critical for the membrane fusion process mediated by gp41, since reintroduction of this motif into the leucine scaffold of the L12 mutant significantly increased the efficiency of cell–cell fusion and infectivity of HIV-1. Moreover, improvement of gp41 fusogenicity was achieved by reintroducing additional F685 and V689 (BH10 isolate numbering). Thus, several of the 12 amino acid residues in the HIV-1 TM core region were implicated in the efficiency of gp41-mediated membrane fusion, consistent with the high conservation of this sequence. Probably, gp41 GGxxG motif can facilitate TM–TM interactions that are necessary for the formation of higher-order fusion pore [59]. Membrane fusion reactions catalyzed by viral fusion proteins require the concerted action of multiple fusion protein trimers [79], and for some HIV-1 isolates between 7 and 14 trimers have been suggested [80]. Lipid mixing precedes Env recruitment [81], and it has been proposed that the merge of the outer leaflets of apposing membranes could initiate with one or few functional trimers at the contact site, with further progress facilitated by the continuous recruitment of adjacent Env subunits [81].
A wealth of information indicates then that HIV fusion proteins mediate membrane fusion by forming a trimeric conformer. It is tempting to speculate that helix–helix interactions between the gp41 TM segments might be initially responsible for the induction of trimer formation, previous to the trimer-of-hairpins folding that is triggered by the heptad repeat regions. As a matter of fact, a chimeric version comprising the gp41 TM domain but lacking the full heptad repeat regions showed that the TM segment constitutes the trimerization domain [57]. Hence, interfering gp41 TM interactions may become an interesting target in AIDS research including development of new and novel anti-HIV inhibitors.
5. HIV-1 Env-mediated membrane fusion (Entry) inhibitors
HIV enters its target cells by means of a sequence of molecular events that provides one of the most attractive targets for inhibitor development. Recent reviews have analyzed CD4-inhibitors and co-receptor-binding inhibitors [4], [10], [11], [82], [83]. On the other hand, receptor recruitment, a prerequisite for fusion, has been shown to be sensitive to lipid modulation, and a number of strategies have been used to alter lipid content of target cells in order to decrease their susceptibility to HIV-1 entry (reviewed in [11]). Other peptide-based inhibitors derived from gp41 NHR and CHR regions such as T21, N36, T20, C34 (Fig. 1A) and chimeric proteins, or small-molecule inhibitors, that interfere with intermediate gp41 structural arrangements have been extensively studied and reviewed elsewhere [10], [83], [84], [85], [86].
Despite all these entry inhibitor developments, at present UK-427857 (Maraviroc, Selzentry (Pfizer)), an attachment inhibitor that blocks the chemokine receptor CCR5, and T20 (Enfuvirtide, Fuzeon (Roche)) are the only FDA-approved HIV inhibitors used for AIDS treatment in patients that fail to respond to antiretroviral therapeutics, but can easily induce drug resistance. Relative to Maraviroc, researchers also question the long-term safety of blocking CCR5, a receptor whose function in healthy individuals is currently not fully understood. In the last years, efforts directed to enhance the biological potency of peptide-based inhibitors, i.e., α-helix-inducible X-EE-XX-KK motifs have been applied to design a CHR-based enfuvirtide analogue that exhibits highly potent anti-HIV activity in vitro [87]. Novel peptide fusion inhibitors have also been designed based on the gp41 fusogenic-core structure involving the upstream region of the binding domain in the CHR region. These peptides have been found highly effective against HIV-1 variants resistant to T20 and C34 [88], [89]. Another anti-HIV peptide, termed sifuvirtide, which is based on the three-dimensional structure of the gp41 fusogenic core, exhibits high potency against infections by a wide range of primary and laboratory-adapted HIV-1 isolates and T20-resistant strains, and is currently in Phase II of clinical studies [90]. Finally, concerning gp41 membranotropic regions, using a synthetic combinatorial library several hexapeptides were identified that inhibited fusion peptide activity in model membranes [91].
6. Amyloid precursor protein and Alzheimer disease
Alzheimer disease (AD) is characterized by the presence of two types of lesions in the brain: neurofibrillary tangles and senile plaques. Amyloid precursor protein (APP) is a ubiquitous-glycosylated type-1 TM protein that plays a central role in the development of extracellular senile plaques, through the generation of a peptide called amyloid-β (Aβ) by proteolysis of the precursor protein (see [92], [93] and references therein).
Full-length APP contains three domains (Fig. 1B): a large ECD that represents around 85% of the total protein mass (for the main neuronal isoform), a single-spanning TM domain, and a small cytoplasmic domain. The ECD consists of several subdomains with functional implications: a cysteine-rich region at the N-terminus with two subdomains, a growth factor-like domain (GFLD), which binds heparin (named also heparin-binding domain 1, HBD1), responsible for neurite outgrowth [94], and a copper-binding domain (CuBD) [95]. Cu2+ binding to CuBD reduces Aβ production, probably through some alterations in signalling mechanisms and/or changes in the APP oligomerization state [96]. A 21-residue disulfide-bonded loop connects the GFLD with the CuBD domain and seems to be crucial for APP homodimerization [97], [98]. The cysteine-rich region is followed by an acidic, random coil, Asp- and Glu-rich region (acidic domain or (DE)n domain), which contains two tyrosine-phosphorylation sites [99], a Kunitz-type protease inhibitor (KPI) domain and an OX2 domain. The KPI and OX2 domains can be spliced out, to produce three main variants: APP770 (770 amino acid residues), APP751 and APP695, with the later being the principal neuronal isoform of human APP. The KPI domain can influence adhesion to extracellular matrix constituents, the activity of secreted APP-degrading proteases, and may act as a ligand for LPR1, a member of the LDL receptor gene family [100], [101].
Following these domains there is a glycosylated domain (referred to as E2) and a largely unstructured juxtamembrane, random coil (RC) region that precedes the TM domain. The E2 domain possesses a RERMS sequence that may be implicated in APP's growth-promoting properties and binds heparin (named also heparin-binding domain 2, HBD2). The α- and β-secretase cleavage sites are located within the RC region and it is possible that this sequence acquires a secondary and/or tertiary structure in the presence of secretases as recently suggested [92]. The APP intracellular domain (AICD) is the center of a complex network of protein–protein interactions whose relevance is highly controversial [102], [103].
Two sites in the ECD seem to be critical for full-length APP homodimerization [97], [104], and a third site localized within the TM sequence of APP determines γ-secretase cleavages [105]. The homophilic binding mechanism of APP is actually a subject of debate, with enormous interest due to putative implications in APP function and regulatory aspects for APP amyloidogenic processing.
7. APP transmembrane domain and membranotropic regions
The TM segment of APP is highly hydrophobic (Fig. 1B) and computer algorithms predict an α-helix of 24 amino acids that can span the plasma membrane. Fifteen point mutations associated with familial Alzheimer disease (FAD) localize in the TM region at positions 705, 713, 714, 715, 716, 717 and 723 according to the Alzheimer Disease Mutation Database [106].
