Abstract
Macro domain is a highly conserved protein domain found in both eukaryotes and prokaryotes. Macro domains are also encoded by a set of positive-strand RNA viruses that replicate in the cytoplasm of animal cells, including coronaviruses and alphaviruses. The functions of the macro domain are poorly understood, but it has been suggested to be an ADP-ribose-binding module. We have here characterized three novel human macro domain proteins that were found to reside either in the cytoplasm and nucleus [macro domain protein 2 (MDO2) and ganglioside-induced differentiation-associated protein 2] or in mitochondria [macro domain protein 1 (MDO1)], and compared them with viral macro domains from Semliki Forest virus, hepatitis E virus, and severe acute respiratory syndrome coronavirus, and with a yeast macro protein, Poa1p. MDO2 specifically bound monomeric ADP-ribose with a high affinity (Kd = 0.15 μM), but did not bind poly(ADP-ribose) efficiently. MDO2 also hydrolyzed ADP-ribose-1″ phosphate, resembling Poa1p in all these properties. Ganglioside-induced differentiation-associated protein 2 did not show affinity for ADP-ribose or its derivatives, but instead bound poly(A). MDO1 was generally active in these reactions, including poly(A) binding. Individual point mutations in MDO1 abolished monomeric ADP-ribose binding, but not poly(ADP-ribose) binding; in poly(ADP-ribose) binding assays, the monomer did not compete against polymer binding. The viral macro proteins bound poly(ADP-ribose) and poly(A), but had a low affinity for monomeric ADP-ribose. Thus, the viral proteins do not closely resemble any of the human proteins in their biochemical functions. The differential activity profiles of the human proteins implicate them in different cellular pathways, some of which may involve RNA rather than ADP-ribose derivatives.
Abbreviations: MDO2, macro domain protein 2; MDO1, macro domain protein 1; ADPR-1″P, ADP-ribose-1″ phosphate; BAL, B-aggressive lymphoma; PARP, poly(ADP-ribose) polymerase; GDAP2, ganglioside-induced differentiation-associated protein 2; MDO3, macro domain protein 3; HEV, hepatitis E virus; SARS-CoV, severe acute respiratory syndrome coronavirus; nsP3, nonstructural protein 3; SFV, Semliki Forest virus; TLC, thin-layer chromatography; PARG, poly(ADP-ribose) glycohydrolase; EGFP, enhanced green fluorescent protein; YFP, yellow fluorescent protein; ZAP, zinc-finger antiviral protein
Keywords: ADP-ribose, poly(ADP-ribose), alphavirus, coronavirus, mitochondrial localization
Introduction
Macro domain is an evolutionarily conserved protein domain, so named because it is found at the C-terminus of large variant core histones, macroH2A.1 MacroH2A is found only in vertebrates and sea urchins, but the macro domain itself is of ancient origin, encoded by the genomes of many, but not all, bacteria and archaea.2 The macro domain of approximately 170 aa folds into a globular mixed α/β-fold containing a deep groove, a potential ligand-binding pocket.3 Recent structural, enzymatic, and binding studies indicate that several macro domains can bind ADP-ribose or its derivates, including poly(ADP-ribose), and that some of them can hydrolyze ADP-ribose-1″ phosphate (ADPR-1″P) to yield ADP-ribose.4, 5, 6 Therefore, macro domain functions may relate to different ADP-ribose metabolites, although many aspects of their physiological roles remain uncertain. The macro domain protein Poa1p of Saccharomyces cerevisiae is quite divergent compared to human homologs, and the main function of the yeast protein has been proposed to be the hydrolysis of ADPR-1″P, which is a by-product of tRNA splicing.7
The human genome contains nine genes encoding macro domain proteins. The two histones macroH2A1 and macroH2A2 have a role in genome silencing, are enriched in female inactive X-chromosomes and other heterochromatin areas, and can also be involved in the regulation of gene expression.8, 9, 10, 11 The gene for an Snf2-like helicase (ALC1), related to the catalytic subunits of ATP-dependent chromatin remodeling complexes, also encodes for a macro domain.12 Three human genes encode the B-aggressive lymphoma (BAL) family of transcription factors.13, 14, 15 BAL proteins include two or three tandem macro domains followed by a poly(ADP-ribose) polymerase (PARP) domain, reinforcing the link of the macro family with ADP-ribose derivatives. The final three human macro domain proteins are poorly understood. LRP16 was originally named during the search for leukemia-relapse-related proteins, but was subsequently found by the authors not to have a connection with that process. Therefore, it is here called macro domain protein 1 (MDO1). The expression of the corresponding gene is activated by estradiol, and it may promote the growth of human breast cancer cells.16, 17, 18 Human locus 140733 encodes a protein [proposed name is macro domain protein 2 (MDO2)] whose macro domain is closely related to that of MDO1. This locus has been linked with a rare developmental disorder, Kabuki syndrome.19 The gene for ganglioside-induced differentiation-associated protein 2 (GDAP2), which could also be called macro domain protein 3 (MDO3), was described as one of the genes induced by ganglioside synthase expression in a mouse neuroblastoma cell line.20
Macro domains are also encoded by several positive-strand RNA viruses: alphaviruses, rubella virus, hepatitis E virus (HEV), and coronaviruses [including the severe acute respiratory syndrome coronavirus (SARS-CoV)].1, 21 These viruses replicate their RNA in the cytoplasm of animal cells in membrane-associated replication complexes.22 The viral macro domains are found in the viral replicase among the more typical replication protein domains, including an RNA-dependent RNA polymerase and RNA helicase. In alphaviruses, the macro domain is essential for virus viability in cell culture, and the macro-domain-containing nonstructural protein 3 (nsP3) is found at the sites of virus replication, on the outer surface of endosomal/lysosomal membranes.23, 24 The viral macro proteins are not known to enter the nucleus. The SARS-CoV macro domain has been shown to bind poly(ADP-ribose) and to hydrolyze ADPR-1″P.6, 25, 26 The viral macro domains have presumably originally been hijacked from the host genome, and certain amino acids in the ADP-ribose binding pocket are highly conserved between viruses and cellular organisms. Thus, the function of viral macro proteins could be similar to the functions of some of the cellular macro domain proteins. Viral macro proteins could act to influence pathways normally regulated by cellular macro domains either to promote virus replication or to inhibit host responses directed against the virus.
We have here studied three small putatively nonnuclear human macro domain proteins with respect to their ligand binding properties and subcellular localization, and compared them directly with the macro domains derived from RNA viruses. We found that each of the three previously uncharacterized human macro proteins has a distinct profile of biochemical activity (also differing from the viral proteins), and that MDO1 is specifically localized in mitochondria.
Results
Domain structure, expression, and purification of macro domain proteins
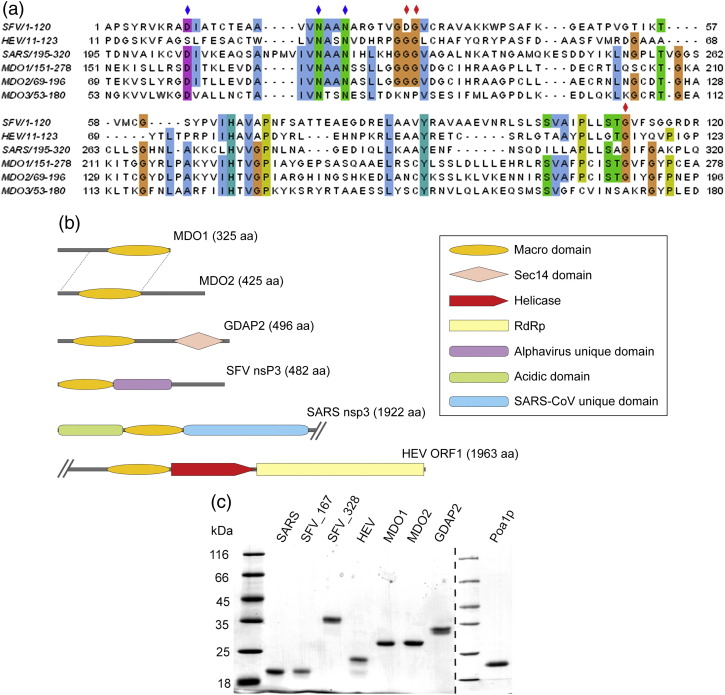
Figure 1a and b shows the domain structure and amino acid sequence alignment of human macro domain proteins MDO1, MDO2, and GDAP2 with the macro domains from Semliki Forest virus (SFV; an alphavirus), HEV (sole member of the Hepeviridae family), and SARS-CoV. MDO1 and MDO2 do not contain any other predicted domains, although they have significant amino acid stretches attached, respectively, to the N-terminus or the C-terminus of the macro domain. However, a high level of sequence homology between them extends for ∼ 230 aa, although the macro domain comprises only ∼ 170 aa of these proteins. GDAP2 includes in its C-terminus a predicted Sec14 domain, which is a putative lipid-binding domain.27 The viral macro proteins form parts of viral replicase polyproteins. SFV macro domain is located at the N-terminus of nsP3, followed by a unique alphavirus domain of unknown function and, after that, a hypervariable phosphorylated region.24 In HEV, the macro domain is immediately preceded by a proline-rich region and a putatively unstructured spacer in the replicase protein, and followed by the viral helicase coding region.28 In coronaviruses, the macro domain is flanked, depending on the virus, by papain-like protease domains or other domains of unknown function that remain in the same mature protein with the macro domain.29
Fig. 1.
