Abstract
The application of optical biosensors, specifically those that use optical fibers and planar waveguides, has escalated throughout the years in many fields, including environmental analysis, food safety and clinical diagnosis. Fluorescence is, without doubt, the most popular transducer signal used in these devices because of its higher selectivity and sensitivity, but most of all due to its wide versatility. This paper focuses on the working principles and configurations of fluorescence-based fiber optic and planar waveguide biosensors and will review biological recognition elements, sensing schemes, as well as some major and recent applications, published in the last ten years. The main goal is to provide the reader a general overview of a field that requires the joint collaboration of researchers of many different areas, including chemistry, physics, biology, engineering, and material science.
Keywords: Fiber optic and planar waveguide biosensors, Optical sensing, Fluorescence techniques, Biorecognition elements, Enzymes, Nucleic acids, Antibodies, Genetically engineered and biomimetic synthetic receptors
Graphical abstract
Highlights
-
•
Principles, configurations and fluorescence techniques using fiber optic and planar waveguide biosensors are discussed.
-
•
The biorecognition elements and sensing schemes used in fiber optic and planar waveguide platforms are reviewed.
-
•
Some major and recent applications of fiber optic and planar waveguide biosensors are introduced.
1. Introduction
Biosensors have been defined [1], [2], [3] as self-integrated devices capable of providing specific quantitative or semiquantitative analytical information on the species of interest using a biological recognition element (biochemical receptor), which is in direct spatial contact with the transducer element. Those biosensors based on the measurement of photons are classified as optical. This review is focused on optical fiber and planar waveguide fluorescence based biosensors, a type of device in which a waveguide (an optical transmitter) is used as a platform for the biochemical receptor as well as to transmit the excitation light and/or the resultant signal to a photodetector that converts the light into an electrical signal.
At present, optical biosensors are not competitive with bulky laboratory instrumentation, such as microplate array systems, for applications where a large number of samples need to be analysed simultaneously. However, they present some desirable features such as potential low cost, small size and ease of use and are well suited for some applications such as on-line monitoring or for the analysis of complex samples, as well as for the measurement of binding events in real time [4]. As a result, this is an active research area and a number of optical biosensing platforms are already in the market for application in specific areas including, environmental analysis, food safety or clinical diagnosis [5], [6], (a), (b).
Fluorescence is, without doubt, the most commonly used transducer signal in biosensors [7], [8]. Several parameters can be recorded and applied for sensing including, fluorescence intensity, that can be measured at the given wavelengths of excitation and emission; decay time or emission anisotropy, which is a function of the fluorescence intensities obtained at two different polarizations, vertical and horizontal [8], [9]. Therefore, a variety of possibilities exists to improve biosensor performance. For example, the excitation or the emission wavelengths of the luminophore can be adequately tuned to improve method selectivity and, in addition, the emission kinetics as well as the anisotropy properties of the luminescent compound may add specificity to the measurement in comparison to other optical methods [10]. Recent biosensor reviews [11], [12], [13], [14], [15] confirm that fluorescence-based transduction, in whichever of its possibilities, is one of the most popular optical detection methods used in biosensing.
This paper focuses on the working principles of fluorescence-based fiber optic and planar waveguides biosensors and will review biological recognition elements, assay formats, as well as selected applications in different areas, published in the last ten years. The main goal is to provide the reader a general overview of this exciting field which requires the joint effort of researchers of many different areas, including optics, biochemistry, electronics and fluidics.
2. Light guiding in optical waveguides and biosensor configurations
Optical waveguides are dielectric structures that transport energy between its two extremes at wavelengths in the UV–Vis and IR region of the electromagnetic spectrum. Depending on their geometry they can be classified in two main groups: cylindrical and planar. Optical fibers are included in the first group and consist of a cylindrical central dielectric core clad by a material of slightly lower refractive index (by ≈ 1%). A planar waveguide is formed from a dielectric slab core sandwiched between two cladding layers with lower refractive indexes [16]. In both, light propagation and guiding along the core is based on the well-known optical phenomenon of total internal reflection (TIR) [5], [16].
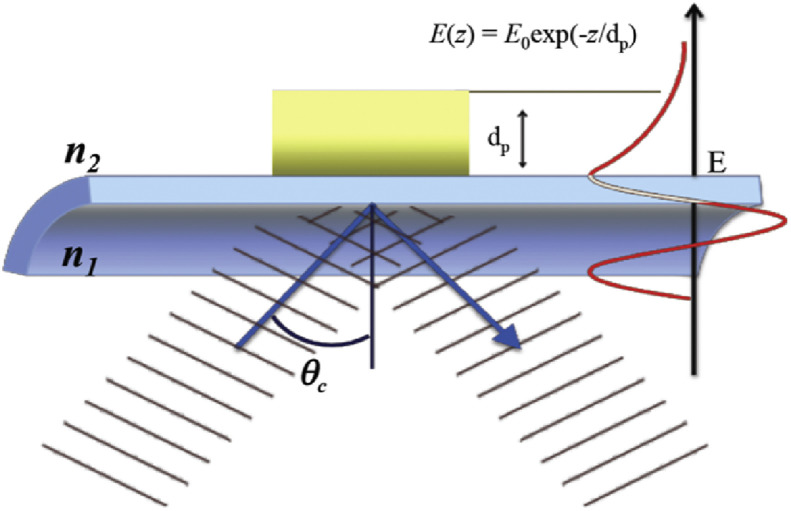
When light propagates along an optical waveguide (Fig. 1 ), such as an optical fiber or a planar waveguide, light is totally reflected at the interface between the optically more dense medium and the optically rare medium if the angle of refraction is larger than a critical angle (θC). At each point of reflection there is a finite decaying electric field across the interface that penetrates some distance into the lower refractive index medium. This field is referred to as the evanescent wave [17].
Fig. 1.
Electric field amplitude E, on both sides of the core/cladding interface of a waveguide. In the lower refractive index medium (n2, cladding) the electric field amplitude of the evanescent wave decays exponentially, with a penetration distance, dp, that depends on l, q, n1 and n2.
In emission spectroscopy, the evanescent wave can be used to generate luminescence or Raman scatter. The wavelength range extends from the UV to the far infrared depending on the quality of the waveguide. Higher order modes, i.e. those that propagate at angles close to the critical angle, contribute in a major extent to the power of the evanescent wave [5]. Therefore, the geometry of the sensing region must be properly designed in fluorescent biosensors based on evanescent field excitation (EFE) so as to increase the excitation power along the probe length, as well as to avoid the coupling of the emitted fluorescence into guided modes that fail to propagate in the clad fiber, what is known as V-number mismatch [5], [18], [19], [20]. Tapering of the sensing region has been proven to be a very effective way to minimize V-number mismatch and increase biosensor sensitivity when using partially clad fibers [5].
Several authors have studied different ways of increasing the performance of evanescent wave fiber optic biosensors. Design characteristics, taper parameters, as well as intrinsic fiber characteristics have been explored in order to increase depth and magnitude of the evanescent field [21], [22]. For optical fibers to be used in evanescence sensing, they must first be de-clad, at least partially, and most times tapered (either the core only, after de-cladding, or the complete fiber maintaining the cladding), so that they can be coated with the sensing element forming a uniform layer and thereby permitting that the evanescent field can interact with the fiber's surroundings [23], [24], [25]. All three processes (etching, tapering and coating) are not simple and require of several physical and chemical operations to be optimized [18], [23], [24], [25], [26]. One of the main advantages of EFE biosensors is the selective excitation of fluorophores attached either to the waveguide surface or within the penetration depth of the evanescent field [82]. The excitation of unbound interfering species present in bulk solution is avoided since the dp is typically in the order of 100 and 200 nm, for visible light, which allows their operation in absorbing, particulate or turbid media, as is the case in many biological samples [27]. This approach works extremely well for the detection of relatively small biomolecules (DNA, proteins) [27]; however the interrogation of micron-size objects, as cells or bacteria, require the use of sensing probes with increased penetration depth such as reverse waveguides, based on low refractive index substrates (below 1.33), or metal-clad waveguides that employ a thin metal layer between the substrate and the thicker waveguide layer to increase the probing depth [28].
Fiber optic (FO) biosensors can be based on two different configurations that rely on TIR for light propagation and guiding [17]: a) the opt(r)ode and, b) the evanescent wave configurations. In the former one, optical fibers are used to guide the excitation light from the light source to the distal end of the fiber where the recognition element is immobilized. In fluorescence based biosensors, the emitted light is guided back through the same fiber, or a different one, to the detector for signal generation. In the second configuration, the biorecognition layer is immobilized onto the longitudinal surface of the optical fiber core and is excited with the evanescent wave. In the latter approach, the penetration depth of the light is much lower, but the interrogated area is larger, than for optodes of the same diameter.
Planar waveguide (PWG) fluorescence biosensors are based on the localized excitation of the fluorophores at the surface of the waveguide upon evanescent field excitation. The emission phenomenon is known in this case as Total Internal Reflection Fluorescence (TIRF), but some papers based on EFE on planar waveguides also call the emission simply evanescent wave fluorescence [29]. PWGs are not usually applied for guiding back the emitted light to the detector but instead, as an advantage over optical fiber devices, they are used in combination with imaging systems (charge-coupled devices (CCDs) or CMOs camera) for multianalyte detection [30].
Planar slab waveguides can be classified in two main groups, bulk waveguides and integrated optical waveguides (IOWs) [29]. The diameter of bulk waveguides, also known as “internal reflection elements” (IREs), is higher than the wavelength of the reflected light and sensing hot spots can be clearly distinguished, at the points of reflection, on the waveguide surface. Integrated optical waveguides (IOWs) are prepared by depositing a very thin layer, usually in the order of the light wavelength or less, of a high refractive index material (usually metal oxides such as SiO2, TiO2, Ta2O5, or Nb2O5) on a glass substrate [29], [30], [31]. The sensitivity of IOWs has shown to be much higher in fluorescence based studies than for IRE-based waveguides, however they are more difficult to prepare and light coupling into the optical waveguide is more difficult requiring the assistance of prism or grating arrangements [29], [32], [33].
Planar waveguides have gained ground on optical fibers, especially when using EFE, for the development of fluorescence-based biosensing platforms in applications where the flexibility and remote capability of optical fibers are not essential. Other advantages of planar waveguides include their mechanical robustness and ease of fabrication, the potential for integration with optoelectronic components for compactness, the relatively easy patterning of the biological reagents needed for the assay, or the possibility to control the penetration depth of the evanescent field by controlling the incident reflection angle [34], [35].
The development of sensing microarrays, which enable multiple and simultaneous analyte detection, using both coherent fiber optic bundles, also referred to as optical imaging fibers [36], [37], [38], as well as integrated optical planar waveguides, hold great potential for fluorescence techniques in biosensing with FO and PWG. Imaging fibers are high density coherent optical arrays, formed by thousands of micron-sized optical fibers, bundled in a coherent way, so that the position of each fiber in one end is identical to the other end. Thus, the light is guided through each individual fiber from one end of the bundle to the other and the fluorescence signal from each region can be spatially resolved, depending on the distance between two adjacent fibers, and detected with a CCD camera. Fiber bundles have been employed for imaging purposes or, for biosensor arrays using encoded bead libraries and a variety of biorecognition elements [39], [40].
3. Fluorescence techniques in biosensing with fiber optics and planar waveguides
The fluorescence phenomenon is multifaceted and has several features that can be the basis of a biosensing design. Its versatility includes intensity-, kinetic-, wavelength- and anisotropy-based measurements [10].
3.1. Fluorescence intensity
After molecular excitation with appropriate light wavelength, spontaneous emission (fluorescence) of excess energy occurs at longer wavelengths, due to an electronic transition between the lowest excited singlet and the ground state. Fluorescence occurs if this deactivation process is favoured by the molecular structure of the compound and by the molecular environment (temperature, solvent characteristics, pH, etc.). Only if these factors make this relaxation mechanism the fastest and most efficient return to the ground state is fluorescence observed. The intensity of the emitted light (I F) at the analytical wavelength is directly related to the concentration of the fluorophore, according to the following equation (Eq. (1)), valid only for diluted solutions where εbc ≤0.02 [41], [42].
| [1] |
where κ is a constant related to the efficiency with which the instrument collects and detects the emitted light, ΦF is the fluorescence quantum yield (ratio of fluorescing to excited molecules; it can also be described in terms of light quanta or rate constants), I 0 is the intensity of the incident light and εbc are the parameters involved in the Lambert-Beer molecular absorption law.
In the case of fluorescence from an (optically) thick polymer film doped with a fluorescent dye, scattering from the solid material has to be also taken into account. Under this situation, very common in fluorescent sensors where the recognition and transducing layers are placed at the distal end of an optical fiber, only the reflectance mode can be used for fluorescence analysis so that the fluorescence intensity as a function of the immobilized fluorophore concentration (c) for an “infinite” layer thickness is given by Eq. (2) [43],
| [2] |
where a and a b are the absorption coefficients of the fluorophore and the “background” material of the film, respectively, at the analytical wavelength; R ∞,b is the diffusely reflected part of the incident radiation by the background, and the other symbols have the same meaning than those in Eq. (1).
For a given fluorophore sensitivity, as well as selectivity, is due mostly to instrumental design. First of all, the detector only views light when the dye in the transducing layer emits; therefore, fluorescence “turn-on” sensors are preferred as long as sufficient discrimination of the emitted and the excitation light is provided (usually with interference filters as monochromators are only used for benchtop spectrofluorometers). Detection limits are in the range of 10−9 and 10−10 M or in the ppt levels [41]. In fact, under very strict conditions, the detection of a single molecule may be attained [44], [45]. On the other hand, the use of the two wavelengths involved in the process (excitation and emission) provides extra selectivity since either of them or both can be controlled [8], [10].
Although not all compounds have intense native fluorescence, this disadvantage can be reverted, sometimes simply by their derivatization into a fluorescent compound, by making them part of a chemical reaction where a fluorescent compound is ultimately produced [42] or by attaching them with a fluorescent label that indicates their presence [46]. Fluorescent dyes, quantum dots (QDs) [47] and green fluorescent protein (GFP) have been used as fluorescent tags that have enabled researchers to study molecular events and the nature and accessibility of binding sites in biological macromolecules [48], [49], [50]. In the field of biosensing, again fluorescence is ideal since there are many naturally fluorescent biochemical substances or they can be easily tagged [51].
3.2. Luminescence lifetime
The luminescence lifetime (τ) represents the reciprocal of the rate constant of the (first order) emission decay that follows an “instantaneous” excitation of the luminophore by a flash of light [10]. Lifetime measurements can be carried out using a pulse light of radiation with a width typically shorter than the decay time of the luminophore. Upon excitation, the digitized emission decay profile is fitted to Eq. (3):
| [3] |
where I o corresponds to the amplitude, i.e. the fluorescence intensity at the end of the “instantaneous” excitation flash [52].
Alternatively, the luminescence lifetime can be evaluated by phase fluorometry (or phosphorimetry) measuring the phase shift between the sinusoidally modulated exciting light and the emitted light [53] (Eq. (4)):
| [4] |
where φ is the measured shift between the excitation and the emission sinusoidal signals and f is the excitation modulation frequency (Hz) [10].
Luminescence lifetime measurements have been extensively applied to the development of gas sensors, especially for oxygen measurements, and also in the development of enzymatic biosensors based on oxygen transduction [8] or in whole cell biosensors for biological oxygen demand (BOD) monitoring [54]. Its application to affinity sensors has been limited by the availability of probes whose decay time changes upon bimolecular interactions.
3.3. Fluorescence quenching
Quenching, in a very simplistic manner, can be described as any bi-molecular interaction by which the fluorescence intensity of the fluorophore molecule is diminished. One of the greatest difficulties of fluorometry is that it suffers from great environment-dependent quenching. Yet, what was originally considered a nuisance, if the fluorophore is the analyte, has been converted into a major application of fluorescence-based biosensors. Many are designed on the basis of the variation the quencher (in this case, the analyte or a third party related to it) produces on the fluorescence of an indicator dye. Although the many mechanisms of fluorescence quenching are beyond the scope of this paper, the process generally follows the Stern-Volmer equation [Eq. (5)] regardless the quenching mechanism [9], [41]:
| [5] |
where I F 0 and I F, and τ F 0 and τ F are, respectively, the fluorescence intensities and the fluorescence lifetimes of the indicator dye in the absence and presence of the quencher (Q) at concentration CQ; K SV is the so-called Stern-Volmer quenching constant which is related to the efficiency with which Q quenches the indicator fluorescence. Eq. (5) is only valid in the case of “dynamic” quenching, i.e. when the fluorophore and the quenching molecules are not pre-associated before the former absorbs a photon and Fick's diffusion laws apply. If quencher and dye are pre-associated to a certain extent, instantaneous “static” quenching occurs within the fluorophore-quencher complex, and Eq. (5) is only valid for the τ F 0/τ F ratio but not for the I F 0/I F ratio because only the free quencher follows the dynamic quenching rules. If there is practically no free quencher (i.e. the association constant is high enough), the emission lifetime ratio will not change with the quencher concentration, but the intensity ratio will obey Eq. (5) with K SV being now the association constant (K as) [10]. Because the fluorophore's signal depends on the quencher concentration, Q (or the third party) can be determined by its quenching action, in a sample that contains, or to which is added, a fluorescent compound.
Numerous procedures based on the quenching process have been devised for biosensing and, in this case, the biorecognition event causes a decrease in fluorescence. In some cases molecular recognition leads to fluorescence quenching of QDs used as markers [55] or of fluorescent dyes, such as certain chelates, which are of common use in this field [56], [57].
Quenching of fluorescent dyes and QD by metallic nanoparticles (NP) can occur at larger distances than those related with molecular quenchers [58], [59], and although NP have increased their application in quenching-based biosensing, there are examples of FO sensors based on the fluorescence enhancement of low-emission-quantum-yield dyes near certain metal surfaces or NPs (“metal-enhanced fluorescence” or MEF) [60]. Several authors have studied, explained and applied to biosensing this phenomenon, which is due to the formation of a mirror dipole on the metallic NP surface plasmon that itself radiates with high efficiency, resulting in enhanced radiative deactivation of the fluorophore excited state [61], [62], [63], [64], [65]. Contribution from the enhanced absorption of light by the fluorophore due to increased electric field between and around the metal NPs has to be taken into account as well. MEF has been shown to be more pronounced for larger particles, more efficient with metals possessing high free electron densities (Ag, Au) and at distances between 2 and 100 nm from the metal surface [60]. At shorter distances, fluorescence quenching due to the competing non-radiative energy or electron transfer to the metal NP surface prevails over the MEF effect [66].