In the absence of full-length APP atomic structure, it has been recently reported the first structural study of the 99-residue C-terminal region (C99) of APP (residues D672–N770, Fig. 1B) that includes the TM domain [107]. NMR data in lysomyristoylphosphatidylglycerol (LMPG) micelles reveals three α-helical segments (Fig. 2B): i) a short surface-associated helix (F690–E693) preceding the TM segment at the extracellular side that may serve as an small anchor to organize the connecting loop to the TM domain, ii) a membrane-spanning region (G700–L723), which is essential for Aβ production, iii) an amphipathic membranotropic helix at the C-terminal end of the cytosolic domain (T761–N770) that plays critical roles in APP trafficking and protein–protein interactions. Three canonical GxxxG motifs are present (Fig. 1B), one located in the juxtamembrane ECD (G696SNKG700), with two others in the TM region (G700AIIG704 and G704LMVG708). It should be noted that a GxxxG-like motif, G709VVIA713, (a glycine residue is substituted by alanine) is also present, where the γ-secretase cleavage sites that generate Aβ peptides are localized (see Fig. 1B). As previously mentioned, GxxxG motifs are responsible for homodimerization of GpA and many other membrane proteins [108], sometimes with repeats in tandem (Fig. 3) [109]. Recent circular dichroism spectroscopy and fluorescence resonance energy transfer studies indicate that synthetic peptides corresponding to TM segments of APP adopted similar highly α-helical structures in sodium dodecyl sulphate (SDS) micelles and phosphatidylglycerol vesicles, and form stable dimers in both systems [110]. Interestingly, C99 was also observed to form dimers in LMPG micelles. In fact, the isolated TM peptides dimerize more avidly than full-length C99, indicating that the native extramembrane domains can influence dimer association [107].
In recent years, at least three models have been proposed for the APP TM dimeric structure, all involving GxxxG or GxxxG-like motifs. In the first model based on site-directed mutagenesis in a neuronal cell system, it was predicted that the interaction that stabilizes homodimer helix–helix contacts is primarily mediated by the GxxxG motif located at the beginning of the TM region [105], i.e. G700AIIG704 in the hAPP770 sequence. Mutations of glycine residues in this motif gradually attenuate the TM dimerization strength. γ-secretase cleavages of APP were shown to be intimately linked to the dimerization strength of the TM substrate and a sequential mechanism for γ-secretase cleavages on dimeric APP TMs (see below) has been postulated [105]. Using synthetic peptides corresponding to the APP TM segment (G700 to L723) and FAD mutant derivatives, a second model for TM dimer formation in SDS micelles has been proposed [110]. This model displays many similarities with the NMR structure of GpA TM helix dimer [111]. According to this second model, the putative dimerization interface relies on the G709VVIA713 sequence (GpA uses G79VMAG83), with similar packing interfaces for both sequences (APP 705–717 and GpA 75–87). It was hypothesized that FAD–APP mutations would destabilize the APP-TM dimer, increasing the population of APP peptide monomers. Thus, it was argued that these mutations are ideally located to disrupt APP dimerization [110]. In the third model [112], supported by recent solid-state NMR data [113], all three canonical GxxxG motifs (the juxtamembrane and the two located in the TM segment) have been proposed to simultaneously participate in the helix–helix interactions that are responsible for homodimerization. In this model, the dimer interface is lined by glycines at positions 696, 700, 704, and 708 (hAPP770 numbering). It was demonstrated that pairwise replacement of glycines by leucines or isoleucines, but not alanines, in the central GxxxG motif results in a decrease in total Aβ production (γ-cleavage) without affecting the yield of AICD (ɛ-cleavage, see below). In this respect, molecular dynamic simulations predicted that all the Gly residues involved in the GxxxG motifs are located at the interface of the wild type TM dimer, and that Gly to Leu or Ile mutations will cause a rotation and the placement of other small residues in the interface (i.e. Gly709 and Ala713), which hinders Aβ generation [112]. In the same direction, replica-exchanged molecular dynamics simulations have predicted that the changes induced by these mutations might be due to the adoption of a different dimer conformation with a shift of some residues relative to the bilayer normal producing a mismatch between the γ-cleavage site and the active site of γ-secretase, which would reduce Aβ secretion [114].
8. APP processing and transmembrane cleavage
APP is subject to alternative pathways of proteolytic processing, leading either to production of the Aβ peptides or to non-amyloidogenic fragments. α- or β-secretase (Fig. 1B) cleavage release its large ECD from the cell surface, a process referred to as ‘shedding’. APP770 processing by α-secretase results in cleavage after K687 and release of an 83-residue C-terminal fragment, C83 [93]. C83 is a TM polypeptide that is further processed by γ-secretase, and the resulting peptide products are not amyloidogenic. Alternatively β-secretase (β-site APP cleaving enzyme 1, BACE1) cleaves after M671 leading to a 99-residue TM C-terminal domain, C99 [115], [116]. The preferential cell surface partitioning of APP to cholesterol-enriched lipid rafts (which can then be internalized to endosomes enriched with β-secretase activity) is believed to be a decisive determinant of the competition between β- and α-secretase for initial proteolysis of APP [107], [117], [118]. Subsequent cleavage of C99 by γ-secretase at membrane-embedded sites leads to release of both the Aβ peptides and the water-soluble AICD domain (V721–N770). Aβ peptides have heterogeneous C-termini due to a somewhat imprecise intramembrane cleavage, and these peptides displayed different propensity to aggregate and to form amyloid deposits in neural tissue [119]. Aβ peptides have been the subject of extensive structural characterization in solution, bound to model membranes or as part of aggregates (reviewed in [120], [121]). It has been proposed that the sequence represented by Aβ may adopt an α-helical structure when present in the full-length APP [112]. After BACE cleavage of APP at the β-position, C99 will likely assume a dimer conformation with the GxxxG motifs in the interface (see above), and will be processed to form Aβ peptides and AICD. Once Aβ is generated, the GxxxG motifs (in other words, the abundance of β-prone residues like glycine) would then promote a conformational change from α-helix to β-strand with the formation of neurotoxic amyloid fibrils [122].
Similar to other intramembrane cleaving proteases, γ-secretase cleaves C99-derived polypeptide at several locations (γ-site, ζ-site and ɛ-site, Fig. 1B) probably through a sequential proteolytic pathway (processive model, reviewed in [115]). This proteolysis would start at the C-terminus of TM α-helix, near the membrane–cytosolic interface, sequentially yielding Aβ49 precursor (ɛ-cleavage), Aβ46 (ζ-cleavage), and Aβ42/40 (γ-cleavage), respectively. Inhibition of C99 dimerization by mutating its first TM GxxxG motif has been shown to reduce the production of Aβ42/40 forms with an increase of non-pathogenic Aβ38/35 shorter forms [105]. Thus, it has been also proposed that γ-secretase removes consecutive fragments from the dimeric helical TM C-terminus, helix turn by helix turn, until cleavage reaches a certain distance from the G700AIIG704 dimerization motif, since this motif sterically hinders γ-secretase proteolysis. Recent structural data support this progressive cleavage mechanism that requires the protease access to the protein backbone to depend on a secondary structure change (here a helix-to-coil transition) at the TM-juxtamembrane interface [113]. In this process, the final cleavages occur after residue V711 and/or A713 and produce mainly Aβ40/42 peptides respectively (Fig. 1B). Perturbed dimers (G700/G704 substituted to alanine) resolve the steric hindrance and allow the γ-secretase to proceed, yielding Aβ38/35 shorter peptides [105]. Membrane retention studies of Aβ segments in microsomal membranes suggests that shorter segments (Aβ 40–45) are not integrated into the membrane, while longer ones (Aβ 46–49) are efficiently retained in the lipid milieu [123]. Since dimer formation seems to be an important feature for γ-secretase activity and this process is likely driven by helix–helix packing, modulating TM interactions arise as a new target for Alzheimer therapeutic intervention.