Macro domain proteins of human and viral origin. (a) Multiple amino acid sequence alignment of macro domain proteins derived from SFV, HEV, and SARS-CoV with human MDO1, MDO2, and GDAP2 (MDO3). Colored bars highlight conserved amino acids with over 51% identity between the aligned proteins. Blue diamonds on top of the sequence indicate the amino acids that were mutated in SFV, and red diamonds indicate the amino acids that were mutated both in SFV and in MDO1. (b) Domain structures of human MDO1, MDO2, and GDAP2, and viral macro proteins from SFV, HEV, and SARS-CoV. Macro-domain-containing proteins from SARS-CoV and HEV are shown only partially, as indicated by slashes, but their full lengths are given by the numbers in parentheses. The region with a high level of homology between MDO1 and MDO2 is indicated by dotted lines. Predicted domains are indicated by colored symbols. RdRp, RNA-dependent RNA polymerase. (c) Purified macro domain proteins were analyzed by SDS-PAGE on 12% gels followed by staining with Coomassie blue. Lanes from left to right: Molecular mass marker, SARS-CoV nsp3 (174 aa), SFV nsP3 (167 aa), SFV nsP3 (328 aa), HEV macro (185 aa), MDO1 (243 aa), MDO2 (243 aa), GDAP2 (231 aa), molecular mass marker, and Poa1p (177 aa). The lengths of the proteins given above exclude the 13-aa to 17-aa N-terminal vector-derived sequence.
To study their biochemical properties, we expressed the macro domains derived from MDO1, MDO2, GDAP2, SFV nsP3, and HEV in Escherichia coli as N-terminally hexahistidine-tagged proteins. The previously characterized yeast protein Poa1p7 was produced as control, and the SARS-CoV macro domain was obtained as previously described.30 The proteins were purified by metal chelate chromatography, in some cases followed by a second purification step (Fig. 1c; see Materials and Methods). The purified proteins were used in enzymatic and binding assays with ADP-ribose and its derivatives.
ADPR-1″P phosphatase activity
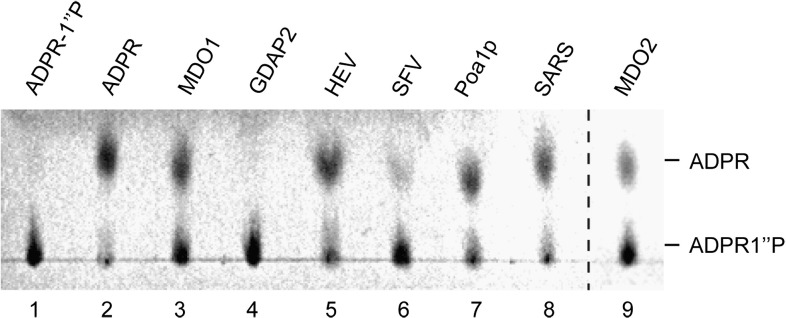
Some macro domain proteins, including Poa1p, have been shown to hydrolyze ADPR-1″P,4, 6 which arises as a by-product during tRNA splicing. The enzymatic activity of the proteins with ADPR-1″P was measured at room temperature, and the reaction products were detected on thin-layer chromatography (TLC). It was observed that the closely related human proteins MDO1 and MDO2 both hydrolyzed ADPR-1″P equally efficiently as yeast Poa1p (Fig. 2 , lanes 3, 7, and 9). HEV and SARS-CoV proteins also demonstrated similar activities (lanes 5 and 8).6 However, SFV nsP3 had only a minor activity, whereas GDAP2 was inactive as an ADPR-1″P phosphoesterase (lanes 6 and 4). The level of activity for SFV nsP3 was independent of the specific construct used, as a protein containing the 167-aa-long macro domain region only and an extended protein of 328 aa had similar activities (data not shown). In parallel with TLC, the reaction products from the hydrolysis of ADPR-1″P by MDO1 were analyzed by HPLC and mass spectrometry. The mass of the reaction product was 558.1 m/z, confirming its identity as ADP-ribose (calculated monoisotopic mass of the protonated molecule, 559.1) and further demonstrating that the reaction involved the removal of the 1″-phosphate and not another type of modification or degradation of the substrate.
Fig. 2.
ADPR-1″P phosphatase activity of macro domains. The indicated macro domain proteins were incubated with ADPR-1″P for 1 h at 28 °C, and reaction products were analyzed by TLC and visualized under UV illumination. Lanes 1 and 2 contain the controls as indicated: ADPR-1″P incubated without the addition of protein, and pure ADP-ribose.
ADP-ribose binding
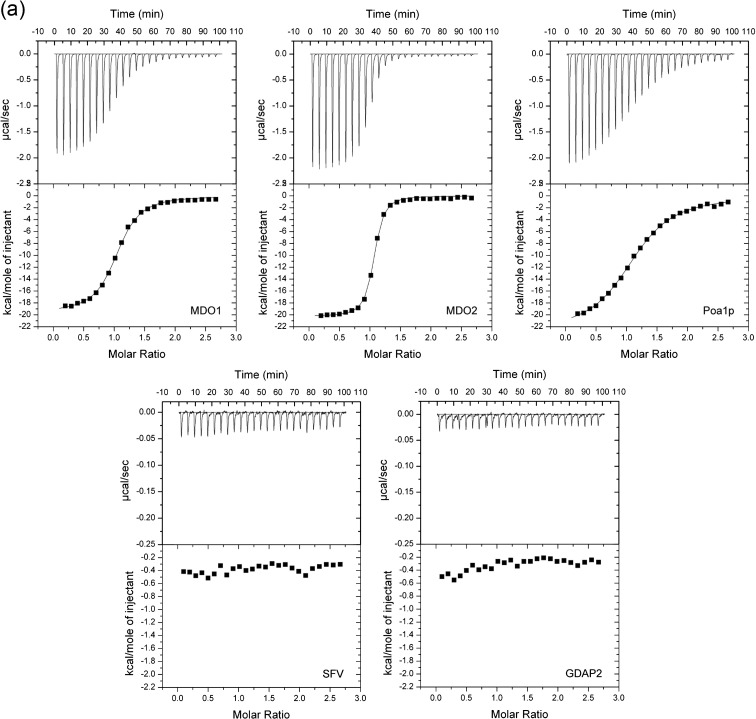
As an alternative to enzymatic function, it has been suggested that cellular and viral macro domains might be ADP-ribose-binding modules.5, 6 We tested the affinity of human, yeast, and viral macro proteins for ADP-ribose by isothermal titration calorimetry (Fig. 3a). MDO1 and MDO2 showed the highest affinity for ADP-ribose, with K d values of 0.9 μM and 0.15 μM, respectively. The yeast protein Poa1p also had a reasonably high affinity (K d = 2.9 μM). Viral macro proteins from SARS-CoV and HEV were previously shown to bind ADP-ribose with a much weaker affinity of ∼ 24 μM or > 50 μM, respectively,6 which was confirmed again in these experiments. We were unable to detect the binding of ADP-ribose to SFV macro. SFV macro domain tended to precipitate in low-salt buffer when the protein concentration was high, as required in calorimetry experiments. Thus, the binding assays for SFV macro protein were repeated in the presence of 500 mM NaCl, where the protein remained soluble. Again, no ADP-ribose binding was detected for SFV macro (both the 167-aa and the 328-aa forms of the protein were tested), whereas MDO1 was able to bind ADP-ribose also in the high-salt buffer, although the dissociation constant measured was approximately twofold higher than that under standard conditions (data not shown). In several independent experiments, human GDAP2 was completely inactive in binding to ADP-ribose (Fig. 3a), although it remained soluble under the assay conditions.
