Abstract
Urgent demand for portable diagnosis has promoted a new sensing strategy that uses personal glucometer (PGM) to detect non-glucose targets. Even though great progresses have been achieved in terms of target range and sensing principle, issues such as low final signal-to-background ratio and hard-to-realize one-tube smart analysis still exist and challenge real-world applications in gene detection. Here we propose a practical solution via coupling isothermal amplification (i.e. LAMP) and three-way amplifiable catalytic hairpin assembly (i.e. CHA) to a PGM. It allows direct transduction from genomic information to commercial portable devices with all of ultra-high sensitivity, specificity and enhanced signal-to-noise ratio. Compared with previous report without signal amplification, the introduction of CHA has successfully improved the signal amplitude by at least 12.5 folds. More importantly, through importing an effective three-way junction based transduction, we also innovatively develop a one-tube logical or multiplex analysis strategy in PGM based detection. Totally four situations of two foodborne bacteria genes, in Cronobacter sakazakii (ompA) and Escherichia coli (malB), could be directly readout using the final PGM signals, with the lowest detection amount down to less than 100 molecular copies (6.6 × 10−18 M). It is believed such a LAMP-CHA-PGM method has been already sensitive, specific, and of great potential for practically portable gene diagnostics.
Keywords: Nucleic acid circuitry, Isothermal amplification, Portable detection, Glucometer, Multi-analysis
Graphical abstract
We report a practical genetic testing method via coupling isothermal amplification and three-way nucleic acid circuit to a personal glucometer. It allows direct transduction from genomic information to commercial portable devices with all of ultra-high sensitivity, specificity and enhanced signal-to-noise ratio. Logical or multiplex analysis can also be realized in one-tube, with only one signal probe, the glucose.
Highlights
-
•
CHA circuit improves the signal-to-background ratio by at least 3.5 folds.
-
•
Misreading frequently induced by signal errors or noises is efficiently reduced.
-
•
The detection limit for pathogen gene is down to 100 molecular copies.
-
•
The method for the first time enables one-tube multiplex assay for PGM based detection.
-
•
Four situations for two pathogen genes can be are directly distinguished using the PGM.
1. Introduction
With the faster population migration and disease spread, more portable or even on-set point-of-care (POC) diagnostic techniques have been urgently called for effective infection control and in-time rescue. Recent advancements in molecular or medical biology are showing increasing possibilities for quickly and accurately diagnosing disease states using nucleic acid or protein biomarkers [[1], [2], [3]]. However, relevant methods are generally energy-consuming or requiring complicated instrumentation restricted within medical laboratories. To accelerate the development of portable, even off-shelf POC diagnosis device, pioneers have opened up a new detection strategy through translating the biomarkers generally to the few types of already commercial POC products. For example, the personal glucose meter (PGM) [4], dipstick [5], blood gas analyser [6], paper chip [[7], [8], [9]], Thermometer [10], and pregnancy test strips [11,12] were recently modified to identify non-original (glucose, H+, trioxypurine, temperature, hCG) targets via aptamer- or DNAzyme-mediated conformational changes or sequence-specific strand exchange. Among these innovations, the transduction of PGM has received especial attention due to its higher accuracy quantitation [4,[13], [14], [15], [16], [17], [18], [19], [20], [21]]. The method uses the invertase as a signal intermediate. Mostly, generation of invertase is designed to be target relative. Through the enzymatic reaction in which invertase oxidases sucrose to glucose, the existence of target could be quantitatively monitored with PGM. Following this principle, drugs, toxic metals, nucleic acids, and various proteins have been successfully detected [4,[12], [13], [14], [15], [16], [17], [18], [19], [20], [21]].
In recent advance, through innovatively coupling loop-mediated isothermal amplification (LAMP) reactions to one-step strand exchange (OSD) signal transduction, we have pushed the PGM based nucleic acid detection further closed to practically sensitive and specific [21]. The lowest detection amount for various pathogen genes, such as Middle-East respiratory syndrome coronavirus (MERS-CoV) or Zaire Ebolavirus (Ebola), could be less than 100 molecular copies (6.6 × 10−18 M), and the false positives frequently induced by the non-specific LAMP reactions have been completely erased. Even through, being limited to a one-to-one transduction principle of OSD, the final signal-to-background ratio, and signal magnitude, may usually be unsatisfied. And because the PGM only have one signal probe, the inverses (glucose), the one-tube smart analysis (logical or multiplex analysis) that is highly demanded by the portable detection is still impossible. This is actually a general challenge for most detection using commercial POC products.
With the purpose to enhance signal-to-background ratio, and at the same time approach the one-tube smart analysis, here we propose a ready-to-use solution that uses amplifiable enzyme-free nucleic acid circuits to replace OSD transduction. In the past decade, we and others have contributed many efforts in developing and optimizing analysis-friendly nucleic acid circuits [[22], [23], [24], [25], [26], [27], [28], [29], [30], [31]]. Among these circuits the catalytic hairpin assembly (CHA) [24,25] has now been proven a “plug-to-use” unit that could transduce various target species to a single-strand reporting oligonucleotide, with tens to hundreds fold signal amplification but almost “zero” non-amplified background [25]. Here in the LAMP-CHA-PGM detection for pathogen genes, the introduction of CHA circuit successfully improved the signal amplitude by at least 12.5 folds, with almost 300 mg/dl increase in signal magnitude. Such a high signal-to-background deviation can efficiently reduce the misreading induced by signal errors or noises, and thus significantly increase the detection reliability. And more importantly, through importing an effective three-way junction based transduction between LAMP and CHA, we innovatively reported a one-tube smart analysis strategy in PGM based detection. Totally four situations for the existences of the genes of two foodborne bacteria, in Cronobacter sakazakii (ompA) and Escherichia coli (malB), could be directly readout using the final PGM signals (glucose concentrations). It is thus believed such a LAMP-CHA-PGM method has been already sensitive, specific, and highly deserving further efforts in the step-integration and final portable device fabrication. After replacing LAMP with another isothermal amplification, the nucleic acid sequence-based amplification (NASBA), the CHA-PGM transduction was effectively enabled to detect RNA amplifications, which further confirmed the universality of the method.