APP processing is affected by other factors. γ-secretase and its substrates are compartmentalized into discrete membrane microdomains and emerging evidence suggest an intimate relationship between cholesterol-containing lipid rafts and APP processing [124], [125]. Recent NMR studies indicate that C99 specifically binds cholesterol, an agent that promotes the amyloidogenic pathway [118], [126], at the loop connecting the short membrane-associated helix to the TM domain [107]. Interestingly, this loop contains a VGSNK sequence immediately preceding the TM region (Fig. 1B) that can be considered as a CRAC-like motif (canonical Tyr in the middle of the motif [37] is substituted by Ser, both Y/S side chains contain a hydroxyl group that can satisfy cholesterol interaction as proposed [36]). Cholesterol binding of this region would be based on H-bond interactions by wrapping and blocking the interfacial cholesterol OH-group. This interaction would give APP the capacity to bind cholesterol-rich domains in biological membranes. On the other hand, protein–protein interactions probably also contributes to APP's propensity to partition into cholesterol-rich domains. In this regard, it has been recently shown that flotillins facilitate clustering of both APP and cholesterol in raft-like membrane domains [127], and LDL receptor-related proteins (LRPs) have been also implicated in raft association, internalization and amyloidogenic processing of APP [101], [107], [128]. Moreover, various cytosolic adaptor proteins have been reported to bind APP and influence its processing [103], [115], and novel insights for APP phosphorylation have been documented [129]. An interactome map has been derived that confirmed eight previously reported interactions of APP and revealed the identity of more than 30 additional proteins that reside in spatial proximity to APP in the brain [130]. The putative role of these proteins in APP processing remains to be determined. Additionally, it has been reported that protein kinases that phosphorylate APP are able to phosphorylate the neuronal protein tau (present in the intraneuronal neurofibrillary tangles). It has been argued that this may be an important factor linking the two characteristic lesions of Alzheimer disease [131].
9. Therapies in Alzheimer disease
Therapeutic approaches for AD are guided by four disease characteristics: amyloid plaques, neurofibrillar tangles, neurodegeneration, and dementia (reviewed in [132], [133]). Current treatments for dementia symptoms are based on the ‘cholinergic deficit hypothesis’ and include FDA-approved, cognition-enhancer, acetylcholinesterase inhibitors (ChEIs) (reviewed in [134], [135]), and an NMDA (N-Methyl-d-Aspartate) receptor antagonist used as adjuvant to ChEI therapy [135]. Since ChEIs have modest efficacy, recent drug developments point towards M1 muscarinic agonists [134] and multi-target-directed ligands as potential disease modifiers [136], [137].
However, the most used hypothesis explaining the pathophysiology of AD is the ‘amyloid hypothesis’ centered in the premise that accumulation of Aβ in the brain leads to oxidative stress, neuronal destruction and finally the clinical syndrome of AD [135]. According to this, a great number of anti-amyloid therapies are currently under investigation and clinical trials are in progress. Efforts are being directed to: i) decreasing Aβ production, including inhibition of β- [138] and γ-secretase, and modulators to produce shorter, non-toxic Aβ fragments [139], as well as activation of α-secretase pathway [135]; ii) increasing Aβ clearance including Aβ immunotherapy [140], active (vaccination) and passive (monoclonal antibodies) immunization [141], and Aβ degradation proteases [142]; iii) inhibition of Aβ aggregation including peptides or peptidomimetics [143], β-sheet packing peptide inhibitors [122], polyphenols [144], intervention of Aβ-metal (Zn/Cu) interactions [145] and glycosaminoglycan inhibitors [146]. At present, the γ-secretase modulator Tarenflurbil or Flurizan [147] and a synthetic glycosaminoglycan (3APS, tramiprosate, Alzhemed) Aβ-aggregation inhibitor [148], are being tested in phase III trials.
10. Concluding remarks
APP and gp41 TM proteins shared astounding structural characteristics and sequence motifs with functional significance, especially in their TM domains and membranotropic regions. The presence of GxxxG motifs that drives protein oligomerization in membranes, and the similarity in the location of a specific cholesterol-binding site (CRAC motif) that can facilitate the clustering of the proteins in cholesterol-rich domains are clearly remarkable (Fig. 2). β-amyloid in Alzheimer disease is related with APP association with cholesterol-enriched microdomains for conversion from non-pathogenic to pathogenic forms. Accumulating evidence suggests that many viruses may hijack these dynamic lipid platforms to be used as an entry portal to the target cell. Even more, similarly to Aβ peptide, the gp41 FP region has been considered amyloidogenic (‘amyloid-like’ peptide) [32], and recent results have suggested that FP bound to membranes as β-sheets may contribute to the cytopathocity of HIV in vivo through an amyloid-type mechanism [33]. Finally, it is worth mentioning that, as previously stated, the gp41 TM domain is essential for HIV fusion activity and APP TM domain is critical for Aβ production. To the extent that in both cases oligomerization of the TM domains play a relevant role, it is tempting to speculate that agents that modulate helix–helix interactions may also be effective new therapies. In the last years efforts are being directed to design peptides and small molecules that can interact with TM helices in a sequence-specific manner [5], [6]. All in all, interfering TM interactions by searching for drugs that selectively modulate helix–helix packing may be a promising new target for HIV inhibitor development and to intervene with APP amyloidogenic processing.
Acknowledgments
We wish to thank Dr. P. Whitley (University of Bath) for critical reading of the manuscript, and the financial support obtained from the Spanish Ministry of Education and Science (MEC) (BFU2006-08542/BMC and BFU2009-08401/BMC to I.M.). S. T. was recipient of a predoctoral fellowship from the University of Valencia (V Segles).
