Abstract
The 63 amino acid polytopic membrane protein, p7, encoded by hepatitis C virus (HCV) is involved in the modulation of electrochemical gradients across membranes within infected cells. Structural information relating to p7 from multiple genotypes has been generated in silico (e.g. genotype (GT) 1a), as well as obtained from experiments in form of monomeric and hexameric structures (GTs 1b and 5a, respectively). However, sequence diversity and structural differences mean that comparison of their channel gating behaviour has not thus far been simulated. Here, a molecular model of the monomeric GT 1a protein is optimized and assembled into a hexameric bundle for comparison with both the 5a hexamer structure and another hexameric bundle generated using the GT 1b monomer structure. All bundles tend to turn into a compact structure during molecular dynamics (MD) simulations (Gromos96 (ffG45a3)) in hydrated lipid bilayers, as well as when simulated at ‘low pH’, which may trigger channel opening according to some functional studies. Both GT 1a and 1b channel models are gated via movement of the parallel aligned helices, yet the scenario for the GT 5a protein is more complex, with a short N-terminal helix being involved. However, all bundles display pulsatile dynamics identified by monitoring water dynamics within the pore.
Keywords: p7 of HCV, Viral channel protein, Genotypes, Channel gating, Molecular dynamics simulation, NMR structure
Graphical abstract
Highlights
-
•
Genotype specific structural features of hexameric bundles of p7 of HCV
-
•
In silico built models of genotype 1a bundle compared with models of genotypes 1b and 5a from NMR
-
•
Genotype 1a and 1b channels are gated via movement of the parallel aligned helices.
-
•
Complex scenario for genotype 5a bundle model
-
•
All bundles show pulsatile dynamics.
1. Introduction
A growing number of viral genomes are being identified as encoding a specialised type of membrane protein which form oligomeric ion channels [1], [2], [3], [4], [5], [6]. These viral channel forming proteins (VCPs), also called viroporins, pose an interesting challenge for the understanding of ion channel mechanics in general, due to their low molecular weight.
Similar to larger host-cell ion channels, the function of the VCPs is to modulate electrochemical gradients in various cellular sub-compartments or at the site of the plasma membrane. This function is essential to the life cycle of many viruses. Some viruses encode bitopic membrane proteins, such as M2 of influenza A [7], Vpu of human immunodeficiency virus type-1 (HIV-1) [8], or 8a of severe acute respiratory syndrome-coronavirus (SARS-CoV) [9]. Other viral genomes harbour the code for polytopic membrane proteins such as p7 of hepatitis C virus (HCV) [10], [11], [12] and 2B of polio virus [13] with 2 transmembrane domains (TMDs), or 3a of SARS-CoV [14] and E5 of human Papilloma-virus type 16 (HPV-16) [15] with 3 TMDs. A common feature of all these proteins is that they have to oligomerise to function as a channel.
Protein p7 of HCV is a 63 amino acid protein [16], [17] essential for virion production ([18], [19], [20], [21], reviewed in [22]). It is a cleavage product from a polyprotein precursor. Its location in the polyprotein follows the two structural proteins E1 and E2 (reviewed in [23]) and is succeeded by non-structural (NS) proteins. Protein p7, together with subsequent protein NS2, is found to act at an early stage of virion morphogenesis [21]. Its function is to support, possibly together with the crosslinked heterodimer E1 and E2 glycoproteins as well as NS2, capsid assembly at the plasma membrane [24]. Experimental results indicate that these proteins initiate the release of RNA-containing core proteins from lipid droplets.
As another function, p7 dissipates proton gradients which has been shown using pH sensitive dyes in cell based fluorescence spectroscopic experiments as well as liposome efflux essays [25]. This function as well as the capability of the protein to render lipid membranes permeable to ions is thought to be due to the protein forming channels, especially when expressed in cells, purified and reconstituted into artificial lipid bilayers [11], [26]. Also, peptides derived from biochemical sources, generate adequate conductance data [10], [12]. Overall, the channel shows weak cation selectivity [11], [12].
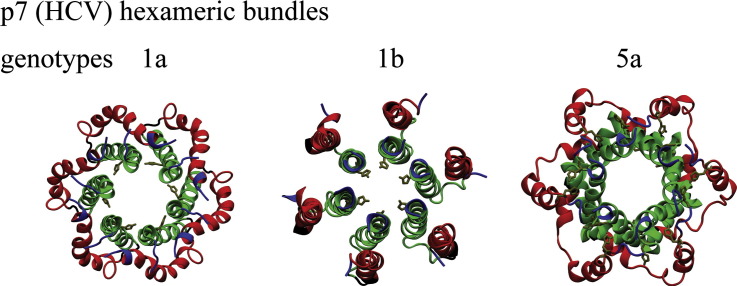
In as much both functions as mentioned come along with different structural features are still under debate. NMR spectroscopic measurements have identified the monomeric protein with two TMDs in a hairpin motif with each of the helices being segmented into two smaller units [27], [28] as well as the protein in an oligomeric state forming a hexameric bundle [29]. A feature of the bundle models built from the hairpin monomers is a ring of histidines (His-17s) pointing into the putative lumen of the pore [30]. The oligomeric architecture suggests an ‘i + 3’ motif in which the C terminal helix of one monomer interacts/links with the N terminal helix of the third monomer. There is currently no information of how the different architectures are related to each other. Negative stain transmission EM data mostly identify a hexameric assembly for the protein [11], [31] as well as GT specific heptameric bundle [26].
The motif of the TMDs of p7, and all of today's known VCPs, is identified to be helical ([4] and references therein). Thus, secondary structure prediction programmes can be applied to identify the length of a putative TMD and the respective amino acids can be translated into a helical conformer. In case of polytopic proteins TMDs need to be assembled to form a monomer using a docking approach [32], [33]. These structures form the basis for simulating functional data, such as ion conductance, which can be compared with those from experiments [34], [35], [36], [37], [38], [39].
The different overall structural features, hairpin motif versus i + 3 motif, sparked the investigations reported in this paper. Bundle models of p7 of GT 1a were generated using the abovementioned protocols [32], [33]. The hexameric bundle of GT 1b was generated using a monomeric NMR structure [40] and positioning six copies around a pseudo six-fold symmetry axis. The bundle of GT 5a derives from NMR spectroscopic data with the protein in an oligomerized (hexamer) form [29]. The three bundles were subjected to multi-nanosecond molecular dynamics (MD) simulations to elucidate structural and dynamic features at both, neutral and acidic pH, with the latter mimicked via protonation of His17 during simulation processing.
2. Materials and methods
The sequence of the TMDs of p7 was taken from the HCV GT 1a, H77 strain [10]: ALENLVILNA10 ASLAGTHGLV20 SFLVFFCFAW30 YLKGRWVPGA40 VYAFYGMWPL50 LLLLLALPQR60 AYA according to Patargias et al. [30]:
-
TMD1:
ALENLVILNA10 ASLAGTHGLV20 SFLVFFCFAW30 YL
-
TMD2:
WVPGA40 VYAFYGMWPL50 LLLLLALPQR60 AYA.
-
Loop (as predicted):
KGR
The model according to GT 1b-J4 uses the following sequence (PDB ID: 3ZD0) [41], [42]:
-
1b (J4):
ALENLVVLNA10 ASVAGAHGIL20 SFLVFFCAAW30 YIKGRLAPGA40 AYAFYGVWPL50 LLLLLALPPR60 AYA.
The following amino acids were found in a helical motif corresponding to the respective TMDs (non-helical amino acids shown in italics):
-
TMD1:
ALENLVVLNA10 ASVAGAHGIL20 SFLVFFCAAW30 YIK
-
TMD2:
GRLAPGA40 AYAFYGVWPL50 LLLLLALPPR60 AYA.
GT 5a model uses (EUH1480; PDB ID: 2M6X) [29]: GAKNVIVLNA10 ASAAGNHGFF20 WGLLVVTLAW30 HVKGRLVPGA40 TYLSLGVWPL50 LLVRLLRPHR60 ALA.