2. Experimental section
2.1. Chemicals and materials
The Bayer Contour Next Blood Glucose Test Strips and Bayer Contour Next Blood Glucose Monitoring System were bought from Amazon.com and used for the tests in this work. Streptavidin-coated magnetic beads (1.5 μM in average diameter) were purchased from Bangs Laboratories Inc. (Fishers, IN, USA) and the Amicon Ultra-2 mL 30 K was purchased from Millipore Inc. (Billerica, MA, USA). Sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC), tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and other chemicals and solvents were purchased from Sigma-Aldrich, Inc. (St. Louis, MO,USA). Bst 2.0 DNA polymerase and 10 × Isothermal Buffer (10 × Iso) were obtained from New England Biolabs (Ipswich, MA, USA). Thermo-stable invertase (TmINV) derived from the hyperthermophilic bacteria, Thermotoga maritima, was home-made according to our previous publication without modification [20,21]. All the oligonucleotides (listed in Table S3) and ompA and malB synthetic DNA templates were synthesized by Sangon Biotech (Shanghai, China). All regents were of analytical grade unless otherwise indicated. The concentrations of the DNA suspensions were measured by UV spectrophotometry using the DeNovix DS-11+ FX spectrophotometer (DeNovix Inc., Wilmington, DE, USA). All fluorescence kinetic curves were monitored with the Coyote Mini-8 portable real-time PCR machine (Beijing, China). Procedures of synthesis of TmINV–F conjugate, preparation of the TmINV–F:Bio-Q-MB conjugate, standard LAMP reaction and fluorescence CHA detection were similar as previously reported [15] and presented in supporting information.
2.2. Detection of mimic target with CHA-to-glucose transduction (3W-CHA-PGM)
In standard 3W-CHA detection, different concentrations of target T (C1, also named with CHA-T, T1 or T2) were mixed with final 80 nM respective TP in 1 × Iso Buffer. Some of this mixture was further mixed with 1.2 μM H1 (in 1 × Iso Buffer), 1.2 μM H2 (in 1 × Iso Buffer) and 1 × Iso Buffer in a 1:1:1:1 vol ratio. H1 and H2 were pre-annealed, separately. The final concentration of each component was as follows. [H1] = [H2] = 300 nM, [TP] = 20 nM. This mixture was incubated for 2.5 h at 55 °C. A series of tubes containing 9 μL aliquots of the 2.1 mg/mL TmINV–F:Bio-Q-MBs were placed close to the magnetic rack for 1 min. The clear solution was discarded and replaced by 10 μL of the above mixture. Then this mixture was incubated for 60 min at 55 °C to allow invertase-mediated catalytic conversion of sucrose to glucose. Subsequently, 1 μL of the reaction solution was transferred to a PGM strip and the amount of glucose was measured by using a commercially available hand-held PGM.
2.3. Detection of mimic target with OSD-to-glucose transduction (OSD-PGM)
A series of tubes containing 9 μL aliquots of the 2.1 mg/mL TmINV–F:Bio-Q-MBs were placed close to the magnetic rack for 1 min. The clear solution was discarded and replaced by 10 μL of the mimic target (OSD-T, in isothermal amplification buffer) at 0 nM and 10 nM concentrations. The OSD reactions were allowed to proceed for 40 min on a vertical rotator at room temperature. The TmINV–F:Bio-Q-MBs were then separated using a magnetic rack and aliquots of the supernatant containing released TmINV–F were transferred into equal volumes of 500 mM sucrose. This mixture was incubated for 60 min at 55 °C to allow invertase-mediated catalytic conversion of sucrose to glucose. Subsequently, 1 μL of the reaction solution was transferred to a PGM strip and the amount of glucose was measured by using a commercially available hand-held PGM.
2.4. Detection of LAMP amplicons (products) with CHA-to-glucose transduction (LAMP-3W-CHA-PGM)
8 μL LAMP amplicons amplified from different copies of ompA synthetic DNA templates were mixed with final 20 nM TP in 1 × Iso Buffer. Some of this mixture was used to replace the mimic target and followed exactly the same 3W-CHA-to-glucose transduction manner as shown in above section.
2.5. Detection of NASBA amplicons (products) with CHA-to-glucose transduction (NASBA-3W-CHA-PGM)
4 μL NASBA amplicons amplified from different copies of Zika RNA templates were mixed with final 40 nM TP in 1 × Iso Buffer. Some of this mixture was used to replace the mimic target and followed exactly the same 3W-CHA-to-glucose transduction manner as shown in above section.
2.6. Detection of multiplex mimic targets with CHA-to-glucose transduction
The procedure of mimic target detection at 55 °C was similar to standard 3W-CHA-to-glucose detection with a little modification. Different target Ts (with or without 10 nM T1 or 10 nM T2) were annealed with corresponding 20 nM TP (T1 with TP1, T2 with TP2) in the same tube in 1 × Iso Buffer. Some of the mixtures were further mixed with H1 mix (final 200 nM H1-T1, 100 nM H1-T2), final 300 nM H2 in 1 × Iso Buffer in a 1:1:1:1 vol ratio. The mixture was incubated for 2.5 h at 55 °C. A series of tubes containing 9 μL aliquots of the 2.1 mg/mL TmINV–F:Bio-Q-MBs were placed close to the magnetic rack for 1 min. The clear solution was discarded and replaced by 10 μL of the above mixture. Then this mixture was incubated for 60 min at 55 °C to allow invertase-mediated catalytic conversion of sucrose to glucose. Subsequently, 1 μL of the reaction solution was transferred to a PGM strip and the amount of glucose was measured by using a commercially available hand-held PGM.