References
- 1.Yin H. Exogenous agents that target transmembrane domains of proteins. Angew Chem. Int. Ed. Engl. 2008;47:2744–2752. doi: 10.1002/anie.200704780. [DOI] [PubMed] [Google Scholar]
- 2.Elofsson A., von Heijne G. Membrane protein structure: prediction versus reality. Annu. Rev. Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- 3.Mackenzie K.R. Folding and stability of alpha-helical integral membrane proteins. Chem. Rev. 2006;106:1931–1977. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 4.Cymer F., Schneider D. Lessons from viruses: controlling the function of transmembrane proteins by interfering transmembrane helices. Curr. Med. Chem. 2008;15:779–785. doi: 10.2174/092986708783955545. [DOI] [PubMed] [Google Scholar]
- 5.Moore D.T., Berger B.W., DeGrado W.F. Protein–protein interactions in the membrane: sequence, structural, and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin H., Slusky J.S., Berger B.W., Walters R.S., Vilaire G., Litvinov R.I., Lear J.D., Caputo G.A., Bennett J.S., DeGrado W.F. Computational design of peptides that target transmembrane helices. Science. 2007;315:1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 7.Zhu P., Liu J., Bess J., Jr., Chertova E., Lifson J.D., Grise H., Ofek G.A., Taylor K.A., Roux K.H. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 8.Wyatt R., Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 9.Colman P.M., Lawrence M.C. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 10.Peisajovich S.G., Shai Y. HIV gp41: a viral membrane fusion machine. In: Fischer W.B., editor. vol. 1. Kluwer Academic/Plenum Publishers; New York: 2005. pp. 35–47. (Viral Membrane Proteins: Structure, Function and Drug Design). [Google Scholar]
- 11.Jacobs A., Garg H., Viard M., Raviv Y., Puri A., Blumenthal R. HIV-1 envelope glycoprotein-mediated fusion and pathogenesis: implications for therapy and vaccine development. Vaccine. 2008;26:3026–3035. doi: 10.1016/j.vaccine.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 13.Melikyan G.B. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melikyan G.B., Markosyan R.M., Hemmati H., Delmedico M.K., Lambert D.M., Cohen F.S. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wexler-Cohen Y., Shai Y. Demonstrating the C-terminal boundary of the HIV 1 fusion conformation in a dynamic ongoing fusion process and implication for fusion inhibition. FASEB J. 2007;21:3677–3684. doi: 10.1096/fj.07-8582com. [DOI] [PubMed] [Google Scholar]
- 16.Lev N., Fridmann-Sirkis Y., Blank L., Bitler A., Epand R.F., Epand R.M., Shai Y. Conformational stability and membrane interaction of the full-length ectodomain of HIV-1 gp41: implication for mode of action. Biochemistry. 2009;48:3166–3175. doi: 10.1021/bi802243j. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Z., Yang R., Bodner M.L., Weliky D.P. Conformational flexibility and strand arrangements of the membrane-associated HIV fusion peptide trimer probed by solid-state NMR spectroscopy. Biochemistry. 2006;45:12960–12975. doi: 10.1021/bi0615902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichert J., Grasnick D., Afonin S., Buerck J., Wadhwani P., Ulrich A.S. A critical evaluation of the conformational requirements of fusogenic peptides in membranes. Eur. Biophys. J. 2007;36:405–413. doi: 10.1007/s00249-006-0106-2. [DOI] [PubMed] [Google Scholar]
- 19.Li Y., Tamm L.K. Structure and plasticity of the human immunodeficiency virus gp41 fusion domain in lipid micelles and bilayers. Biophys. J. 2007;93:876–885. doi: 10.1529/biophysj.106.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salom D., Perez-Paya E., Pascal J., Abad C. Environment- and sequence-dependent modulation of the double-stranded to single-stranded conformational transition of gramicidin A in membranes. Biochemistry. 1998;37:14279–14291. doi: 10.1021/bi980733k. [DOI] [PubMed] [Google Scholar]
- 21.Gabrys C.M., Weliky D.P. Chemical shift assignment and structural plasticity of a HIV fusion peptide derivative in dodecylphosphocholine micelles. Biochim. Biophys. Acta. 2007;1768:3225–3234. doi: 10.1016/j.bbamem.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaroniec C.P., Kaufman J.D., Stahl S.J., Viard M., Blumenthal R., Wingfield P.T., Bax A. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 23.Han X., Bushweller J.H., Cafiso D.S., Tamm L.K. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 24.Gomara M.J., Mora P., Mingarro I., Nieva J.L. Roles of a conserved proline in the internal fusion peptide of Ebola glycoprotein. FEBS Lett. 2004;569:261–266. doi: 10.1016/j.febslet.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 25.White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nieva J.L., Agirre A. Are fusion peptides a good model to study viral cell fusion? Biochim. Biophys. Acta. 2003;1614:104–115. doi: 10.1016/s0005-2736(03)00168-8. [DOI] [PubMed] [Google Scholar]
- 27.Sackett K., Shai Y. The HIV fusion peptide adopts intermolecular parallel beta-sheet structure in membranes when stabilized by the adjacent N-terminal heptad repeat: a 13C FTIR study. J. Mol. Biol. 2005;350:790–805. doi: 10.1016/j.jmb.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 28.Qiang W., Weliky D.P. HIV fusion peptide and its cross-linked oligomers: efficient syntheses, significance of the trimer in fusion activity, correlation of beta strand conformation with membrane cholesterol, and proximity to lipid headgroups. Biochemistry. 2009;48:289–301. doi: 10.1021/bi8015668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiang W., Bodner M.L., Weliky D.P. Solid-state NMR spectroscopy of human immunodeficiency virus fusion peptides associated with host-cell-like membranes: 2D correlation spectra and distance measurements support a fully extended conformation and models for specific antiparallel strand registries. J. Am. Chem. Soc. 2008;130:5459–5471. doi: 10.1021/ja077302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiang W., Yang J., Weliky D.P. Solid-state nuclear magnetic resonance measurements of HIV fusion peptide to lipid distances reveal the intimate contact of beta strand peptide with membranes and the proximity of the Ala-14-Gly-16 region with lipid headgroups. Biochemistry. 2007;46:4997–5008. doi: 10.1021/bi6024808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callebaut I., Tasso A., Brasseur R., Burny A., Portetelle D., Mornon J.P. Common prevalence of alanine and glycine in mobile reactive centre loops of serpins and viral fusion peptides: do prions possess a fusion peptide? J. Comput. Aided Mol. Des. 1994;8:175–191. doi: 10.1007/BF00119866. [DOI] [PubMed] [Google Scholar]
- 32.Saez-Cirion A., Nieva J.L., Gallaher W.R. The hydrophobic internal region of bovine prion protein shares structural and functional properties with HIV type 1 fusion peptide. AIDS Res. Hum. Retroviruses. 2003;19:969–978. doi: 10.1089/088922203322588323. [DOI] [PubMed] [Google Scholar]