This sequence contains 5 mutations of unconserved amino acids (T1G, C2A, A12S, C27T, C44S). The three helices extend from
-
H1:
VIVLNA10 ASAAGN
-
H2:
F20 WGLLVVTLAW30 HVKGRLVPGA40 T
-
H3:
WPL50 LLVRLLRP.
2.1. Generation of the protein models
The computational p7 bundles were generated in a stepwise manner according to protocols described previously [32], [33]. Briefly, the two GT 1a TMDs are assembled into a monomer (monomer-1a) first, followed by assembling six copies of monomer-1a around a pseudo six-fold symmetry axis into a bundle (bundle-1a). The conformational space for both of the assemblies, the monomer and the bundle, is screened by varying the degrees of distance, rotational angle and tilt stepwise as follows: (i) inter helical distance in steps of 0.25 Å covering 7.5 Å to 13.0 Å for monomers and 11.75 Å to 20 Å for hexameric bundles; (ii) rotational angles around the helical axis in steps of 5° covering 360°; and (iii) tilt in steps of 2° covering − 36° to + 36°. At each position, the conformation of the side chains was introduced choosing the most likely conformation from an internal library of the MOE software suit (www.chemcomp.com). At this stage, 15 steps of steepest descent minimization followed to remove ‘bad’ (overlaps) van-der-Waals contacts. Any extensive minimization is compensated by a fine grained, in terms of step width of the three degrees of freedom such as distance, rotational and tilt angle, search of the conformational space. Potential energy of each conformer was evaluated, using AMBER94. The docking was done using a dielectric constant of ε = 2 to mimic the environment of the bilayer.
The structure of the bundle according to GT 1b (bundle-1b) was generated from a FLAG-tagged monomer (PDB ID: 3ZD0) [40]. The FLAG-tag was removed and the respective monomer used for pore assembly as described for bundle-1a. Bundle of the geneotype 5a (PDB ID: 2M6X, bundle-5a) was taken without modification for the MD simulations.
2.2. MD simulations
The monomers and the bundles were individually inserted into pre-equilibrated patches of POPC lipids (16:1–18:1 diester PC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) for which parameters of Chandrasekhar et al. [43] were adopted. Lipids which overlapped with the helices were removed. The protein–lipid system was hydrated and, after steps of minimization (2000 steps of steepest decent and 5000 steps of conjugated gradient), it was equilibrated for a total of 1.9 ns. Equilibration was achieved by gradually increasing the temperature from 100 K to 200 K and 310 K, whilst keeping the peptide fully restrained with k = 1000 kJ mol− 1 nm− 2. The first two simulations (100 K and 200 K) were run for 200 ps each, the third simulation (310 K) was run for 500 ps. Holding the system at 310 K, the restraints, imposed by a force constant k on the peptide, were released in 2 steps (k = 500 kJ mol− 1 nm− 2, k = 250 kJ mol− 1 nm− 2), running each of the steps for 500 ps. MD simulation of the system is carried out using Gromos96 (ffG45a3) force field. The temperature of the peptide, lipid, and the water molecules were separately coupled to a velocity rescaling thermostat [44] with a coupling time of 0.1 ps and a Berendsen barostat with a coupling time of 2.0 ps during the early equilibration at temperatures 100 K, 200 K and 310 K. During the production run, the system was coupled to a Nosé–Hoover thermostat with a coupling time of 0.1 ps [45], [46].
The simulation systems of the individual TMDs consisted of 122 lipids (6344 atoms) and 3655 water molecules (10,965 atoms), for simulating monomers 276 lipids (14,352 atoms) and 8746 water molecules (26,238 atoms), the bundle systems consisted of 488 lipids (25,376 atoms) and 14,640 water molecules (43,920 atoms).
2.3. Data analysis and hardware equipment
The software ‘Fluxer’ developed and supplied by Dr. Manuel N. Melo, University of Groningen, NL, was used to count the number of water molecules passing through the pore (http://cgmartini.nl/cgmartini/index.php/tools2/proteins-and-bilayers/223). The central membrane embedded region of the bundle was chosen from residues 14 to 24 of each of the individual monomers.
The simulations were run on an Acer i7-2600 workstation with 8 cores and submitted to the ALPS-Acer AR585 F1 Cluster in the National Center for High-Performance Computing (NCHC), Hsinchu, TW. Plots and pictures were made with Origin 8.5.1 and VMD 1.9, respectively
3. Results
3.1. Generation of GT 1a channel complexes in silico (bundle-1a)
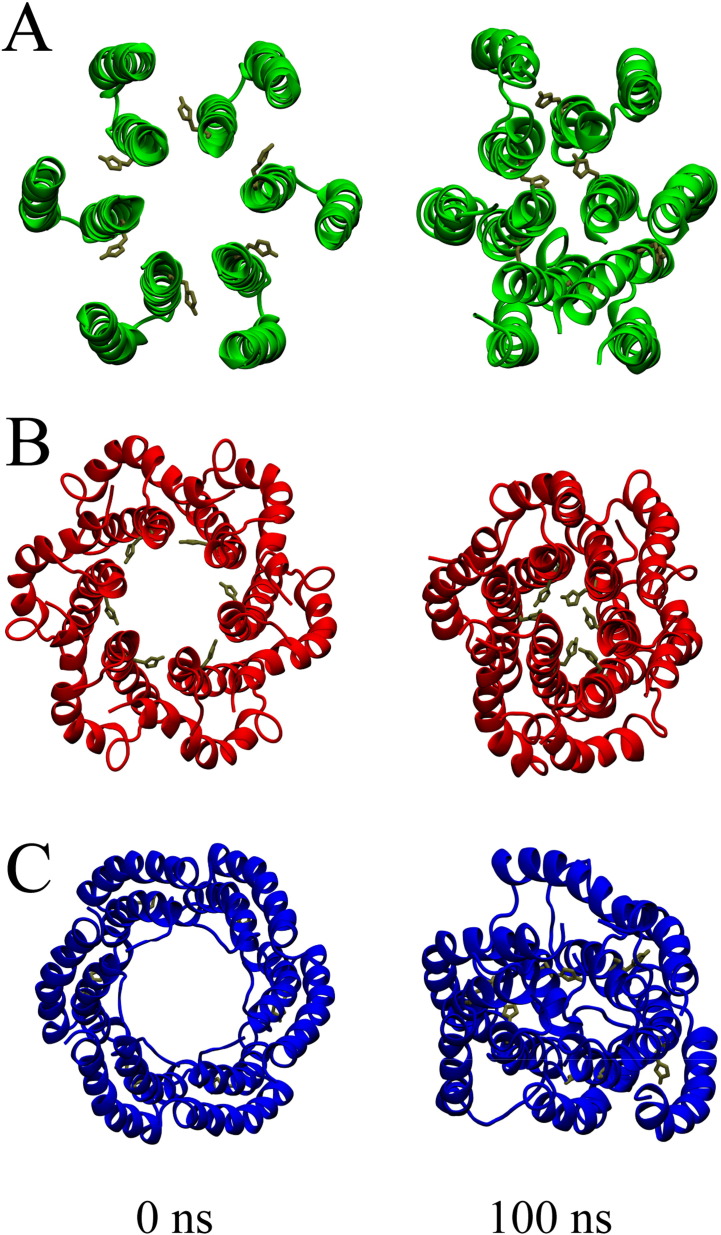
Three different protocols are used to generate hexameric assemblies of p7 monomers (Fig. 1 ). In the first protocol (P1, Fig. 1A) two ‘ideal’ helical TMDs are generated (ϕ and ψ angles of − 65° and − 39°, respectively). The TMDs are assembled into a monomer. The monomer is copied six times and a hexameric bundle is generated. In the second protocol (P2, see Fig. 1B) the monomer is equilibrated in a fully hydrated lipid bilayer during a 100 ns MD simulation prior to the assembly into a bundle. In the third protocol (P3, see Fig. 1C), the individual TMDs as well as the monomer have been simulated for 100 ns, prior to the assembly into a bundle. Whenever the assembly protocol is used, the lowest energy structures are used for any of the consecutive step.