2.7. Detection of multiplex LAMP amplicons (products) with CHA-to-glucose transduction
The detection was carried out in the same manner as the detection of multiplex mimic targets with 3W-CHA-to-glucose transduction, except that mimic targets were replaced by LAMP amplicons amplified from samples with or without 2000 copies of ompA or malB synthetic DNA templates.
3. Results and discussion
3.1. Principle of LAMP-3W-CHA-PGM detection
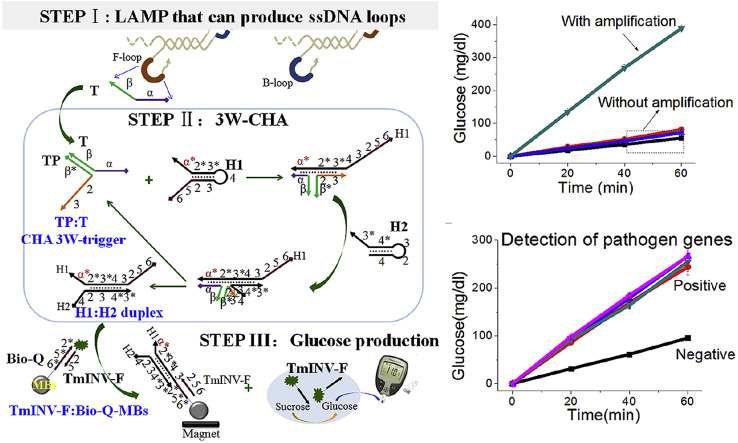
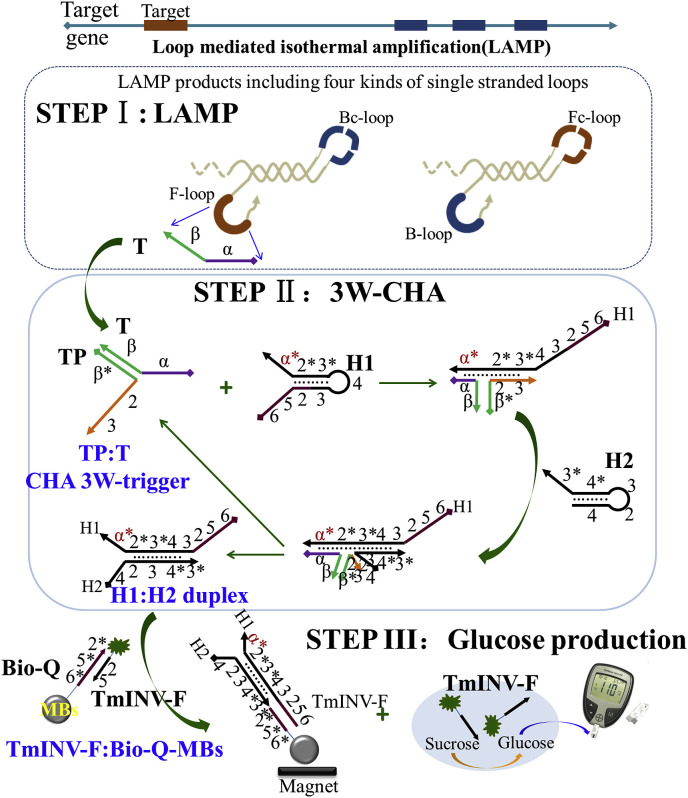
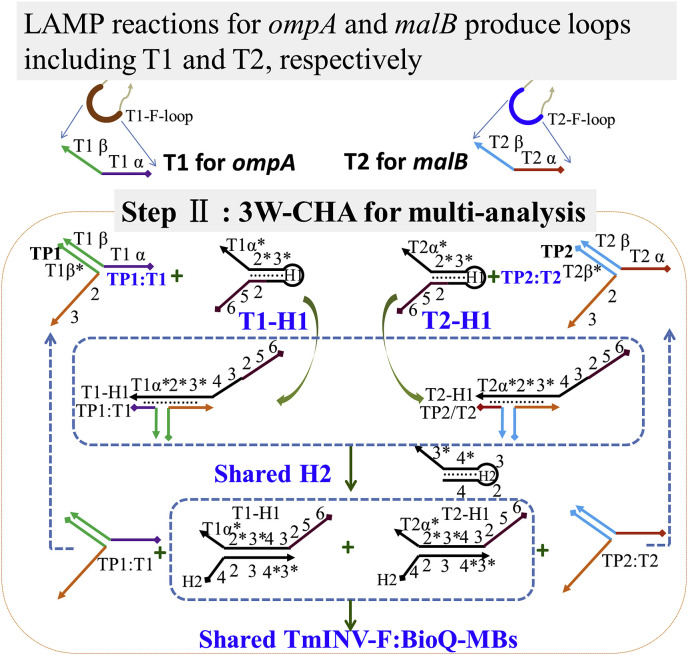
The detailed pathway of gene detection is shown in Fig. 1 , including three sub-steps allowed happening step by step or together. The first step (STEP I) is loop mediated isothermal amplification, the LAMP [32]. At a constant temperature (60±5 °C), LAMP can amplify a segment of pathogen gene from as few as 10 copies for up to 108–109 folds, until the products reach a detectable concentration. The reaction products include cauliflower-like structures with four types of single-stranded nucleic acid loops. In the second step (STEP II), one of the ssDNA loop sequence (here, F-loop) is sequence specifically designed as an associative trigger to activate a downstream CHA circuit detector via a strand displacement reaction across the three-way junction. In details, the F-loop is separated into two sequence domains, α and β. Domain β is designed to hybridize with a transducer probe (TP). Through this process domain α will be close enough to the domain 2-3 in the TP, finally forming an associative trigger to start downstream CHA circuit (thus also called 3W-CHA). The 3W-CHA circuit pathway should be similar to any classic one [31]. The associative trigger opens H1 and then H2 through two successive OSD reactions, forming hybrid H1:H2 as the final product. And the trigger itself will be released into solution free to open another H1, as a catalyst. Third step (STEP III) is signal reporting step. During the H1:H2 hybrid formation, domain 2 previously embedded into the stem of H1 will be released into free linear stage together with domain 5-6. The domain 2-5-6 could start another downstream OSD reaction to displace an inverse-modified oligonucleotide (TmINV–F) previously hybridized onto magnetic beads (TmINV–F:Bio-Q-MBs). After simply magnet separation, the TmINV–F will be left in supernatant and trigger the catalytic transduction of sucrose to glucose. The concentration of glucose could be directly read with a commercially personal PGM, as the indicator to the existence of targeting gene.