3.4. Förster Resonance Energy Transfer (FRET)
Förster Resonance Energy Transfer, commonly (but incorrectly) called “Fluorescence” Resonance Energy Transfer, is a particular manifestation of fluorescence quenching that occurs when two different species, one (a donor) with a fluorescence spectrum that overlaps the excitation spectrum of the other (the acceptor), are close enough, usually less than 80 nm [67]. Thereby, the radiation-excited donor can transfer energy non-radiatively to the acceptor; so that the fluorescence intensity of the former is partially quenched, no matter if the acceptor is fluorescent or not. The electronic energy exchange is due to the dipole-dipole resonance interaction between the two molecules, not requiring a collision. Nevertheless, the FRET rate constant (k t) not only depends strongly on the donor-acceptor distance and on the extent of the spectral overlap between their respective spectra, but also on the relative orientation of the donor and the acceptor species [Eq. (6)] [68].
| [6] |
where κ 2 is the orientation factor, ΦD is the donor fluorescence quantum yield, F D(ν) is the spectral distribution of the donor fluorescence (normalized to unity in the wavenumber scale), ε A(ν) is the molar absorption coefficient (spectral distribution) of the acceptor, N A is the Avogadro's number, n is the solvent refractive index, τ 0 is the excited state lifetime of the donor in the absence of any quencher (including the acceptor), and R is the distance between the donor and the acceptor (in cm). Eq. (6) if often displayed as Eq. (7) after defining R 0 as the so-called “Förster distance”, i.e., the donor-acceptor distance at which k t = 1/τ 0, so that at that particular distance, the probability of emission of the excited donor is equal to the probability of energy transfer to the acceptor.
| [7] |
Evidently, the molecular recognition of the analyte must alter in some way the FRET process, thus producing a change that signals its presence. The most straightforward sensing application is obtained when the distance or orientation between the donor and acceptor changes in the presence of the target.
Several strategies have been used in FRET biosensing; the simplest, when the target itself is the acceptor-quenching moiety. Another one is that this second species also be fluorescent and thereby uses the transferred energy to emit its own fluorescence at a much longer wavelength, thereby dramatically increasing the emission Stokes shift. In this way, the primary excitation of the donor, whose fluorescence is quenched, will give rise to the fluorescence of the acceptor. When this is the case, it is possible to advantageously use a two-wavelength ratiometric method to sense the binding event [69], [70], [71], [72].
According to some authors [9], only in rare, lucky cases is one of the host/guest partners fluorescent. Therefore, labelling them with one or two organic dyes (or other type of label) is usually necessary. Also, FRET mechanism is not limited to the use of fluorescent organic dyes, QD and noble metal NP are now being used as either donors or acceptors, respectively [73], [74], [75], [76].
Intramolecular FRET in single stranded nucleic acids that form a stem-and-loop structure is the basis of molecular beacons [77], also known as molecular hairpins (see section 4.3). Very recently, FRET between luminescently-labelled (donor) NPs and a fluorescently-tagged (acceptor) analyte has been successfully used in a competitive assay format based on molecularly-imprinted polymer nanoshell coatings on the NPs [78].
4. Applications of fluorescence based fiber optic and planar waveguide biosensors
Recognition elements have become an extraordinary tool in recent years playing a successful role in the implementation of optical biosensors. Typical recognition elements are macromolecules or molecular assemblies that provide specific analytical information upon the recognition of the target molecule (ligand or analyte) in the course of a reaction.
The chemical information derived from the driven interactions is optically trapped and transformed into an electrical signal for data acquisition and interpretation. The recognition receptors feature inherent binding capabilities that determine the specificity and sensitivity of any proposed biosensor. In general, based on the nature of this binding event, recognition elements can be classified in [79], [80], [81]:
-
(A)
Catalytic Recognition Elements: The metabolic properties of these molecules, with natural or artificial origin, rely on catalyzed reactions whose progress is related to the concentration of the analyte, which can be measured by monitoring the changes in the product rate formation, reactant rate consumption or the inhibition of the reaction. The first biosensors described in literature were based on these recognition elements. Examples of catalytic recognition elements are: isolated enzymes, subcellular organelles, whole cells, tissues and/or organisms, etc. Moreover, engineered DNAzymes and catalytic antibodies (Abzymes) should also be included.
-
(B)
Affinity Recognition Elements: The recognition is typically driven by many weak interactions between the analyte and the receptor in a non-catalytically controlled process. It covers natural recognition elements such as antibodies, receptors, nucleic acids, bacteriophages, whole cells, etc. and genetically engineered or artificial recognition elements, such as recombinant antibodies, aptamers, engineered whole cells and/or organisms, as well as molecularly imprinted polymers (MIPs). Although most devices based on affinity interactions cannot work in a fully reversible way, they have been termed as biosensors for decades by the scientific community [51].
Biological, engineered and biomimetic recognition elements, employed for fiber optic and planar waveguide (bio)sensor development, have been described in detail and classified accordingly [8], [82], [83]. We will highlight herein those supported by a remarkable number of publications during the last years. Besides complementing the description of the (bio)recognition element, sensing schemes typically applied in optical fiber and planar waveguide based (bio)sensors will be reviewed.
4.1. Enzymes in biosensing with FO and PWG
Enzymes are the gold standard of the catalytic recognition elements used in biosensing development. Enzymes were the first receptors applied in biosensors [84] and they are still the basis of many devices in combination with different types of transducers (optical, electrochemical, etc). Enzyme-based biosensors proceed with extraordinary catalytic efficiency at small concentration levels. Interestingly, although major efforts to develop non-enzymatic sensors have been made, still there is a significant number of optical enzyme-biosensors developed in recent years [51], [85].
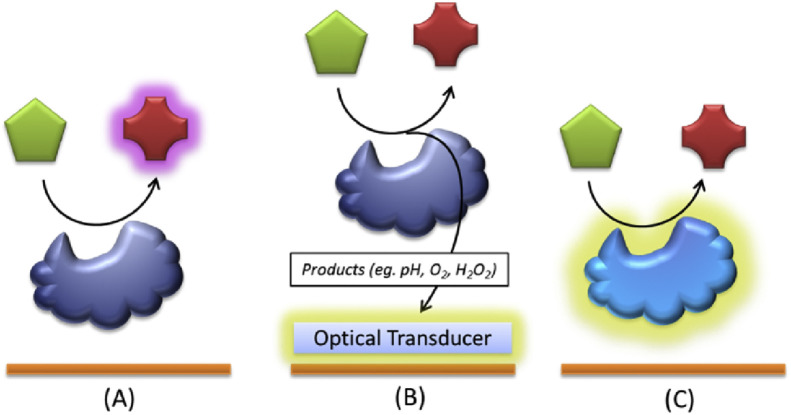
Three different sensing schemes are applied in enzyme-based FO and PWG biosensors, considering the following principles (Fig. 2 ) [8], [86]: (i) the analyte is assessed directly by monitoring the rate of formation of a product or the disappearance of a reagent with characteristic fluorescent properties in the course of the enzymatic reaction; (ii) the concentration of the target analyte is indirectly related to the signal generated by a fluorescent indicator (optochemical transducer) which responds to the changes in the biocatalytic process and (iii) the enzyme itself reports the presence of the analyte by variations in its luminescence properties.
Fig. 2.
Classification of enzymatic assays: (A) Analyte/product is assessed directly, (B) The analyte is assessed by the monitoring of an optochemical transducer, (C) the enzyme reports itself the presence of the analyte by variations in its luminescence.
Based on the first sensing scheme, Orellana et al. [87] have developed a fiber optic dosimeter for the analysis of carbamate pesticides. The authors have synthesized a red emitting Ru(II)polypyridyl complex containing a 4-acetoxyphenantroline ligand whose luminescence is strongly decreased upon hydrolysis by the enzyme acetylcholinesterase (AChE). The pesticide inhibits the enzymatic activity, and the luminescence intensity or the lifetime of the indicator dye can be used to monitor its concentration. The reduced form of nicotinamide adenine dinucleotide (NADH), a coenzyme involved in many oxidation or hydrogen transfer reactions, can be monitored via its emission at λ max 450 nm. For example, Kudo et al. [88], [89] have developed a fiber optic set-up for formaldehyde sensing (bio-sniffer) via the increase in the fluorescence of NADH using either formaldehyde dehydrogenase (FALDH) or the system aldehyde dehydrogenase (ALDH)/formate dehydrogenase (FDH) immobilized in a polytetrafluoroethylene (PTFE) membrane coated with a phospholipid polymer at the tip of an optical fiber. The reported biosensor enables real-time formaldehyde monitoring with a limit of detection of only 0.75 μg L−1.
As mentioned above, the second sensing scheme, mostly used for these enzymatic biosensors, is based on the indirect detection of the target analyte by monitoring the signal generated by a fluorescent indicator (optochemical transducer). In most cases, the concentration of the target compound is related to the oxygen consumption, pH change, hydrogen peroxide or ammonia production in the course of a biocatalytic reaction that is measured with the corresponding optochemical transducer near the immobilized enzyme. Recent advances in this field include the development of miniaturized devices that are applied to in vivo analysis. For example, an optical fiber microsensor, using fluorescence lifetime measurements, has been applied to the continuous measurement of glucose in subcutaneous tissues at concentrations up to 20 mM [90]. The hybrid sensor system consists of two oxygen optodes, one of them containing immobilized GOx to monitor glucose, and the second compensating for oxygen fluctuations in the intercellular fluid as well as for slight temperature changes. In a different approach, Pospiskova et al. [91] have developed a fiber optic biosensor with incorporated magnetic microparticles for the determination of biogenic amines. The device consisted of a lens coated with a UV-cured hybrid polymer that incorporates magnetic microparticles whose cores contain an optical oxygen indicator, Ruthenium tris-(bathophenatroline) dichloride [Ru (BaPhen)3]2+ 2Cl−, and their shells are modified with diamine oxidase. The measurement is performed by obtaining the quenched fluorescence lifetime of the ruthenium complex. The proposed system features a limit of detection of 25 μmol L−1 for putrescine and 30 μmol L−1 for cadaverine.
Stein et al. [92] have proposed an in vivo biochemical monitoring platform using what they call “smart tattoo” sensors. The biosensors consist of fluorescent microspheres that can be implanted intradermal and interrogated noninvasively using a custom optical fiber. The sensor particles were prepared using Pt(II) octaethylporphine (PtOEP), as oxygen indicator, incorporated into hybrid mesoporous alginate-silicate microspheres of ca. 10 μm, followed by loading and subsequent covalent immobilization of glucose oxidase. The surface of the nanoparticles was assembled with multilayer nanofilms doped with rhodamine B to provide a reference signal. The particles showed a fast (t95 = 84 s) and reversible response to glucose levels in the range of 2 and 10 mg dL−1, which is below the accepted clinical range of 40–350 mg dL−1 required for in vivo monitoring. However, the adequate selection of the surface-adsorbed nanofilm thickness, ionic strength of assembly conditions and capping layer allowed to tune the sensor response to cover the hypo- (0–80 mg dL−1), normo- (80–120 mg dL−1), and hyperglycemic levels (>120 mg dL−1) [93].
Zheng et al. [94] have described an optical fiber based nanosensor for the analysis of extracellular lactate at single cell level. The nanobiosensor that contains covalently immobilized lactate dehydrogenase at the fiber tip is parked on the plasma membrane of the cell and the evanescent field from the nanotip illuminates a spatially confined region. The enzyme catalyzes the conversion of lactate into pyruvate, and the fluorescence of the byproduct NADH is monitored as analytical signal. The device has been applied to the analysis of extracellular lactate concentrations in single HeLa, MCF-7 and human fetal osteoblast cells, allowing to distinguish the high extracellular lactate levels of cancer cell lines as compared to the normal cell line. The same instrumental set up has also been applied to the detection of telomerase, a general cancer biomarker, at single cell level, but using selective antibodies as recognition elements [95]. Single cell analysis using fiber optics and different recognition elements has been recently reviewed by Vo Dinh and Zhang [96].
Environmental monitoring has also been a growing area for biosensor application in the last decade, as these devices allow a simple, rapid and reliable measurement of various contaminants. For example, the analysis of 1,2-dichloroethane (DCA), a widely used halogenated organicreleased into the environment in large quantities, has been accomplished using haloalkane dehalogenase, DhlA, in whole cells of Xanthobacter autotrophicus GJ10 immobilized in calcium alginate on the tip of a fiber optic fluoresceinamine-based pH optode [97]. The biosensor had a response time of 8–10 min, mainly limited by DCA transport across the cell membrane, and provided a rapid, reagentless, continuous and simple measurement of DCA in water. In a further development [98] Rhodoccocus sp GJ70 expressing DhlA were immobilized on the tip of the optical fiber, along with the fluoresceinamine dye, allowing the detection of 1,2-dibromoethane in the range of 1–10 μg L−1.
Fluorescent nanoparticles, such as quantum dots (QDs), are being increasingly applied in combination with enzymes for biosensor development. QDs exhibit unique photophysical properties reflected by size-controlled fluorescence, high fluorescence quantum yields, stability against photobleaching and sensitivity to surface ligands. Therefore they offer an interesting alternative to common fluorophores for biosensor interrogation. In a recent example they have been applied to the development of microarray-based biosensors for the determination of phenol [99] fabricated by photopatterning of a solution containing PEG diacrylate (PEG-DA), tyrosinase, and CdSe/ZnS QDs. The entrapped enzyme catalyzes the oxidation of phenol to produce quinones, which subsequently quench the fluorescence of QDs within hydrogel microarray allowing 1 μM detection limits. Other applications of QDs are based on their photoluminescence quenching by H2O2 [100]. The production of this compound during the GOx biocatalysed reaction allowed glucose monitoring with a detection limit of 1.8 μM and the sensing platform has been adapted to 96-well plates for blood and urine multisample analysis.
Following the third sensing mechanism, Portaccio et al. [101] have characterized the intrinsic fluorescence of GOx immobilized in monolithic silica gel supports, in the UV and visible regions, using steady state and time resolved measurements, and they have further developed a fiber optic biosensor for glucose monitoring in the range from 0.2 to 10 mM. De Marcos et al. [102] have proposed a chemical modification of the enzymes using fluorescent labels, such as fluorescein derivatives, to increase the sensitivity and selectivity of direct enzyme based biosensors. GOx has been labelled with fluorescein-5(6)-carboxamidocaproic acid N-succinimidyl ester (GOx-FS) and entrapped in a polyacrylamide gel. The fluorescence intensity of GOx-FS is increased in the presence of glucose, with a linear response range between 300 and 2000 mg L−1 that has been attributed to an inner filter effect [8]. New trends using this approach deal with the synthesis of more sensitive devices by application of molecular genetics techniques, although their application in combination with fiber optics or planar waveguides is still limited. For example, a fluorescent molecular biosensor for the analysis of methyl parathion (MP), an organophosphate pesticide, has been described based on the construction of the fusion protein between a green fluorescent protein mutant (E2GFP) and the enzyme methyl parathion hydrolase (MPH). In the presence of the pesticide, the H+ ions released during the enzymatic reaction decrease the fluorescence intensity of E2GFP. Biosensor sensitivity was greatly enhanced by the integration of the E2GFP-MPH fusion protein biosensor structure into protein nanowires through self-assembly of Sup35 protein of Saccharomyces cerevisiae. Concentrations as low as 0.26 ng mL−1 MP, have been detected [103].
Enzyme immobilization on the transducer surface is of paramount importance for biosensor performance and several physical or chemical immobilization techniques can be applied to obtain a sensitive, robust and stable sensing layer [86], [104]. Doong and Shih [105] have proposed the preparation of titanium dioxide sol-gel matrices by vapor deposition for the co-immobilization of a fluorescent pH indicator, carboxy seminaphthorhodamine-1-dextran (SNARF-1-dextran), and glutamate dehydrogenase (GLDH). The sensing layers were deposited by pin-printing onto a glass-slide and evaluated using fluorescence microscopy allowing application to the determination of glutamate in water and biological samples. In a further development, the same approach has been applied to the development of an optical array biosensor for the simultaneous monitoring of glucose, urea and glutamate in serum samples [106]. Urease, glucose dehydrogenase (GDH), and GLDH have been coupled with the pH indicator SNARF-1-dextran and the change in the solution pH was evaluated by measuring the fluorescence ratio of the dye at 585 and 630 nm. Dynamic ranges of 2–3 orders of magnitude, with LODs of 3.1–7.8 μM, were obtained in serum samples without cross-interference from other species. Additional representative examples of fluorescence based enzymatic biosensors are depicted in Table 1 .
Table 1.
Fiber optics and planar waveguide fluorescence based enzymatic biosensors.
| Analyte(s) | Material | Sensor platform | Scheme | LOD | Analytical performance | Ref. |
|---|---|---|---|---|---|---|
| Direct sensors | ||||||
| Glucose | Glucose oxidase chemically modified with fluorescein-5(6)-carboxamidocaproic acid N-succinimidyl ester (GOx-FS) |
|
FI | – | Working range: 300–2000 mg L−1 Response time: 80 to 240 s |
[102] |
| Lactate | Lactate dehydrogenase |
|
FI | – | Working range: 0.06–1 mM Response time: 1 s |
[94] |
| l-Leucine | Leucine dehydrogenase immobilized in a PTFE membrane with PMEH polymer |
|
FI | 1 μM | Working range: 1-10000 mol L−1 |
[201] |
| Sorbitol | Sorbitol dehydrogenase from Flavimonas immobilized in a PMEH membrane |
|
FI | – | Working range: 1-1000 μmol L−1 Response time: 1.5 min |
[202] |
| pH transduction | ||||||
| Glutamate, urea and glucose | Enzymes: glutamate dehydrogenase, urease and glucose dehydrogenase Indicator: SNARF-1-dextran |
|
FI | Glutamate: 7.8 μM Urea: 3.1 μM Glucose: 5.4 μM |
Serum samples Glutamate: 40–8000 μM Urea: 2–300 μM Glucose: 20–10000 μM |
[106] |
| Organophosphate pesticides | Organophosphate hydrolase and carboxynaphthofluorescein |
|
EW | Paraoxon 2.5 μM Parathion 10 μM |
Working range: 0.00125–0.1 mM |
[203] |
| Halogenated aliphatic hydrocarbons | Haloalkane dehalogenase co-immobilized with 5(6)-carboxyfluorescein conjugated to bovine serum albumin |
|
FI | 0.133 mM for DBE 0.014 mM for CCMP |
30 min incubation | [204] |
| O2transduction | ||||||
| Uric acid | Uricase co-immobilized with Ru(dpp)3TMS2 (B1) or Ir(ppy)3 (B2) |
|
FI | B1: 50 μM B2: 20 μM |
B1: 5–600 μM B2: 5–800 μM Response time: 1.5 min |
[205] |
APTES = (3-aminopropyl)triethoxysilane; CCMP = 3-Chloro-2-(chloromethyl)-1-propene; CNF = carboxynaphthofluorescein; DBE = 1,2-Dibromoethane; EW = evanescence wave; FI = fluorescence intensity; Ir(ppy)3 = Tris(2-phenylpyridyl)iridium(III)complex; PVA = Polyvinylalcohol; PMEH = 2-methacryloyloxyethyl phosphorylcholine; PTFE = polytetrafluoroethylene; Ru(dip)32+ = Tris(4,7-diphenyl-1,10-phenanthroline)ruthenium(II) cation; Ru(dpp)3TMS2 = Tris(4,7-diphenyl-1,10-phenanthroline)ruthenium(II) trimethyl 3-(trimethylsilyl)-1-propanesulfonate; SNARF-1-dextran = Carboxy seminaphthorhodamine-1-dextran.