- 33.Gordon L.M., Nisthal A., Lee A.B., Eskandari S., Ruchala P., Jung C.L., Waring A.J., Mobley P.W. Structural and functional properties of peptides based on the N-terminus of HIV-1 gp41 and the C-terminus of the amyloid-beta protein. Biochim. Biophys. Acta. 2008;1778:2127–2137. doi: 10.1016/j.bbamem.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorizate M., Huarte N., Saez-Cirion A., Nieva J.L. Interfacial pre-transmembrane domains in viral proteins promoting membrane fusion and fission. Biochim. Biophys. Acta. 2008;1778:1624–1639. doi: 10.1016/j.bbamem.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzwedel K., West J.T., Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Epand R.F., Thomas A., Brasseur R., Vishwanathan S.A., Hunter E., Epand R.M. Juxtamembrane protein segments that contribute to recruitment of cholesterol into domains. Biochemistry. 2006;45:6105–6114. doi: 10.1021/bi060245+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epand R.M. Proteins and cholesterol-rich domains. Biochim. Biophys. Acta. 2008;1778:1576–1582. doi: 10.1016/j.bbamem.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 38.Vishwanathan S.A., Thomas A., Brasseur R., Epand R.F., Hunter E., Epand R.M. Large changes in the CRAC segment of gp41 of HIV do not destroy fusion activity if the segment interacts with cholesterol. Biochemistry. 2008;47:11869–11876. doi: 10.1021/bi8014828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vishwanathan S.A., Thomas A., Brasseur R., Epand R.F., Hunter E., Epand R.M. Hydrophobic substitutions in the first residue of the CRAC segment of the gp41 protein of HIV. Biochemistry. 2008;47:124–130. doi: 10.1021/bi7018892. [DOI] [PubMed] [Google Scholar]
- 40.Luo C., Wang K., Liu de Q., Li Y., Zhao Q.S. The functional roles of lipid rafts in T cell activation, immune diseases and HIV infection and prevention. Cell Mol. Immunol. 2008;5:1–7. doi: 10.1038/cmi.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A.A. Waheed, E.O. Freed, Lipids and membrane microdomains in HIV-1 replication, Q5 Virus Res. (in press), doi:10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed]
- 42.Tenchov B.G., MacDonald R.C., Siegel D.P. Cubic phases in phosphatidylcholine–cholesterol mixtures: cholesterol as membrane “fusogen”. Biophys. J. 2006;91:2508–2516. doi: 10.1529/biophysj.106.083766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vishwanathan S.A., Hunter E. Importance of the membrane-perturbing properties of the membrane-proximal external region of human immunodeficiency virus type 1 gp41 to viral fusion. J. Virol. 2008;82:5118–5126. doi: 10.1128/JVI.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S.S., Yang P., Ke P.Y., Li H.F., Chan W.E., Chang D.K., Chuang C.K., Tsai Y., Huang S.C. Identification of the LWYIK motif located in the human immunodeficiency virus type 1 transmembrane gp41 protein as a distinct determinant for viral infection. J. Virol. 2009;83:870–883. doi: 10.1128/JVI.01088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter G.C., Bernstone L., Sangani D., Bee J.W., Harder T., James W. HIV entry in macrophages is dependent on intact lipid rafts. Virology. 2009;386:192–202. doi: 10.1016/j.virol.2008.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorizate M., de la Arada I., Huarte N., Sanchez-Martinez S., de la Torre B.G., Andreu D., Arrondo J.L., Nieva J.L. Structural analysis and assembly of the HIV-1 Gp41 amino-terminal fusion peptide and the pretransmembrane amphipathic-at-interface sequence. Biochemistry. 2006;45:14337–14346. doi: 10.1021/bi0612521. [DOI] [PubMed] [Google Scholar]
- 47.Lorizate M., Gomara M.J., de la Torre B.G., Andreu D., Nieva J.L. Membrane-transferring sequences of the HIV-1 Gp41 ectodomain assemble into an immunogenic complex. J. Mol. Biol. 2006;360:45–55. doi: 10.1016/j.jmb.2006.04.056. [DOI] [PubMed] [Google Scholar]
- 48.Noah E., Biron Z., Naider F., Arshava B., Anglister J. The membrane proximal external region of the HIV-1 envelope glycoprotein gp41 contributes to the stabilization of the six-helix bundle formed with a matching N' peptide. Biochemistry. 2008;47:6782–6792. doi: 10.1021/bi7023139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bellamy-McIntyre A.K., Lay C.S., Baar S., Maerz A.L., Talbo G.H., Drummer H.E., Poumbourios P. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J. Biol. Chem. 2007;282:23104–23116. doi: 10.1074/jbc.M703485200. [DOI] [PubMed] [Google Scholar]
- 50.Miller M.A., Cloyd M.W., Liebmann J., Rinaldo C.R., Jr., Islam K.R., Wang S.Z., Mietzner T.A., Montelaro R.C. Alterations in cell membrane permeability by the lentivirus lytic peptide (LLP-1) of HIV-1 transmembrane protein. Virology. 1993;196:89–100. doi: 10.1006/viro.1993.1457. [DOI] [PubMed] [Google Scholar]
- 51.Srinivas S.K., Srinivas R.V., Anantharamaiah G.M., Segrest J.P., Compans R.W. Membrane interactions of synthetic peptides corresponding to amphipathic helical segments of the human immunodeficiency virus type-1 envelope glycoprotein. J. Biol. Chem. 1992;267:7121–7127. [PubMed] [Google Scholar]
- 52.Kliger Y., Shai Y. A leucine zipper-like sequence from the cytoplasmic tail of the HIV-1 envelope glycoprotein binds and perturbs lipid bilayers. Biochemistry. 1997;36:5157–5169. doi: 10.1021/bi962935r. [DOI] [PubMed] [Google Scholar]
- 53.Moreno M.R., Perez-Berna A.J., Guillen J., Villalain J. Biophysical characterization and membrane interaction of the most membranotropic region of the HIV-1 gp41 endodomain. Biochim. Biophys. Acta. 2008;1778:1298–1307. doi: 10.1016/j.bbamem.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 54.Jiang J., Aiken C. Maturation-dependent human immunodeficiency virus type 1 particle fusion requires a carboxyl-terminal region of the gp41 cytoplasmic tail. J. Virol. 2007;81:9999–10008. doi: 10.1128/JVI.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viard M., Ablan S.D., Zhou M., Veenstra T.D., Freed E.O., Raviv Y., Blumenthal R. Photoinduced reactivity of the HIV-1 envelope glycoprotein with a membrane-embedded probe reveals insertion of portions of the HIV-1 Gp41 cytoplasmic tail into the viral membrane. Biochemistry. 2008;47:1977–1983. doi: 10.1021/bi701920f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonigo P., Alizon M., Staskus K., Klatzmann D., Cole S., Danos O., Retzel E., Tiollais P., Haase A., Wain-Hobson S. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985;42:369–382. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- 57.Lenz O., Dittmar M.T., Wagner A., Ferko B., Vorauer-Uhl K., Stiegler G., Weissenhorn W. Trimeric membrane-anchored gp41 inhibits HIV membrane fusion. J. Biol. Chem. 2005;280:4095–4101. doi: 10.1074/jbc.M411088200. [DOI] [PubMed] [Google Scholar]