Fig. 1.
Scheme of LAMP-3W-CHA-PGM. According to common rules, each oligonucleotide component is separated into several domains named with numbers or characters. Complementarity between numbered domains is denoted by an asterisk (*). Each domain may contain 4-10 bases. Arrow denotes 3’end of an oligonucleotide.
There are several key points that should be noted for deeper understanding the importance of each step. LAMP is specially chosen because it contributes the main the ultra-sensitivity and an isothermal, constant temperature (60±5 °C) reaction condition that is more easily to connect portable device [33]. 3W-CHA here provides the amplified signal magnitude as well as the ultra-selectivity to merely discriminate correct LAMP reaction, with up to 100-fold resolution to single mismatch [31,[33], [34], [35]]. The whole reaction components are high temperature-resistant. Until now CHA has been the only type of enzyme-free circuitry that is well engineered to execute at very large temperature window, through 4 °C to as high as 60 °C [34,35]. Also we have previously engineered a thermo-stable invertase that can keep efficient at even 95 °C [21]. The coupling with LAMP should thus be very flexible, either at end point or in real-time at 60±5 °C. Therefore, through the LAMP-3W-CHA-PGM detection, sensitive, selective, flexible, and portable gene detection could be realized at the same time [21].
3.2. LAMP-3W-CHA-PGM detection of mimic and synthetic gene targets
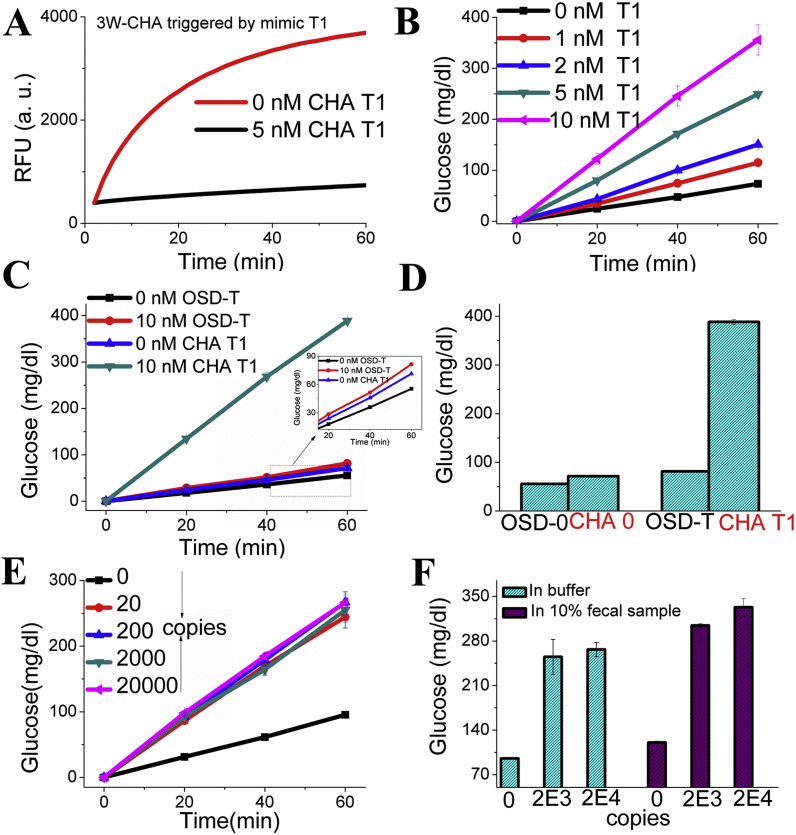
Before carrying out gene detection, the success of LAMP and 3W-CHA has been demonstrated separately. The ompA gene has been selected as our model target. Its five primer set for LAMP has been self-designed and proven very efficient in our previous publications [31]. They can amplify down to 20 copies of selected 236-mer gene segment within 1.5 h (Fig. S1). The F-loop of LAMP product contains 33 nucleotides. This 33-mer has been in-vitro synthesized as the mimic target T (CHA T1 or T1) of the CHA reaction. As shown in Fig. 1, the basic principle says that only the domain β in TP, and domain α in CHA H1 were relevant to the T1. Therefore, except domain β* of TP, and α* of H1, we directly borrow all other CHA component sequences from a well-performed CHA circuit previously designed for other purpose [31,35]. Classic fluorescence resonance energy transfer (FRET) experiments have been firstly performed to test the efficiency of 3W-CHA reaction (scheme shown in Fig. S2). As shown in Fig. 2 A, the sample with T1 clearly shows higher and faster fluorescence emission compared with the one without T1. To demonstrate the amplification efficiency of 3W-CHA over OSD, domain 2-5-6 of H1 has been synthesized as the control target (OSD-T) for the OSD reaction on magnetic beads. It is observed when OSD-T and CHA T1 is used as respective input, under the same reaction buffer condition, 10 nM OSD-T shows much slower glucose production rate compared the one with 10 nM CHA T1 (Fig. 2C). While, the OSD background (0 nM OSD-T) is kept closed to that of CHA background (0 nM CHA T1, and 10 nM non-specific sequences). Fig. 2D provides the bar graph indicates the glucose concentration at the 60 min invertase-sucrose reaction. It clearly shows compared with OSD reaction CHA reaction produces 3.5 fold higher signal-to-background ratio as well as 12.5 fold higher signal magnitude.