Fig. 3.
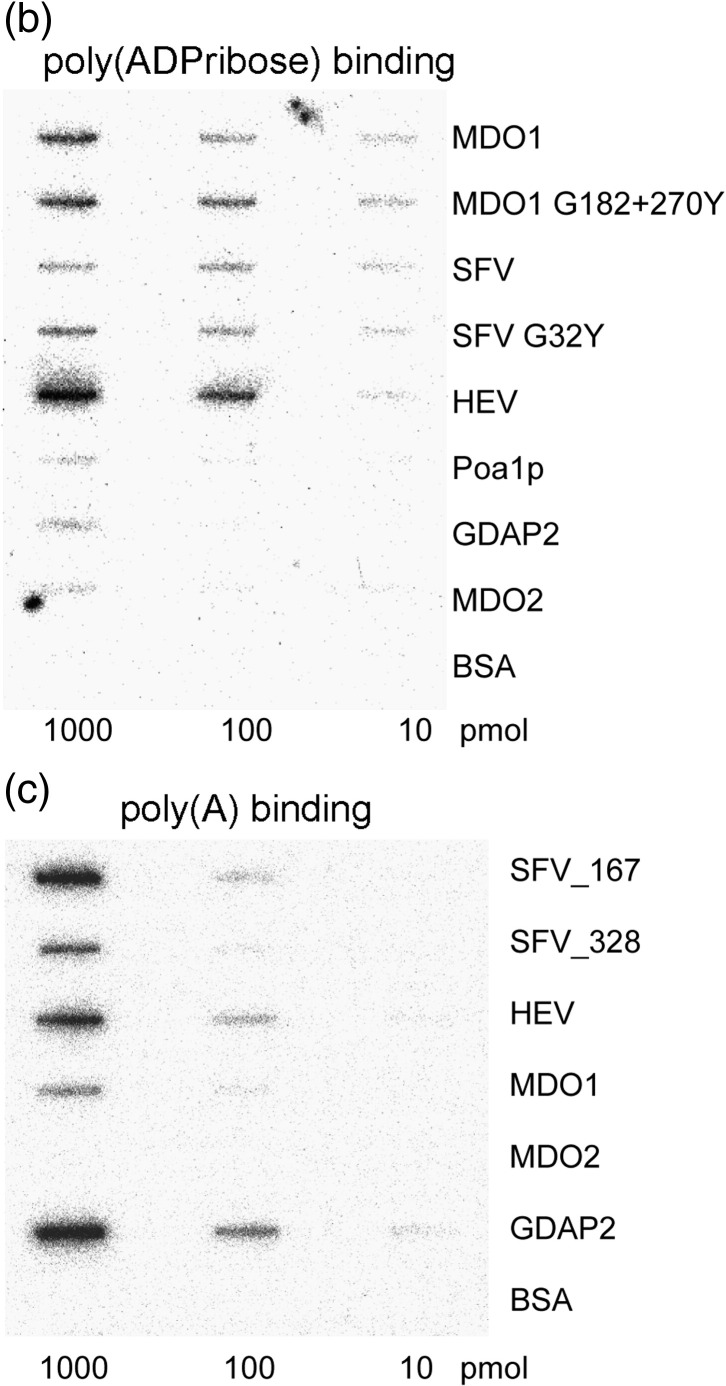
Macro domain proteins of different origins have different affinities for ADP-ribose monomer, poly(ADP-ribose), and poly(A). (a) Isothermal titration calorimetry of ADP-ribose binding. Titration curves show the stepwise addition of ADP-ribose into a solution containing the purified MDO1, MDO2, Poa1p, SFV, and GDAP2 macro domains in the presence of 100 mM NaCl (500 mM NaCl for SFV macro) at 30 °C. The lower parts of the figures show the fit of the measured data (first data point omitted) to an equilibrium binding isotherm. The protein assayed is indicated on the bottom-right corner of each panel. (b) Binding of 32P-labeled poly(ADP-ribose) by macro domain proteins blotted onto a nitrocellulose membrane. About 10 pmol, 100 pmol, or 1000 pmol of each protein was immobilized onto the membrane, followed by blocking with milk proteins. After 1 h of incubation with [32P]poly(ADP-ribose), the membrane was washed carefully, and bound radioactivity was detected with PhosphorImager. Bovine serum albumin was used as negative control. In addition to the wild-type macro proteins, a double mutant (G182Y + G270Y) of MDO1 and a single mutant (G32Y) of SFV nsP3 are illustrated. (c) Binding of 32P-labeled poly(A) by macro proteins was tested similarly to the poly(ADP-ribose) binding experiment.
Binding of poly(ADP-ribose)
Most of the ADP-ribose in the cells is in the form of poly(ADP-ribose), a polymer produced by PARPs. It has been shown that some macro domains bind poly(ADP-ribose).5, 6 Thus, we wanted to examine if this represents a common property for all the macro domains. For this assay, poly(ADP-ribose) was produced from radioactively labeled NAD + by auto-ADP-ribosylation of PARP-1 (see Materials and Methods). Macro domain proteins immobilized on a nitrocellulose filter were incubated with a protein-free purified poly(ADP-ribose) preparation or with a preparation in which the labeled polymer was still bound to PARP-1. Overall, the experiments showed that the binding of poly(ADP-ribose) with PARP-1 in the preparation and the binding of poly(ADP-ribose) without PARP-1 in the preparation were practically identical. Incubation of macro domains with radioactive NAD + alone produced no radioactive signal.
The viral macro domains from HEV and SFV bound poly(ADP-ribose) effectively (Fig. 3b),6 although they bound monomeric ADP-ribose with low affinity (HEV) or no affinity at all (SFV) in previous experiments. In complete contrast, macro domains derived from MDO2 and Poa1p did not show effective binding to poly(ADP-ribose) (Fig. 3b), although they bound monomeric ADP-ribose tightly. MDO1 bound poly(ADP-ribose) and also the monomer, and GDAP2 was inefficient in binding poly(ADP-ribose) and ADP-ribose (Table 1 ). Thus, the binding of poly(ADP-ribose) showed striking differences between macro domains and was not directly related to monomer binding.
Table 1.
Binding and hydrolysis activities of macro domain proteins
| Protein | ADPR-1″P hydrolysis | ADP-ribose binding, Kd (μM) | Poly(ADP-ribose) binding | Poly(A) binding |
|---|---|---|---|---|
| MDO1 | ++ | 0.9 | + | + |
| MDO2 | ++ | 0.15 | − | − |
| GDAP2 | − | NB | − | + |
| Poa1p | ++ | 2.9 | − | ND |
| SFV macro | + | NB | + | + |
| HEV macro | ++ | > 50 | + | + |
| SARS macro | ++ | 24 | +a | ND |
ADPR-1″P hydrolysis was tested at 28 °C and detected by TLC. Hydrolysis activity was classified as moderate (++), poor (+), or none (−). ADP-ribose binding was measured by isothermal titration calorimetry at 30 °C. Binding of poly(ADP-ribose) and poly(A) was detected by filter binding assays with a 32P-labeled ligand and classified as efficient (+) or inefficient (−).
NB, nonbinding; ND, not determined.