Fig. 1.
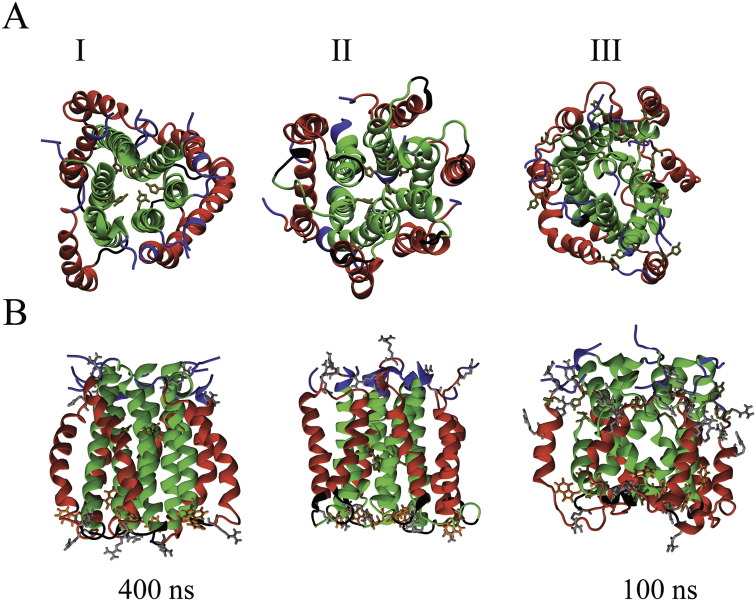
Graphical representation of the hexameric bundles of p7 (GT 1a) using protocol 1(A), protocol 2 (B) and protocol 3 (C) as outlined in the text. The structures are shown at the initial stage of 0 ns of the MD simulation and at the end at 100 ns. Unprotonated His-17 is highlighted in tan. The view is from the side of the termini towards the loop region.
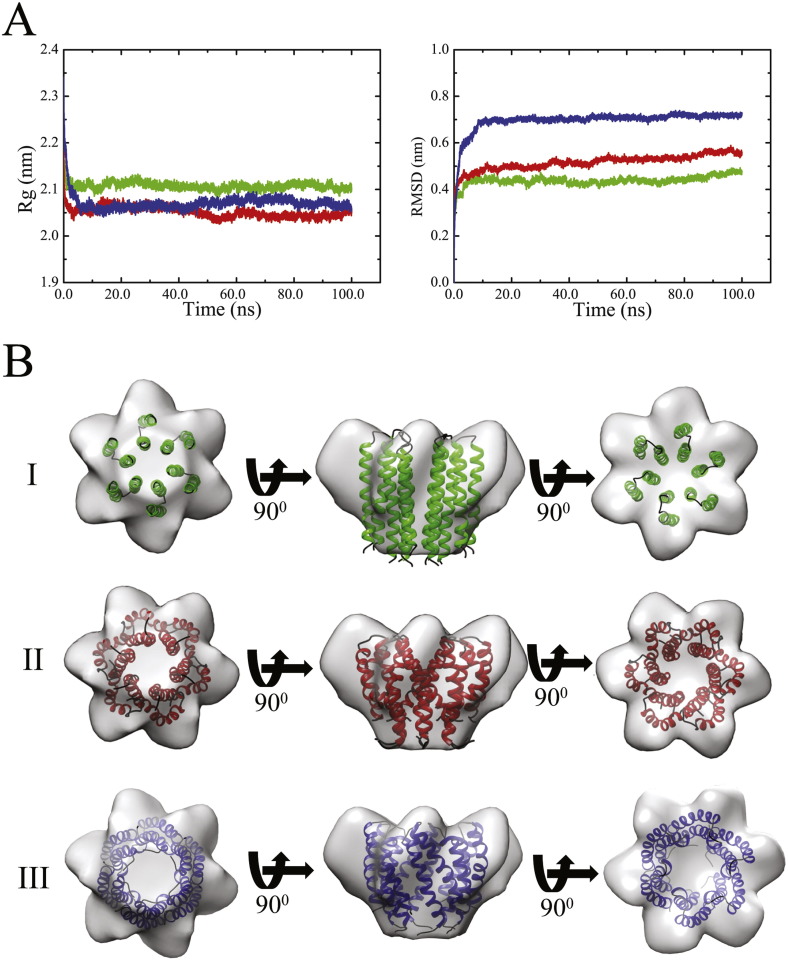
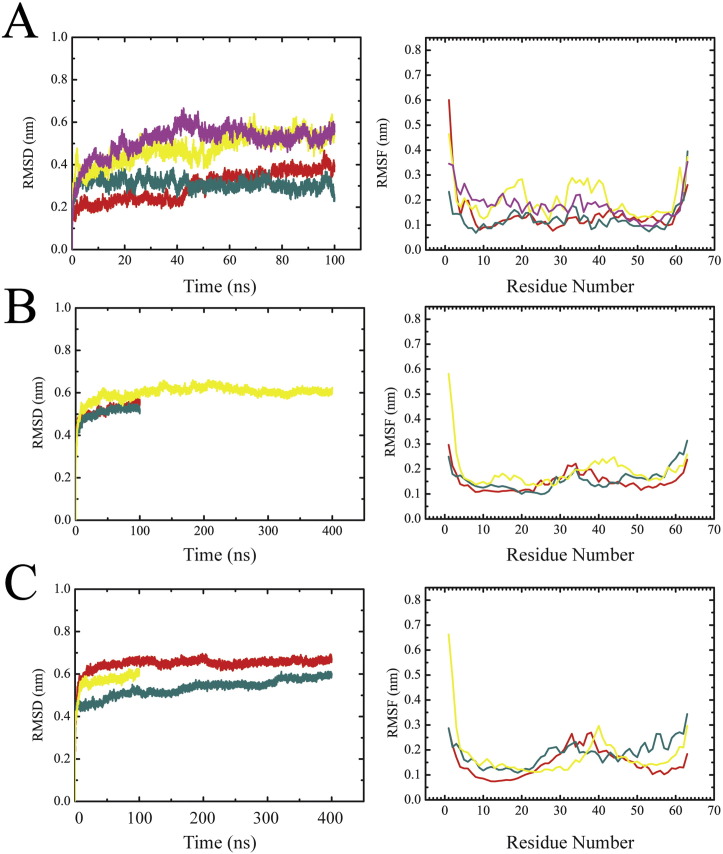
During 100 ns MD simulations, all bundles develop a compact structure (Fig. 1A–C) as indicated by a decline of the radius of gyration (Fig. 2A, left). Considering the levelling-phase of the root mean square deviation (RMSD) values (Fig. 2A, right), the bundle generated using P3 adopts the highest values of around 0.7 nm (blue curve) whilst the other two bundles level off at around 0.45 nm (P1, green curve) and 0.5 nm (P2, red curve). Levelling off suggests that the bundles have deviated from the starting structure and reached a reasonably stable structure.
Fig. 2.
Data of the radius of gyration (Rg) (A, left) and the root mean square fluctuation (RMSD) of the Cα-atoms (A, right) from 100 ns MD simulations of p7 bundles (GT 1a) using protocols 1 (green), 2 (red) and 3 (blue). Bundle models generated using protocols 1 (green, B,I), 2 (red, B,II) and 3 (blue, B,III) embedded into a 16 A° EM map (EM database ID 1661) [31] at various angles of observation.
The assembled lowest energy bundles from all three protocols are fitted into the 16 Å EM map (EM database ID 1661) of a hexameric p7 of GT 2a [31] (Fig. 2B). The lowest calculated fitting correlations are 0.88 (P1, Fig. 2B,I), 0.92 (P2, Fig. 2B,II) and 0.89 (P3, Fig. 2B, III) and resulting in an ‘upside down’ fit of the bundles (also reported in [29]) compared to the model reported in [31]. Based on the largest fitting correlation with the EM map, bundle of P2 is considered further for data evaluation. In this bundle, His-17 is pointing towards the lumen of the pore. In the other two bundles His-17 is pointing towards the helix–helix interface.