Fig. 2.
LAMP-3W-CHA-PGM detection of mimic and synthetic gene targets. (A) Kinetic fluorescence curves of 3W-CHA reactions with and without mimic T1. (B) Kinetic glucose production curves after 3W-CHA reactions with different concentrations of T1. (C) Kinetic glucose production curves after OSD and 3W-CHA reactions with and without respective target. (D) Bar-graph of glucose concentration of OSD and 3W-CHA reactions with and without respective target at 60 min glucose production. The signal amplitude of CHA and OSD detection could be represented as “[Glucose]CHA-T1-[Glucose]CHA-0” and “[Glucose]OSD-T-[Glucose]OSD-0”, respectively. (E) Kinetic glucose production curves after LAMP-3W-CHA with different copies of ompA synthetic gene. (F) Bar-graph of glucose concentration of LAMP-3W-CHA reactions with different copies of ompA synthetic gene, at 60 min glucose production. The concentrations of TP, H1, H2, and Reporter were present in Table S4 of supporting information. Note: here for proof-of-concept we don’t use real genes exacted from pathogens, but only adapt a 236 bp gene segment in-vitro synthesized by companies. It is because in previous studies we have proven that the LAMP efficiency will be very similar between real genes and synthetic segments [21].
During the concentration dependence for the CHA-T (Fig. 2B), even 1 nM CHA T1 can produce more glucose than that of 10 nM OSD-T, indicating at current experimental condition, at least 10 times more invertase can be released compared with same concentration of OSD-T. The reliability of these control experiments and following tests came from the high reproducibility of the whole reaction system, including making TmINV–F:Bio-Q duplex modified MBs and the OSD reaction on these TmINV–F:Bio-Q-MBs (Fig. S3).
After above demonstration, the detection for ompA gene is ready to be carried out. For ompA gene detection, the T1 was replaced with LAMP products generated from different concentrations of synthetic ompA genes, after 1.5 h LAMP amplification. Clearly deviation of kinetic curves away from the negative control have been shown for all the gene-positive samples, including the one with as few as 20 copies (aM molar concentration, Fig. 2E). Notably, here the similar kinetic curves generated for different concentrations of ompA genes indicates 1.5 h LAMP reaction could be enough to consume up all the primers, no matter how few the template is. This is consistent with previous studies [21,31]. While, when we execute the LAMP with the ompA embedded in 10% fecal samples (Fig. 2F), the non-target induced leakage of CHA reaction can be slightly increased. Even through, in assistance of CHA amplification, the 20 copies ompA still generated enough LAMP products that could be reliably detected with PGM.
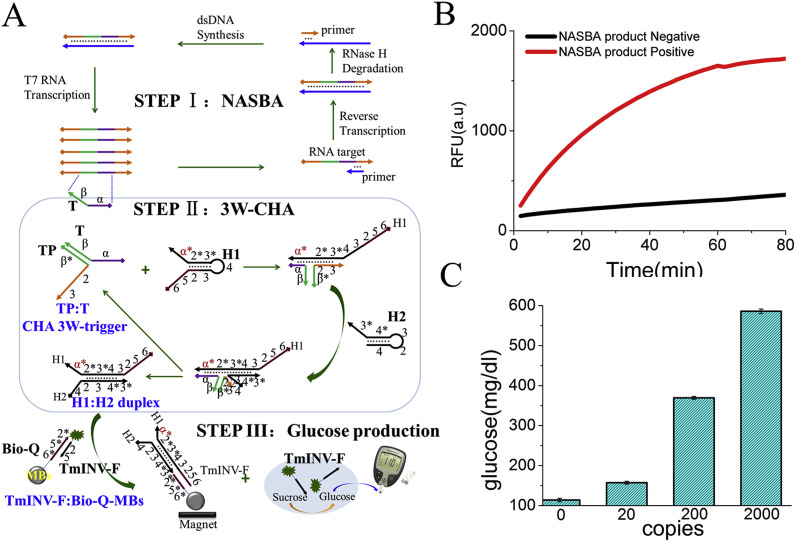
As well known, LAMP is usually used for the amplification of DNAs. To further prove the universality of this method for RNA detection, here we replaced LAMP with another power isothermal amplification, nucleic acid sequence-based amplification (NASBA, Fig. 3 A Step I) [36]. Zika RNA is used as the model-target. TP and H1 sequences were designed according to 33-mer within synthetic Zika RNA template, named with TP-T3 and H1-T3, respectively. Fluorescence kinetic curves prove the success of NASBA and 3W-CHA reactions (Fig. 3B). And PGM detection exhibit very clear signal-to-background deviations for all the Zika RNA positive samples (Fig. 3C), which proves the universality and flexibility of the detection.
Fig. 3.
NASBA-3W-CHA-PGM detection of synthetic gene targets. (A) Scheme of NASBA-3W-CHA-PGM. (B) Kinetic fluorescence curves of 3W-CHA reactions with and without NASBA products amplified from 2000 copies of Zika synthetic RNA. (C) Bar-graph of glucose concentration of NASBA-3W-CHA reactions with different copies of Zika synthetic RNA.