Data from Egloff et al.6
The binding of poly(ADP-ribose) appeared to be specific in the case of MDO1, SFV macro, and HEV macro. When the filter-bound proteins were incubated with a large molar excess of poly(A) (∼ 400-fold) or monomeric ADP-ribose (1500-fold excess), together with poly(ADP-ribose), no competition for the binding of poly(ADP-ribose) was noted (data not shown). Competition of ADP-ribose with poly(ADP-ribose) was also tested in experiments in which the proteins were first preincubated with the monomer, followed by addition of the polymer, with similar results. When we tested the binding of labeled poly(A) alone, however, all the proteins that bound poly(ADP-ribose) were also able to bind poly(A) (Fig. 3c; Table 1). MDO2 did not bind poly(A); however, surprisingly, GDAP2, which was inactive in all of the ADP-ribose-related assays, bound poly(A) in this assay, leading to a hypothesis that sequence alterations in the substrate-binding pocket of GDAP2, as compared to other macro domain proteins, might be related to different substrate specificities [nucleic acid binding versus poly(ADP-ribose) binding].
So far, only one enzyme, poly(ADP-ribose) glycohydrolase (PARG), has been known to hydrolyze poly(ADP-ribose) to monomeric ADP-ribose. We also tested the macro proteins' poly(ADP-ribose) hydrolysis activity. The proteins were incubated with poly(ADP-ribose) in hydrolysis buffer, and the reactions were analyzed by TLC for separation of the monomer from the polymer. None of the macro proteins degraded poly(ADP-ribose) under these conditions, whereas cell lysates containing abundant PARG yielded clear evidence of ADP-ribose generation (data not shown). Thus, the macro domains derived from HEV, SFV, and human MDO1—rather than hydrolytic enzymes acting on poly(ADP-ribose)—are specific poly(ADP-ribose)-binding modules.
Mutagenesis of the active site
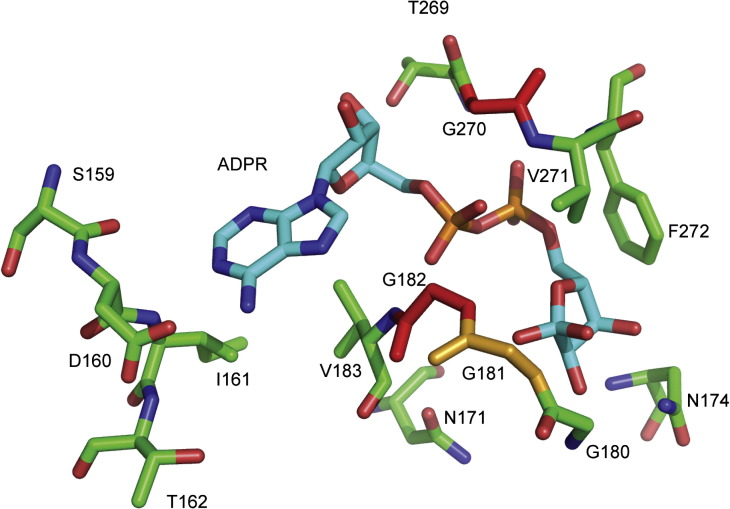
Based on the existing macro domain structures,3, 5, 6 we constructed mutations to the suggested ligand-binding pockets of human MDO1 and SFV macro. The sites for two mutations in both proteins were selected so that by replacing highly conserved glycine residues (which flank the pocket) with a larger amino acid (tyrosine), the narrow binding pocket might become physically blocked (Fig. 4 ). In addition, some of the other conserved amino acids in SFV macro ligand-binding pocket were mutated (Fig. 1a). The mutant proteins were tested for ADPR-1″P hydrolysis and for binding of ADP-ribose and poly(ADP-ribose). Several of the single-point mutations were sufficient to inactivate ADP-ribose binding and ADPR-1″P hydrolysis. Mutations G182Y and G270Y in MDO1 inactivated ADP-ribose binding and ADPR-1″P hydrolysis reactions, and the corresponding mutations in SFV macro (G32Y and G112Y) also totally abolished ADPR-1″P hydrolysis (Table 2 ). The ADP-ribose binding of SFV macro mutants was not tested, as no binding was detected for the wild-type protein. Most macro domains contain a stretch of three glycines (residues 180–182 in MDO1), but SFV macro has an aspartate in the central position (Fig. 1a). An attempt to return to the consensus sequence by mutation of this aspartate to glycine further reduced the small ADPR-1″P hydrolysis activity of SFV macro, and, for MDO1, an acidic residue (glutamate) was detrimental in this position (Table 2). Mutations in the other tested positions of SFV macro abolished the enzymatic activity of the protein (Table 2).
Fig. 4.
Structure prediction of the MDO1 ligand binding site. The ADP-ribose bound to the MDO1 ligand-binding pocket is shown as predicted, using SARS-CoV macro domain as model. Sites of mutations inactivating ADP-ribose binding (residues G182 and G270) are highlighted in red, and the site of mutation reducing binding (G181) is shown in orange. In the SFV macro sequence, positions 31 and 32 correspond to MDO1 residues 181–182, and position 112 corresponds to MDO1 residue 270. The residues corresponding to the additionally mutated sites in SFV are also seen in this rendering: residues D10, N21, and N24 in the SFV sequence correspond to MDO1 D160, N171, and N174, respectively.
Table 2.
Binding and hydrolysis activities of MDO1 and SFV macro protein mutants
| Protein | ADPR-1″P hydrolysis | ADP-ribose binding, Kd (μM) | Poly(ADP-ribose) binding |
|---|---|---|---|
| MDO1 wild type | ++ | 0.9 | + |
| G181E | + | 1.7 | + |
| G182Y | − | NB | + |
| G270Y | ± | NB | + |
| G182Y + G270Y | − | ND | + |
| SFV macro wild type | + | NB | + |
| D10A | − | ND | + |
| N21A + N24A | − | ND | + |
| D31G | ± | ND | + |
| G32Y | − | ND | + |
| G112Y | − | ND | + |
ADPR-1″P hydrolysis was tested at 28 °C and detected by TLC. Hydrolysis activity was classified as moderate (++), poor (+), very weak (±), or none (−). ADP-ribose binding was measured by isothermal titration calorimetry at 30 °C. Poly(ADP-ribose) binding was detected by filter binding assays with 32P-labeled poly(ADP-ribose) and classified as efficient (+) or inefficient (−).
NB, nonbinding; ND, not determined.
None of the mutations affected poly(ADP-ribose) binding (Table 2; two mutants are also illustrated in Fig. 3b). This could be explained by the interaction of poly(ADP-ribose) with additional extensive sites on the surface of the macro domain outside of the deep pocket that binds monomeric ADP-ribose, as suggested.6 However, it was surprising that none of the mutants could even noticeably reduce poly(ADP-ribose) binding, as examined by filter binding assay. For instance, mutating either of the glycines (G182 and G270 in MDO1) flanking the binding pocket to tyrosine residues totally abolished ADP-ribose binding, possibly by physically blocking the binding pocket. Furthermore, combining these two mutations (G182Y + G270Y) did not have an effect on poly(ADP-ribose) binding (Fig. 3b), even when the filter was washed under more stringent conditions (1 M NaCl) (not shown). Thus, it seems that poly(ADP-ribose) binding is a relatively robust property of those macro domains that show an affinity for it.
Subcellular localization of human macro proteins
The macroH2A and BAL families of proteins are known to localize to the nucleus and to function in transcriptional regulation.8, 10, 14 As the virus-encoded macro domain proteins function in the cytosol, we were interested to know whether some of their human counterparts would also reside in the cytosolic compartment.