3.2. Comparison of the individual monomers
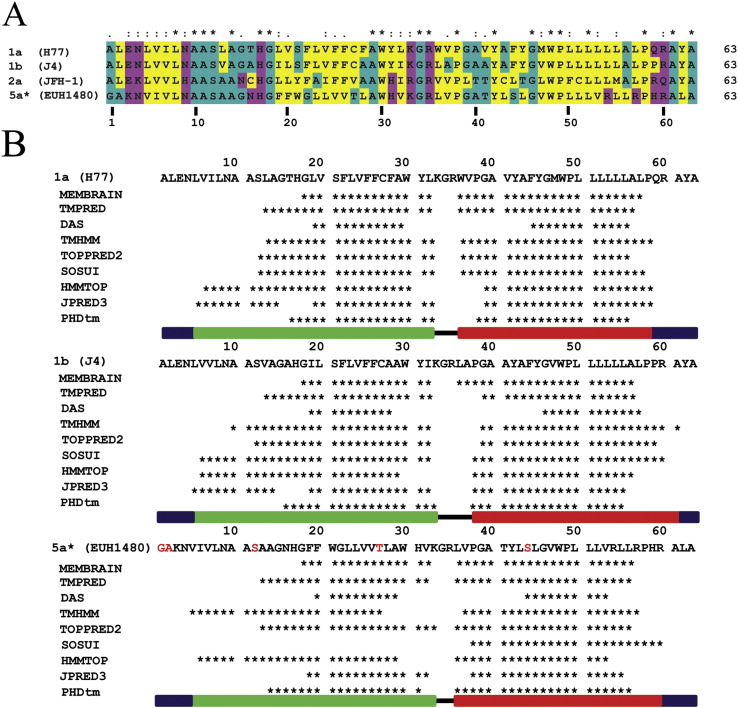
Despite significant (~ 50%) variation in amino acid sequence (Fig. 3A), computer predictions of secondary structure support that two helical segments are present for all three GTs, 1a, 1b and 5a (Fig. 3B). As this differs from the conformation of monomer-5a in the complete bundle structure, the fold of monomer-5a is assessed using protocol P2, as employed for the 1a protein. As expected from secondary structure predictions, the resultant model, named monomer-5a′, comprised a hairpin fold, reminiscent of the GT 1a and 1b proteins.
Fig. 3.
Multiple alignment (ClustalX) of the sequences of GTs 1a (H77), 1b (J4), 2a (JFH-1) and 5a (EUH1480, contains 5 mutations of unconserved amino acids (T1G, C2A, A12S, C27T, C44S marked in red)) (A). The colour scheme identifies hydrophobic amino acids in yellow, hydrophilic amino acids in magenta and neutral amino acids in cyan. The similarity index (ClustalX, top row) is as follows: ‘*’ = invariant (49%); ‘:’ = highly similar (16%); ‘.’ = similar (10%). Alignment of the results predicting helical motifs (indicated as ‘*’) from a series of secondary structure prediction programmes (B). The green and red bars indicate that the sequence taken for simulation of TMD1 and TMD2, respectively, has a helix. The black line indicates the amino acids of the loop-region, the blue bars indicate the extramembrane parts.
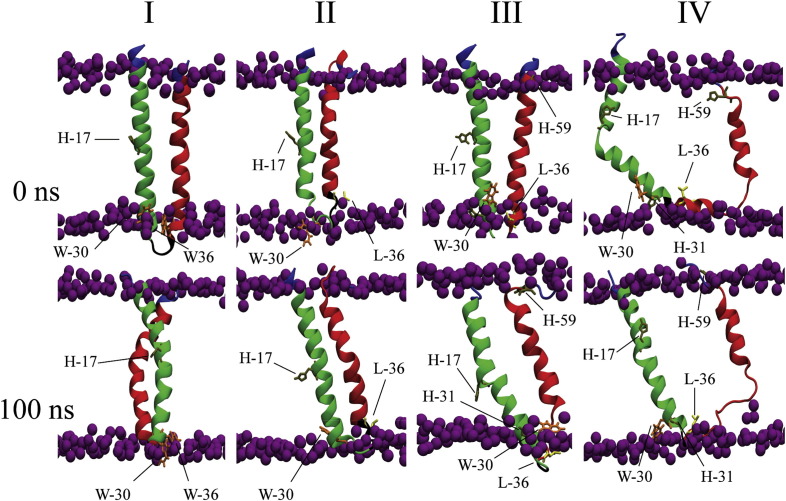
Next the structural integrity of all the possible monomeric forms (Fig. 4 , I: monomer-1a, II: monomer-1b (PDB ID: 3ZD0), III: monomer-5a′ and IV: monomer-5a from PDB ID: 2M6X) for each GT is investigated when inserted into a lipid bilayer, during a 100 ns MD simulation. For monomer-1b, RMSD values levelled off after the first few ns (Suppl. Fig. 1A, left), whereas other monomers show plateauing values after about 50 ns. This indicates that the structures do not change within the remaining simulation time. In the region 33–43, which includes the loop and the amino-N terminal side of TMD2, both monomer-5a and -5a′ show identical fluctuations of the respective residues (Suppl. Fig. 1A, right). The TMDs of 1a, 1b and 5a′ maintain their helical motif (Fig. 4, I–III, lower panel). Helix1 (H1, notation according to [29]: Val-5 to Asn-16) and the N terminal side of helix2 (H2) up to Gly-34 of monomer-5a form a continuous TMD1 with the C terminal side of H2 unwinding within the lipid head group region (Fig. 4,IV, lower panel).
Fig. 4.
Structural models of the monomers of GTs 1a (I), 1b (II), 5a, modelled according to protocol 2 (P2) (III) and 5a, taken from the NMR structure, (IV). Structures are shown at 0 ns (upper panel) and 100 ns (lower panel). The backbones of TMD1 and TMD2 are shown in green and red, respectively. Side chains of residues H17, − 31, − 59, W-30, − 36, L-36 are highlighted. The phosphorous atoms are shown in van der Waals representation (purple) to indicate the boundaries of the lipid bilayer. Lipids and water molecules are omitted for clarity.
3.3. Comparison of the computational GT 1a model with GT 1b and 5a bundles generated using experimental data
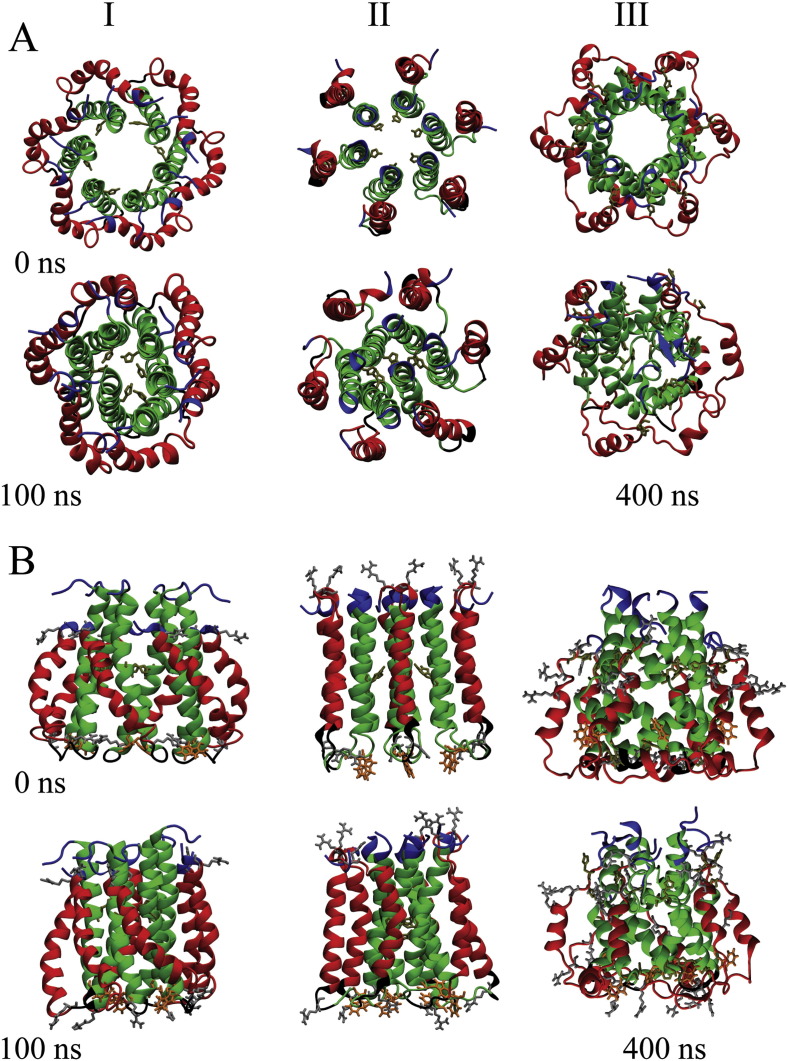
Structural and functional features are compared amongst three bundle models: (i) GT 1a bundles derived from protocol P2, (ii) GT 1b bundles (PDB ID: 3ZD0) (Fig. 5A, II), and (iii) the GT 5a channel structure (PDB ID: 2M6X) (Fig. 5A, III).