3.3. Principle of one-tube smart assay based on LAMP-3W-CHA-PGM detection
Traditionally, smart analysis (logical or multiplex analysis) is mostly realized following a one-tube multi-channel detection, in which multiple analytes are transduced into multiple signal probes with different color emissions or redox potentials [37,38]. However, such a method is no longer applicable to the sensing strategy using commercial POC devices because each POC device only fits to a single analyte (signal probe). There is thus always a high technical barrier for the development of smart analysis in one tube. Here, we proposed a potential solution being assisted by the CHA circuit using three-way associative trigger. As demonstrated above, to detect any new target oligonucleotide, only the sequences of domain α* of CHA-H1, and domain β of TP have to be changed according to the target sequence. The other sequences and all components with probe label can be remained unchanged. As shown in the pathway show in Fig. 4 . When we aim to target two gene segments from two pathogens, for example, ompA and malB, respective LAMP reaction can generate the F-loop sequence for both ompA (T1-F-loop, shortened as T1) and malB (T2-F-loop, shortened as T2). According to the design shown in Fig. 1, TP-T1 and TP-T2 are respectively designed with β* coming from T1 and T2. Similarly, CHA H1-T1 and CHA H1-T2 were respectively designed with α* coming from T1 and T2. All other sequences, H2, TmINV–F, and Bio-Q are shared, remaining the same as those used in Fig. 2. In this way, the multiple signal probes can be replaced by different concentrations of H1s for each different T.
Fig. 4.
Scheme of multi-analysis based on LAMP-3W-CHA-PGM.
3.4. One-tube smart assay of mimic targets and LAMP amplicons based on LAMP-3W-CHA-PGM detection
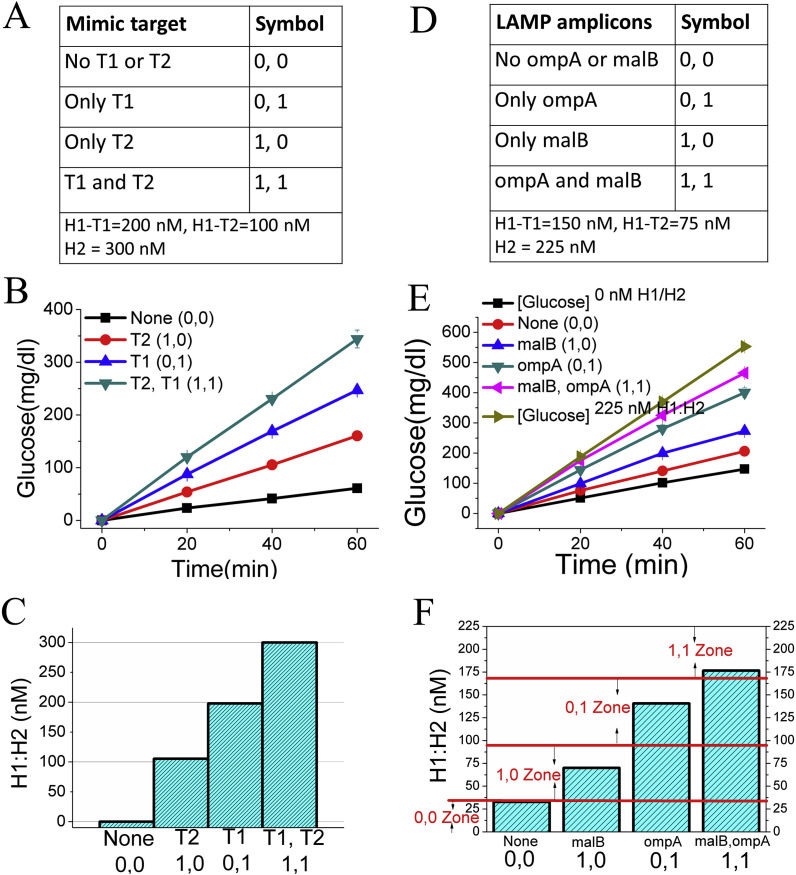
Before coupling with LAMP reactions, we used the T1 and T2 as the mimic targets to demonstrate the ideal situation of the mechanism. For example, if the concentration of H1-T1 is 200 nM, while H1-T2 is 100 nM, these H1s together share 300 nM H2. Such a design allows T1 and T2 to release up to 200 nM and 100 nM H1–H2 duplex, respectively; no matter how much each T was added. As shown in Fig. 5 A and B, the target in each of 4 situations (no target, T1, T2, and T1 + T2) generates four clearly distinguished glucose generated curves. If we define the glucose concentration at 60 min produced by target T1+T2 and no target is respectively contributed by the 300 nM H1:H2 and 0 nM H1:H2, after the calculation the glucose concentration at 60 min of T1 and T2 well fit the values generated from 200 nM, and 100 nM H1:H2 (Fig. 5C). Therefore, the recognition of the target in each of 4 situations (no target, T1, T2, and T1 + T2) could be directly readout through the concentrations of H1:H2 generated. When T1 and T2 are replaced using respective LAMP reaction products, similar results are still achieved. But due to the amounts of LAMP products might vary for each reaction, sometimes the final loops may be not enough to finish downstream CHA reaction during the defined time zone. In other words, the actual glucose generation curve may more or less deviate from the ideal situation, as shown in Fig. 5D and E. Even through, to generally provide an accurate recognition during multiplex analysis, here we derive a simple calculation that helps to confirm the signal (glucose or H1:H2 duplex concentration) zone which a specific target may sit into, as shown in Fig. 5F. The detail for the calculation is provided in the supporting information. Through aligning the actual detection results to the signal zones, each of 4 situations (no target, ompA, malB, and ompA + malB) can still be discriminated (Fig. 5F), no matter whether the CHA reaction has been finished or not. Therefore, following this method, logical or multiplex analysis can be easily realized in one-tube, with the only one signal probe, glucose. It is notable that Fig. 5D–F is gotten with LAMP amplicons generated from equal copy numbers of ompA and malB. But it actually doesn’t matter once the copy ratio between ompA and malB is changed. It is because in most cases different template copies will generate very similar amounts of amplicons after 1.5 h LAMP reactions. Therefore, the method developed here may be generalized to most targets after isothermal amplifications.