4.2. Antibodies in biosensing with FO and PWG
Antibodies have been for years one of the most used affinity recognition elements in areas as clinical medicine or the environmental and biotechnological fields. The combination of high affinity and high specificity together with relatively good stability makes antibodies suitable biosensor probes and thus, they have been applied to the development of fluorescent FO and PWG immunosensors. These devices exploit the principles of solid-phase immunoassays, such as ELISA, where selective bio-affinity interactions between an antibody and a specific compound are commonly observed on a solid support. Table 2 shows some examples of the trends in recent years in fluorescence immunosensing using FO and PWG.
Table 2.
Fiber optics and planar waveguide fluorescence based immunosensors.
| Analyte(s) | Material | Sensor platform | Scheme | LOD | Analytical performance | Ref. |
|---|---|---|---|---|---|---|
| C reactive protein, interleukin-6 (IL 6) and procalcitonin, in hospitalized patients | Specific type of antibody for each analyte on glass substrate coated with thin high index waveguide (Ta2O5) |
|
FI (EW) |
For IL-6: 0.08 ng mL−1; for PCT: 1 ng mL−1; for CRP: 0.35 ng mL−1 |
35 min assay | [32] |
| Capalstatin, in beef samples |
Capture antibodies, conjugated to donor fluorophore, on tips of tapered OF |
|
FRET | – | – | [71] |
| Telomerase, in MCF-7 breast cancer single cell nucleus | OF silver-coated nanotip probe immobilized with capture antibody |
|
FI | – | – | [95] |
| Escherichia coli O157:H7, in spiked food matrices | Stripes of capture antibodies, in a 12-channel PDMS template over the surface of glass microscope slides |
|
FI (EW) | 5 × 103 cells mL−1 | <30 min assay | [110] |
| SARS coronavirus nucleocapsid protein, in human serum | Capture antibody on surface of declad portion of plastic OFs |
|
FI (SP coupled) | ∼0.1 pg mL−1 | Working range: 0.1 pg mL−1–1 ng mL−1 | [116] |
| Factor V leiden (FVL) and FV, in blood plasma | Dual quartz FO system, with capture antibodies (FV preferred and FVL preferred) on surface |
|
FI | – | Working range: 0–12 μg mL−1 <10 min assay | [118] |
| VEGF; IP-10; IL-8 EGF; MMP-9; IL-1β in saliva samples |
Capture antibody-functionalized microspheres loaded in microwell array etched on the distal end of imaging OF bundle |
|
FI | VEGF (6 pg mL−1), IP-10 (26 pg mL−1), IL-8 (4 pg mL−1), EGF (3 pg mL−1), MMP-9 (1311 pg mL−1), IL-1β (5 pg mL−1) | Working range: VEGF (202–16000 pg mL−1), IP-10 (99–16000 pg mL−1), IL-8 (24–1000 pg mL−1), EGF (6–10000 pg mL−1), MMP-9 (1311–800000 pg mL−1), IL-1β (378–4000 pg mL−1) | [124] |
| Carcinoembryon antigen, in serum and nipple aspirate fluid samples | Capture Ab attached to phospholipid bilayer film joint to surface of a thin SiO2 deposited on a PWG |
|
FI (EW) | <0.5 pM | 15 min assay | [206] |
| Paralytic shellfish toxins: saxitoxin (STX), neosaxitoxin (NEO) produced by marine dinoflagellates in coastal waters | Capture antibodies anti-STX and anti-NEO labelled with Alexa Fluor® 647 carboxylic acid |
|
FI (EW) | STX: 12 pg mL−1 | STX: (CCβ: 20 pg mL−1) (%CV: 11%) total time to test a sample is 20 min | [207] |
| Microcystin LR (MCLR) | Planar waveguide (borosilicate microscope slides) patterned with dextran-BTL2/MCLR conjugates for increased loading capability |
|
FI (EW) | For MCLR: 0.007 ± 0.001 ng L−1 | Working range: 0.09–136.6 ng L−1 ∼3 h assay |
[208] |
| Microcystins in freshwater samples | Capture antibodies anti-STX and anti-NEO labelled with Alexa Fluor® 647 carboxylic acid |
|
FI (EW) | MCLR: 0.78 ng mL−1 |
MCLR: (CCβ: 1 ng mL−1) (%CV: 12.6%) Dynamic range (IC10–IC90) of 0.22–5.12 ng mL−1 | [209] |
|
Yersinia pestis, in infected mice |
Capture antibodies on polystyrene FO probes |
|
FI (EW) | – | Working range: 6.101–6.107 CFU mL−1 20 min assay | [210] |
| C reactive protein (CRP) | Polypeptide with tailored binder on bulk glass slide |
|
FI (EW) | – | With low affinity binder: 1.9×10−9– 5×10−8 M With high affinity binder: 3.4×10−10–2×10−8 M |
[211] |
| Progesterone, in bovine milk |
Progesterone-11- hemisuccinate on glass slide with bound aminodextran layer |
|
FI (EW) | 0.04 ng mL−1 | 5 min assay | [212] |
| 2,4-dichloro-phenoxyacetic acid (2,4-D) and microcystin-LR (MC-LR) |
Hapten–carrier conjugate 2,4-D-BSA or MC-LR-OVA on plastic-clad step-index silica optical fiber, uncladded and tapered on distal end. |
|
FI (EW) | for 2,4-D: 0.09 μg L−1 (0.07) for MC-LR: 0.03 μg L−1 | – | [213], [214] |
| Melamine, in milk products |
BSA-mel crosslinked by GMBS to a mercaptosilanized glass. |
|
EW | 6.6 μg L−1 | 26.6–517.5 μg L−1 20 min assay | [215] |
FI = fluorescence intensity; EW = evanescence wave; SP = surface plasmon; BTL2 = Geobacillus thermocatenulatus lipase 2; VEGF = human vascular endothelial growth factor; IP-10 = interferon gamma-induced protein 10; IL-8 = interleukin-8; EGF = epidermal growth factor; MMP-9 = matrix metalloproteinase 9; interleukin-1 beta (IL-1β); BSA = bovine serum albumin; GMBS = N-(4-maleimidobutyryloxy)succinimide.
The most widely used schemes of the aforementioned optical platforms can be classified according to four different assay formats [107], [108] (Fig. 3 ): (i) direct immunoassay – the antigen is incubated with excess amounts of immobilized antibody and the interaction is detected through, for example, a luminescence property of the antigen; (ii) competitive immunoassay – the preferred format for detecting small target molecules which contain only one epitope. Two modes of competitive assay are widely used: in one mode, the competition for the binding sites of the immobilized antibody is carried out between the target molecule and a labelled target. In the second mode, a target derivative is immobilized and the antibody is labelled so any target molecule in the sample is able to inhibit the binding of the antibody to the immobilized target; this mode is also known as the “binding-inhibition” assay; (iii) sandwich immunoassay – the preferred format for detecting targets with, at least, two epitopes in their structure. The assay involves the capture of the target molecules in solution by an immobilized primary antibody. Afterwards, a secondary labelled antibody (detection or reporter antibody) is added and binds the target molecule. Finally, the resultant complex is measured after washing the excess of immunoregeants; (iv) displacement immunoassay – in this flow assay configuration, the active sites of an immobilized antibody are previously saturated with an excess of labelled antigen. After a stable signal is achieved, the addition of the target molecule produces the displacement of the labelled antigen whose signal is proportional to the amount of target molecule added.
Fig. 3.
Classification of (bio)mimetic assays: (A) Direct immunoassay, (B) Competitive immunoassay, (C) Sandwich immunoassay and (D) Displacement immunoassay.
Several simple conclusions can be drawn from the information presented in literature. Although the binding-inhibition configuration has been broadly used, the sandwich type format has been, by far, the preferred detection set up [107]. On the other hand, most of the fluorescent labels used for marking of reporter antibodies are either organic fluorophores, such as Alexa®Fluor or Dylight® dyes, or fluorescent proteins, such as R-phycoerythrin. The advantages presented by QD labelling are barely used as yet, and the phenomenon of fluorescence enhancement near metal structures, such as NPs, has not been sufficiently exploited in FO and PWG biosensors [51].
One representative and commercial example of FO immunosensor is RAPTOR device. The field portable system was used to detect simultaneously biological agents, such as B. anthracis, Ricin, Cholera toxin or Salmonella, thanks to the four individual channels corresponding to one optical fiber [8]. Two representative examples of PWG immunosensors are based on a binding-inhibition format [109]: the first prototype was RIver ANAlyser platform (RIANA) and the second, built to overcome the drawbacks of RIANA, was the Automated Water Analyser Computer Supported System (AWACSS) [8]. Both were used to determine pesticides, pollutants and hormones in different liquid samples (waters and milk). The AWACSS platform employs a fiber-pigtailed chip, driven by a semiconductor laser that consists of a waveguide circuit which distributes excitation light to 32 separate sensing patches on the chip surface. Besides, a micro-fluidic system handles automatically the sample injection over the sensor surface and a fiber-coupled detection array monitors the fluorescence signals.
Ligler and coworkers developed a multi-analyte TIRF biosensor (National Research lab (NRL) array biosensor), which is fully automated and extensively applied in different fields [110], [111], [112], [113]. According to these authors [114], [115], the system consists of a two-dimensional array of immobilized capture molecules on the surface of an optical waveguide (glass slide), forming parallel patterns of either stripes or spots; the assays are performed within the channels of a multiple-channel flow cell placed on the PWG and multiple samples can be simultaneously applied. This biosensor uses mostly a sandwich type format and has been used in screening environmental, food and clinical matrices, as well as in the investigation of binding kinetics.
The three abovementioned platforms, take advantage of the qualities of PWG (their ability to define separate patterns of immobilized sensing elements on a single surface and the ease of integration into microfluidic platforms) as well as all the advantages provided by the evanescent field excitation (basically, surface selectivity). Moreover, the success of these platforms is due to the possibility of multiplexing and semi or fully automated analysis.
Chou and co-workers [64], [116], [117] developed another group of immunosensors that combine localized surface plasmon techniques, to increase the sensitivity of protein immunoassays, and EW excitation, to perform protein binding kinetics. Lastly, the group of K. Kang and colleagues [118] have determined several analytes of clinical interest and have studied the surface plasmon enhancement of cardiac marker detection.
In all the FO biosensing platforms that are presented in Table 2, waveguides are used as solid supports activated with the appropriate antibody to capture the target molecule during the assay. However, other works in the field of fluorescence immunosensing use optical fibres, usually in the form of bundles or arrays, simply to collect and transport emitted light [119], [120], [121], [122], [123] from antibody-based microspheres that report the presence of target analytes in the sample. In particular, microarrays which combine microsphere-technology and randomly ordered arrays have grown rapidly. For example, Nie and coworkers [124] reported a multiplexed protein microarray based on the fiber-optic bundles and microsphere-based antibody arrays for the detection of six salivary biomarkers for asthma and cystic fibrosis diagnosis. The potential to use the multiplexed protein array for respiratory disease diagnosis was fully demonstrated by the construction of an analyser, SDReader, which combines non-invasive sample collection and fully automated analysis [125]. Using the fiber-optic bundles, the Walt group has also developed a single molecule immunoarray, which they called digital ELISA [126], that can detect subfemtomolar concentrations of proteins in serum. Digital ELISA, also known as single molecule array (or SiMoA), features not only greater sensitivity but also enables faster assays, minimizes (by dilution) interferences and allows smaller sample volumes to be tested [127].
Although monoclonal and polyclonal antibodies are sensitive to temperature and have a limited self-life, they remain to be the main probes for immunosensor development. It is interesting to note that, despite the increasing production of recombinant antibodies engineered to improve their affinity and stability (e.g. Fab, Fv, single chain Fv, VH or VHH fragments), scarce number of works are published operating them in combination with either FO or PWG platforms. For example, Murphy et al. [128] produced a novel biotinylated single chain fragment (scFv) recombinant anti-microcystin avian antibody for the detection of microcystin-LR. This antibody fragment was applied in a fluorescence inhibition immunoassay on a planar waveguide detection system, where microcystin LR (MC-LR)-OVA conjugates were printed onto portable optical-planar waveguide cartridges and Streptavidin-Alexa-647® was used to detect the presence of biotinylated-anti-MC-LR. The immunoassay allows the detection of free MC-LR with a limit of detection of 0.19 ng mL−1 and a detection range of 0.21–5.9 ng mL−1.
4.3. Nucleic acids in biosensing with FO and PWG
Single-core optical fiber sensors, fiber optic microarrays and PWG sensors become powerful platforms by virtue of using nucleic acids as affinity biorecognition elements [129], [130], [131]. Recent applications are based on diagnostic tests of infections [132], detection of genetic mutations such as single nucleotide polymorphisms (SNPs) [133] or fast microbial pathogen detectors [134]; some of them are illustrative examples and will be reviewed in this section. Besides, Table 3 presents the general characteristics of other fluorescence-based Nucleic Acid FO and PWG biosensors (see Table 3).
Table 3.
Fiber optic and planar waveguide fluorescence based nucleic acid biosensors.
| Analyte(s) | Material | Sensor platform | Scheme | LOD | Analytical performance | Ref. |
|---|---|---|---|---|---|---|
| HABs (Harmful Algal Bloom) A. fundyense, A. ostenfeldii, and Pseudo-nitzschia australis |
ssDNA capture probes (<23 mer) covalently attached to encoded microspheres |
|
FI | <100 fM of rRNA | 45 min | [216] |
| Peanut allergens, Ara H1 in food matrices | Use as bioreceptor Ara H1 DNA aptamer, selected by CE-SELEX |
|
FI (FO-SPR) | 75 nM | Concentration tested from 0 nM to 634 nM | [217] |
| uidA gene of Escherichia coli | ssDNA-coated fiber probe (30 mer) |
|
FI (EW) | 3.2 amol of bound target DNA | 10 min/assay | [218] |
| mRNA abundances in FFPE tissue samples: Expression control of 14 genes markers of breast cancer | Spotted DNA array on slides coated with epoxysilane or polyvinylamine. DNA capture probes ranged between 53 and 71 mer. |
|
FI (EW) |
<10 fM. No target amplification is necessary | 7–18 h/24 samples | [219] |
| 17β-estradiol | DNA aptamer probe (76 mer) specific to 17β-estradiol |
|
FI (EW) | 2.1 nM (0.6 ng mL−1) | 10 min, 50 min/assay | [220] |
| Simultaneous detection of proteins (IL-6, IL-8) and gen encoding membrane protein, P6, of Haemophilus influenzae | A fiber-optic protein-DNA microarray using microsphere- immobilized capture antibodies specific to IL-6 and IL-8 |
|
FI | 100 fM for IL-6 and IL-8 1 pM for target DNA sequence |
3.4 h/assay | [221] |
EW = evanescence wave; FI = fluorescence intensity; FFPE = formalin-fixed paraffin-embedded; Phen = 1,10-phenanthroline and PHPIP = p-hydroxyphenylimidazo[f]1,10-phenanthroline; FO-SPR = Fiber optic surface plasmon resonance; CE = Capillary electrophoresis; SELEX = Systematic evolution of ligands by exponential enrichment.
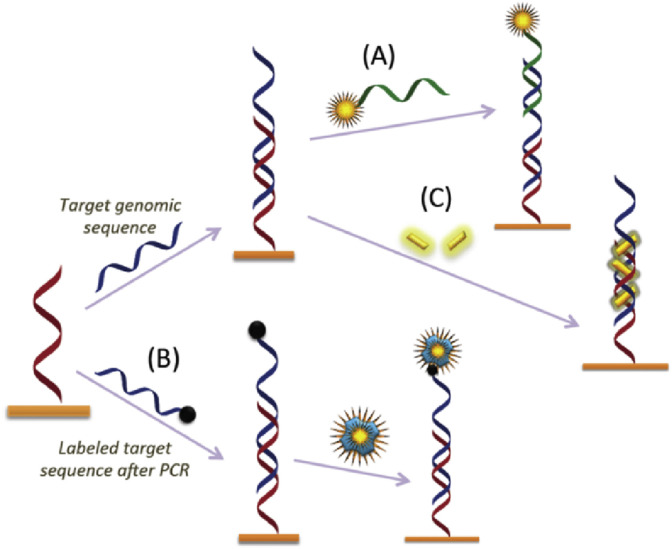
The typical transduction employed in this class of biosensors requires immobilization of a single strand oligonucleotide probe to the surface of the optical substrate and capture or hybridization of the target sequence of interest. Afterwards, the most frequent scheme of detection is based on the well characterized sandwich assay, in which a captured target sequence is allowed to hybridize with a labelled complementary target sequence (Fig. 4 A). A second indirect transduction format for detection is the use of luminescence reporter molecules that bind target nucleic acids previously labelled (e.g. biotinylated) by PCR amplification with labelled PCR primers or by a post-amplification labeling process (Fig. 4B) [39]. In a third approach, the hybridization between the capture probe and the target sequence is queried by a fluorogenic dye whose emission is enhanced by its binding to the double strand oligonucleotide structure (Fig. 4C).
Fig. 4.
Most frequent sensing schemes for nucleic acid detection: (A) Sandwich assay; a captured target sequence is allowed to hybridize with a labelled complementary target sequence. (B) Post-amplification labeling process. (C) Signal is queried by a fluorogenic dye whose emission is enhanced upon intercalation in the double strand oligonucleotide structure.
During the last years, the number of publications describing nucleic acid fluorescence biosensors based on single core fibers has decreased notoriously since microarray architectures on fibers and other substrates, such as planar guides, have become powerful tools for multiplexed sensing and fundamental studies of single molecules and cells [135]. However, several recent publications demonstrate that single core optical fibers are still the preferred platform to perform nucleic acid biosensing. For example, Massey and Krull [136] have recently explored a novel strategy for nucleic acid detection based on scaffolding duplexes bound to a silica fused optical fiber. The tandem consists of a capture DNA probe and a long-chain thiazole orange (TO) which switches on its emission when dsDNA is nearby. As a proof of concept, the biosensor was immersed in a solution containing 1 μM of the target gene sequence and a fivefold signal increment was observed. The authors claim that the suggested platform can be further used for simple nanoscale biosensors in analytical applications. In a different approach, the same group reported a multiplexed solid-phase assay for the detection of nucleic acid hybridization using optical fibers modified with QDs and co-immobilized with two different capture DNA probes. A sandwich assay is performed to detect the target gene sequence and the signal response is achieved upon hybridization through FRET-sensitized acceptor emission. To that aim, the detection probes are labelled at the 3′-end with Cy3 and Rhodamine Red-X dyes which are suitable acceptors of the emission upcoming from the QD film. Although the LOD of the proposed DNA-sensor is not sensitive enough, 10 nM, this fiber biosensor opens a new insight in the development of FRET multiplexed biosensors [137].