Fig. 5.
Graphical representation of the bundle models of GT 1a (I), 1b (II) and 5a (III) from NMR at 0 ns (upper panel) and 100 ns (lower panel) (A). The structure shown for 5a is taken at 400 ns. The segments belonging to TMD1 and 2 are highlighted in green and red, respectively. The side chains of His-17 (bundle-1a, -1b, -5a) as well as His-31 and -59 of bundle-5a are highlighted. The view is from the side of the termini to the loop region. A side view of the respective structures is highlighting specific residues: His-17, -31, -59, Trp-30, Arg-35, -54, -57, -60 (B). Lipids and water molecules are omitted for clarity.
RMSD values calculated from 100 ns (400 ns in case of bundle-5a) MD simulations of each bundle level around 0.5 nm after 10 ns for bundle-1a and -1b, and around 0.6 nm for bundle-5a (Suppl. Fig. 1B). Looking along the central axis of the unprotonated bundles shows that all bundles lose their circular arrangement (Fig. 5). In bundles-1a and -1b structural rearrangements are due to a movement of the entire TMD1, independent of the protonation state of His-17 (Fig. 5A, I, II). In bundle-5a several H1s are moving so that the pore adopts a kind of closed structure (Fig. 5A, III). The movement of one H1 domain is so that the segment linking to H2 (His-17,Gly-18,Phe-19) points into the lumen of the pore.
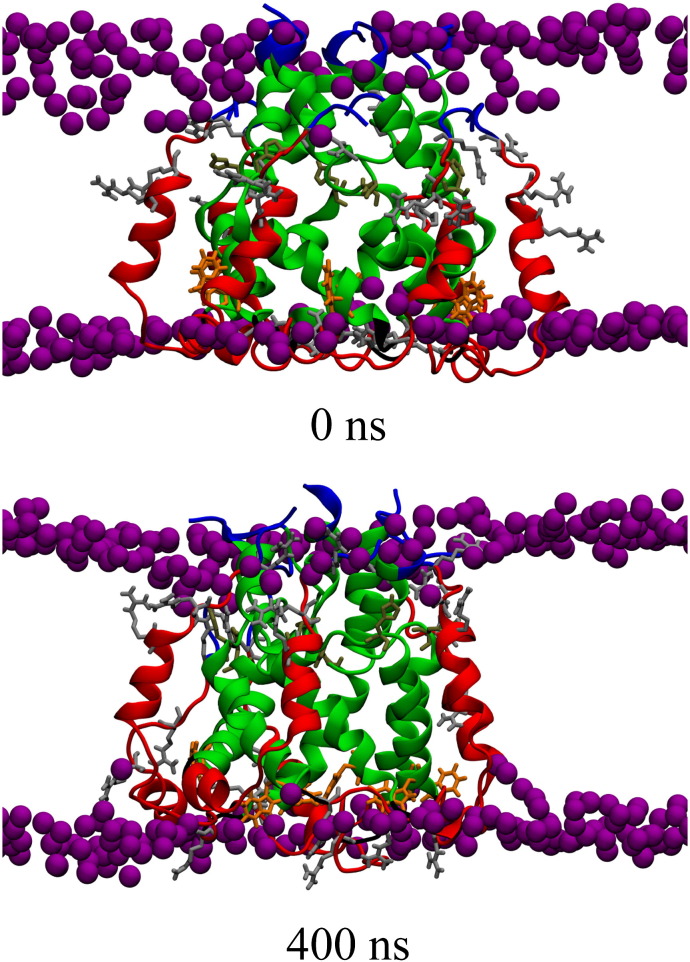
Visible inspection of the bundles from the side (within the membrane) of bundle-1a indicates a straightening of TMD2, an almost unchanged shape of bundle-1b and a strong rearrangement of bundle-5a H3 (Fig. 5B,I–III). The rearrangement of the latter is so that especially Arg-60 interacts with the head groups of the opposing leaflet of the lipid bilayer, which leads to a thinning of the bilayer (Fig. 6 ). The movement of the helices in bundle-5a can be best described as a swinging of H1 into the pore and a movement of the arginine rich H3 (Trp-48 to Pro-58) to the opposing head group region, leaving H2 almost unaffected. These features are observed independent of the protonation state of the bundles.
Fig. 6.
Side view of the unprotonated bundle-5a highlighting the phosphorous atoms (van der Walls representation) to identify the boundaries of the lipid membrane. Specific amino acids of the protein are highlighted as follows: His-17, -31, -59, Trp-30, Arg-35, -54, -57, -60. Lipids and water molecules are omitted for clarity.
Averaged tilt angles of the TMD1s ((24.2 ± 1.5) ° unprotonated; (12.5 ± 1.1) ° protonated) of bundle-1a are smaller than the values for the respective TMD2 ((27.7 ± 1.3) ° unprotonated; (18.1 ± 1.4) ° protonated) (Table 1 ). Upon protonation of His-17 in the bundles, the kink is smaller for TMD2 ((159.2 ± 1.7) °) than in the unprotonated bundle ((148.8 ± 1.4) °). This pattern regarding tilt and kink is also found in bundle-1b except for the tilt in the protonated 1b bundle. The outer TMD2s of bundle-1a and -1b are stronger tilted than the inner TMD1s and respond to the protonation by repulsion and straightening of the inner TMD1s.
Table 1.
Averaged tilt and kink angles of the individual TMDs of p7 GTs 1a and 1b.
| GT | Tilt | Kink | |
|---|---|---|---|
| 1a | |||
| Unprotonated | TMD1 | 24.2 ± 1.5 | 159.6 ± 1.2 |
| TMD2 | 27.7 ± 1.3 | 148.4 ± 1.4 | |
| Protonated | TMD1 | 12.5 ± 1.1 | 158.7 ± 1.2 |
| TMD2 | 18.1 ± 1.4 | 159.2 ± 1.7 | |
| 1b | |||
| Unprotonated | TMD1 | 12.0 ± 1.6 | 166.6 ± 1.1 |
| TMD2 | 16.5 ± 1.1 | 153.7 ± 1.7 | |
| Protonated | TMD1 | 16.4 ± 1.1 | 166.7 ± 1.2 |
| TMD2 | 16.3 ± 1.0 | 163.8 ± 2.6 | |
3.4. Features based on individual amino acids
Upon protonation of bundle-1a as well as for the unprotonated bundle-1b one of the protonated His-17 turns so that a circular arrangement of the His-17 is perturbed. Generally, the His-17s adopt a staggered-like arrangement as reported earlier [47].
The side chains of Arg-35 in unprotonated bundle-5a face the aqueous phase allowing a circular arrangement of His-31 at the mouth of the pore. Over the duration of the simulation Cl-ions enter the lumen of the pore freely. The lumen of the pore is maintained by H2. In the NMR structure His-17 and His-59 face each other and are exposed to the lipids. They diverge from each other upon a 400 ns simulation. This feature is independent of the simulated protonation state of His-17.