Fig. 5.
One-tube smart assay of mimic targets and LAMP amplicons based on LAMP-3W-CHA-PGM detection. (A) True value table and concentrations of CHA components used for detecting two mimic targets, T1 and T2. (B) Kinetic glucose production curves after 3W-CHA reactions with and without T1 or T2. (C) Bar-graph of concentration of 3W-CHA product (H1:H2) with and without T1 or T2 at 60 min glucose production. The value of reaction without any target has been defined as the background. (D) True value table and concentrations of CHA components used for detecting two kinds of LAMP amplicons, ompA and malB. (E) Kinetic glucose production curves after LAMP-3W-CHA reactions with and without 2000 copies of malB or ompA synthetic gene. [Glucose]0 nM H1:H2 represents the background with only LAMP reagents but no 3W-CHA components. [Glucose]225 nM H1:H2 represents the reaction containing enough LAMP loops that may consume all of 225 nM H2 after 3W-CHA reaction. (F) Bar-graph of concentration of LAMP-3W-CHA product (H1:H2) with and without 2000 copies of malB or ompA synthetic gene, at 60 min glucose production. Note: the calculation of the recognition zone for each of 4 situations was present in the supporting information.
4. Conclusions
In conclusion, here we proposed a smart connection between isothermal amplifications and personal glucometer detection via enzyme-free nucleic acid, the catalytic hairpin assembly. Through combining the advantage of each component, ultrasensitive, ultra specific and portable gene detection was realized. Being assisted by the design of three-way associative trigger, detection of a new target could be simply realized through chancing two sequence domains on TP probe and CHA H1. The sequences critical to signal out or required chemical label could remain unchanged. More importantly, through reading the different concentrations of H1:H2 generated for different target, we proposed a new multiplex detection strategy, which may potentially solve the traditional challenge encountered by most methods using commercial POC devices. Even though with these successes, many optimizations in experimental details are still required until a mature detection kit can be manufactured. And a device or chip that integrates all the three steps of this method is still anticipated to further simplify the detection.
CRediT authorship contribution statement
Lulu Guo: Writing - original draft. Yan Du: Conceptualization, Writing - original draft. Bingling Li: Conceptualization, Writing - original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the Harbin Scentia Biotechnology Corporation for the support of buffers and DNA sequences. This work was financially supported by the Natural Science Foundation of China (21605138, 21874129), K. C. Wong Education Foundation, Cooperation of Province and the Institution of Heilong Jiang province (YS17C21), Heilong Jiang Technology-based SME Technology Innovation Foundation (2017FK3GJ023), and Open Project of State Key Laboratory of Supramolecular Structure and Materials (sklssm2019013).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aca.2020.01.068.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wong D.T. Salivary diagnostics powered by nanotechnologies, proteomics and genomics. J. Am. Dent. Assoc, JADA. 2006;137:313–321. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 2.Vo-Dinh T., Cullum-Fresenius B. Biosensors and biochips: advances in biological and medical diagnostics. J. Anal. Chem. 2000;366:540–551. doi: 10.1007/s002160051549. [DOI] [PubMed] [Google Scholar]
- 3.Cheng M.M.C., Cuda G., Bunimovich Y.L., Gaspari M., Heath J.R., Hill H.D., Mirkin C.A., Nijdam A.J., Terracciano R., Thundat T., Ferrari M. Nanotechnologies for biomolecular detection and medical diagnostics. Curr. Opin. Chem. Biol. 2006;10:11–19. doi: 10.1016/j.cbpa.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Xiang Y., Lu Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J., Mazumdar D., Lu Y. A simple and sensitive “dipstick” test in serum based on lateral flow separation of aptamer-linked nanostructures. Angew. Chem. Int. Ed. 2006;45:7955–7959. doi: 10.1002/anie.200603106. [DOI] [PubMed] [Google Scholar]
- 6.Liu G., Chen H., Peng H., Song S., Gao J., Lu J., Ding M., Li L., Ren S., Zou Z., Fan C. A carbon nanotube-based high-sensitivity electrochemical immunosensor for rapid and portable detection of clenbuterol. Biosens. Bioelectron. 2011;28:308–313. doi: 10.1016/j.bios.2011.07.037. [DOI] [PubMed] [Google Scholar]
- 7.Nie Z., Deiss F., Liu X., Akbulut O., Whitesides G.M. Integration of paper-based microfluidic devices with commercial electrochemical readers. Lab Chip. 2010;10:3163–3169. doi: 10.1039/c0lc00237b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sardesai N.P., Barron J.C., Rusling J.F. Carbon nanotube microwell array for sensitive electrochemiluminescent detection of cancer biomarker proteins. Anal. Chem. 2011;83:6698–6703. doi: 10.1021/ac201292q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J., Li Y., Gu H., Xia F., Zuo X. Recent development of sandwich assay based on the nanobiotechnologies for proteins, nucleic acids, small molecules, and ions. Chem. Rev. 2014;114:7631–7677. doi: 10.1021/cr300248x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J.J., Xing H., Lu Y. Translating molecular detections into a simple temperature test using target-responsive smart thermometer. Chem. Sci. 2018;9:3906–3910. doi: 10.1039/c7sc05325h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du Y., Pothukuchy A., Gollihar J.D., Nourani A., Li B., Ellington A.D. Coupling sensitive nucleic acid amplification with commercial pregnancy test strips. Angew. Chem. Int. Ed. 2017;56:992–996. doi: 10.1002/anie.201609108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Ma C.B., Yang M.T., Pothukuchy A., Du Y. Point-of-care testing of various analytes by means of a one-step competitive displacement reaction and pregnancy tests trips. Sensor. Actuator. B Chem. 2019;288:163–170. [Google Scholar]