Since Ferguson and co-workers pioneered in the creation of a fiber optic array biosensor for the detection of seven DNA sequences [138], numerous FO microarray biosensors have been reported. Remarkable is the work of Walt and co-workers based on an innovative high density fiber optic bead-array platform, which has been applied for a variety of DNA and RNA biosensors in the last five years, highlighting very low LODs as well as low non-specific binding [39]. For example, Song [139] uses this platform for the simultaneous detection of Bacillus Anthracis, Yersinia pestis, Francisella tularensis and four more biological warfare agents (BWA) by a RNA assay where 50-mer capture probes are bound to uniquely identified microspheres. The authors reported LODs between 10 and 100 fM, and multiplexed assay with mixed samples indicated high specificity on the target detection. Gunderson and co-workers performed a DNA assay for SNPs detection using high density fiber optic array and beads decorated with highly specific ssDNA capture probes. The assay, which is based on the well-known Illumina GoldenGate Genotyping Assay [140], is able to interrogate up to 1536 SNPs per sample, minimizing time, reagent volumes, and material requirements of the process [141].
Rindorf and co-workers proposed a novel optical detection system incorporating a microstructured optical fiber (MOF) into an optic-compatible fluidic coupler chip. This hybrid biosensor operates towards the capture of specific DNA targets by functionalizing the MOF with an oligonucleotide capture probe in an optical sensing layer [134].
A novel emerging tool for bioassays is the use of single-molecule detection (SMD) techniques that, apart from the fascination of looking at individual biomolecules at work, offer the possibility of developing highly sensitive assays. Although SMD techniques were at the beginning a challenging task that required complex microscope apparatus, recent studies have been published about single molecule detection using optical fiber arrays [142], [143]. A nucleic acid assay with SMD is described by Z. Li and co-workers [144] using arrays of femtoliter-sized reaction vessels and a fluorescent enzymatic detection system. By a sandwich assay format, the simple binary read-out based scheme allows DNA detection down to 1 fM and exhibits a high signal-to-noise ratio.
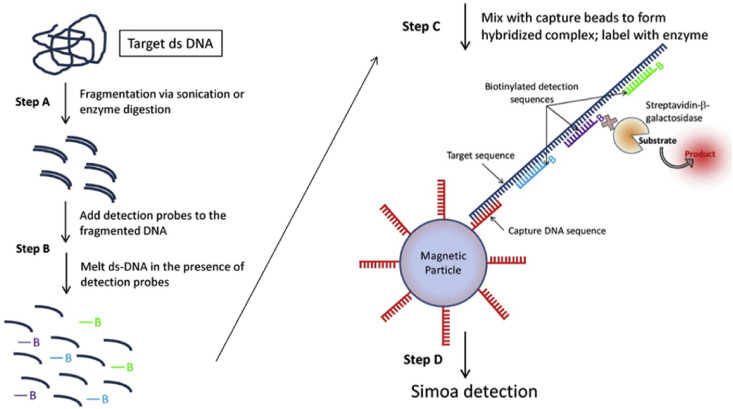
The ability of single molecule (fiber-optic) arrays (SiMoA) to literally count the number of protein molecules in an assay has also been exploited to detect bacterial genomic DNA at sub-femtomolar concentrations (Fig. 5 ). The SiMoA DNA assay, reported by Song et al. [145], directly detects target DNA molecules without molecular replication, such as the polymerase chain reaction (PCR). Furthermore, the method enables the quantification of DNA in river water without purification and authors claimed that it would be suitable for measuring bacteria in many other samples, such as bloodstream.
Fig. 5.
Workflow of the process for detecting genomic DNA using single molecule arrays (SiMoA). The target DNA, fragmented by the use of restriction enzymes, is mixed with biotinylated detection probes at an elevated temperature to form single-stranded DNA. The DNA-probe mixture is then incubated with magnetic beads decorated with DNA probes. Hybridized DNA complexes are then labelled with beta-galactosidase via biotin-streptavidin interaction. These beads are then loaded into arrays of femtoliter wells, and single DNA fragments are detected. Reproduced with permission from Ref. [145], http://dx.doi.org/10.1021/ac303426b.
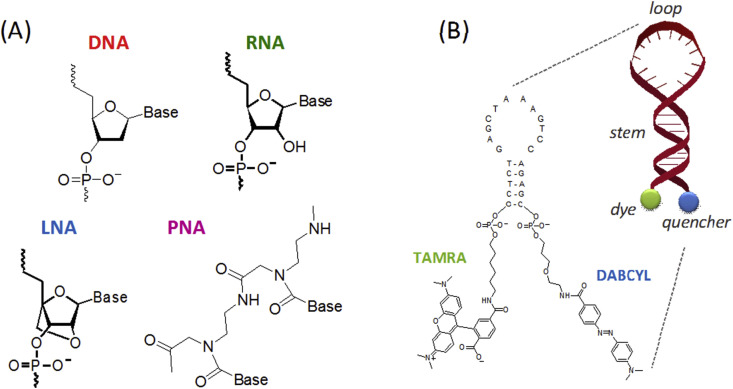
For significant progress in the development of DNA biosensors, it is essential to achieve rapid and accurate detections of specific DNA and RNA sequences. To that aim, it is important to obtain stable DNA/DNA duplex (or DNA/RNA) structures upon hybridization [146]. Thus, synthetic nucleic acids, such as Peptide Nucleic Acid (PNA) and Locked Nucleic Acid (LNA) (Fig. 6 A), are being used as alternatives, for example, to the classical ssDNA-based capture probes [147], [148]. Briefly, PNAs are synthetic DNA analogs whose sugar phosphate backbone has been replaced by an uncharged N-(2-aminoethyl) glycine scaffold. The uncharged nature of PNAs is responsible for a better thermal stability of PNA-DNA [149], [150]. LNAs are tailor-made nucleotides in which the ribose moiety is locked by an extra bridge connecting the 2′-oxygen and 4′-carbon. The novel nucleic acid architecture enhances base stacking and backbone pre-organization. LNA was initially prepared as an ideal oligomer for recognition of RNA, but LNA also displays high affinity toward DNA single strands [146].
Fig. 6.
(A) Chemical structure of DNA, RNA and LNA, PNA analogs. (B) Example of a molecular beacon probe.
The first publications of PNA polymers as novel recognition elements predicted high impact in biosensor development; however, it has still not been observed. PNAs exhibit disadvantages related to the cost of their synthesis and the high occurrence of unspecific binding events [151]. If these problems can be solved, PNAs will represent an interesting alternative for the development of the next generation of nucleic acid biosensors. For example, Sozzi et al. recently report a label free DNA sensor based on the use of capture PNA probes immobilized on the surface of a grating-based fiber optic platform [152].
In a similar way as PNAs, the use of LNAs has increased in the last years knowing their improved affinity and specificity with respect to their DNA and RNA targets. However, the new reported biosensors based on LNA capture probes are mostly based on non-optical techniques and their use in optical biosensors has not extended so far [153]. Optical platforms demonstrating the usefulness of LNA-modified oligonucleotides are mostly chip-based biosensing platforms constructed by photolithography of glass surfaces which eventually attach LNA capture probes arrayed by a combinatorial chemistry approach [154].
Molecular beacons (MBs) are particular dual labelled single strand oligonucleotide probes, DNA or RNA, that have self-complementary ends to form a stem-loop structure (hairpin) in their native state. The hairpin forces reporter and quencher into contact. Upon hybridization of a matching target to the loop portion, reporter and quencher are separated through conformational reorganization and the molecular beacon becomes bright (Fig. 6B). The use of MBs as immobilized capture probes on the optical substrate show two sensing schemes in which the signal is directly obtained upon both hybridization to the target sequence or ssDNA binding protein (Fig. 6B) [155]. The use of MBs has the advantage that no additional labelled species are necessary [156], [157], [158], [159]. In the last years, the employment of MBs in the development of solid-surface based biosensors has not fully matured. The results, which initially underscored the potential of MBs as smart biorecognition elements in fluorescence based FO and PWG biosensors, never overcame some of their critical limitations, such as the poor stability of the stem-and-loop structure once a MB nucleotide is immobilized onto a solid substrate. Some examples of these biosensors are depicted in Table 3.
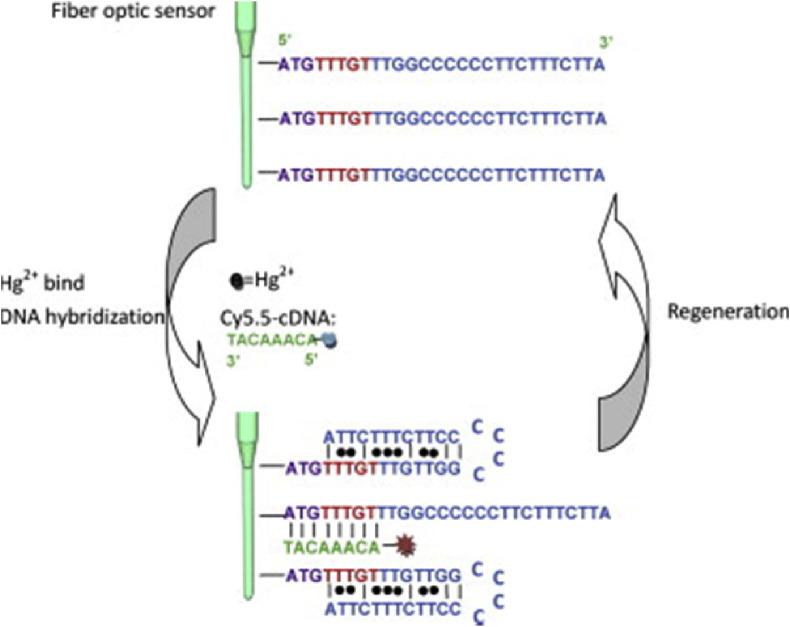
Biosensors using aptamers as biorecognition elements are referred to as aptasensors. In the same way as immunosensors, aptasensors can be applied in several sensing schemes to transduce the recognition process, such as, direct, competitive and sandwich assay formats. There are some interesting examples of fluorescence based optical aptasensors that have been reviewed recently [160]. One example of aptasensor, combined with PWG and fluorescence detection, is the evanescent wave DNA-aptamer-based biosensor for Hg (II) detection [161]. In this work, a hybrid DNA-aptamer probe is covalently immobilized onto an optical fiber. These DNA-aptamer probes have two different parts (Fig. 7 ): (i) a short sequence in 5′ for the hybridization of a complementary chain labelled with Cy5.5 and (ii) a sequence of T–T mismatch pairs that, in presence of Hg (II), promotes the folding of the probe into a hairpin structure forming a T-Hg(II)-T complex. The sensing scheme is a competitive assay, between the Hg (II) of the sample and a complementary DNA fluorescently labelled with Cy5.5. This platform has a detection limit of 2.1 nM and can be regenerated with a 0.5% SDS solution (pH 1.9) more than 100 times. The authors also evaluated the response of other metal ions and proved the high specificity of the sensor.
Fig. 7.
Schematic representation of a structure-competitive sensing mechanism of Hg (II) detection. Reproduced with permission from Ref. [161]http://dx.doi.org/10.1016/j.bios.2011.03.022.
Aptazymes (RNAzymes and DNAzymes) are bioinspired materials with allosteric properties that transduce the recognition event of their targets into catalytically generated reporting signals. An advantage of using aptazymes is their stability to repeated denaturation processes without losing neither catalytic nor binding capabilities. Furthermore, their production is not expensive and they are suitable for a wide range of analytes [162]. Although these recognition elements seem to be smart tools for optical sensing, they have not been applied up to date in fluorescence based FO or PWG sensing. The only publication we have found is the work of Yildirim et al. [163] who developed a portable and rapid DNAzyme based sensor to monitor Pb (II). The presence of the metal produces the cleave of the DNAzyme and a fluorescent labelled fragment is released that hybridizes with the complementary strand immobilized on the optic fiber sensor surface. Fluorescent signals of the hybridized fluorescent labelled fragment allows the detection of Pb (II) in a range from 2 to 75 nM, with a detection limit of 1.03 nM (0.21 ng mL−1). The sensor demonstrated good selectivity against other metal ions, and can be regenerated by treatment with a 1% solution of SDS (pH 1.9) to be used over 100 times.
4.4. Whole cells in biosensing with FO and PWG
Whole cells, including microorganisms (eukaryotic and prokaryotic cells), animal or plant tissues and cell receptors [164], [165], [166], [167] are the choice biorecognition elements to obtain information regarding the bioavailability, general toxicity and genotoxicity of target analytes, as well as to understand the interaction between cells and target compounds. Non-genetically engineered whole cells have been applied to the analysis of the total amount of pollutants and hazardous substances, and sensors for the analysis of the biological oxygen demand (BOD) are already on the market [168].
Peña-Vázquez et al. [169] developed an optical fiber sensor for the detection of simazine using three microalgae (Dictyosphaerium chlorelloides (D.c.), Scenedesmus intermedius (S.i.) and Scenedesmus sp. (S.s.)) The three microbial species were entrapped in a silicate sol-gel membrane placed at the tip of an optical fiber to create the sensor probe that showed response to those herbicides that inhibit the photosynthesis at photosystem II (PSII). The intensification in the chlorophyll fluorescence signal was measured, and the results showed that D.c. has a lower limit of detection (3.6 μg L−1) than S.s. (48. μg L−1) and S.i. (31.0 μg L−1). This biosensor is stable for at least 3 weeks. In a similar approach, Nguyen-Ngoc and co-workers prepared an optical biosensor using Chlorella vulgaris to detect the pesticide diuron [170]. Again, the distal end of an optical fiber is modified with a sol-gel membrane, containing the microalga, and an increase of fluorescence is observed when the pesticide inhibits the photosynthesis of PSII. The detection limit of the biosensor is 1 μg L−1 and maintains the activity after 5 weeks.
Another approach relies on the ability of genetically engineered whole cells to be used as sensing systems. In this case, both prokaryotic or eukaryotic cells could be modified by introducing a plasmid prepared by coupling a reporter gene with a sensing element, generally formed by an operon promoter region and a regulatory gene that encodes for a regulatory protein responsible of target binding (e.g. luxCDABE operon) [171]. Upon formation, the regulatory protein-target complex binds the promoter, and triggers the expression of the reporter gene responsible for the production of a luminescent reporter protein such as bacterial and firefly luciferases. These biosensors can be applied to the detection of a broad range of analytes including metals, organic pollutants, sugars, drugs, etc. [165]. For example, Ivask et al. [172] prepared two luminescent fiber-optic biosensors for the detection of Hg and As. The biosensors consist of alginate-immobilized recombinant bacteria emitting light specifically in the presence of bioavailable Hg or As in a dose-dependent manner. The limits of quantification of the developed sensors were 0.0026 mg L−1 for Hg and 0.012 and 0.14 mg L−1 for As(III) and As(V), respectively, which are good enough to allow their use in environmental analysis.
In a different approach, Futra and coworkers [173] propose an approach for monitoring the heavy metal toxicity. The biosensor is based on a recombinant E. coli modified with the green fluorescence protein (GFP) gene, which is directly immobilized on a cellulose nitrate membrane. The fluorescence response of the immobilized GFP-E. coli is monitored by a fiber-optic system, where the sensor membrane has been located in the distal end of the fiber. The response of the biosensor was stable for at least 3 weeks, and the detection limits were of 0.04, 0.32, 0.46, 2.80, 100, 250, 400, 720 and 2600 μg L−1 for Cu(II), Cd(II), Pb(II), Zn(II), Cr(VI), Co(II), Ni(II), Ag(I) and Fe(III), respectively.
4.5. Other receptors in biosensing with FO and PWG
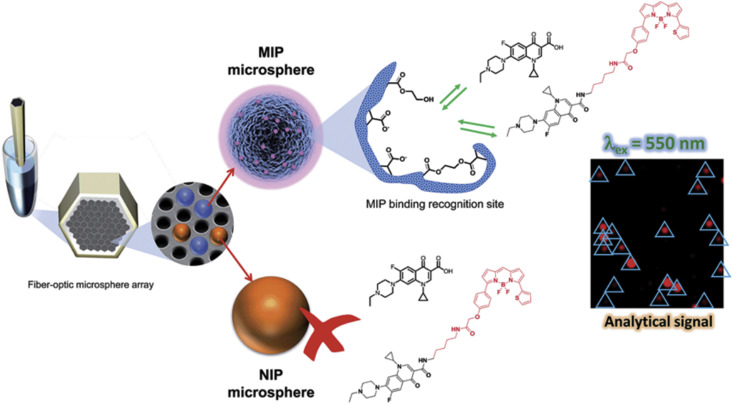
The nature of the receptors is very diverse, thus, we will focus our attention to those used in either optical fiber or planar waveguide platforms. In the last decade, four types of natural receptors have been used as affinity biorecognition elements in FO and PWG platforms combined with fluorescence detection: saccharide and glycoprotein receptors, toxin receptors, antimicrobial peptides, bacteriophages and molecularly imprinted polymers [8], [51], [174] (Table 4 ).
Table 4.