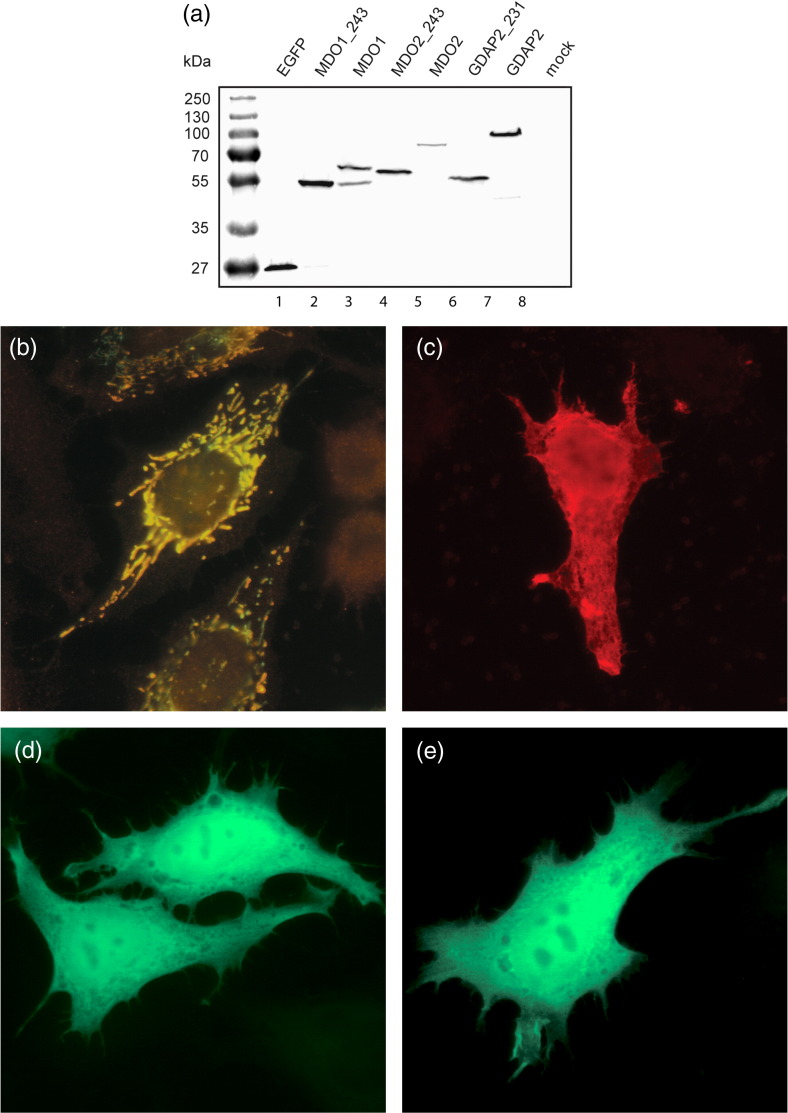
We expressed MDO1, MDO2, and GDAP2 in HeLa cells as C-terminally myc-tagged or enhanced green fluorescent protein (EGFP)-tagged proteins. Two constructs, encoding either the full-length protein or the macro domain only, were made for each (see Materials and Methods). The myc antibodies failed to recognize the expressed proteins on Western blot analysis. However, the macro proteins fused to EGFP were readily expressed, and proteins of the expected size were detected by Western blot analysis with EGFP antibodies (Fig. 5a). In the case of full-length MDO1-EGFP, besides the protein of expected size (62 kDa), an additional smaller protein of ∼ 56 kDa—corresponding to roughly 29 kDa for the MDO1 portion when the mass of EGFP was subtracted—was also detected. Similarly, two bands of ∼ 36 and ∼ 30 kDa were detected when MDO1-myc was expressed and detected by anti-MDO1 antibodies (not shown).
Fig. 5.
Expression and localization of the human macro proteins MDO1, MDO2, and GDAP2 in HeLa cells. (a) Western blot analysis of EGFP-tagged proteins, as detected by antibodies against EGFP. An EGFP coding vector without fusion partner was used as positive control (lane 1), and mock-transfected cells were used as negative controls (lane 8). Either the full-length protein (lanes 3, 5, and 7) or the macro domain with flanking sequences (length indicated by the numbers following the name of the protein; lanes 2, 4, and 6) was expressed, as indicated on top. Molecular mass marker is shown on the left. (b) MDO1 fused to a myc epitope was cotransfected with YFP linked to a mitochondrial localization signal (mt-YFP) and detected by indirect immunofluorescence using anti-MDO1 antibodies. Localization of mt-YFP was detected in the green channel, and anti-MDO1 staining was detected in the red channel. Colocalization of the signals is illustrated in yellow. (c) The 243-aa C-terminal of MDO1 detected by anti-MDO1 did not show mitochondrial localization. Full-length MDO2 (d) and GDAP2 (e) were expressed as C-terminally EGFP-tagged recombinant proteins in HeLa cells and were detected by EGFP fluorescence both in the cytosol and in the nucleus.
For immunofluorescence, HeLa cells were transfected with EGFP fusion constructs and, in the case of MDO1, also with myc fusion for detection by anti-MDO1 antibody. The transfected cells were studied by immunofluorescence at 6 h and 18 h posttransfection. At both time points, MDO1 showed bright staining of specific and abundant organelles, which were confirmed as mitochondria, using anti-MDO1 antibody together with yellow fluorescent protein (YFP) linked to a mitochondrial localization signal (Fig. 5b). This localization was identical independently of the vector used (EGFP fusion and myc-tagged). Notably, a faint staining of the mitochondria was also observed when untransfected HeLa cells were stained with anti-MDO1 antibodies (not shown), suggesting that endogenous MDO1 is expressed and present in the mitochondria at low levels.
The smaller band detected for MDO1 by Western blot analysis could indicate either translation initiation from the second in-frame methionine (Met83) in the MDO1 gene (yielding a 243-aa protein) or proteolytic cleavage of the full-length protein. The localization of MDO1 in mitochondria led us to suspect the latter possibility, as it is common that mitochondrial import sequence is cleaved after transport to the mitochondria. The MDO1 protein sequence was analyzed with the MitoProt program,31 which predicted export to the mitochondria with a probability of 0.99. The predicted import sequence was 77 aa long, the cleavage of which would result in a mature protein of 248 aa in the mitochondria. To support the presence of a mitochondrial import sequence, we additionally expressed in HeLa cells a construct comprising the last 243 aa of MDO1. Expression in this construct was initiated from the second in-frame methionine and resulted in an evenly distributed cytoplasmic protein (Fig. 5c), which was sometimes concentrated in large aggregate-like structures at later time points (not shown), and no colocalization with the mitochondria was detected. Therefore, it is likely that MDO1 is initially expressed in the cells as a 325-aa protein, which is transported to the mitochondria and cleaved by mitochondrial enzymes to result in the shorter protein.
MDO2-EGFP and GDAP2-EGFP proteins showed an even distribution throughout the cytoplasm and nucleus (Fig. 5d and e). This localization was identical for the full-length protein and the smaller macro core units fused to EGFP (not shown). Thus, the localization of the proteins was not affected by the predicted Sec14 domain in GDAP2 or the ∼ 190-aa unique sequence in the MDO2 C-terminus. Based on these results, MDO2 and GDAP2 are the first macro domain proteins to be found in the cytosol and the first macro domain proteins that could potentially be shuttling to and from the nucleus.
Discussion
In this study, we characterized the biochemical functions of small human macro domain proteins, which turned out to have rather distinct properties compared to one another. This study revealed new types of human macro domain proteins: two cytosolic/nuclear macro proteins MDO2 and GDAP2, one of which (GDAP2) contains the first macro domain binding poly(A), but not ADP-ribose derivatives. Furthermore, MDO1 was the first macro protein found to localize in mitochondria. We also compared the viral macro domain proteins with their human homologs, expecting that the viral proteins would resemble some of their cellular counterparts and thus putatively influence the same cellular pathways. However, the viral macro proteins from SFV, HEV, and SARS-CoV did not show a clear relatedness to any of the cellular proteins studied, but resembled each other in their biochemical properties. Based on this study, it seems likely that the functions of the macro domain proteins are more diversified than has been thought before.