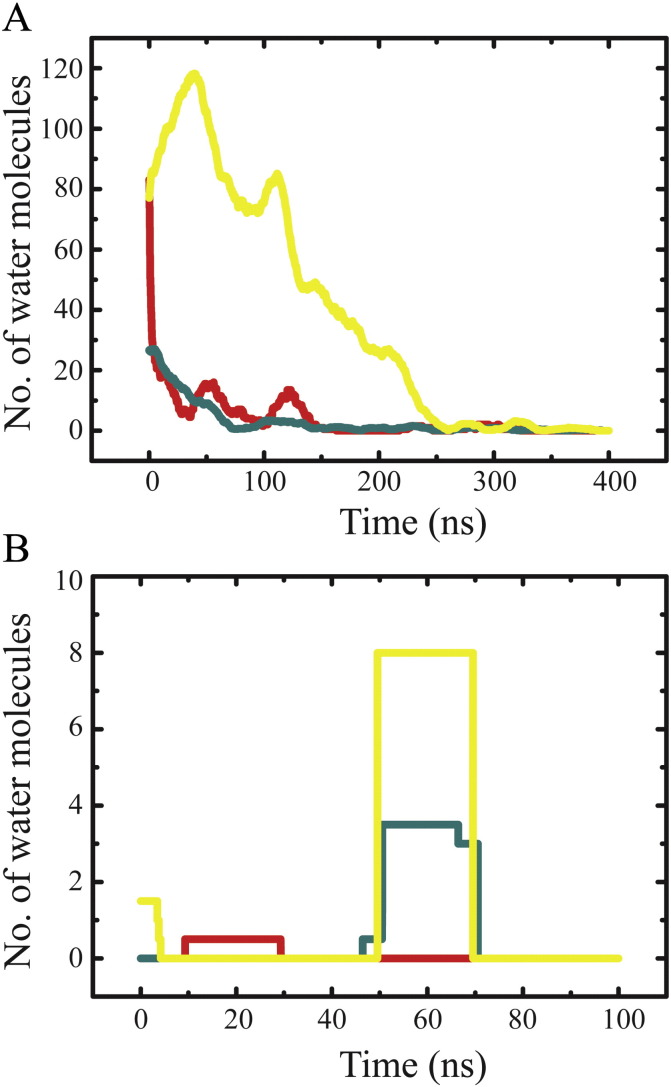
3.5. Pore open-state and water permeation
In its unprotonated state, only bundle-5a allows water molecules to pass through the pore (Fig. 7A, yellow trace). Neither bundle-1a nor -1b are sufficiently long in an ‘open’ conformation to allow for a similar observation (Fig. 7B, red and cyan, respectively). Upon protonation (for RMSD of the protonated bundles see Suppl. Fig. 1C), bundle-1a and -1b allow water molecules to pass through (Fig. 7A, red and cyan), but bundle-5a water permeation is reduced (Fig. 7B, yellow trace), since just 8 water molecules are observed for about 29 ns during a 100 ns simulation. Simulations over 400 ns of the protonated bundle-1a and -1b as well as the unprotonated 5a bundles reveal that the quantity of water molecules crossing fluctuates periodically on top of an overall decrease.
Fig. 7.
(A) Number of the water traversing the protonated hexameric bundles of GTs 1a (red) and 1b (cyan) as well as the unprotonated bundle of GT 5a from NMR (yellow). (B) Number of water molecules traversing unprotonated bundles of GT 1a (red), 1b (cyan) and protonated bundle-5a (yellow).
4. Discussion
4.1. GT specific structural differences
The protocol applied in this study has been used, with slight variations, to suggest structural motifs of other assemblies of viral channel proteins [9], [30], [32], [33], [48], [49]. As a general feature, the protocol delivers monomers and bundles of parallel aligned TMDs. Experimental data from NMR spectroscopy confirm the model [28], [42], [50], [51]. It is suggested, that the two TMDs range from Val-6 to Cys-27 and Pro-38 to Pro-58 [50], [51]. Each of the two helices consists of two segments which are separated by Ala-16 to His-17 of TMD1 and Trp-48 to Pro-49 of TMD2. In another study helical regions are suggested from Leu-2 to Ala-14 and Leu-19 to Ile-32, as well as Ala-40 to Ala-56 [28]. Using DPC, p7 is found to exist in an oligomeric state adopting the hairpin motif [28], as well as in the ‘unusual architecture’ [29]. In p7 of GT 5a, it seems that two adjacent phenylalanines which are at positions 25 and 26 in GT 1a/1b, are shifted by one helical turn to positions 19 and 20. Thus, a phenylalanine is missing for the suggested π–π stacking motif comprised of Phe-26, Trp-30, Tyr-45 reported to support the hairpin motif and putatively responsible for the formation of the middle helix, H2, in bundle-5a [28]. In bundle-5a instead, Phe-19 and -20 are part of a hydrophobic pocket accessible from the outside of the bundle [29], with Phe-19 being reported to contribute to harbour amantadine. Another striking feature of the 5a-sequence is the two arginines (Arg-54, -57) at the C terminus. They are not observed in the other GTs and confess most likely another reason for the structural difference between bundle-1a/1b and -5a [52]. At this stage it is considered, that the experimental conditions used in various NMR-experiments support either the monomeric appearance of the protein (bundles-1a/1b) or the oligomeric state (bundles-2a/5a). When simulating the monomer out of bundle-5a it tends to develop into an almost parallel alignment as shown in this study. Consequently, aforementioned phylogenetic differences between 1a/1b and 5a strains can cause considerable differences in structural features.
HCV enters the host cells via interaction of its glycoproteins E1/E2 with four receptor proteins one of which is CD81 [53]. Mutations in the viral glycoproteins renders HCV susceptible to mouse CD81 binding [54]. This is explained by structural changes enabled upon mutation. A single amino acid mutation in the core protein of HCV GT3 leads to an increase in GT 3-specific steatosis [55]. These features underline further, that the change of a single amino acid or just a few can alter conformational dynamics and structural features which result in highly diverse GT specific features despite a moderate difference of ~ 30% at the nucleotide level [56].
Given the genetic distance between 1a and 2a [52], the only one available EM structure of assembled GT 2a p7 is used as a selection criteria (Fig. 2B). Due to the genetic distance between the GTs, a pedal like structure seems to be likely for bundles of GTs 2a and 5a. In the light of sequence identity between 1a and 2a (Fig. 3A) however, it is assumed to still be reasonable to use the overall structure envelope of the EM map as a selection criterion. At this stage, a reason for ‘upside down’ fitting other than simply the mathematical shape-matching algorithm cannot be given.
4.2. Equilibration of the monomers
Three plausible, ‘in-silico generated’ bundle models are presented of the polytopic p7. Generally, the protocol requires the interrogation of secondary structure prediction programmes to suggest TMD motifs, followed by the assembly of the TMDs in various ways. Computationally derived models, based on a docking approach, are screened by fitting respective assembled lowest energy bundle structures into an existing EM structure available for a proposed hexameric p7 bundle. Fitting the initial bundle to experimental data, is reasonable in as much an experiment is the ultimate validation of computational data. At this stage, simulating the monomeric model prior to further assembly into larger oligomers is a reasonable and essential step to generate bundle models which match experimental finding. The protocol suggested, matches the biologically assumed pathway to generate assembled channel bundles as well as larger pores, e.g. those of toxins [57], [58].
Comparison of all monomers based on computational tools suggests that they would possibly adopt the same structural feature, including monomer-5a. In the light of the experimentally derived structure of bundle-5a, it is suggested that there are GT specific differences in the structure.