- 58.West J.T., Johnston P.B., Dubay S.R., Hunter E. Mutations within the putative membrane-spanning domain of the simian immunodeficiency virus transmembrane glycoprotein define the minimal requirements for fusion, incorporation, and infectivity. J. Virol. 2001;75:9601–9612. doi: 10.1128/JVI.75.20.9601-9612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shang L., Yue L., Hunter E. Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell–cell fusion and virus infection. J. Virol. 2008;82:5417–5428. doi: 10.1128/JVI.02666-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 61.Armstrong R.T., Kushnir A.S., White J.M. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 2000;151:425–437. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Helseth E., Olshevsky U., Gabuzda D., Ardman B., Haseltine W., Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J. Virol. 1990;64:6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Owens R.J., Burke C., Rose J.K. Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J. Virol. 1994;68:570–574. doi: 10.1128/jvi.68.1.570-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salzwedel K., Johnston P.B., Roberts S.J., Dubay J.W., Hunter E. Expression and characterization of glycophospholipid-anchored human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 1993;67:5279–5288. doi: 10.1128/jvi.67.9.5279-5288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyauchi K., Komano J., Yokomaku Y., Sugiura W., Yamamoto N., Matsuda Z. Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion. J. Virol. 2005;79:4720–4729. doi: 10.1128/JVI.79.8.4720-4729.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Welman M., Lemay G., Cohen E.A. Role of envelope processing and gp41 membrane spanning domain in the formation of human immunodeficiency virus type 1 (HIV-1) fusion-competent envelope glycoprotein complex. Virus Res. 2007;124:103–112. doi: 10.1016/j.virusres.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 67.Miyauchi K., Curran R., Matthews E., Komano J., Hoshino T., Engelman D.M., Matsuda Z. Mutations of conserved glycine residues within the membrane-spanning domain of human immunodeficiency virus type 1 gp41 can inhibit membrane fusion and incorporation of Env onto virions. Jpn. J. Infect Dis. 2006;59:77–84. [PubMed] [Google Scholar]
- 68.Lemmon M.A., Flanagan J.M., Treutlein H.R., Zhang J., Engelman D.M. Sequence specificity in the dimerization of transmembrane α-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 69.Mingarro I., Whitley P., Lemmon M.A., von Heijne G. Ala-insertion scanning mutagenesis of the glycophorin A transmembrane helix. A rapid way to map helix–helix interactions in integral membrane proteins. Protein Sci. 1996;5:1339–1341. doi: 10.1002/pro.5560050712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Melnyk R.A., Kim S., Curran A.R., Engelman D.M., Bowie J.U., Deber C.M. The affinity of GXXXG motifs in transmembrane helix–helix interactions is modulated by long-range communication. J. Biol. Chem. 2004;279:16591–16597. doi: 10.1074/jbc.M313936200. [DOI] [PubMed] [Google Scholar]
- 71.Schneider D., Engelman D.M. Motifs of two small residues can assist but are not sufficient to mediate transmembrane helix interactions. J. Mol. Biol. 2004;343:799–804. doi: 10.1016/j.jmb.2004.08.083. [DOI] [PubMed] [Google Scholar]
- 72.Orzaez M., Salgado J., Gimenez-Giner A., Perez-Paya E., Mingarro I. Influence of proline residues in transmembrane helix packing. J. Mol. Biol. 2004;335:631–640. doi: 10.1016/j.jmb.2003.10.062. [DOI] [PubMed] [Google Scholar]
- 73.Orzaez M., Perez-Paya E., Mingarro I. Influence of the C-terminus of the glycophorin A transmembrane fragment on the dimerization process. Protein Sci. 2000;9:1246–1253. doi: 10.1110/ps.9.6.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orzaez M., Lukovic D., Abad C., Perez-Paya E., Mingarro I. Influence of hydrophobic matching on association of model transmembrane fragments containing a minimised glycophorin A dimerisation motif. FEBS Lett. 2005;579:1633–1638. doi: 10.1016/j.febslet.2005.01.078. [DOI] [PubMed] [Google Scholar]
- 75.Ciczora Y., Callens N., Penin F., Pecheur E.I., Dubuisson J. Transmembrane domains of hepatitis C virus envelope glycoproteins: residues involved in E1E2 heterodimerization and involvement of these domains in virus entry. J. Virol. 2007;81:2372–2381. doi: 10.1128/JVI.02198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., Nilsson I., White S.H., von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 77.Hessa T., Meindl-Beinker N.M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S.H., von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 78.Martinez-Gil L., Perez-Gil J., Mingarro I. The surfactant peptide KL4 sequence is inserted with a transmembrane orientation into the endoplasmic reticulum membrane. Biophys. J. 2008;95:L36–38. doi: 10.1529/biophysj.108.138602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bentz J. Minimal aggregate size and minimal fusion unit for the first fusion pore of influenza hemagglutinin-mediated membrane fusion. Biophys. J. 2000;78:227–245. doi: 10.1016/S0006-3495(00)76587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chertova E., Bess J.W., Jr., Crise B.J., Sowder I.R., Schaden T.M., Hilburn J.M., Hoxie J.A., Benveniste R.E., Lifson J.D., Henderson L.E., Arthur L.O. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 2002;76:5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chien M.P., Lin C.H., Chang D.K. Recruitment of HIV-1 envelope occurs subsequent to lipid mixing: a fluorescence microscopic evidence. Retrovirology. 2009;6:20. doi: 10.1186/1742-4690-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Clercq E. The design of drugs for HIV and HCV. Nat. Rev. Drug Discov. 2007;6:1001–1018. doi: 10.1038/nrd2424. [DOI] [PubMed] [Google Scholar]
- 83.Melby T., Westby M. Inhibitors of viral entry. Handb. Exp. Pharmacol. 2009:177–202. doi: 10.1007/978-3-540-79086-0_7. [DOI] [PubMed] [Google Scholar]
- 84.Qian K., Morris-Natschke S.L., Lee K.H. HIV entry inhibitors and their potential in HIV therapy. Med. Res. Rev. 2009;29:369–393. doi: 10.1002/med.20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Debnath A.K. Prospects and strategies for the discovery and development of small-molecule inhibitors of six-helix bundle formation in class 1 viral fusion proteins. Curr. Opin. Investig. Drugs. 2006;7:118–127. [PubMed] [Google Scholar]
- 86.Kaushik-Basu N., Basu A., Harris D. Peptide inhibition of HIV-1: current status and future potential. BioDrugs. 2008;22:161–175. doi: 10.2165/00063030-200822030-00003. [DOI] [PubMed] [Google Scholar]
- 87.Oishi S., Ito S., Nishikawa H., Watanabe K., Tanaka M., Ohno H., Izumi K., Sakagami Y., Kodama E., Matsuoka M., Fujii N. Design of a novel HIV-1 fusion inhibitor that displays a minimal interface for binding affinity. J. Med. Chem. 2008;51:388–391. doi: 10.1021/jm701109d. [DOI] [PubMed] [Google Scholar]
- 88.He Y., Cheng J., Lu H., Li J., Hu J., Qi Z., Liu Z., Jiang S., Dai Q. Potent HIV fusion inhibitors against Enfuvirtide-resistant HIV-1 strains. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16332–16337. doi: 10.1073/pnas.0807335105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He Y., Cheng J., Li J., Qi Z., Lu H., Dong M., Jiang S., Dai Q. Identification of a critical motif for the human immunodeficiency virus type 1 (HIV-1) gp41 core structure: implications for designing novel anti-HIV fusion inhibitors. J. Virol. 2008;82:6349–6358. doi: 10.1128/JVI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.He Y., Xiao Y., Song H., Liang Q., Ju D., Chen X., Lu H., Jing W., Jiang S., Zhang L. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 2008;283:11126–11134. doi: 10.1074/jbc.M800200200. [DOI] [PubMed] [Google Scholar]