Other fiber optic and planar waveguide fluorescence biosensors.
| Analyte(s) | Material | Sensor platform | Scheme | LOD | Analytical performance | Ref. |
|---|---|---|---|---|---|---|
| Glucose |
Natural Receptors Glucose/Galactose binding protein-Badan conjugate (GBP-Badan) attached to either polystyrene or agarose beads via oligo His-Tag. |
|
FLT TC-SPC |
100 mM |
Kd = 13 mM (agarose) Kd = 20 mM (polystyrene) |
[182] |
| OPs |
Genetically Engineering Whole Cells OPH and EGFP co-displaying yeast. |
|
PW | – | 0.02–20 mM of paraoxon Stability at least a month at 4 °C |
[222] |
| Cocaine |
MIPs FITC-MIP. |
|
FI | – | 0–500 μM | [223] |
| BoNT serotype A light chain protease (LcA) |
Natural Receptors LcA peptide substrate sequence labelled with 6His-Tag and Cy3. |
|
FI-FRET | 350 pM | – | [185] |
| Theophylline |
Genetically Engineering Whole Cell Theophylline-sensitive riboswitch (TSR) placed upstream of the TEV protease coding sequence. |
|
FI-FRET | – | 11-fold increase in cellular extract fluorescence in the presence of theophylline Dynamic range: 0.01–2.5 mM |
[224] |
|
Triazines: simazine, atrazine, propazine, terbuthylazine; Urea based herbicides: linuron |
Whole Cell Three microalgal species (Dictyosphaeriumchlorelloides (D.c.), Scenedesmusintermedius (S.i.) and Scenedesmus sp. (S.s.) were encapsulated in silicate sol–gel matrices. |
|
FI | Simazine (3.6 μg L−1), atrazine (13.5 μg L−1), propazine (7.6 μg L−1), terbuthylazine (3.3 μg L−1), linuron (4.1 μg L−1) | Dynamic range: Simazine (19–860 μg L−1), atrazine (28–282 μg L−1), propazine (20–540 μg L−1), terbuthylazine (6–55 μg L−1), linuron (9–149 μg L−1). Stability: 3 weeks |
[169] |
| EA2192, VX, sarin and soman |
MIPs Thin films coated onto chemically tapped optical fiber. |
|
FI | EA2192 (11 ppt), Sarin (24 ppt), Soman (33 ppt), VX (21 ppt) | Linear dynamic ranges: ppt-ppm 15 min sensor response |
[225] |
| Pathogenic bacterium sources of HAI |
Bacteriophage RB encoding GFP genomic sequence |
|
FI | – | – | [226] |
| Heavy metals |
Genetically Engineering Whole Cell Cellulose nitrate membrane with recombinant GFP - E. coli immobilized. |
|
FI | Cu(II) (0.04 μg L−1); Cd(II) (0.32 μg L−1); Pb(II) (0.46 μg L−1); Zn(II) (2.80 μg L−1); Cr(VI) (100 μg L−1); Co(II) (250 μg L−1); Ni(II) (400 μg L−1); Ag(I) (720 μg L−1); Fe(III) (2600 μg L−1). | Dynamic range: Cu(II) (0.05–1 μg L−1); Cd(II) (0.50–10 μg L−1); Pb(II) (0.70–20 μg L−1); Zn(II) (5–100 μg L−1); Cr(VI) (0.10–5 mg L−1); Co(II) (0.50–7 mg L−1); Ni(II) (0.70–10 mg L−1); Ag(I) (1.00–20 mg L−1); Fe(III) (5.00–70 mg L−1). Sensor response stable for at least five weeks. |
[173] |
| Bisphenol A (BPA) |
MIPs Thin films on the flow cell, in contact with an optical fiber. |
|
EW | 1.7 μg L−1 | Dynamic range: 0.003–5 mg L−1 | [227] |
4.5.1. Saccharide and glycoprotein-based biosensors
Lectins constitute a broad family of protein receptors involved in numerous biological processes. They are very specific receptors and exhibit strong binding for saccharide moieties via multivalent interactions. Concanavalin A (ConA) [175] is the most widely used lectin in optical sensors. Several sensing schemes have been applied in the development of optical biosensors based on ConA but competitive and sandwich assays are the most used [176], [177]. Besides, the majority are associated to the use of advanced fluorescence techniques such as FRET and fluorescent nanomaterials [178].
Other protein receptors, with no lectin origin, are also used as recognition elements including the rivoflavin binding protein (RBP) [179], the maltose binding proteins (MBPs) [180], or the glucose binding protein [181], etc. A glucose/galactose-binding protein (GBP), labelled with the fluorophore 6-Bromoacetyl-2-Dimethylaminonaphthalene (Badan), was used by Saxl et al. [182] for the development of a glucose sensor. GBP-Badan was attached to the surface of NTA-agarose or polystyrene beads via Histag, and loaded into a porous chamber at the end of an optical fiber. Glucose responses of the immobilized sensor are reversible and stable, showing the best sensor response using Ni–NTA functionalized agarose beads (Kd of 13 mM (agarose) and 20 mM (polystyrene)).
4.5.2. Toxin receptor-based biosensors
Many bacterial toxins and viruses bind specifically to carbohydrate moieties on the cell surfaces to facilitate their invasion. Consequently, sugars and gangliosides have been used as recognition elements due to their specific ability of recognizing pathogens and toxins [183]. One of the most used toxin-receptor biosensor is based on the fluorescently labelled ganglioside GM1 receptor for the detection of cholera toxin [184]. In this case, the sensing scheme is based on a direct assay. The presence of the target molecule, with multiple binding sites, aggregates the labelled GM1 receptors and fluorescence self-quenching is observed.
In a different approach, Sapsford et al. [185] investigated the use of QDs to construct a peptide-FRET probe to detect botulinum neurotoxins (BoNTs), one of the most poisonous protein toxins known. The developed sensor monitors the BoNT serotype A light chain protease (LcA) activity, using a LcA substrate that combines: a LcA recognition/cleavage moiety, a specific residue to allow labeling a Cy3 acceptor dye, a linker-spacer sequence and a terminal oligohistidine to assemble the peptide on the QDs surface. The proteolytic assay is performed following an indirect pre-exposure of LcA-peptide-Cy3 conjugate to LcA, before the assembly on QDs. In this format, QDs are essentially utilized as a visualization reagent, demonstrating a limit of detection of 17.5 ng mL−1 for LcA (Fig. 8 ).
Fig. 8.
QD-FRET assay design components. (A) The QD-FRET-based assay format used to investigate LcA enzymatic activity. (B) Peptide sequences of the LcA substrate used in this study. Colors are used to highlight the different functional moieties. Partially reproduced with permission from Ref. [185], http://dx.doi.org/10.1021/nn102997b.
4.5.3. Antimicrobial peptides - based biosensors
Antimicrobial peptides (AMPs) represent an ancient and efficient innate defense mechanism, which protects species from infection with pathogenic microorganisms [186]. AMPs have the ability to associate with bacterial membranes so they are an alternative to some protein-based receptors for detecting their natural targets such as bacteria, fungi, viruses and also toxins such as botulinium toxins A, B and vaccinia virus [187], [188], [189]. The analytical characteristics of AMP-based biosensors were better, in some cases, than those obtained with antibodies as target captures, whereas sensing schemes are similar to those used for immunoassays (section 4.2.).
An example of the use of AMPs as affinity recognition elements is described in the study of Taitt's group [188] for detecting botulinum neurotoxoids A and B in an array biosensor using a sandwich immuno-like assay where the AMP and fluorophore-labelled detection antibody were simultaneously bound to different epitopes on the antigen (target toxoid A and B). Toxoid A could be detected with a limit of detection of 1 ng mL−1 in a 75 min assay, whereas toxoid B was detected at 10 ng mL−1.
4.5.4. Bacteriophages in biosensing with FO and PWG
Bacteriophages, as well as plant and some animal viruses [190], [191], have been used as probes for sensing and imaging [192], [193], [194]. Bacteriophages, shortened to phages, can be easily conjugated, they are robust, ubiquitous in nature, harmless to humans and their production is easy and economical. Currently, bacteriophage-based optical biosensors are well established thanks to their easy integration into transduction devices, comparable to antibodies and nucleic acids. They are typically used for the analysis of pathogens in food and water samples, reporting better response than classical microbiological methods [195].
The advances in phage-based biosensors have been impressive in the last few years. In this regard, engineered phages, and their combination with emerging materials and nanostructures, have boosted their analytical applications to the same level as other common biorecognition elements. However, their integration in fluorescence based FO and PWG platforms is still minimal and few examples are available in the literature. For example, Phillips and co-workers [196] describes a device for determining different bacteria, including: Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa using recombinant bacteriophages. In this case, the phage is used to deliver, to a targeted bacterial pathogen, either genetic material of a fluorescent detectable biomarker (e.g. GFP) or an operon promoter region and a regulatory gene that encodes the production of an enzyme, such as bacterial luciferase or aequorin. The device consists of a portable system that carries a power source, a fiber-optical detector, a signal processor, and a signaling device for emitting a signal upon detection of the pathogenic bacterium in an environment.
4.5.5. Molecularly imprinted polymers-based biosensors-like
Molecularly imprinted polymers (MIPs) have been proposed as alternatives to antibodies for the development of biomimetic sensors. During the imprinting process recognition sites are formed in the sensing material that enables selective binding of the target molecule even in complex mixtures [197].
Bio(mimetic) optical fiber and planar waveguide fluorescence sensors using MIPs as recognition elements, are usually based on the following sensing schemes [198]: (i) direct, based on the evaluation of the selective binding to the polymer of a naturally or labelled fluorescent target derivative. Alternatively, the polymer itself can be labelled and binding can induce a change in the fluorescent properties of the probes incorporated into the polymer backbone; (ii) competitive, as in immunosensors, are based on the competition of the target and a fluorescently labelled derivative for the polymer selective binding sites. Alternatively, the MIP binding sites can be saturated with the fluorescent labelled analogue that will be displaced by the target compound.
One of the main limitations for the development of MIP sensors has been the difficulty of integrating them with the transducer element. To overcome this shortcoming, Carrasco et al. [199] reported a fiber-optic microarray platform prepared by random self-assembly of MIP microspheres into a fiber optic microwell array. The device has been applied to the analysis of enrofloxacin (ENRO), a fluoroquinolone antibiotic in sheep blood serum samples. The sensing mechanism is based on a fluorescent competitive assay between the target antibiotic and a novel highly fluorescent labelled ENRO (Fig. 9 ). Strong fluorescence is observed in the absence of free analyte. The limit of detection was 40 nM. The selectivity of the assay was evaluated by measuring the cross-reactivity of other fluoroquinolones and non-related antibiotics. Only ciprofloxacin yielded an 8% inhibition of labelled ENRO binding at 10 μM concentration. No response was observed for the other antimicrobials tested.
Fig. 9.
Workflow of the assay protocol. ENRO quantification was based on a competitive assay in which the target analyte competes with the labelled ENRO (BODIFLOXACIN) to bind to specific binding sites on the MIP microspheres. Reproduced with permission from Ref. [199]http://dx.doi.org/10.1039/c5sc00115c - Published by The Royal Society of Chemistry.
5. Conclusions
Optical biosensors are increasingly finding their application in highly diversified areas such as medical and veterinary diagnosis, environmental testing, homeland security or food safety and several commercial devices are already found on the market [5]. This evolution has been the result of the combined efforts of specialists in very different disciplines, as well as the implementation of discoveries in other areas related to biosensor design, such as biochemistry, optics, fluidics or electronics, that in the end allowed the transfer of these devices from laboratory controlled environments, where they were initially applied, to the point of use.
Several factors have been identified that will play an important role in the future of biosensors, including [4], [200]: a) the availability of new immobilization strategies to enhance the performance and long-term stability of biomolecules; b) signal amplification schemes allowing higher sensitivities; c) the application of nanotechnology for geometric control of the biochemistry and signal enhancement; d) microfluidics for automation, reaction control and minimization of analysis time; e) the availability of optical elements to facilitate system integration and portability; f) new methodologies that facilitate the development of self-powered devices for the application of biosensors outside the lab walls.
The search of new recognition elements and sensing schemes has always received a great deal of attention. In fact, research in the search of bio and biomimetic selective recognition elements is an area of continuous research. However, although some applications require the use of very specific probes for each target of interest, new efforts are being focused on the application of more generic recognition approaches in combination with chemometric techniques that allow multiplexing capabilities with simpler designs.
Multianalyte sensing is not always a must but for some applications it represents a clear advantage over other detection techniques. DNA arrays and enzyme arrays, especially for clinical analysis of glucose, urea, cholesterol and lactate, are perfectly established. However protein arrays are most complex and still require further development [8].
Nanoparticles, QDs and metallic nanoparticles are being increasingly used for biosensing purposes including in vivo applications, although instrumentation is still a limiting factor in this area. Coded particles are an alternative as well for multiplexed analysis, but a large effort is being focused on the application of magnetic coded nanoparticles that enable sample treatment and target preconcentration with the corresponding increase in the sensitivity. Easy manipulation is another advantage of the application of such nanoparticles, especially in combination with microfluidic devices. Nevertheless, there is a great concern regarding the environmental toxicity of nanomaterials that must not be overlooked.
Another area that requires future attention is related to the synthesis of new dyes, with long-term stability and brightness, especially near infrared and long lifetime luminophores, as well as materials and surface chemistries that allow higher sensitivities, lower costs and favor mass production [200]. The final goal would be the production of affordable, easy to use, customer driven fully automated biosensors.
In conclusion, although advances in this field have been impressive in the last thirty years, the future of optical biosensors based on fluorescent measurements requires a close collaboration between experts in chemistry, physics, biology, engineers, material scientists and, very important, of end-users to get devices that can facilitate our everyday life.
ConA = Concanavalin A; DES = diethylstilbestrol; EGFP = Enhanced green fluorescence protein, ERE = Estrogen response element (ERE); EW = evanescence wave: FI = fluorescence intensity; FITC = fluorescein; FLT = fluorescence lifetime; GFP = Green fluorescence protein; HAI = Hospital acquired infection; MIPs: molecularly imprinted polymers; OPH = Organaphosphorus hydrolase; OPs = Organophosphorus compounds; PBac = Pathogenic bacterium; PEG = polyethylene glycol; PW = Planar Waveguide; QD = Quantum dots; RB = Recombinant bacteriophage; REACh = a non fluorescent YFP mutant (for resonance energy-accepting chromoprotein); rhERa and rhERb = recombinant human estrogen receptor a and b; TC-SPC = time correlated single-photon counting; TEV = Tobacco Etch Virus, YFP = yellow fluorescent protein, BPA = bisphenol A.
Acknowledgements
This work has been funded by the Spanish Ministry of Economy and Competitiveness (MINECO, grant CTQ2015-69278-C2-1R). B. Glahn-Martínez thanks MINECO for a Grant (“Promoción de Empleo Joven e Implantación de la Garantía Juvenil en I + D + i”). The authors thank Prof. G. Orellana for helpful discussions on the fundamentals of fluorescence spectroscopy.
Biographies

Elena Benito-Peña received her Ph.D in Chemistry from Complutense University (Madrid) in 2006. In 2005 she moved to the private sector working at the Chromatography Lab of Interlab Group (now Labaqua). In 2006, she returned to Complutense University where she is currently a Postdoctoral Research Associate with the Department of Analytical Chemistry. In 2009 she was awarded a Postdoctoral fellowship from the Spanish Government (MEC-FECYT) to work with Prof. David Walt at Tufts University (2009–2011) in the development of multiplexed optical-fiber bundle based biosensor microarrays and InfoBiology. Her research interests are optical sensors and biosensors, microarray biosensing platforms, synthesis of biomimetic elements for sensor and separation purposes, and their application to food, clinical and environmental analysis.

Mayra Granda-Valdés received her BSc (1971) and PhD (1987) degrees in Chemistry from Havana University (Cuba), and her MSc (1975) from Dalhousie University (Halifax, Canada). Since graduating, she has been in the teaching staff of the Department of Analytical Chemistry at the Faculty of Chemistry and is currently a Professor there. For a 5 year period she was the director of the department and for over 20 years member of the National Commission of Chemistry. Her primary research interests cover optical (bio)sensing, FIA, molecular imprinted materials, luminescence spectroscopy, pre-concentration methods and education in chemistry.

Bettina Glahn-Martínez obtained her degree in Chemistry in 2014 and a Master degree in Chemical Science and Technology in 2015 at Complutense University. In October 2013, she began her research activity as undergraduate student at GSOLFA (Group of Optosensors and Applied Photochemistry Lab) of Prof. Guillermo Orellana at Complutense University. In 2015, she was awarded an Erasmus scholarship to work with Prof. Homola at the Institute of Photonics and Electronics of the CAS in Prague on the development of SPR-based biosensors. In December 2015, she start her Ph.D under supervision of Prof. Moreno-Bondi and Dr. Benito-Peña. She is current research involves the development of biosensors for the detection of immunosuppressants.

Maria C. Moreno-Bondi received her Ph.D. in Analytical Chemistry (1990) at Complutense University of Madrid (Spain) where she was promoted to full professor in 2008. Her research interests lie in the development and applications of optochemical sensors and biosensors as well as on molecularly imprinted polymers for sensing and separation purposes (www.gsolfa.info/en/). She received the Young Researcher's Award (Spanish Society of Analytical Chemistry, 1993) and the Research Award in Analytical Chemistry from the Royal Spanish Society of Chemistry in 2010. She is currently Department Chair of the Analytical Chemistry Department at UCM and President of the Society of Applied Spectroscopy.