4.3. Functional and mechanical features of p7 bundles from different GTs
With the diverse structures at hand, it is hypothesised that some functions of p7, e.g. proton conductance would be highly GT specific. Assuming narrow passages are necessary for proton conductance through proteins, the pore of bundle-5a is too large and the putatively His-17 relevant for proton translocation, is not in a similar position as reported for His-37 in M2 of influenza A. Thus, bundle-5a would be the channel least selective for protons. However, it needs to be mentioned that mutations of His-17 is reported to play an important role in the infectivity cycle of HCV [59]. His-17 of bundle-5a could still be a key residue to guarantee necessary structural and dynamic features.
The development of more compact structures, which could be synonymous with a closed structure of bundle-1a and -1b, is possible due to the dynamics of more or less straight helices. It is proposed that, the mechanical features upon alteration of the protonation state of the pore lining histidines (His-17) are a straightening of the TMD2s of bundle-1a and -1b in respect to the membrane normal (Fig. 5B, I, II and Suppl. Fig. 2, I, II). In contrast, bundle-5a shows versatile mechanics, best described by movements of ‘outer helix (H3)’ and the ‘inner helix’ H1. This would turn the region between the first two helices into a hinge involved in potentially voltage sensitive gating. The ‘closure’ of 5a is occurring in parallel with a thinning of the lipid bilayer due to the motion of H3. Most striking is, that any gating or structural changes come with an overall oscillating behaviour within the protein.
5. Conclusions
A two-step in silico protocol, assembling the monomer prior to bundle assembly, delivers plausible p7 bundles for GTs 1a and 1b. Ultimately, differences in mechanical features due to the specific bundle architectures are observed as well as common features such as periodicity in the dynamics of the bundles.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbamem.2015.03.006.
Conflict of interest
There is no conflict of interest.
The following are the supplementary data related to this article.
Suppl. Fig. 1.
RMSD (left) and RMSF values (right) of the simulations of the monomeric structures (A), as well as the unprotonated (B) and protonated (C) bundle models. The data show the monomeric structure according to GT 1a (red), 1b (dark cyan), 5a from protocol P2 (magenta) and 5a from NMR (yellow). The data of the bundle structures are shown in red (1a), dark cyan (1b) and yellow (5a).
Suppl. Fig. 2.
Graphical representation of the protonated bundle models of GT 1a (I), 1b (II), both at 400 ns, and 5a from NMR (III) at 100 ns (A). The segments belonging to TMD1 and 2 are highlighted in green and red, respectively. The side chains of His-17 (bundle-1a, -1b, -5a) as well as His-31 and -59 of bundle-5a are highlighted. The view is from the side of the termini to the loop region. A side view of the respective structures is highlighting specific residues: His-17, -31, -59, Trp-30, Arg-35, -54, -57, -60 (B). Lipids and water molecules are omitted for clarity.
Acknowledgement
WBF thanks the National Science Council Taiwan (NSC-101-2112-M-010-002-MY3) for financial support and the National Center for High-performance Computing (NCHC), Hsinchu, TW, for technical support. MMK acknowledges a student-fellowship of National Yang-Ming University. We thank M.-H. Lin, Y.-T. Wang, and R. D. Mahato (Taipei, TW) as well as R. Schilling (Heidelberg, D) for their helpful discussions.
References
- 1.Fischer W.B., Sansom M.S.P. Viral ion channels: structure and function. Biochim. Biophys. Acta. 2002;1561:27–45. doi: 10.1016/s0304-4157(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 2.Gonzales M.E., Carrasco L. Viroporins. FEBS Lett. 2003;552:28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- 3.Wang K., Xie S., Sun B. Viral proteins function as ion channels. Biochim. Biophys. Acta. 2010;1808:510–515. doi: 10.1016/j.bbamem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer W.B., Wang Y.-T., Schindler C., Chen C.-P. Mechanism of function of viral channel proteins and implications for drug development. Int. Rev. Cell Mol. Biol. 2012;294:259–321. doi: 10.1016/B978-0-12-394305-7.00006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nieva J.L., Madan V., Carrasco L. Viroporins: structure and biological functions. Nat. Rev. Microbiol. 2012;10:563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.OuYang B., Chou J.J. The minimalist architectures of viroporins and their therapeutic implications. Biochim. Biophys. Acta. 2014;1838:1058–1067. doi: 10.1016/j.bbamem.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duff K.C., Ashley R.H. The transmembrane domain of influenza A M2 protein forms amantadine-sensitive proton channels in planar lipid bilayers. Virology. 1992;190:485–489. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- 8.Schubert U., Ferrer-Montiel A.V., Oblatt-Montal M., Henklein P., Strebel K., Montal M. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398:12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- 9.Chen C.-C., Krüger J., Sramala I., Hsu H.-J., Henklein P., Chen Y.-M.A., Fischer W.B. ORF 8a of severe acute respiratory syndrome coronavirus forms an ion channel: experiments and molecular dynamics simulations. Biochim. Biophys. Acta. 2011;1808:572–579. doi: 10.1016/j.bbamem.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavlović D., Neville D.C.A., Argaud O., Blumberg B., Dwek R.A., Fischer W.B., Zitzmann N. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin S.D.C., Beales L.P., Clarke D.S., Worsfold O., Evans S.D., Jäger J., Harris M.P.G., Rowlands D.J. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 2003;535:34–38. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 12.Premkumar A., Wilson L., Ewart G.D., Gage P.W. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557:99–103. doi: 10.1016/s0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- 13.van Kuppeveld F.J.M., Melchers W.J.G., Kirkegaard K., Doedens J.R. Structure–function analysis of coxsackie B3 virus protein 2B. Virology. 1997;227:111–118. doi: 10.1006/viro.1996.8320. [DOI] [PubMed] [Google Scholar]
- 14.Lu W., Zheng B.-J., Xu K., Schwarz W., Du L., Wong C.K.L., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wetherill L.F., Holmes K.K., Verow M., Müller M., Howell G., Harris M., Fishwick C., Stonehouse N., Foster R., Blair G.E., Griffin S., Macdonald A. High-risk human papillomavirus E5 oncoprotein displays channel-forming activity sensitive to small-molecule inhibitors. J. Virol. 2012;86:5341–5351. doi: 10.1128/JVI.06243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C., Lindenbach B.D., Pragai B.M., McCourt D.W., Rice C.M. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbers K., Tautz N., Becher P., Stoll D., Rumenapf T., Thiel H.J. Processing in the pestivirus E2-NS2 region: identification of proteins p7 and E2p7. J. Virol. 1996;70:4131–4135. doi: 10.1128/jvi.70.6.4131-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann V., Korner F., Koch J., Herian U., Theilmann L., Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 19.Sakai A., St. Claire M., Faulk K., Govindarajan S., Emerson S.U., Purcell R.H., Bukh J. The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11646–11651. doi: 10.1073/pnas.1834545100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinmann E., Penin F., Kallis S., Patel A.H., Bartenschlager R., Pietschmann T. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3:962–971. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones C.T., Murray C.L., Eastman D.K., Tassello J., Rice C.M. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinmann E., Pietschmann T. Hepatitis C virus p7 — a viroporin crucial for virus assembly and emerging target for antiviral therapy. Viruses. 2010;2:2078–2095. doi: 10.3390/v2092078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vieyres G., Dubuisson J., Pietschmann T. Incorporation of hepatitis C virus E1 and E2 glycoproteins: the keystones on a peculiar virion. Viruses. 2014;6:1149–1187. doi: 10.3390/v6031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentzsch J., Brohm C., Steinmann E., Friesland M., Menzel N., Vieyres G., Perin P.M., Frentzen A., Kaderali L., Pietschmann T. Hepatitis C virus p7 is critical for capsid-assembly and envelopment. PLoS Pathog. 