- 13.Zhang J.J., Xiang Y., Wang M., Basu A., Lu Y. dose-dependent response of personal glucose meters to nicotinamide coenzymes: applications to point-of-care diagnostics of many non-glucose targets in a single step. Angew. Chem. Int. Ed. 2016;55:732–736. doi: 10.1002/anie.201507563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J.J., Lu Y. Biocomputing for portable, resettable, and quantitative point-of-care diagnostics: making the glucose meter a logic-gate responsive device for measuring many clinically relevant targets. Angew. Chem. Int. Ed. 2018;57:9702–9706. doi: 10.1002/anie.201804292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Y., Zhang Y.T., Huang Z., Hu S.W., Zhao W., Xu J., Chen Y.H. An exploration of nucleic acid liquid biopsy using a glucose meter. Chem. Sci. 2018;9:3517–3522. doi: 10.1039/c8sc00627j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hun X., Xu Y., Xie G., Luo X. Aptamer biosensor for highly sensitive and selective detection of dopamine using ubiquitous personal glucose meters. Sensor. Actuator. B Chem. 2015;209:596–601. [Google Scholar]
- 17.Joo J., Kwon D., Shin H.H., Park K.-H., Cha H.J., Jeon S. A facile and sensitive method for detecting pathogenic bacteria using personal glucose meters. Sensor. Actuator. B Chem. 2013;188:1250–1254. [Google Scholar]
- 18.Gu C.M., Lan T., Shi H.C., Lu Y. Portable detection of melamine in milk using a personal glucose meter based on an in vitro selected structure-switching aptamer. Anal. Chem. 2015;87:7676–7682. doi: 10.1021/acs.analchem.5b01085. [DOI] [PubMed] [Google Scholar]
- 19.Xiang Y., Lu Y. An Invasive DNA Approach toward a general method for portable quantification of metal ions using a personal glucose meter. Chem. Commun. 2013;49:585–587. doi: 10.1039/c2cc37156a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong Q., Liu Q.Y., Guo L.L., Li D., Shang X.D., Li B.L., Du Y. A signal-flexible gene diagnostic strategy coupling loop-mediated isothermal amplification with hybridization chain reaction, Anal. Chim. Acta. ASAP. 2019 doi: 10.1016/j.aca.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Y., Hughes R.A., Bhadra S., Jiang Y.S., Ellington A.D., Li B. A sweet spot for molecular diagnostics: coupling isothermal amplification and strand exchange circuits to glucometers. Sci. Rep. 2015;5:11039. doi: 10.1038/srep11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robert M.D., Pierce N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D.Y., Turberfield A.J., Yurke B., Winfree E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- 24.Yin P., Choi H.M.T., Calvert C.R., Pierce N.A. Programming biomolecular self-assembly pathways. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 25.Li B., Ellington A.D., Chen X. Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods. Nucleic Acids Res. 2011;39:e110. doi: 10.1093/nar/gkr504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D.Y., Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 27.Stojanovic M.N., Stefanovic D., Rudchenko S. Exercises in molecular computing. Acc. Chem. Res. 2014;47:1845–1852. doi: 10.1021/ar5000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung C., Ellington A.D. Diagnostic applications of nucleic acid circuits. Acc. Chem. Res. 2014;47:1825–1835. doi: 10.1021/ar500059c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bi S., Yue S., Zhang S. Hybridization chain reaction: a versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem. Soc. Rev. 2017;46:4281–4298. doi: 10.1039/c7cs00055c. [DOI] [PubMed] [Google Scholar]
- 30.Wang F., Lu C.H., Willner I. From cascaded catalytic nucleic acids to enzyme. Chem. Rev. 2014;114:2881–2941. doi: 10.1021/cr400354z. [DOI] [PubMed] [Google Scholar]
- 31.Tang Y., Lu B., Zhu Z., Li B. Establishment of a universal and rational gene detection strategy through three-way junction-based remote transduction. Chem. Sci. 2018;9:760–769. doi: 10.1039/c7sc03190d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Notomi T., Okayama H., Masubuchi H. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B., Chen X., Ellington A.D. Adapting enzyme-free DNA circuits to the detection of loop-mediated isothermal amplification reactions. Anal. Chem. 2012;84:8371–8377. doi: 10.1021/ac301944v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B., Jiang Y., Chen X., Ellington A.D. Real-time detection of isothermal amplification reactions with thermostable catalytic hairpin assembly. J. Am. Chem. Soc. 2013;135:7430–7433. doi: 10.1021/ja4023978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Z., Tang Y., Jiang Y.S., Bhadra S., Du Y., Ellington A.D., Li B. Strand-exchange nucleic acid circuitry with enhanced thermo-and structure-buffering abilities turns gene diagnostics ultra-reliable and environmental compatible. Sci. Rep. 2016;6:36605. doi: 10.1038/srep36605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 37.Craw P., Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12:2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 38.Vashist S.K., Luppa P.B., Yeo L.Y., Ozcan A., Luong J.H.T. Emerging technologies for next-generation point-of-care testing. Trends Biotechnol. 2015;33:692–705. doi: 10.1016/j.tibtech.2015.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.