References
- 1.Hulanicki A., Geab S., Ingman F., Toth K., Durst R.A., Wilson G.S. Chemical sensors: definitions and classification. Pure Appl. Chem. 1991;63:1247–1250. [Google Scholar]
- 2.Thevenot D.R., Toth K., Durst R.A., Wilson G.S. Electrochemical biosensors: recommended definitions and classification. Anal. Lett. 2001;34:635–659. doi: 10.1016/s0956-5663(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 3.Thevenot D.R., Toth K., Durst R.A., Wilson G.S. Electrochemical biosensors: recommended definitions and classification. Biosens. Bioelectron. 2001;16:121–131. doi: 10.1016/s0956-5663(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 4.Taitt C.R., Anderson G.P., Ligler F.S. Evanescent wave fluorescence biosensors: advances of the last decade. Biosens. Bioelectron. 2016;76:103–112. doi: 10.1016/j.bios.2015.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ligler F.S., Taitt C.R., editors. Optical Biosensors: Today and Tomorrow. second ed. Elsevier; Netherlands: 2008. [Google Scholar]
- 6.(a) Bayer Technology Services, http://www.bayertechnology.com/es/.(accessed 01.06.16).; (b) Illumina, www.illumina.com (accessed 30.11.15)
- 7.White N.S., Errington R.J. Fluorescence techniques for drug delivery research: theory and practice. Adv. Drug Deliv. Rev. 2005;57:17–42. doi: 10.1016/j.addr.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Borisov S.M., Wolfbeis O.S. Optical biosensors. Chem. Rev. 2008;108:423–461. doi: 10.1021/cr068105t. [DOI] [PubMed] [Google Scholar]
- 9.Demchenko A.P. Springer; Netherlands: 2009. Introduction to Fluorescence Sensing. [Google Scholar]
- 10.Orellana G. Fluorescence-based sensors. In: Baldini F., Chester A.N., Homola J., Martellucci S., editors. Optical Chemical Sensors. Springer; Netherlands: 2006. pp. 99–116. [Google Scholar]
- 11.Medintz I.L., Delehanty J.B. Fluorescence-based intracellular sensing. In: Ligler F.S., R Taitt C., editors. Optical Biosensors: Today and Tomorrow. second ed. Elsevier; Netherlands: 2008. pp. 623–657. [Google Scholar]
- 12.Wark A.W., Lee J., Kim S., Faisal S.N., Lee H.J. Bioaffinity detection of pathogens on surfaces. J. Ind. Eng. Chem. 2010;16:169–177. doi: 10.1016/j.jiec.2010.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suri C.R., Boro R., Nangia Y., Gandhi S., Sharma P., Wangoo N., Rajesh K. Immunoanalytical techniques for analyzing pesticides in the environment. Trends Anal. Chem. 2009;28:29–39. [Google Scholar]
- 14.Farré M., Kantiani L., Pérez S., Barceló D. Sensors and biosensors in support of EU Directives. Trends Anal. Chem. 2009;28:170–185. [Google Scholar]
- 15.Ince R., Narayanaswamy R. Analysis of the performance of interferometry, surface plasmon resonance and luminescence as biosensors and chemosensors. Anal. Chim. Acta. 2006;569:1–20. [Google Scholar]
- 16.Banica F.G. Wiley; United Kingdom: 2012. Chemical Sensors and Biosensors. Fundamentals and Applications. [Google Scholar]
- 17.Taitt C.R., Anderson G.P., Ligler F.S. Evanescence wave fluorescence biosensors. Biosens. Bioelectron. 2005;20:2470–2487. doi: 10.1016/j.bios.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Rijal K., Leung A., Shankar P.M., Mutharasan R. Detection of pathogen Escherichia coli O157:H7 AT 70 cells/mL using antibody-immobilized biconical tapered fiber sensors. Biosens. Bioelectron. 2005;21:871–880. doi: 10.1016/j.bios.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Leung A., Shankar P.M., Mutharasan R. Label-free detection of DNA hybridization using gold-coated tapered fiber optic biosensors (TFOBS) in a flow cell at 1310 nm and 1550 nm. Sens. Actuators B-Chem. 2008;131:640–645. [Google Scholar]
- 20.Tao S., Gong S., Fanguy J.C., Hu X. The application of a light guiding flexible tubular waveguide in evanescent wave absorption optical sensing. Sens. Actuators B-Chem. 2007;120:724–731. [Google Scholar]
- 21.Ahmad M., Hench L.L. Effect of taper geometries and launch angle on evanescent wave penetration depth in optical fibers. Biosens. Bioelectron. 2005;20:1312–1319. doi: 10.1016/j.bios.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 22.Leung A., Shankar P.M., Mutharasan R. A review of fiber-optic biosensors, Sens. Actuators B-Chem. 2007;125:688–703. [Google Scholar]
- 23.Taitt C.R., Ligler F.S. Evanescent wave fiber optic biosensors. In: Ligler F.S., Taitt C.R., editors. Optical Biosensors: Present and Future. Elsevier Science; 2002. pp. 57–94. [Google Scholar]
- 24.Kasik I., Matejec V., Chomat M., Hayer M., Berkova D. Optical fibers for optical sensing. In: Baldini F., Chester A.N., Homola J., Martellucci S., editors. Optical Chemical Sensors. Springer; Netherlands: 2006. pp. 59–76. [Google Scholar]
- 25.El-Sherif M. Fiber-optic chemical and biosensors. In: Zourob M., Lakhtakia A., editors. Optical Guided-wave Chemical and Biosensors II. Springer; Berlin Heidelberg: 2010. pp. 109–150. [Google Scholar]
- 26.Ren H., Jiang C., Hu W., Gao M., Wang J., Wang H., He J., Liang E. The preparation of optical fiber nanoprobe and its application in spectral detection. Opt. Laser Technol. 2007;39:1025–1029. [Google Scholar]
- 27.Zourob M., Skivesen N., Horvath R., Mohr S., Goddard N.J. Deep-probe optical waveguides for chemical and biosensors. In: Xudong F., editor. Advanced Photonic Structures for Biological and Chemical Detection. Springer; Berlin Heidelberg: 2009. pp. 395–441. [Google Scholar]
- 28.Skivesen N., Horvath R., Thinggaard S., Larsen N.B., Pedersen H.C. Deep-probe metal-clad waveguide biosensors. Biosens. Bioelectron. 2007;22:1282–1288. doi: 10.1016/j.bios.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 29.Sapsford K.E. Total-internal-reflection platforms for chemical and biological sensing applications. In: Zourob M., Lakhtakia A., editors. Optical Guided-wave Chemical and Biosensors I. Springer; Berlin Heidelberg: 2009. pp. 3–20. [Google Scholar]
- 30.Sapsford K.E., R Taitt C., Ligler F.S. Planar waveguides for fluorescent biosensors. In: Ligler F.S., R Taitt C., editors. Optical Biosensors: Today and Tomorrow. second ed. Elsevier; Netherlands: 2008. pp. 139–184. [Google Scholar]
- 31.Gründler P. Springer; Berlin Heidelberg: 2007. Chemical Sensors: an Introduction for Scientists and Engineers. [Google Scholar]
- 32.Kemmler M., Koger B., Sulz G., Ursula Sauer, Schleicher E., Preininger C., Brandenburg A. Compact point-of-care system for clinical diagnostics. Sens. Actuators B-Chem. 2009;139:44–51. [Google Scholar]
- 33.Sagarzazu G., Bedu M., Martinelli L., Pelletier N., Safarov V.I., Weisbuch C., Gacoin T., Benisty H. Quantitative analysis of enhanced light irradiance in waveguide-based fluorescent microarrays. Biosens. Bioelectron. 2009;24:2281–2284. doi: 10.1016/j.bios.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Bernini R., Cennamo N., Minardo A., Zeni L. Planar waveguides for fluorescence-based biosensing: optimization and analysis. IEEE Sensors J. 2006;6:1218–1226. [Google Scholar]
- 35.Scully P.J., Merchant D.F. Optical sensors and biosensors for environmental monitoring. In: Butterworth F.M., Gunatilaka A., Bonaparte M.E.G., editors. Vol. 2. Plenum Publishing Coperation; New York: 2001. pp. 175–201. (Biomonitors and Biomarkers as Indicators of Environmental Change). [Google Scholar]
- 36.Epstein J.R., Walt D.R. Fluorescence-based fiber optic arrays: a universal platform for sensing. Chem. Soc. Rev. 2003;32:203–214. doi: 10.1039/b300617d. [DOI] [PubMed] [Google Scholar]
- 37.Biran I., Yu X., Walt D.R. Optrode-based fiber optic biosensors (bio-optode) In: Ligler F.S., Taitt C.R., editors. Optical Biosensors: Today and Tomorrow. second ed. Elsevier; Netherlands: 2008. pp. 3–83. [Google Scholar]
- 38.Espinosa Bosch M., Ruiz Sánchez A., Sánchez Rojas F., Bosch Ojeda C. Recent development in optical fiber biosensors. Sensors. 2007;7:797–859. [Google Scholar]
- 39.Walt D.R. Fiber optic microarrays. Chem. Soc. Rev. 2010;39:38–50. doi: 10.1039/b809339n. [DOI] [PubMed] [Google Scholar]
- 40.Biran I., Walt D.R. Optrode-based fiber optic biosensors. In: Ligler F.S., Taitt C.R., editors. Optical Biosensors: Present and Future. Elsevier Science; 2002. pp. 5–56. [Google Scholar]
- 41.Valeur B., Berberan-Santos M.N. second ed. Wiley-VCH; Weinheim: 2012. Molecular Fluorescence: Principles and Applications. [Google Scholar]
- 42.Harvey D. McGraw-Hill; Boston: 2000. Modern Analytical Chemistry. [Google Scholar]
- 43.Oelkrug D., Mammel U., Brun M., Günther R., Uhl S. In: Fluorescence Spectroscopy: New Methods and Applications. Wolfbeis O.S., editor. Springer; Berlin Heidelberg: 1993. pp. 65–78. [Google Scholar]
- 44.Walt D.R. Optical methods for single molecule detection and analysis. Anal. Chem. 2013;85:1258–1263. doi: 10.1021/ac3027178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kunzelmann S., Solscheid C., Webb M.R. Fluorescent biosensors: design and application to motor proteins. EXS. 2014;105:25–47. doi: 10.1007/978-3-0348-0856-9_2. [DOI] [PubMed] [Google Scholar]
- 46.Mikkelsen S.R., Cortón E. John Wiley and Sons; New Jersey: 2004. Bioanalytical Chemistry. [Google Scholar]
- 47.Algar W.R., Massey M., Krull U.J. The application of quantum dots, gold nanoparticles and molecular switches to optical nucleic-acid diagnostics. Trends Anal. Chem. 2009;28:292–306. [Google Scholar]
- 48.Hine A.V., Chen X., Hughes M.D., Zhou K., Davies E., Sugden K., Bennion I., Zhang L. Optical fiber-based detection of DNA hybridization, 2009. Biochem. Soc. Trans. 2009;37:445–449. doi: 10.1042/BST0370445. [DOI] [PubMed] [Google Scholar]
- 49.Chen X., Zhang L., Zhou K., Davies E., Sugden K., Bennion I., Hughes M., Hine A. Real-time detection of DNA interactions with long-period fiber-grating-based biosensor. Opt. Lett. 2007;32:2541–2543. doi: 10.1364/ol.32.002541. [DOI] [PubMed] [Google Scholar]
- 50.Simpson-Stroot J.M., Kearns E.A., Stroot P.G., Magaña S., Lim D.V. Monitoring biosensor capture efficiencies: development of a model using GFP-expressing Escherichia coli O157:H7. J. Microbiol. Methods. 2008;72:29–37. doi: 10.1016/j.mimet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 51.Wang X.D., Wolfbeis O.S. Fiber-optic chemical sensors and biosensors (2013−2015) Anal. Chem. 2016;88:203–227. doi: 10.1021/acs.analchem.5b04298. [DOI] [PubMed] [Google Scholar]
- 52.Carraway E.R., Demas J.N., DeGraff B.A. Luminescence quenching mechanism for microheterogeneous systems. Anal. Chem. 1991;63:332–336. [Google Scholar]
- 53.McNaught A.D., Wilkinson A. second ed. Blackwell Scientific Publications; Oxford: 1997. IUPAC. Compendium of Chemical Terminology (The “Gold Book”) [Google Scholar]
- 54.Orellana G., Haigh D. New trends in fiber-optic chemical and biological sensors. Curr. Anal. Chem. 2008;4:273–295. [Google Scholar]
- 55.Aoyagi S., Kudo M. Development of fluorescence change-based, reagent-less optic immunosensor. Biosens. Bioelectron. 2005;20:1680–1684. doi: 10.1016/j.bios.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 56.Lin L., Xiao L.L., Huang S., Zhao L., Cui J.S., Wang X.H., Chen X. Novel BOD optical fiber biosensor based on co-immobilized microorganisms in ormosils matrix. Biosens. Bioelectron. 2006;21:1703–1709. doi: 10.1016/j.bios.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 57.Kwok N.Y., Dong S., Lo W., Wong K.Y. An optical biosensor for multi-sample determination of biochemical oxygen demand (BOD) Sens. Actuators B-Chem. 2005;110:289–298. [Google Scholar]
- 58.Dulkeith E., Ringler M., Klar T.A., Feldmann J., Muñoz Javier A., Parak W.J. Gold nanoparticles quench fluorescence by phase induced radiative rate suppression. Nano Lett. 2005;5:585–589. doi: 10.1021/nl0480969. [DOI] [PubMed] [Google Scholar]
- 59.Pons T., Medintz I.L., Sapsford K.E., Higashiya S., Grimes A.F., English D.S., Mattoussi H. On the quenching of semiconductor quantum dot photoluminescence by proximal gold nanoparticles. Nano Lett. 2007;7:3157–3164. doi: 10.1021/nl071729+. [DOI] [PubMed] [Google Scholar]
- 60.Geddes C.D., editor. Metal-enhanced Fluorescence. Wiley; Hoboken, New Jersey: 2010. [Google Scholar]
- 61.Hong B., Kang K.A. Biocompatible, nanogold-particle fluorescence enhancer for fluorophore mediated, optical immunosensor. Biosens. Bioelectron. 2006;21:1333–1338. doi: 10.1016/j.bios.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 62.Bharadwaj P., Novotny L. Spectral dependence of single molecule fluorescence enhancement. Opt. Express. 2007;15:14266–14274. doi: 10.1364/oe.15.014266. [DOI] [PubMed] [Google Scholar]
- 63.Ng M.Y., Liu W.C. Fluorescence enhancements of fiber-optic biosensor with metallic nanoparticles. Opt. Express. 2009;17:5867–5878. doi: 10.1364/oe.17.005867. [DOI] [PubMed] [Google Scholar]
- 64.Chang Y.F., Chen R.C., Lee Y.J., Chao S.C., Su L.C., Li Y.C., Chou C. Localized surface plasmon coupled fluorescence fiber-optic biosensor for alpha-fetoprotein detection in human serum. Biosens. Bioelectron. 2009;24:1610–1614. doi: 10.1016/j.bios.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 65.Abdulhalim I. Nanophotonic and subwavelength structures for sensing and biosensing. In: Zourob M., Lakhtakia A., editors. Optical Guided-wave Chemical and Biosensors II. Springer; Berlin Heidelberg: 2010. pp. 73–106. [Google Scholar]
- 66.Goulet P.J.G., Aroca R.F. In: Geddes C., Lakowicz J.R., editors. Vol. 8. Springer; New York: 2005. pp. 223–247. (Topics in Fluorescence Spectroscopy: Radiative Decay Engineering). [Google Scholar]
- 67.Clegg R.M. Förster resonance energy transfer −FRET− what is it, why do it, and how it's done. In: Gadella T.W.J., editor. vol. 33. Elsevier; Amsterdam, The Netherlands: 2009. pp. 1–57. (Laboratory Techniques in Biochemistry and Molecular Biology). [Google Scholar]
- 68.Medintz I., Hildebrandt N., editors. FRET - Förster Resonance Energy Transfer: from Theory to Applications. Wiley-VCH; Weinheim, Germany: 2014. [Google Scholar]
- 69.Demchenko A.P. Optimization of fluorescence response in the design of molecular biosensors. Anal. Biochem. 2005;343:1–22. doi: 10.1016/j.ab.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 70.Ko S., Grant S.A. A novel FRET-based optical fiber biosensor for rapid detection of Salmonella typhimurium. Biosens. Bioelectron. 2006;21:1283–1290. doi: 10.1016/j.bios.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 71.Bratcher C.L., Grant S.A., Stringer R.C., Lorenzen C.L. Correlation between a novel calpastatin biosensor and traditional calpastatin assay techniques. Biosens. Bioelectron. 2008;23:1429–1434. doi: 10.1016/j.bios.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 72.Bratcher C.L., Grant S.A., Vasalli J.T., Lorenzen C.L. Enhanced efficiency of a capillary-based biosensor over an optical fiber biosensor for detecting calpastatin. Biosens. Bioelectron. 2008;23:1674–1679. doi: 10.1016/j.bios.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 73.Algar W.R., Tavares A.J., Krull U.J. Beyond labels: a review of the application of quantum dots as integrated components of assays, bioprobes and biosensors utilizing optical transduction. Anal. Chim. Acta. 2010;673:1–25. doi: 10.1016/j.aca.2010.05.026. [DOI] [PubMed] [Google Scholar]
- 74.Teles F.R.R., Fonseca L.P. Trends in DNA biosensors. Talanta. 2008;77:606–623. [Google Scholar]
- 75.Algar W.R., Krull U.J. Towards multi-colour strategies for the detection of oligonucleotide hybridization using quantum dots as energy donors in fluorescence resonance energy transfer (FRET) Anal. Chim. Acta. 2007;581:193–201. doi: 10.1016/j.aca.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 76.Cady N.C., Strickland A.D., Batt C.A. Optimized linkage and quenching strategies for quantum dot molecular beacons. Mol. Cell Probes. 2007;21:116–124. doi: 10.1016/j.mcp.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 77.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 78.Descalzo A.B., Somoza C., Moreno-Bondi M.C., Orellana G. Luminescent core–shell imprinted nanoparticles engineered for targeted Förster resonance energy transfer-based sensing. Anal. Chem. 2013;85:5316–5320. doi: 10.1021/ac400520s. [DOI] [PubMed] [Google Scholar]
- 79.Marazuela M.D., Moreno-Bondi M.C. Fiber-optic biosensors – an overview. Anal. Bioanal. Chem. 2002;372:664–682. doi: 10.1007/s00216-002-1235-9. [DOI] [PubMed] [Google Scholar]
- 80.Monk D.J., Walt D.R. Optical fiber-based biosensors. Anal. Bioanal. Chem. 2004;379:931–945. doi: 10.1007/s00216-004-2650-x. [DOI] [PubMed] [Google Scholar]
- 81.Pisoschi A.M. Biosensors as bio-based materials in chemical analysis: a review. J. Biobased Mater. Bioenergy. 2013;7:19–38. [Google Scholar]
- 82.Gasparac R., Walt D.R. Fiber-optic array biosensors. In: Marks R.S., Cullen D.C., Karube I., Lowe C.R., Weetall H.H., editors. Handbook of Biosensors and Biochips. John Wiley and Sons; New York: 2007. pp. 895–916. [Google Scholar]
- 83.Wang X.D., Wolfbeis O.S. Fiber-optic chemical sensors and biosensors (2008–2012) Anal. Chem. 2013;85:487–508. doi: 10.1021/ac303159b. [DOI] [PubMed] [Google Scholar]
- 84.Clark L.C., Jr., Lyons C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962;102:29–45. doi: 10.1111/j.1749-6632.1962.tb13623.x. [DOI] [PubMed] [Google Scholar]
- 85.Reardon K.F., Zhong Z., Lear K.L. Environmental applications of photoluminescence-based biosensors. Adv. Biochem. Eng. Biotechnol. 2009;116:99–123. doi: 10.1007/10_2008_51. [DOI] [PubMed] [Google Scholar]
- 86.Leca-Bouvier B.D., Blum L.J. Enzyme for biosensing application. In: Zourob M., editor. Recognition Receptors in Biosensors. Springer; New York: 2010. pp. 177–220. [Google Scholar]
- 87.Orellana G., Garcia-Fresnadillo D., Moreno-Bondi M.C. Carbamate pesticides sensing with a catalytic biosensor and molecularly engineered luminescent dyes. Afinidad. 2007;64:257–264. [Google Scholar]
- 88.Kudo H., Wang X., Suzuki Y., Ye M., Yamashita T., Gessei T., Miyajima K., Arakawa T., Mitsubayashi K. Fiber-optic biochemical gas sensor (bio-sniffer) for sub-ppb monitoring of formaldehyde vapor. Sens. Actuator B-Chem. 2012;161:486–492. [Google Scholar]
- 89.Kudo H., Yamashita T., Miyajima K., Arakawa T., Mitsubayashi K. NADH-fluorometric biochemical gas sensor (Bio-Sniffer) for evaluation of indoor air quality. IEEE Sensors J. 2013;13:2828–2833. [Google Scholar]
- 90.Pasic A., Koehler H., Klimant I., Schaupp L. Miniaturized fiber-optic hybrid sensor for continuous glucose monitoring in subcutaneous tissue. Sens. Actuator B-Chem. 2007;122:60–68. [Google Scholar]
- 91.Pospiskova K., Safarik I., Sebela M., Kuncova G. Magnetic particles–based biosensor for biogenic amines using an optical oxygen sensor as a transducer. Microchim. Acta. 2013;180:311–318. [Google Scholar]
- 92.Stein E.W., Grant P.S., Zhu H., McShane M.J. Microscale enzymatic optical biosensors using mass transport limiting nanofilms. 1. Fabrication and characterization using glucose as a model analyte. Anal. Chem. 2007;79:1339–1348. doi: 10.1021/ac061414z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stein E.W., Singh S., McShane M.J. Microscale enzymatic optical biosensors using mass transport limiting nanofilms. 2. Response modulation by varying analyte transport properties. Anal. Chem. 2008;80:1408–1417. doi: 10.1021/ac701738e. [DOI] [PubMed] [Google Scholar]
- 94.Zheng X.T., Yang H.B., Li C.M. Optical detection of single cell lactate release for Cancer metabolic analysis. Anal. Chem. 2010;82:5082–5087. doi: 10.1021/ac100074n. [DOI] [PubMed] [Google Scholar]