- 91.Gomara M.J., Lorizate M., Huarte N., Mingarro I., Perez-Paya E., Nieva J.L. Hexapeptides that interfere with HIV-1 fusion peptide activity in liposomes block GP41-mediated membrane fusion. FEBS Lett. 2006;580:2561–2566. doi: 10.1016/j.febslet.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 92.Gralle M., Ferreira S.T. Structure and functions of the human amyloid precursor protein: the whole is more than the sum of its parts. Prog. Neurobiol. 2007;82:11–32. doi: 10.1016/j.pneurobio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 93.Thinakaran G., Koo E.H. Amyloid precursor protein trafficking, processing, and function. J. Biol. Chem. 2008;283:29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ohsawa I., Takamura C., Kohsaka S. The amino-terminal region of amyloid precursor protein is responsible for neurite outgrowth in rat neocortical explant culture. Biochem. Biophys. Res. Commun. 1997;236:59–65. doi: 10.1006/bbrc.1997.6903. [DOI] [PubMed] [Google Scholar]
- 95.Kong G.K., Adams J.J., Harris H.H., Boas J.F., Curtain C.C., Galatis D., Masters C.L., Barnham K.J., McKinstry W.J., Cappai R., Parker M.W. Structural studies of the Alzheimer's amyloid precursor protein copper-binding domain reveal how it binds copper ions. J. Mol. Biol. 2007;367:148–161. doi: 10.1016/j.jmb.2006.12.041. [DOI] [PubMed] [Google Scholar]
- 96.Kong G.K., Miles L.A., Crespi G.A., Morton C.J., Ng H.L., Barnham K.J., McKinstry W.J., Cappai R., Parker M.W. Copper binding to the Alzheimer's disease amyloid precursor protein. Eur. Biophys. J. 2008;37:269–279. doi: 10.1007/s00249-007-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kaden D., Munter L.M., Joshi M., Treiber C., Weise C., Bethge T., Voigt P., Schaefer M., Beyermann M., Reif B., Multhaup G. Homophilic interactions of the amyloid precursor protein (APP) ectodomain are regulated by the loop region and affect beta-secretase cleavage of APP. J. Biol. Chem. 2008;283:7271–7279. doi: 10.1074/jbc.M708046200. [DOI] [PubMed] [Google Scholar]
- 98.Gralle M., Oliveira C.L., Guerreiro L.H., McKinstry W.J., Galatis D., Masters C.L., Cappai R., Parker M.W., Ramos C.H., Torriani I., Ferreira S.T. Solution conformation and heparin-induced dimerization of the full-length extracellular domain of the human amyloid precursor protein. J. Mol. Biol. 2006;357:493–508. doi: 10.1016/j.jmb.2005.12.053. [DOI] [PubMed] [Google Scholar]
- 99.Gralle M., Botelho M.M., de Oliveira C.L., Torriani I., Ferreira S.T. Solution studies and structural model of the extracellular domain of the human amyloid precursor protein. Biophys. J. 2002;83:3513–3524. doi: 10.1016/S0006-3495(02)75351-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Menendez-Gonzalez M., Perez-Pinera P., Martinez-Rivera M., Calatayud M.T., Blazquez Menes B. APP processing and the APP-KPI domain involvement in the amyloid cascade. Neurodegener. Dis. 2005;2:277–283. doi: 10.1159/000092315. [DOI] [PubMed] [Google Scholar]
- 101.Jaeger S., Pietrzik C.U. Functional role of lipoprotein receptors in Alzheimer's disease. Curr. Alzheimer Res. 2008;5:15–25. doi: 10.2174/156720508783884675. [DOI] [PubMed] [Google Scholar]
- 102.Muller T., Meyer H.E., Egensperger R., Marcus K. The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics-relevance for Alzheimer's disease. Prog. Neurobiol. 2008;85:393–406. doi: 10.1016/j.pneurobio.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 103.McLoughlin D.M., Miller C.C. The FE65 proteins and Alzheimer's disease. J. Neurosci. Res. 2008;86:744–754. doi: 10.1002/jnr.21532. [DOI] [PubMed] [Google Scholar]
- 104.Scheuermann S., Hambsch B., Hesse L., Stumm J., Schmidt C., Beher D., Bayer T.A., Beyreuther K., Multhaup G. Homodimerization of amyloid precursor protein and its implication in the amyloidogenic pathway of Alzheimer's disease. J. Biol. Chem. 2001;276:33923–33929. doi: 10.1074/jbc.M105410200. [DOI] [PubMed] [Google Scholar]
- 105.Munter L.M., Voigt P., Harmeier A., Kaden D., Gottschalk K.E., Weise C., Pipkorn R., Schaefer M., Langosch D., Multhaup G. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. EMBO J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.M. Cruts, http://www.molgen.ua.ac.be/ADMutations, (2009).
- 107.Beel A.J., Mobley C.K., Kim H.J., Tian F., Hadziselimovic A., Jap B., Prestegard J.H., Sanders C.R. Structural studies of the transmembrane C-terminal domain of the amyloid precursor protein (APP): does APP function as a cholesterol sensor? Biochemistry. 2008;47:9428–9446. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Senes A., Gerstein M., Engelman D.M. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J. Mol. Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 109.Sulistijo E.S., MacKenzie K.R. Sequence dependence of BNIP3 transmembrane domain dimerization implicates side-chain hydrogen bonding and a tandem GxxxG motif in specific helix–helix interactions. J. Mol. Biol. 2006;364:974–990. doi: 10.1016/j.jmb.2006.09.065. [DOI] [PubMed] [Google Scholar]
- 110.Gorman P., Kim S., Guo M., Melnyk R., McLaurin J., Fraser P., Bowie J., Chakrabartty A. Dimerization of the transmembrane domain of amyloid precursor proteins and familial Alzheimer's disease mutants. BMC Neuroscience. 2008;9:17. doi: 10.1186/1471-2202-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.MacKenzie K.R., Prestegard J.H., Engelman D.M. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 112.Kienlen-Campard P., Tasiaux B., Van Hees J., Li M., Huysseune S., Sato T., Fei J.Z., Aimoto S., Courtoy P.J., Smith S.O., Constantinescu S.N., Octave J.N. Amyloidogenic processing but not amyloid precursor protein (APP) intracellular C-terminal domain production requires a precisely oriented APP dimer assembled by transmembrane GXXXG motifs. J. Biol. Chem. 2008;283:7733–7744. doi: 10.1074/jbc.M707142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sato T., Tang T.C., Reubins G., Fei J.Z., Fujimoto T., Kienlen-Campard P., Constantinescu S.N., Octave J.N., Aimoto S., Smith S.O. A helix-to-coil transition at the epsilon-cut site in the transmembrane dimer of the amyloid precursor protein is required for proteolysis. Proc. Natl. Acad. Sci. U. S. A. 2009;106:1421–1426. doi: 10.1073/pnas.0812261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Miyashita N., Straub J.E., Thirumalai D., Sugita Y. Transmembrane structures of amyloid precursor protein dimer predicted by replica-exchange molecular dynamics simulations. J. Am. Chem. Soc. 2009;131:3438–3439. doi: 10.1021/ja809227c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beel A.J., Sanders C.R. Substrate specificity of gamma-secretase and other intramembrane proteases. Cell Mol. Life Sci. 2008;65:1311–1334. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Steiner H., Fluhrer R., Haass C. Intramembrane proteolysis by gamma-secretase. J. Biol. Chem. 2008;283:29627–29631. doi: 10.1074/jbc.R800010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stefani M., Liguri G. Cholesterol in Alzheimer's disease: unresolved questions. Curr. Alzheimer Res. 2009;6:15–29. doi: 10.2174/156720509787313899. [DOI] [PubMed] [Google Scholar]