The human cytosolic macro proteins MDO2 and GDAP2 bound ADP-ribose monomer and poly(A), respectively, but did not bind poly(ADP-ribose). These proteins seem to reside also in the nucleus (EGFP-tagged proteins were detected in both the cytoplasm and the nucleus) and could potentially also participate in nuclear functions such as RNA processing, in addition to cytosolic metabolic/signaling pathways. The high binding affinity of MDO2 for ADP-ribose, its inability to bind poly(ADP-ribose), and its hydrolysis activity towards ADPR-1″P are all properties that it shares with the yeast Poa1p (Table 1). If mammalian cells harbor an enzymatic activity scavenging the ADPR-1″P by-product of tRNA splicing, this role could be fulfilled by MDO2, as previously suggested for Poa1p in yeast.7 The unusual cytoplasmic splicing event of XBP-1 mRNA during the unfolded protein response caused by endoplasmic reticulum stress may also proceed via a tRNA-like pathway.32 Interestingly, disruption of the gene encoding MDO2 was suggested to be linked to a rare developmental disorder in one patient,19 thus implying an important developmental role for this protein. This could be related to ADP-ribose-mediated cellular signaling, as mono(ADP-ribosyl)ation has an important role in the signaling and metabolic pathways of eukaryotic cells (reviewed by Di Girolamo et al. 33). Alternatively, accumulation of metabolic products such as ADPR-1″P could lead to developmental disturbances.
GDAP2 was originally found in a search for ganglioside-induced neuronal differentiation factors, and its expression was found to be developmentally regulated in mouse brain.20 GDAP2 contains in its C-terminus a predicted Sec14 domain—a lipid-binding domain that is known to function in intracellular trafficking. In our experiments, the inclusion of the Sec14 domain did not affect the subcellular localization of GDAP2 when the protein was overexpressed in HeLa cells. However, it remains possible that, under specific circumstances, the Sec14 domain could have targeting effects on the protein. The GDAP2 macro did not possess the ability to interact with ADP-ribose and poly(ADP-ribose), but we demonstrated the binding of poly(A) (Fig. 3b and c). Clarification of the nucleic acid binding properties and specificities of GDAP2 macro domain requires further studies.
MDO1 is a relatively potent ADP-ribose binding protein and is also able to bind poly(ADP-ribose) and to hydrolyze ADPR-1″P (Table 1). MDO1 is the first ADP-ribose binding protein that has been shown to localize in mitochondria. The role of poly(ADP-ribosyl)ation in mitochondrial regulatory events has been under discussion (reviewed by Scovassi34). Recently, two newly discovered poly(ADP-ribose)-hydrolyzing PARG isoforms were detected in the mitochondria.35 Specific PARG activity was shown to be higher in the mitochondria than in the nucleus, although PARP activity in the mitochondria was low.35 This could indicate transport of poly(ADP-ribose) from the nucleus to the mitochondria. In another study, nuclear–mitochondrial signaling was studied under apoptotic stress, and no poly(ADP-ribose) transport to the mitochondria was detected.36 Rather, it was proposed that ADP-ribose might serve as messenger molecule in the activation of mitochondrial apoptosis mechanisms after nuclear poly(ADP-ribosyl)ation. The localization of MDO1 in the mitochondria may support a role for ADP-ribosylation (either in polymeric or in monomeric form) also in the mitochondria, although the possibility of MDO1 functioning in ADPR-1″P hydrolysis cannot be excluded.
The viral macro domain proteins have been adopted as an essential part of viral genome, but are present only in a small subset of positive-strand RNA viruses that replicate in the cytosolic compartment. It has been suggested that viral macro domains might function as poly(ADP-ribose)-binding modules.6 The present study confirms that viral macro domain have substantial affinity for poly(ADP-ribose), but not for monomeric ADP-ribose. Poly(ADP-ribose) binding, coupled with poly(A)/nucleic acid binding, further suggests that viral macro domains could have a role in viral RNA replication and/or transcription. The macro domains of HEV and SFV can also bind poly(ADP-ribose) in the presence of a poly(A) competitor. This suggests that they could recruit poly(ADP-ribosyl)ated cellular factors to the replication complex while bound to RNA. It is also possible that the function of poly(ADP-ribose) binding would facilitate the disassembly of macro domain from the viral genome. This possibility could not be tested directly here because poly(ADP-ribose) could not be produced in amounts high enough to be tested in competition experiments against poly(A). Although the viral proteins can—to a higher or a lower extent—hydrolyze ADPR-1″P (Fig. 2; Putics et al. 25, 37), it is not easy to understand how this activity might be linked to the advancement of viral RNA replication. This function has been also shown to be dispensable in a cell culture for coronavirus.25 Whether poly(ADP-ribose) or poly(A) binding by viral macro domains is important for virus replication remains to be established.
One possible pathway targeted by viral macro proteins involves a cellular protein called zinc-finger antiviral protein (ZAP), which has been shown to inhibit alphavirus growth in cell culture.38, 39 ZAP consists of repeated zinc-fingers, a TiPARP homology domain, a WWE domain, and a PARP-like domain.40 The zinc-finger domain of human ZAP was found to be sufficient for alphavirus inhibition; however, in a recent study, it was shown that the PARP-like domain increased the inhibition by fivefold. In the same study, the PARP-like domain was also found to have been under positive evolutionary selection, unlike other ZAP domains.40 Thus, SFV nsP3 and other viral macro proteins could aim to sequester ZAP, or other proteins modified by the putative PARP activity of ZAP, to prevent the normal function of ZAP and the inhibitory event. Also other cytosolic PARP-like proteins or proteins modified by these PARPs could interact with viral macro proteins, but even less is known about these proteins. Altogether, 17 PARP-like proteins, some of which are likely to function in the cytosolic compartment, have been identified in humans.41 Some of the already characterized PARP proteins have been shown to localize at least partly outside the nucleus, including PARP-3 and PARP-4, tankyrase-1 and tankyrase-2, and probably even PARP-1, the main PARP (reviewed by Virág42). Among the cellular macro domain proteins studied, only MDO1 was able to bind poly(ADP-ribose); however, due to its mitochondrial localization, MDO1 could only indirectly interact with the putative networks influenced by the viral macro proteins. Thus, in contrast to our initial assumptions, it seems that functional studies of cellular macro proteins might not directly illuminate the functions of the related viral proteins.
We show here that different macro domains can have a variety of binding properties, and that the binding of ADP-ribose monomer and polymer is not necessarily interrelated, as some of the examined proteins only bound one, but not the other, ligand. These results are in accordance with experiments on the histone splicing variant mH2a1.1, which binds O-acetyl-ADP-ribose and ADP-ribose, but not poly(ADP-ribose).43 The gene mH2A1 results in two differently spliced proteins, of which mH2A1.1 binds ADP-ribose, but mH2A1.2 does not. The reported changes in the mH2A1.2 splicing variant were replacements of G223 and G224 by K and D, respectively, and a ∼ 90° flip in F348 stacking the adenine residue.43 In GDAP2, the amino acids corresponding to G223 and G224 in histone mH2A1.1 are also replaced by larger amino acids, K82 and N83. In most macro domains, this region contains a stretch of three glycines (Fig. 1). We also observed in our experiments that mutating the last of the three glycines (G182 in MDO1) to tyrosine resulted in loss of ADP-ribose binding and ADPR-1″P hydrolysis. It seems that the flexible glycine stretch at the edge of the binding pocket (aa 180–182 in MDO1; Fig. 4) is required for ADP-ribose binding. The conservation of this stretch could be one hallmark for the binding of ADP-ribose-type ligands by selected macro domains. In GDAP2, the glycine residue on the opposite side of the binding pocket (G270 in MDO1) has also been lost.