- 95.Zheng X.T., Li C.M. Single living cell detection of telomerase over-expression for cancer detection by an optical fiber nanobiosensor. Biosens. Bioelectron. 2010;25:1548–1552. doi: 10.1016/j.bios.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 96.Vo-Dinh T., Zhang Y. Single-cell monitoring using fiberoptic nanosensors. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2011;3:79–85. doi: 10.1002/wnan.112. [DOI] [PubMed] [Google Scholar]
- 97.Campbell D.W., Müller C., Reardon K.F. Development of a fiber optic enzymatic biosensor for 1,2-dichloroethane. Biotechnol. Lett. 2006;28:883–887. doi: 10.1007/s10529-006-9014-x. [DOI] [PubMed] [Google Scholar]
- 98.Reardon K.F., Campbell D.W., Müller C. Optical fiber enzymatic biosensor for reagentless measurement of ethylene dibromide. Eng. Life Sci. 2009;9:291–297. [Google Scholar]
- 99.Jang E., Son K.J., Kim B., Koh W.G. Phenol biosensor based on hydrogel microarrays entrapping tyrosinase and quantum dots. Analyst. 2010;135:2871–2878. doi: 10.1039/c0an00353k. [DOI] [PubMed] [Google Scholar]
- 100.Hu M., Tian J., Lu H.T., Weng L.X., Wang L.H. H2O2-sensitive quantum dots for the label-free detection of glucose. Talanta. 2010;82:997–1002. doi: 10.1016/j.talanta.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 101.Portaccio M., Lepore M., Della Ventura B., Stoilova O., Manolova N., Rashkov I., Mita D.G. Fiber-optic glucose biosensor based on glucose oxidase immobilised in a silica gel matrix. J. Sol-Gel. Sci. Technol. 2009;50:437–448. [Google Scholar]
- 102.De Marcos S., Sanz V., Andreu Y., Galbán J. Comparative study of polymeric supports as the base of immobilisation of chemically modified enzymes. Microchim. Acta. 2006;153:163–170. [Google Scholar]
- 103.Leng Y., Wei H.P., Zhang Z.P., Zhou Y.F., Deng J.Y., Cui Z.Q., Men D., You X.Y., Yu Z.N., Luo M., Zhang X.E. Integration of a fluorescent molecular biosensor into self-assembled protein nanowires: a large sensitivity enhancement. Angew. Chem. Int. Ed. 2010;49:7243–7246. doi: 10.1002/anie.201002452. [DOI] [PubMed] [Google Scholar]
- 104.Gupta R., Chaudhury N.K. Entrapment of biomolecules in sol–gel matrix for applications in biosensors: problems and future prospects. Biosens. Bioelectron. 2007;22:2387–2399. doi: 10.1016/j.bios.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 105.Doong R.A., Shih H.M. Glutamate optical biosensor based on the immobilization of glutamate dehydrogenase in titanium dioxide sol–gel matrix. Biosens. Bioelectron. 2006;22:185–191. doi: 10.1016/j.bios.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 106.Doong R.A., Shih H.M. Array-based titanium dioxide biosensors for ratiometric determination of glucose, glutamate and urea. Biosens. Bioelectron. 2010;25:1439–1446. doi: 10.1016/j.bios.2009.10.044. [DOI] [PubMed] [Google Scholar]
- 107.Wild D., editor. The Immunoassay Handbook: Theory and Applications of Ligand Binding, ELISA and Related Techniques. fourth ed. Elsevier; Oxford: 2013. [Google Scholar]
- 108.Ligler F.S. Fluorescence-based optical biosensors. In: Pavesi L., Fauchet P.M., editors. Biophotonics. Springer; New York: 2008. pp. 199–215. [Google Scholar]
- 109.Barceló D., Hansen P.D., editors. Biosensors for the Environmental Monitoring of Aquatic Systems. Springer; Berlin Heidelberg: 2009. [Google Scholar]
- 110.Shriver-Lake L.C., Turner S., Taitt C.R. Rapid detection of Escherichia coli O157:H7 spiked into food matrices. Anal. Chim. Acta. 2007;584:66–71. doi: 10.1016/j.aca.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 111.Moreno-Bondi M.C., Taitt C.R., Shriver-Lake L.C., Ligler F.S. Multiplexed measurement of serum antibodies using an array biosensor. Biosens. Bioelectron. 2006;21:1880–1886. doi: 10.1016/j.bios.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 112.Sapsford K.E., Ngundi M.M., Moore M.H., Lassman M.E., Shriver-Lake L.C., Taitt C.R., Ligler F.S. Rapid detection of foodborne contaminants using an array biosensor. Sens. Actuators B-Chem. 2006;113:599–607. [Google Scholar]
- 113.Herranz S., Marazuela M.D., Moreno-Bondi M.C. Automated portable array biosensor for multisample microcystin analysis in freshwater samples. Biosens. Bioelectron. 2012;33:50–55. doi: 10.1016/j.bios.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 114.Ngundi M.M., Taitt C.R. An array biosensor for detection of bacterial and toxic contaminants of foods. In: O'Connor L., editor. Diagnostic Bacteriology Protocols. second ed. Humana Press; New York: 2006. pp. 53–68. [DOI] [PubMed] [Google Scholar]
- 115.Taitt C.R., Shriver-Lake L.C., Ngundi M.M., Ligler F.S. Array biosensor for toxin detection: continued advances. Sensors. 2008;8:8361–8377. doi: 10.3390/s8128361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang J.C., Chang Y.F., Chen K.H., Su L.C., Lee C.W., Chen C.C., Chen Y.M.A., Chou C. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2009;25:320–325. doi: 10.1016/j.bios.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin C.H., Chen H.Y., Yu C.J., Lu P.L., Hsieh C.H., Hsieh B.Y., Chang Y.F., Chou C. Quantitative measurement of binding kinetics in sandwich assay using a fluorescence detection fiber-optic biosensor. Anal. Biochem. 2009;385:224–228. doi: 10.1016/j.ab.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 118.Kang K.A., Ren Y., Sharma V., Peiper S.C. Near real-time immuno-optical sensor for diagnosing single point mutation (a model system: sensor for factor V Leiden diagnosis) Biosens. Bioelectron. 2009;24:2785–2790. doi: 10.1016/j.bios.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rubenstein R., Chang B., Gray P., Piltch M., Bulgin M.S., Sorensen-Melson S., Miller M.W. A novel method for preclinical detection of PrPSc in blood. J. Gen. Virol. 2010;91:1883–1892. doi: 10.1099/vir.0.020164-0. [DOI] [PubMed] [Google Scholar]
- 120.Warner M.G., Grate J.W., Tyler A., Ozanich R.M., Miller K.D., Lou J., Marks J.D., Bruckner-Lea C.J. Quantum dot immunoassays in renewable surface column and 96-well plate formats for the fluorescence detection of botulinum neurotoxin using high-affinity antibodies. Biosens. Bioelectron. 2009;25:179–184. doi: 10.1016/j.bios.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Grate J.W., Warner M.G., Ozanich R.M., Jr., Miller K.D., Colburn H.A., Dockendorff B., Antolick K.C., Anheier N.C., Jr., Lind M.A., Lou J., Marks J.D., Bruckner-Lea C.J. Renewable surface fluorescence sandwich immunoassay biosensor for rapid sensitive botulinum toxin detection in an automated fluidic format. Analyst. 2009;134:987–996. doi: 10.1039/b900794f. [DOI] [PubMed] [Google Scholar]
- 122.Lucas L.J., Chesler J.N., Yoon J.Y. Lab-on-a-chip immunoassay for multiple antibodies using microsphere light scattering and quantum dot emission. Biosens. Bioelectron. 2007;23:675–681. doi: 10.1016/j.bios.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 123.Baldini F., Carloni A., Giannetti A., Mencaglia A., Porro G., Tedeschi L., Trono C. Optical PMMA chip for multianalyte detection. IEEE Sensors J. 2008;8:1305–1309. [Google Scholar]
- 124.Nie S., Benito-Peña E., Zhang H., Wu Y., Walt D.R. Multiplexed salivary protein profiling for patients with respiratory diseases using fiber-optic bundles and fluorescent antibody-based microarrays. Anal. Chem. 2013;85:9272–9280. doi: 10.1021/ac4019523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nie S., Henley W.H., Miller S.E., Zhang H., Mayer K.M., Dennis P.J., Oblath E.A., Alarie J.P., Wu Y., Oppenheim F.G., Little F.F., Uluer A.Z., Wang P., Ramsey J.M., Walt D.R. An automated integrated platform for rapid and sensitive multiplexed protein profiling using human saliva samples. Lab. Chip. 2014;14:1087–1098. doi: 10.1039/c3lc51303c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rissin D.M., Kan C.W., Campbell T.G., Howes S.C., Fournier D.R., Song L., Piech T., Patel P.P., Chang L., Rivnak A.J., Ferrell E.P., Randall J.D., Provuncher G.K., Walt D.R., Duffy D.C. Single-molecule enzyme-linked immunosorbent assay detects serum proteins. Nat. Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Quanterix, www.quanterix.com (accessed May 31, 2016).
- 128.Murphy C., Stack E., Krivelo S., McPartlin D.A., Byrne B., Greef C., Lochhead M.J., Husar G., Devlin S., Elliott C.T., O'Kennedy R.J. Detection of the cyanobacterial toxin, microcystin-LR, using a novel recombinant antibody-based optical-planar waveguide platform. Biosens. Bioelectron. 2015;67:708–714. doi: 10.1016/j.bios.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 129.Sassolas A., Leca-Bouvier B.D., Blum L.J. DNA biosensors and microarrays. Chem. Rev. 2008;108:109–139. doi: 10.1021/cr0684467. [DOI] [PubMed] [Google Scholar]
- 130.Gorris H.H., Blicharz T.M., Walt D.R. Optical-fiber bundles. FEBS J. 2007;274:5462–5470. doi: 10.1111/j.1742-4658.2007.06078.x. [DOI] [PubMed] [Google Scholar]
- 131.Wang Y., Pang X., Zhang Y. Recent advances in fiber-optic DNA biosensors. J. Biomed. Sci. Eng. 2009;2:312–317. [Google Scholar]
- 132.Massey M., Piunno P.A.E., Krull U.J. Challenges in the design of optical DNA biosensors. In: Orellana G., Moreno-Bondi M.C., editors. Frontiers in Chemical Sensors: Novel Principles and Techniques. Springer; Berlin Heidelberg: 2005. pp. 227–260. [Google Scholar]
- 133.Wang X., Krull U.J. Synthesis and fluorescence studies of thiazole orange tethered onto oligonucleotide: development of a self-contained DNA biosensor on a fiber optic surface. Bioorg. Med. Chem. Lett. 2005;15:1725–1729. doi: 10.1016/j.bmcl.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 134.Rindorf L., Høiby P.E., Jensen J.B., Pedersen L.H., Bang O., Geschke O. Towards biochips using microstructured optical fiber sensors. Anal. Bioanal. Chem. 2006;385:1370–1375. doi: 10.1007/s00216-006-0480-8. [DOI] [PubMed] [Google Scholar]
- 135.Whitaker R.D., Walt D.R. Fiber-based single cell analysis of reporter gene expression in yeast two-hybrid systems. Anal. Biochem. 2007;360:63–74. doi: 10.1016/j.ab.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 136.Massey M., Krull U.J. Towards a fluorescent molecular switch for nucleic acid biosensing. Anal. Bioanal. Chem. 2010;398:1605–1614. doi: 10.1007/s00216-010-3794-5. [DOI] [PubMed] [Google Scholar]
- 137.Algar W.R., Krull U.J. Multiplexed interfacial transduction of nucleic acid hybridization using a single color of immobilized quantum dot donor and two acceptors in fluorescence resonance energy transfer. Anal. Chem. 2010;82:400–405. doi: 10.1021/ac902221d. [DOI] [PubMed] [Google Scholar]
- 138.Ferguson J.A., Boles T.C., Adams C.P., Walt D.R. A fiber-optic DNA biosensor microarray for the analysis of gene expression. Nat. Biotechnol. 1996;14:1681–1684. doi: 10.1038/nbt1296-1681. [DOI] [PubMed] [Google Scholar]
- 139.Song L., Ahn S., Walt D.R. Fiber-optic microsphere-based arrays for multiplexed biological warfare agent detection. Anal. Chem. 2006;78:1023–1033. doi: 10.1021/ac051417w. [DOI] [PubMed] [Google Scholar]
- 140.Illumina, www.illumina.com (accessed 30.11.15).
- 141.Gunderson K.L., Kruglyak S., Graige M.S., Garcia F., Kermani B.G., Zhao C., Che D., Dickinson T., Wickham E., Bierle J., Doucet D., Milewski M., Yang R., Siegmund C., Haas J., Zhou L., Oliphant A., Fan J.B., Barnard S., Chee M.S. Decoding randomly ordered DNA arrays. Genome Res. 2004;14:870–877. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rissin D.M., Walt D.R. Digital concentration readout of single enzyme molecules using femtoliter arrays and Poisson statistics. Nano Lett. 2006;6:520–523. doi: 10.1021/nl060227d. [DOI] [PubMed] [Google Scholar]
- 143.Aouani H., Deiss F., Wenger J., Ferrand P., Sojic N., Rigneault H. Optical-fiber-microsphere for remote fluorescence correlation spectroscopy. Opt. Express. 2009;17:19085–19092. doi: 10.1364/OE.17.019085. [DOI] [PubMed] [Google Scholar]
- 144.Li Z., Hayman R.B., Walt D.R. Detection of single-molecule DNA hybridization using enzymatic amplification in an array of femtoliter-sized reaction vessels. J. Am. Chem. Soc. 2008;130:12622–12623. doi: 10.1021/ja8053018. [DOI] [PubMed] [Google Scholar]
- 145.Song L., Shan D.,, Zhao M., Pink B.A., Minnehan K.A., York L., Gardel M., Sullivan S., Phillips A.F., Hayman R.B., Walt D.R., Duffy D.C. Direct detection of bacterial genomic DNA at sub-femtomolar concentrations using single molecule arrays. Anal. Chem. 2013;85:1932–1939. doi: 10.1021/ac303426b. [DOI] [PubMed] [Google Scholar]
- 146.Sze P.-S., Bergstrom D.E. Alternative nucleic acid analogues for programmable assembly: hybridization of LNA to PNA. NanoLetters. 2005;5:107–111. doi: 10.1021/nl048246f. [DOI] [PubMed] [Google Scholar]
- 147.Kubota K., Ohashi A., Imachi H., Harada H. Improved in situ hybridization efficiency with locked-nucleic-acid-incorporated DNA probes. Appl. Environ. Microbiol. 2006;72:5311–5317. doi: 10.1128/AEM.03039-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Astakhova I.K., Pasternak K., Campbell M.A., Gupta P., Wengel J. A locked nucleic acid-based nanocrawler: Designed and reversible movement detected by multicolor fluorescence. J. Am. Chem. Soc. 2013;135:2423–2426. doi: 10.1021/ja311250w. [DOI] [PubMed] [Google Scholar]
- 149.Egholm M., Buchardt O., Christensen L., Behrens C., Freier S.M., Driver D.A., Berg R.H., Kim S.K., Norden B., Nielsen P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 150.Gao F., Lei J., Ju H. Label-free surface-enhanced Raman spectroscopy for sensitive DNA detection by DNA-mediated silver nanoparticle growth. Anal. Chem. 2013;85:11788–11793. doi: 10.1021/ac4032109. [DOI] [PubMed] [Google Scholar]
- 151.Brandt O., Hoheisel J.D. Peptide nucleic acids on microarrays and other biosensors. Trends Biotechnol. 2004;22:617–622. doi: 10.1016/j.tibtech.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 152.Sozzi M., Cucinotta A., Corradini R., Marchelli R., Konstantaki M., Pissadakis S., Selleri S. Modification of a long period grating-based fiber optic for DNA biosensing. Proc. SPIE. 2011;7894 [Google Scholar]
- 153.Chen J., Zhang J., Wang K., Lin X., Huang L., Chen G. Electrochemical biosensor for detection of BCR/ABL fusion gene using locked nucleic acids on 4-aminobenzenesulfonic acid-modified glassy carbon electrode. Anal. Chem. 2008;80:8028–8034. doi: 10.1021/ac801040e. [DOI] [PubMed] [Google Scholar]
- 154.Kauppinen S., Alsbo C., Nielsen P., Jeffares D., Mourier T., Mork S., Arctander P., Tommerup N., Tolstrup N., Vissing H. 2007. Oligonucleotides useful for detecting and analyzing nucleic acids of interest. US20070117144 A1. [Google Scholar]
- 155.Chambers J.P., Arulanandam B.P., Matta L.L., Weiss A., Valdes J.J. Biosensor recognition elements. Curr. Issues Mol. Biol. 2008;10:1–12. [PubMed] [Google Scholar]
- 156.Dong H., Hao K., Tian Y., Jin S S., Lu H., Zhou S.F., Zhang X. Label-free and ultrasensitive microRNA detection based on novel molecular beacon binding readout and target recycling amplification. Biosens. Bioelectron. 2014;53:377–383. doi: 10.1016/j.bios.2013.09.061. [DOI] [PubMed] [Google Scholar]
- 157.Li Y., Zhou X., Ye D. Molecular beacons: An optimal multifunctional biological probe. Biochem. Biophys. Res. Commun. 2008;373:457–461. doi: 10.1016/j.bbrc.2008.05.038. [DOI] [PubMed] [Google Scholar]
- 158.Marras S.A., Tyagi S., Kramer F.R. Real-time assays with molecular beacons and other fluorescent nucleic acid hybridization probes. Clin. Chim. Acta. 2006;363:48–60. doi: 10.1016/j.cccn.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 159.Strohsahl C.M., Du H., Miller B.L., Krauss T.D. Towards single-spot multianalyte molecular beacon biosensors. Talanta. 2005;67:479–485. doi: 10.1016/j.talanta.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 160.Šmuc T., Ahn I.Y., Ulrich H. Nucleic acid aptamers as high affinity ligands in biotechnology and biosensorics. J. Pharm. Biomed. Anal. 2013;81–82:210–217. doi: 10.1016/j.jpba.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 161.Long F., Gao C., Shi H.C., He M., Zhu A.N., Klibanov A.M., Gu A.Z. Reusable evanescent wave DNA biosensor for rapid, highly sensitive, and selective detection of mercury ions. Biosens. Bioelectron. 2011;26:4018–4023. doi: 10.1016/j.bios.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 162.Yin X.B. Functional nucleic acids for electrochemical and electrochemiluminescent sensing applications. Trends Anal. Chem. 2012;33:81–94. [Google Scholar]
- 163.Yildirim N., Long F., He M., Gao C., Shi H.C., Gu A.Z. A portable DNAzyme-based optical biosensor for highly sensitive and selective detection of lead (II) in water sample. Talanta. 2014;129:617–622. doi: 10.1016/j.talanta.2014.03.062. [DOI] [PubMed] [Google Scholar]
- 164.Wong L.S., Lee Y.H., Surif S. Performance of a Cyanobacteria Whole Cell-Based Fluorescence Biosensor for Heavy Metal and Pesticide Detection. Sensors. 2013;13:6394–6404. doi: 10.3390/s130506394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Struss A.K., Pasini P., Daunert S. Biosensing Systems Based on Genetically Engineered Whole Cells. In: Zourob M., editor. Recognition Receptors in Biosensors. Springer; New York: 2010. pp. 565–598. [Google Scholar]
- 166.Belkin S., Gu M.B., editors. Whole Cell Sensing Systems I: Reporter Cells and Devices. Springer; Berlin Heidelberg: 2010. [Google Scholar]
- 167.Belkin S., Gu M.B., editors. Whole Cell Sensing Systems II: Reporter Cells and Devices. Springer; Berlin Heidelberg: 2010. [Google Scholar]
- 168.Bedoya-Gutiérrez M., Delgado-Alonso J., García-Ares E., García J.L., Orellana G., Moreno-Bondi M.C. 2009. Measuring cell, analyser, process, and computer program product for measuring biochemical oxygen demand (bod) EP2110430 A2. [Google Scholar]
- 169.Peña-Vázquez E., Maneiro E., Pérez-Conde C., Moreno-Bondi M.C., Costas E. Microalgae fiber optic biosensors for herbicide monitoring using sol–gel technology. Biosens. Bioelectron. 2009;24:3538–3543. doi: 10.1016/j.bios.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 170.Nguyen-Ngoc H., Tran-Minh C. Fluorescent biosensor using whole cells in an inorganic translucent matrix. Anal. Chim. Acta. 2007;583:161–165. doi: 10.1016/j.aca.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 171.Michelini E., Roda A. Staying alive: new perspectives on cell immobilization for biosensing purposes. Anal. Bioanal. Chem. 2012;402:1785–1797. doi: 10.1007/s00216-011-5364-x. [DOI] [PubMed] [Google Scholar]
- 172.Ivask A., Green T., Polyak B., Mor A., Kahru A., Virta M., Marks R. Fiber-optic bacterial biosensors and their application for the analysis of bioavailable Hg and As in soils and sediments from Aznalcollar mining area in Spain. Biosens. Bioelectron. 2007;22:1396–1402. doi: 10.1016/j.bios.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 173.Futra D., Heng L.Y., Ahmad A., Surif S., Ling T.L. An Optical Biosensor from Green Fluorescent Escherichia coli for the Evaluation of Single and Combined Heavy Metal Toxicities. Sensors. 2015;15:12668–12681. doi: 10.3390/s150612668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Subrahmanyam S., Piletsky S.A., Turner A.P.F. Application of Natural Receptors in Sensors and Assays. Anal. Chem. 2002;74:3942–3951. doi: 10.1021/ac025673+. [DOI] [PubMed] [Google Scholar]
- 175.RCSB Protein Data Bank, www.rcsb.org/(accessed 30.11. 2015).