- 118.Ehehalt R., Keller P., Haass C., Thiele C., Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J. Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Irvine G.B., El-Agnaf O.M., Shankar G.M., Walsh D.M. Protein aggregation in the brain: the molecular basis for Alzheimer's and Parkinson's diseases. Mol. Med. 2008;14:451–464. doi: 10.2119/2007-00100.Irvine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q. Rev. Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 121.Glabe C.G. Structural classification of toxic amyloid oligomers. J. Biol. Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sato T., Kienlen-Campard P., Ahmed M., Liu W., Li H., Elliott J.I., Aimoto S., Constantinescu S.N., Octave J.N., Smith S.O. Inhibitors of amyloid toxicity based on beta-sheet packing of Abeta40 and Abeta42. Biochemistry. 2006;45:5503–5516. doi: 10.1021/bi052485f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lundin C., Johansson S., Johnson A.E., Naslund J., von Heijne G., Nilsson I. Stable insertion of Alzheimer Abeta peptide into the ER membrane strongly correlates with its length. FEBS Lett. 2007;581:3809–3813. doi: 10.1016/j.febslet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cheng H., Vetrivel K.S., Gong P., Meckler X., Parent A., Thinakaran G. Mechanisms of disease: new therapeutic strategies for Alzheimer's disease-targeting APP processing in lipid rafts. Nat. Clin. Pract. Neurol. 2007;3:374–382. doi: 10.1038/ncpneuro0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Reid P.C., Urano Y., Kodama T., Hamakubo T. Alzheimer's disease: cholesterol, membrane rafts, isoprenoids and statins. J. Cell Mol. Med. 2007;11:383–392. doi: 10.1111/j.1582-4934.2007.00054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cordy J.M., Hussain I., Dingwall C., Hooper N.M., Turner A.J. Exclusively targeting beta-secretase to lipid rafts by GPI-anchor addition up-regulates beta-site processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11735–11740. doi: 10.1073/pnas.1635130100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schneider A., Rajendran L., Honsho M., Gralle M., Donnert G., Wouters F., Hell S.W., Simons M. Flotillin-dependent clustering of the amyloid precursor protein regulates its endocytosis and amyloidogenic processing in neurons. J. Neurosci. 2008;28:2874–2882. doi: 10.1523/JNEUROSCI.5345-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fuentealba R.A., Barria M.I., Lee J., Cam J., Araya C., Escudero C.A., Inestrosa N.C., Bronfman F.C., Bu G., Marzolo M.P. ApoER2 expression increases Abeta production while decreasing Amyloid Precursor Protein (APP) endocytosis: possible role in the partitioning of APP into lipid rafts and in the regulation of gamma-secretase activity. Mol. Neurodegener. 2007;2:14. doi: 10.1186/1750-1326-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Suzuki T., Nakaya T. Regulation of amyloid beta-protein precursor by phosphorylation and protein interactions. J. Biol. Chem. 2008;283:29633–29637. doi: 10.1074/jbc.R800003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bai Y., Markham K., Chen F., Weerasekera R., Watts J., Horne P., Wakutani Y., Bagshaw R., Mathews P.M., Fraser P.E., Westaway D., St George-Hyslop P., Schmitt-Ulms G. The in vivo brain interactome of the amyloid precursor protein. Mol. Cell Proteomics. 2008;7:15–34. doi: 10.1074/mcp.M700077-MCP200. [DOI] [PubMed] [Google Scholar]
- 131.Pierrot N., Octave J.N. Processing of amyloid precursor protein and amyloid peptide neurotoxicity. Curr. Alzheimer Res. 2008;192:92–99. doi: 10.2174/156720508783954721. [DOI] [PubMed] [Google Scholar]
- 132.Barten D.M., Albright C.F. Therapeutic strategies for Alzheimer's disease. Mol. Neurobiol. 2008;37:171–186. doi: 10.1007/s12035-008-8031-2. [DOI] [PubMed] [Google Scholar]
- 133.Forette F., Hauw J.J. [Alzheimer's disease: from brain lesions to new drugs] Bull. Acad. Natl. Med. 2008;192:363–378. discussion 378–380. [PubMed] [Google Scholar]
- 134.Fisher A. Cholinergic treatments with emphasis on m1 muscarinic agonists as potential disease-modifying agents for Alzheimer's disease. Neurotherapeutics. 2008;5:433–442. doi: 10.1016/j.nurt.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van Marum R.J. Current and future therapy in Alzheimer's disease. Fundam. Clin. Pharmacol. 2008;22:265–274. doi: 10.1111/j.1472-8206.2008.00578.x. [DOI] [PubMed] [Google Scholar]
- 136.Munoz-Torrero D. Acetylcholinesterase inhibitors as disease-modifying therapies for Alzheimer's disease. Curr. Med. Chem. 2008;15:2433–2455. doi: 10.2174/092986708785909067. [DOI] [PubMed] [Google Scholar]
- 137.Pepeu G., Giovannini M.G. Cholinesterase inhibitors and beyond. Curr. Alzheimer Res. 2009;6:86–96. doi: 10.2174/156720509787602861. [DOI] [PubMed] [Google Scholar]
- 138.Shimmyo Y., Kihara T., Akaike A., Niidome T., Sugimoto H. Flavonols and flavones as BACE-1 inhibitors: structure–activity relationship in cell-free, cell-based and in silico studies reveal novel pharmacophore features. Biochim. Biophys. Acta. 2008;1780:819–825. doi: 10.1016/j.bbagen.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 139.Wolfe M.S. Gamma-secretase inhibition and modulation for Alzheimer's disease. Curr. Alzheimer Res. 2008;5:158–164. doi: 10.2174/156720508783954767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nitsch R.M., Hock C. Targeting beta-amyloid pathology in Alzheimer's disease with Abeta immunotherapy. Neurotherapeutics. 2008;5:415–420. doi: 10.1016/j.nurt.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wisniewski T., Konietzko U. Amyloid-beta immunisation for Alzheimer's disease. Lancet. Neurol. 2008;7:805–811. doi: 10.1016/S1474-4422(08)70170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Leissring M.A. The AbetaCs of Abeta-cleaving proteases. J. Biol. Chem. 2008;283:29645–29649. doi: 10.1074/jbc.R800022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Doig A.J. Peptide inhibitors of beta-amyloid aggregation. Curr. Opin Drug Discov. Devel. 2007;10:533–539. [PubMed] [Google Scholar]
- 144.Bastianetto S., Krantic S., Quirion R. Polyphenols as potential inhibitors of amyloid aggregation and toxicity: possible significance to Alzheimer's disease. Mini. Rev. Med. Chem. 2008;8:429–435. doi: 10.2174/138955708784223512. [DOI] [PubMed] [Google Scholar]
- 145.Bush A.I., Tanzi R.E. Therapeutics for Alzheimer's disease based on the metal hypothesis. Neurotherapeutics. 2008;5:421–432. doi: 10.1016/j.nurt.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wright T.M. Tramiprosate. Drugs Today (Barc) 2006;42:291–298. doi: 10.1358/dot.2006.42.5.973584. [DOI] [PubMed] [Google Scholar]
- 147.Wilcock G.K., Black S.E., Hendrix S.B., Zavitz K.H., Swabb E.A., Laughlin M.A. Efficacy and safety of tarenflurbil in mild to moderate Alzheimer's disease: a randomised phase II trial. Lancet Neurol. 2008;7:483–493. doi: 10.1016/S1474-4422(08)70090-5. [DOI] [PubMed] [Google Scholar]
- 148.Aisen P.S., Gauthier S., Vellas B., Briand R., Saumier D., Laurin J., Garceau D. Alzhemed: a potential treatment for Alzheimer's disease. Curr. Alzheimer Res. 2007;4:473–478. doi: 10.2174/156720507781788882. [DOI] [PubMed] [Google Scholar]
- 149.Vilar M., Charalampopoulos I., Kenchappa R.S., Simi A., Karaca E., Reversi A., Choi S., Bothwell M., Mingarro I., Friedman W.J., Schiavo G., Bastiaens P.I., Verveer P.J., Carter B.D., Ibanez C.F. Activation of the p75 neurotrophin receptor through conformational rearrangement of disulphide-linked receptor dimers. Neuron. 2009;62:72–83. doi: 10.1016/j.neuron.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]