In conclusion, it seems likely that most macro domain proteins have evolved to bind a variety of metabolites related to ADP-ribose, but some macro domains such as GDAP2 have evolved to a different direction to interact, for instance, with nucleic acids. This divergence might perhaps not be so surprising, as it has been suggested that poly(ADP-ribose) could assume helical conformations similar to those of nucleic acids.44 Very recently, PBZ, another poly(ADP-ribose) binding domain that consists of motifs (zinc-fingers) originally characterized for nucleic acid binding, was identified.45
Materials and Methods
Protein expression and purification
Macro domain encoding sequences derived from two viral genes, one yeast gene, and three human genes were cloned to a pHAT expression vector that encodes an N-terminal hexahistidine tag.46 The cloned regions, obtained by PCR and verified by sequencing, encoded the following amino acids: aa 775–960 from HEV open reading frame 1 protein (Burmese strain, described earlier47); aa 1–167 or aa 1–328 from SFV nsP3 (prototype strain SFV4); aa 1–177 (entire protein) for S. cerevisiae Poa1p; aa 1–231 for Homo sapiens GDAP2; aa 83–325 for MDO1; and aa 1–243 for MDO2. Clones for MDO1 (IRALp962M206) and GDAP2 (IRAKp961B043) were obtained from the RZPD German Resource Center for Genome Research (sequences correspond to the Entrez nucleotide database with accession numbers BC000270 and BC013132), and the clone for MDO2 was obtained from Origene (accession number NM_080676.5). The POA1 gene was cloned in-house from S. cerevisiae, and Ser2 was mutated to Ala for cloning purposes. HEV sequence was cloned to pHAT using SpeI and BamHI sites, and the others were cloned using NcoI and HindIII sites. Point mutations were made to the E. coli expression constructs of SFV nsP3 (328 aa) and human MDO1 (243 aa) using the XL mutagenesis kit (Promega) in accordance with the manufacturer's instructions. In nsP3, the following mutations were created: D10A, D31G, G32Y, N21A + N24A, G112Y; and in MDO1, the following mutations were created: G181E, G182Y, G270Y, and G182Y + G270Y. All constructs were fully sequenced.
Proteins were expressed in E. coli BL21(DE3) and purified by nickel affinity chromatography (HiTrap; GE Healthcare) in 20 mM sodium phosphate (pH 8.0) and 0.5 M NaCl. Protein binding was carried out in 20 mM imidazole, followed by washes with 40–100 mM imidazole and elution with 500 mM imidazole in sodium phosphate buffer. Affinity purification was followed by ion exchange chromatography (Resource S; GE Healthcare) for MDO2. The ion exchange chromatography was run in 20 mM Hepes and 100 mM NaCl, and the proteins were eluted with a NaCl gradient from 100 mM to 500 mM (1 ml/min for 15 min). The proteins were stored in a protein storage buffer containing 25 mM Hepes (pH 8.0), 250 mM NaCl, 1 mM dithiothreitol (DTT), and 20% glycerol at − 70 °C. SARS-CoV macro protein was kindly provided by Dr. Bruno Canard (CNRS, Marseille, France).30
Human macro protein encoding sequences were also cloned to vectors for expression in mammalian cell lines. Two constructs, encoding either the full-length protein or a truncated derivative (macro domain with some flanking amino acids), were made for each protein. The MDO1 gene was cloned to pEGFP-N1 (EcoRI-AgeI; Clonetech) and pcDNA4/TO (EcoRI-XbaI; Invitrogen) to create a C-terminally myc-tagged (EQKLISEEDL) variant. Two different constructs were made for both vectors to express either the full-length (325 aa) protein or a 243-aa “macro core unit” (aa 83–325). Corresponding constructs were made for GDAP2 and MDO2 (encoding aa 1–239 or aa 1–496, and aa 1–243 or aa 1–425, respectively) in pEGFP-N1 (MDO2: HindIII-SacII; GDAP2: SacI-SacII) and pcDNA4/TO (GDAP2: EcoRI-XbaI).
Enzyme assays
ADPR-1″P was produced from ADP-ribose-1″2″ cyclic phosphate by cyclic phosphodiesterase (reagents were kindly provided by Dr. Witold Filipowicz, Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland), and ADPR-1″P hydrolysis activity of the proteins was detected essentially as previously described.6 The proteins (15 μM) were incubated with 2.3 mM ADPR-1″P [stored in 50 mM Tris (pH 7.0) and 0.1% Triton X-100] in the reaction mixture at pH 6.0 (1:1:1, protein storage buffer, ADPR-1″P buffer, and 60 mM 4-morpholineethanesulfonic acid, pH 5.0) at 28 °C for 1 h or 2 h. The longer incubation time was used for GDAP2 and in studies of mutants of the less active SFV nsP3. The produced ADP-ribose was separated from the substrate by TLC on PEI-F cellulose plates in 0.15 M NaCl and 0.15 M sodium formate (pH 3.0), and detected under UV illumination. To determine the mass of the reaction product, 6 μl of the reaction mixture was run on reverse-phase HPLC with an RP-18 column (2.1 mm × 100 mm, Spherisorb C18, 5 μm, 120 Å) over a linear gradient of 50 min at 180 μl/min [buffer 1: 0.2 M triethyl-ammoniumbicarbonate (pH 7); buffer 2: 50% acetonitrile]. The peak fraction was studied with an Ultraflex TOF/TOF mass spectrometer (Bruker-Daltonik GmbH, Bremen, Germany) using alpha-cyano-4-hydroxycinnamic acid matrix in linear negative mode.
PARG activity was tested by incubating 140 pmol of macro domain or HeLa cell nuclear extract with 0.6 pmol (calculated as NAD + equivalents) of [32P]poly(ADP-ribose) in 5 μl of glycohydrolase buffer [50 mM NaPO4 (pH 7.4), 50 mM KCl, and 10 mM β-mercaptoethanol] for 20 min at 37 °C. The reaction was stopped by adding 0.5 μl of 1% SDS, and products were analyzed on PEI-TLC with 0.3 M LiCl and 0.9 M acetic acid. The cleaved ADP-ribose was detected with BAS-1500 PhosphorImager (Fujifilm).
ADP-ribose binding
Isothermal titration calorimetry binding assays were carried out at 30 °C using a VP-ITC instrument (Microcal). Binding reactions were performed in 25 mM Hepes (pH 7.0) and 100 mM NaCl, using 25–45 μM protein and 350–500 μM ADP-ribose (Sigma) as the injected ligand. The binding assays were also performed in a stabilizing high-salt buffer of 25 mM Hepes (pH 7.0) and 500 mM NaCl, with SFV nsP3, GDAP2, and MDO1 as reference. Data analysis was conducted using Origin software (OriginLab).
Poly(ADP-ribose) synthesis
Poly(ADP-ribose) was synthesized by auto-ADP-ribosylation of PARP-1 as previously described.48 A 100-μl reaction mixture containing 500 ng of PARP-1 (Alexis), 2.5 μg of activated DNA (Sigma), and 150 μM NAD +, including 7.5 μCi of [32P]NAD + (GE Healthcare), was incubated in 25 mM Tris (pH 8.0), 10 mM MgCl2, 0.5 mM DTT, and 100 mM NaCl. After 1 h at 24 °C, the reaction mixture was used directly in a binding assay. Alternatively, to prepare purified poly(ADP-ribose), the reaction mixture was treated with proteinase K, and the protein-free polymer was collected by phenol–chloroform extraction as previously described.5
Poly(ADP-ribose) and poly(A) binding assay
Poly(ADP-ribose) binding assay was performed essentially as previously described,49 using either the polymer bound to PARP-1 or a protein-free preparation. Proteins (10 pmol, 100 pmol, or 1000 pmol) were blotted onto a nitrocellulose membrane using Minifold II slot blot apparatus (Schleichter and Schuell). The membrane was blocked for 1 h in TBS-T [10 mM Tris (pH 7.4), 150 mM NaCl, and 0.05% Tween] containing 5% nonfat milk powder. Subsequently, the membrane was incubated for 1 h with the poly(ADP-ribose) preparation (100 μl) diluted in 10 ml of TBS-T. The membrane was washed as described5 and autoradiographed after drying.
Poly(A) was labeled with 32P using [γ-32P]ATP (GE Healthcare) and T4 polynucleotide kinase (New England Biolaboratories). A 50-μl reaction mixture contained 4.2 μg poly(A) (Sigma), 140 nM ATP (corresponding to 24 μCi), 20 U of T4 polynucleotide kinase, and 4.8% polyethylene glycol 8000 in exchange buffer (0.1 mM spermidine, 5 mM DTT, 18 mM MgCl2, 0.1 mM ethylenediaminetetraacetic acid, and 50 mM imidazole–HCl, pH 7.0). The mixture was incubated for 45 min at 37 °C, and then the reaction was stopped by adding 2 μl of 0.5 M ethylenediaminetetraacetic acid and incubating for 10 min at 70 °C. Labeled poly(A) was purified with a Sephadex-25 column (PD-10; GE Healthcare), and binding assay was performed as described above for poly(ADP-ribose).