- 176.Fu Y., Lu D., Lin B., Sun Q., Liu K., Xu L., Zhang S., Hu C., Wang C., Xu Z., Zhang W. Fluorescence assay for glycan expression on living cancer cells based on competitive strategy coupled with dual-functionalized nanobiocomposites. Analyst. 2013;138:7016–7022. doi: 10.1039/c3an01226c. [DOI] [PubMed] [Google Scholar]
- 177.Chevolot Y., Vidal S., Laurenceau E., Morvan F., Vasseur J.J., Souteyrand E. Carbohydrates as recognition receptors in biosensing applications. In: Zourob M., editor. Recognition Receptors in Biosensors. Springer; New York: 2010. pp. 275–341. [Google Scholar]
- 178.Ballerstadt R., Evans C., Gowda A., McNichols R. In vivo performance evaluation of a transdermal near-infrared fluorescence resonance energy transfer affinity sensor for continuous glucose monitoring. Diabetes Technol. Ther. 2006;8:296–311. doi: 10.1089/dia.2006.8.296. [DOI] [PubMed] [Google Scholar]
- 179.Ogasawa F.K., Wang Y., Bobbitt D.R. Dynamically modified, biospecific optical fiber sensor for riboflavin binding protein based on hydrophobically associated 3-octylriboflavin. Anal. Chem. 1992;64:1637–1642. doi: 10.1021/ac00039a003. [DOI] [PubMed] [Google Scholar]
- 180.Medintz I.L., Clapp A.R., Mattoussim H., Goldman E.R., Fisher B., Mauro J.M. Self-assembled nanoscale biosensors based on quantum dot FRET donors. Nat. Mater. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 181.Siegrist J., Kazarian T., Ensor C., Joel S., Madou M., Wang P., Daunert S. Continuous glucose sensor using novel genetically engineered binding polypeptides towards in vivo applications. Sens. Actuator B-Chem. 2010;149:51–58. [Google Scholar]
- 182.Saxl T., Khan F., Ferla M., Birch D., Pickup J. A fluorescence lifetime-based fiber-optic glucose sensor using glucose/galactose-binding protein. Analyst. 2011;136:968–972. doi: 10.1039/c0an00430h. [DOI] [PubMed] [Google Scholar]
- 183.Campbell K., Rawn D.F.K., Niedzwiadek B., Elliott C.T. Paralytic shellfish poisoning (PSP) toxin binders for optical biosensor technology: problems and possibilities for the future: a review. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2011;28:711–725. doi: 10.1080/19440049.2010.531198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Song X., Swanson B.I. Direct, Ultrasensitive, and Selective Optical Detection of Protein Toxins Using Multivalent Interactions. Anal. Chem. 1999;71:2097–2107. doi: 10.1021/ac981145f. [DOI] [PubMed] [Google Scholar]
- 185.Sapsford K.E., Granek J., Deschamps J.R., Boeneman K., Blanco-Canosa J.B., Dawson P.E., Susumu K., Stewart M.H., Medintz I.L. Monitoring Botulinum Neurotoxin A Activity with Peptide-Functionalized Quantum Dot Resonance Energy Transfer Sensors. ACS Nano. 2011;5:2687–2699. doi: 10.1021/nn102997b. [DOI] [PubMed] [Google Scholar]
- 186.Lai Y., Gallo R.L. AMPed Up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shriver-Lake L.C., North S.H., Dean S.N., Taitt C.R. Antimicrobial peptides for detection and diagnostic assays. In: Piletsky S.A., M.J Whitcombe, editors. Designing Receptors for the Next Generation of Biosensors. Springer; Berlin Heidelberg: 2013. pp. 85–104. [Google Scholar]
- 188.Kulagina N.V., Anderson G.P., Ligler F.S., Shaffer K.M., Taitt C.R. Antimicrobial peptides: new recognition molecules for detecting botulinum toxins. Sensors. 2007;7:2808–2824. doi: 10.3390/s7112808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kulagina N.V., Shaffer K.M., Ligler F.S., Rowe-Taitt C. Antimicrobial peptides as new recognition molecules for screening challenging species. Sens. Actuator B-Chem. 2007;121:150–157. doi: 10.1016/j.snb.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Goicochea N.L., De M., Rotello V.M., Mukhopadhyay S., Dragnea B. Core-like particles of an enveloped animal virus can self-assemble efficiently on artificial templates. Nano Lett. 2007;7:2281–2290. doi: 10.1021/nl070860e. [DOI] [PubMed] [Google Scholar]
- 191.Petrenko V.A. Evolution of phage display: from bioactive peptides to bioselective nanomaterials. Expert Opin. Drug Deliv. 2008;5:825–836. doi: 10.1517/17425247.5.8.825. [DOI] [PubMed] [Google Scholar]
- 192.Fernandes E., Martins V.C., Nóbrega C., Carvalho C.M., Cardoso F.A., Cardoso S., Dias J., Deng D., Kluskens L.D., Freitas P.P., Azeredo J. A bacteriophage detection tool for viability assessment of Salmonella cells. Biosens. Bioelectron. 2014;52:239–246. doi: 10.1016/j.bios.2013.08.053. [DOI] [PubMed] [Google Scholar]
- 193.Souza G.R., Christianson D.R., Staquicini F.I., Ozawa M.G., Snyder E.Y., Sidman R.L., Miller J.H., Arap W., Pasqualini R. Networks of gold nanoparticles and bacteriophage as biological sensors and cell-targeting agents. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1215–1220. doi: 10.1073/pnas.0509739103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Petrenko V.A., Sorokulova I.B. Detection of biological threats. A challenge for directed molecular evolution. J. Microbiol. Methods. 2004;58:147–168. doi: 10.1016/j.mimet.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 195.Peltomaa R.J., López-Perolio I., Benito-Peña E., Barderas R., Moreno-Bondi M.C. Application of bacteriophages in sensor development. Anal. Bioanal. Chem. 2016;408:1805–1828. doi: 10.1007/s00216-015-9087-2. [DOI] [PubMed] [Google Scholar]
- 196.Phillips E.M., Hantke R., Baird D., Rainone M., Plowman T.E., Presley T. 2012. Recombinant bacteriophage for detection of nosocomial infection. US 20120143024 A1. [Google Scholar]
- 197.Moreno-Bondi M.C., Navarro Villoslada F., Benito-Peña E., Urraca J.L. Molecularly imprinted polymers as selective recognition elements in optical sensing. Curr. Anal. Chem. 2008;4:316–340. [Google Scholar]
- 198.Moreno-Bondi M.C., Benito-Peña M.E., Urraca J.L., Orellana G. Immuno-like assays and biomimetic microchips. Top. Curr. Chem. 2012;325:111–164. doi: 10.1007/128_2010_94. [DOI] [PubMed] [Google Scholar]
- 199.Carrasco S., Benito-Peña E., Walt D.R., Moreno-Bondi M.C. Fiber-optic array using molecularly imprinted microspheres for antibiotic analysis. Chem. Sci. 2015;6:3139–3147. doi: 10.1039/c5sc00115c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wolfbeis O.S. Probes, sensors and labels: Why is real progress slow? Angew. Chem. Int. Ed. 2013;52:9864–9865. doi: 10.1002/anie.201305915. [DOI] [PubMed] [Google Scholar]
- 201.Koshida T., Arakawa T., Gessei T., Takahashi D., Kudo H., Saito H., Yano K., Mitsubayashi K. Fluorescence biosensing system with a UV-LED excitation for L-leucine detection. Sens. Actuator B-Chem. 2010;146:177–182. [Google Scholar]
- 202.Gessei T., Arakawa T., Kudo H., Mitsubayashi K. A fiber-optic sorbitol biosensor based on NADH fluorescence detection toward rapid diagnosis of diabetic complications. Analyst. 2015;140:6335–6342. doi: 10.1039/c4an01593b. [DOI] [PubMed] [Google Scholar]
- 203.Ramanathan M., Simonian A.L. Array biosensor based on enzyme kinetics monitoring by fluorescence spectroscopy: Application for neurotoxins detection. Biosens. Bioelectron. 2007;22:3001–3007. doi: 10.1016/j.bios.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 204.Bidmanova S., Chaloupkova R., Damborsky J., Prokop Z. Development of an enzymatic fiber-optic biosensor for detection of halogenated hydrocarbons. Anal. Bioanal. Chem. 2010;398:1891–1898. doi: 10.1007/s00216-010-4083-z. [DOI] [PubMed] [Google Scholar]
- 205.Schrenkhammer P., Wolfbeis O.S. Fully reversible optical biosensors for uric acid using oxygen transduction. Biosens. Bioelectron. 2008;24:994–999. doi: 10.1016/j.bios.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 206.Mukundan H., Kubicek J.Z., Holt A., Shively J.E., Martinez J.S., Grace K., Grace W.K., Swanson B.I. Planar optical waveguide-based biosensor for the quantitative detection of tumor markers. Sens. Actuator B-Chem. 2009;138:453–460. [Google Scholar]
- 207.Meneely J.P., Campbell K.C., Greef M.J., Lochhead C.T., Elliott Development and validation of an ultrasensitive fluorescence planar waveguide biosensor for the detection of paralytic shellfish toxins in marine algae. Biosens. Bioelectron. 2013;41:691–697. doi: 10.1016/j.bios.2012.09.043. [DOI] [PubMed] [Google Scholar]
- 208.Herranz S., Marciello M., Olea D., Hernández M., Domingo C., Vélez M., Gheber L.A., Guisán J.M., Moreno-Bondi M.C. Dextran-lipase conjugates as tools for low molecular weight ligand immobilization in microarray development. Anal. Chem. 2013;85:7060–7068. doi: 10.1021/ac400631t. [DOI] [PubMed] [Google Scholar]
- 209.Devlin S., Meneely J.P., Greer B., Greef C., Lochhead M.J., Elliott C.T. Next generation planar waveguide detection of microcystins in freshwater and cyanobacterial extracts, utilising a novel lysis method for portable sample preparation and analysis. Anal. Chim. Acta. 2013;769:108–113. doi: 10.1016/j.aca.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 210.Wei H., Zhao Y., Bi Y., Liu H., Guo Z., Song Y., Zhai J., Huang H., Yang R. Direct detection of Yersinia pestis from the infected animal specimens by a fiber optic biosensor. Sens. Actuators B-Chem. 2007;123:204–210. [Google Scholar]
- 211.Albrecht C., Fechner P., Honcharenko D., Baltzer L., Gauglitz G. A new assay design for clinical diagnostics based on alternative recognition elements. Biosens. Bioelectron. 2010;25:2302–2308. doi: 10.1016/j.bios.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 212.Käppel N.D., Pröll F., Gauglitz G. Development of a TIRF-based biosensor for sensitive detection of progesterone in bovine milk. Biosens. Bioelectron. 2007;22:2295–2300. doi: 10.1016/j.bios.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 213.Long F., He M., Shi H.C., Zhu A.N. Development of evanescent wave all-fiber immunosensor for environmental water analysis. Biosens. Bioelectron. 2008;23:952–958. doi: 10.1016/j.bios.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 214.Long F., Shi H.C., He M., Zhu A.N. Sensitive and rapid detection of 2,4-dicholorophenoxyacetic acid in water samples by using evanescent wave all-fiber immunosensor. Biosens. Bioelectron. 2008;23:1361–1366. doi: 10.1016/j.bios.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 215.Guo H., Zhou X., Zhang Y., Song B., Liu L., Zhang J., Shi H. Highly sensitive and rapid detection of melamine in milk products by planar waveguide fluorescence immunosensor (PWFI) Sens. Actuators B-Chem. 2014;194:114–119. [Google Scholar]
- 216.Ahn S., Kulis D.M., Erdner D.L., Anderson D.M., Walt D.R. Fiber-optic microarray for simultaneous detection of multiple harmful algal bloom species. Appl. Environ. Microbiol. 2006;72:5742–5749. doi: 10.1128/AEM.00332-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tran D.T., Knez K., Janssen K.P., Pollet J., Spasic D., Lammertyn J. Selection of aptamers against Ara h 1 protein for FO-SPR biosensing of peanut allergens in food matrices. Biosens. Bioelectron. 2013;43:245–251. doi: 10.1016/j.bios.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 218.Long F., Wu S., He M., Tong T., Shi H. Ultrasensitive quantum dots-based DNA detection and hybridization kinetics analysis with evanescent wave biosensing platform. Biosens. Bioelectron. 2011;26:2390–2395. doi: 10.1016/j.bios.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 219.Schwers S., Reifenberger E., Gehrmann M., Izmailov A., Bohmann K. A high-sensitivity, medium-density, and target amplification–free planar waveguide microarray system for gene expression analysis of formalin-fixed and paraffin-embedded tissue. Clin. Chem. 2009;55:1995–2003. doi: 10.1373/clinchem.2009.128215. [DOI] [PubMed] [Google Scholar]
- 220.Yildirim N., Long F., Gao C., He M., Shi H.C., Gu A.Z. Aptamer-based optical biosensor for rapid and sensitive detection of 17 β-estradiol in water samples. Environ. Sci. Technol. 2012;46:3288–3294. doi: 10.1021/es203624w. [DOI] [PubMed] [Google Scholar]
- 221.Konry T., Hayman R.B., Walt D.R. Microsphere-based rolling circle amplification microarray for the detection of DNA and proteins in a single assay. Anal. Chem. 2009;81:5777–5782. doi: 10.1021/ac900694y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tsuchiya K., Ueda M., Mulchandani A., Enami Y., Suye S. International Chemical Congress of Pacific Basin Societies; Honolulu, United States: December 15-20, 2010. Combination of Optical Waveguide Device and Arming Yeast for Organophosphorus Compounds Fluorescence Sensing System. [Google Scholar]
- 223.Nguyen T.H., Sun T., Grattan K.T.V., Hardwick S.A. A fiber optic chemical sensor for the detection of cocaine. Proc. SPIE. 2010;7653 [Google Scholar]
- 224.Harbaugh S., Kelley-Loughnane N., Davidson M., Narayanan L., Trott S., Chushak Y.G., Stone M.O. FRET-based optical assay for monitoring Riboswitch activation. Biomacromolecules. 2009;10:1055–1060. doi: 10.1021/bm801117f. [DOI] [PubMed] [Google Scholar]
- 225.Jenkins A.L., Bae S.Y. Molecularly imprinted polymers for chemical agent detection in multiple water matrices. Anal. Chim. Acta. 2005;542:32–37. [Google Scholar]
- 226.Rainone M., Plowman T.E., Baird D., Hantke R., Phillips E.M., Presley T. 2009. Recombinant bacteriophage for detection of nosocomial infection. WO2009074893 A2. [Google Scholar]
- 227.Xiong Y., Ye Z., Xu J., Liu Y., Zhang H. A microvolume molecularly imprinted polymer modified fiber-optic evanescent wave sensor for bisphenol A determination. Anal. Bioanal. Chem. 2014;406:2411–2420. doi: 10.1007/s00216-014-7664-4. [DOI] [PubMed] [Google Scholar]