2013;9:e1003355. doi: 10.1371/journal.ppat.1003355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wozniak A.L., Griffin S., Rowlands D., Harris M., Yi M.-K., Lemon S.M., Weinman S.A. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 2010;6:e1001087. doi: 10.1371/journal.ppat.1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke D., Griffin S., Beales L., Gelais C.S., Burgess S., Harris M., Rowlands D. Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 2006;281:37057–37068. doi: 10.1074/jbc.M602434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook G.A., Opella S.J. NMR studies of the p7 protein from hepatitis C virus. Eur. Biophys. J. 2010;39:1097–1104. doi: 10.1007/s00249-009-0533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montserret R., Saint N., Vanbelle C., Salvay A.G., Simorre J.P., Ebel C., Sapay N., Renisio J.-G., Böckmann A., Steinmann E., Pietschmann T., Dubuisson J., Chipot C., Penin F. NMR structure and ion channel activity of the p7 protein from hepatitis C virus. J. Biol. Chem. 2010;285:31446–31461. doi: 10.1074/jbc.M110.122895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.OuYang B., Xie S., Berardi M.J., Zhao X., Dev J., Yu W., Sun B., Chou J.J. Unusual architecture of the p7 channel from hepatitis C virus. Nature. 2013;498:521–525. doi: 10.1038/nature12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patargias G., Zitzmann N., Dwek R., Fischer W.B. Protein–protein interactions: modeling the hepatitis C virus ion channel p7. J. Med. Chem. 2006;49:648–655. doi: 10.1021/jm050721e. [DOI] [PubMed] [Google Scholar]
- 31.Luik P., Chew C., Aittoniemi J., Chang J., Wentworth P., Jr., Dwek R., Biggin P.C., Vénien-Bryan C., Zitzmann N. The 3-dimensional structure of the hepatitis C virus p7 ion channel by electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12712–12716. doi: 10.1073/pnas.0905966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krüger J., Fischer W.B. Assembly of viral membrane proteins. J. Chem. Theory Comput. 2009;5:2503–2513. doi: 10.1021/ct900185n. [DOI] [PubMed] [Google Scholar]
- 33.Hsu H.-J., Fischer W.B. In silico investigations of possible routes of assembly of ORF 3a from SARS-CoV. J. Mol. Model. 2011;18:501–514. doi: 10.1007/s00894-011-1092-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grice A.L., Kerr I.D., Sansom M.S.P. Ion channels formed by HIV-1 Vpu: a modelling and simulation study. FEBS Lett. 1997;405:299–304. doi: 10.1016/s0014-5793(97)00198-1. [DOI] [PubMed] [Google Scholar]
- 35.Fischer W.B., Pitkeathly M., Wallace B.A., Forrest L.R., Smith G.R., Sansom M.S. Transmembrane peptide NB of influenza B: a simulation, structure, and conductance study. Biochemistry. 2000;39:12708–12716. doi: 10.1021/bi001000e. [DOI] [PubMed] [Google Scholar]
- 36.Allen T.W., Andersen O.S., Roux B. Molecular dynamics — potential of mean force calculations as a tool for understanding ion permeation and selectivity in narrow channels. Biophys. Chem. 2006;124:251–267. doi: 10.1016/j.bpc.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 37.Hub J.S., de Groot B.L. Mechanism of selectivity in aquaporins and aquaglycoporins. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1198–1203. doi: 10.1073/pnas.0707662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kutzner C., Grubmüller H., de Groot B.L., Zachariae U. Computational electrophysiology: the molecular dynamics of ion channel permeation and selectivity in atomistic detail. Biophys. J. 2011;101:809–817. doi: 10.1016/j.bpj.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandler D.E., Penin F., Schulten K., Chipot C. The p7 protein of hepatitis C virus forms structurally plastic, minimalist ion channels. PLoS Comput. Biol. 2012;8:e1002702. doi: 10.1371/journal.pcbi.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foster T.L., Verow M., Wozniak A.L., Bentham M.J., Thompson J., Atkins E., Weinmann S.A., Fishwick C., Foster R., Harris M., Griffin S. Resistance mutations define specific antiviral effects for inhibitors of the hepatitis C virus p7 ion channel. Hepatology. 2011;54:79–90. doi: 10.1002/hep.24371. [DOI] [PubMed] [Google Scholar]
- 41.Yanagi M., St Claire M., Shapiro M., Emerson S.U., Purcell R.H., Bukh J. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 42.Foster T.L., Thompson G.S., Kalverda A.P., Kankanala J., Bentham M., Wetherill L.F., Thompson J., Barker A.M., Clarke D., Noerenberg M., Pearson A.R., Rowlands D.J., Homans S.W., Harris M., Foster R., Griffin S. Structure-guided design affirms inhibitors of hepatitis C virus p7 as a viable class of antivirals targeting virion release. Hepatology. 2014;59:408–422. doi: 10.1002/hep.26685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chandrasekhar I., Kastenholz M., Lins R.D., Oostenbrink C., Schuler L.D., van Gunsteren W.F. A consistent potential energy parameter set for lipids: dipalmitoylphosphatidylcholine as a benchmark of the GROMOS96 45A3 force field. Eur. Biophys. J. 2003;32:67–77. doi: 10.1007/s00249-002-0269-4. [DOI] [PubMed] [Google Scholar]
- 44.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity-rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 45.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 46.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A. 1985;31 doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y.-T., Schilling R., Fink R.H.A., Fischer W.B. Ion-dynamics in HCV p7 hexameric bundles — a molecular dynamics simulation study. Biophys. Chem. 2014;192:33–40. doi: 10.1016/j.bpc.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Cordes F.S., Tustian A.D., Sansom M.S., Watts A., Fischer W.B. Bundles consisting of extended transmembrane segments of Vpu from HIV-1: computer simulations and conductance measurements. Biochemistry. 2002;41:7359–7365. doi: 10.1021/bi025518p. [DOI] [PubMed] [Google Scholar]
- 49.Patargias G., Barke T., Watts A., Fischer W.B. Model generation of viral channel forming 2B protein bundles from polio and coxsackie viruses. Mol. Membr. Biol. 2009;26:309–320. doi: 10.1080/09687680903164101. [DOI] [PubMed] [Google Scholar]
- 50.Cook G.A., Opella S.J. Secondary structure, dynamics, and architecture of the p7 membrane protein from hepatitis C virus by NMR spectroscopy. Biochim. Biophys. Acta. 2011;1808:1448–1453. doi: 10.1016/j.bbamem.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cook G.A., Dawson L.A., Tian Y., Opella S.J. Three-dimensional structure and interaction studies of hepatitis C virus p7 in 1,2-dihexanoyl-sn-glycero-3-phosphatidylcholine by solution nuclear magnetic resonance. Biochemistry. 2013;52:5295–5303. doi: 10.1021/bi4006623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith D.B., Bukh J., Kuiken C., Muerhoff A.S., Rice C.M., Stapleton J.T., Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindenbach B.D., Rice C.M. The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 2013;11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haid S., Windisch M.P., Bartenschlager R., Pietschmann T. Mouse-specific residues of claudin-1 limit hepatitis C virus genotype 2a infection in a human hepatocyte cell line. J. Virol. 2010;84:964–975. doi: 10.1128/JVI.01504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hourioux C., Patient R., Morin A., Blanchard E., Moreau A., Trassad S., Giraudeau B., Roingeard P. The genotype 3-specific hepatitis C virus core protein residue phenylalanine 164 increases steatosis in an in vitro cellular model. Gut. 2007;56:1302–1308. doi: 10.1136/gut.2006.108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartenschlager R., Frese M., Pietschmann T. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 2004;63:71–180. doi: 10.1016/S0065-3527(04)63002-8. [DOI] [PubMed] [Google Scholar]
- 57.Müller M., Grauschopf U., Maier T., Glockshuber R., Ban N. The structure of a cytolytic alpha-helical toxin pore reveals its assembly mechanism. Nature. 2009;459:726–730. doi: 10.1038/nature08026. [DOI] [PubMed] [Google Scholar]
- 58.Bayley H. Piercing insights. Nature. 2009;459:651–652. doi: 10.1038/459651a. [DOI] [PubMed] [Google Scholar]
- 59.Meshkat Z., Audsley M., Beyer C., Gowans E.J., Haqshenas G. Reverse genetic analysis of a putative, influenza virus M2 HXXXW-like motif in the p7 protein of hepatitis C virus. J. Viral Hepat. 2009;16:187–194. doi: 10.1111/j.1365-2893.2008.01064.x. [DOI] [PubMed] [Google Scholar]