For competition binding assays, the amount of poly(ADP-ribose) in the protein-free purified preparation was calculated as NAD + equivalents, based on the specific activity of [32P]NAD + and on the amount of 32P in the product, as measured by liquid scintillation. The protein-containing membrane was incubated with 1500 pmol NAD + equivalents of poly(ADP-ribose) and 580 nmol of poly(A) in 10 ml of TBS-T for 1 h at room temperature, followed by washing, as before. For competition with ADP-ribose monomer, the membrane was preincubated in 10 ml of TBS-T with 3750 nmol of ADP-ribose for 10 min, then 1500 pmol NAD equivalents of purified and labeled poly(ADP-ribose) was added. The mixture was incubated for 1 h, and the membrane was washed.
Prediction of MDO1 structure
Structure prediction for MDO1 was performed using the SARS-CoV macro domain as model (Protein Data Bank code FAV2). FAV2 was chosen for the model because its structure includes an ADP-ribose ligand. Alignment of MDO1 with FAV2 was obtained from Phyre† (E = 6.4 × 10− 18) and sent to SwissModel‡ . The prediction obtained from SwissModel was used as the MDO1 structure model.
Protein expression and detection in human cells
For localization studies, the macro domain expression constructs (in pEGFP-N1 or pcDNA4/TO) were transfected into HeLa cells using Exgen (Fermentas) in accordance with the manufacturer's instructions. For detection of mitochondria, the mitochondrial localization marker mt-YFP containing a mitochondrial localization signal fused to YFP (construct designed by Dr. A. Miyawaki, Riken Brain Science Institute, Japan) was cotransfected with the macro domain construct. The cells were incubated at 37 °C for 6 h or 18 h, fixed with 4% paraformaldehyde, and stained with an antibody (anti-MDO1, produced in-house against aa 83–325, as earlier described for HEV antibodies)50 or detected directly by EGFP or YFP fluorescence. In parallel, cells were collected in 1% SDS, and the proteins were separated by SDS-PAGE on 12% gels and detected by Western blot analysis with the appropriate antibodies (anti-MDO1 or anti-EGFP; kindly provided by Dr. Andres Merits, University of Tartu, Estonia) as described previously.51
Acknowledgements
We thank Dr. Tuula Nyman for help with mass spectrometry, Pia Salomaa for cloning and protein purification in the initial phase of the project, and Riikka Väyrynen for technical assistance. Financial support from the Sigrid Jusélius Foundation and the Academy of Finland (grant 211121) is gratefully acknowledged.
Edited by F. Schmid
Footnotes
References
- 1.Pehrson J.R., Fuji R.N. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 1998;26:2837–2842. doi: 10.1093/nar/26.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anantharaman V., Koonin E.V., Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen M.D., Buckle A.M., Cordell S.C., Löwe J., Bycroft M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 2003;330:503–511. doi: 10.1016/s0022-2836(03)00473-x. [DOI] [PubMed] [Google Scholar]
- 4.Martzen M.R., McCraith S.M., Spinelli S.L., Torres F.M., Fields S., Grayhack E.J., Phizicky E.M. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- 5.Karras G.I., Kustatscher G., Buhecha H.R., Allen M.D., Pugieux C., Sait F., et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., et al. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shull N.P., Spinelli S.L., Phizicky E.M. A highly specific phosphatase that acts on ADP-ribose 1″-phosphate, a metabolite of tRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:650–660. doi: 10.1093/nar/gki211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pehrson J.R., Fried V.A. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- 9.Costanzi C., Pehrson J.R. MACROH2A2, a new member of the MARCOH2A core histone family. J. Biol. Chem. 2001;276:21776–21784. doi: 10.1074/jbc.M010919200. [DOI] [PubMed] [Google Scholar]
- 10.Costanzi C., Pehrson J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 11.Agelopoulos M., Thanos D. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J. 2006;25:4843–4853. doi: 10.1038/sj.emboj.7601364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaus A., Martin D.M., Barton G.J., Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguiar R.C., Yakushijin Y., Kharbanda S., Salgia R., Fletcher J.A., Shipp M.A. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood. 2000;96:4328–4433. [PubMed] [Google Scholar]
- 14.Aguiar R.C., Takeyama K., He C., Kreinbrink K., Shipp M.A. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- 15.Goenka S., Boothby M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proc. Natl Acad. Sci. USA. 2006;103:4210–4215. doi: 10.1073/pnas.0506981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han W.D., Mu Y.M., Lu X.C., Xu Z.M., Li X.J., Yu L., et al. Up-regulation of LRP16 mRNA by 17beta-estradiol through activation of estrogen receptor alpha (ERalpha), but not ERbeta, and promotion of human breast cancer MCF-7 cell proliferation: a preliminary report. Endocr. Relat. Cancer. 2003;10:217–224. doi: 10.1677/erc.0.0100217. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y.L., Han W.D., Li Q., Mu Y.M., Lu X.C., Yu L., et al. Mechanism of transcriptional regulation of LRP16 gene expression by 17-beta estradiol in MCF-7 human breast cancer cells. J. Mol. Endocrinol. 2005;34:77–89. doi: 10.1677/jme.1.01628. [DOI] [PubMed] [Google Scholar]
- 18.Meng Y.G., Han W.D., Zhao Y.L., Huang K., Si Y.L., Wu Z.Q., Mu Y.M. Induction of the LRP16 gene by estrogen promotes the invasive growth of Ishikawa human endometrial cancer cells through the downregulation of E-cadherin. Cell Res. 2007;17:869–880. doi: 10.1038/cr.2007.79. [DOI] [PubMed] [Google Scholar]
- 19.Maas N.M., Van de Putte T., Melotte C., Francis A., Schrander-Stumpel C.T., Sanlaville D., et al. The C20orf133 gene is disrupted in a patient with Kabuki syndrome. J. Med. Genet. 2007;44:562–569. doi: 10.1136/jmg.2007.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H., Nakagawa T., Kanematsu T., Uchida T., Tsuji S. Isolation of 10 differentially expressed cDNAs in differentiated Neuro2a cells induced through controlled expression of the GD3 synthase gene. J. Neurochem. 1999;72:781–790. doi: 10.1046/j.1471-4159.1999.0721781.x. [DOI] [PubMed] [Google Scholar]
- 21.Gorbalenya A.E., Koonin E.V., Lai M.M. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salonen A., Ahola T., Kääriäinen L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kujala P., Ikäheimonen A., Ehsani N., Vihinen H., Auvinen P., Kääriäinen L. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 2001;75:3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kääriäinen L., Ahola T. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:187–222. doi: 10.1016/S0079-6603(02)71044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Putics A., Filipowicz W., Hall J., Gorbalenya A.E., Ziebuhr J. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005;79:12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., et al. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Angelo I., Welti S., Bonneau F., Scheffzek K. A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein. EMBO Rep. 2006;7:174–179. doi: 10.1038/sj.embor.7400602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koonin E.V., Gorbalenya A.E., Purdy M.A., Rozanov M.N., Reyes G.R., Bradley D.W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl Acad. Sci. USA. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malet H., Dalle K., Brémond N., Tocque F., Blangy S., Campanacci V., et al. Expression, purification and crystallization of the SARS-CoV macro domain. Acta Crystallogr. Sect. F. 2006;62:405–408. doi: 10.1107/S1744309106009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Claros M.G. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput. Appl. Biosci. 1995;11:441–447. doi: 10.1093/bioinformatics/11.4.441. [DOI] [PubMed] [Google Scholar]
- 32.Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 33.Di Girolamo M., Dani N., Stilla A., Corda D. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2005;272:4565–4575. doi: 10.1111/j.1742-4658.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 34.Scovassi A.I. Mitochondrial poly(ADP-ribosylation): from old data to new perspectives. FASEB J. 2004;18:1487–1488. doi: 10.1096/fj.04-1841rev. [DOI] [PubMed] [Google Scholar]
- 35.Meyer R.G., Meyer-Ficca M.L., Whatcott C.J., Jacobson E.L., Jacobson M.K. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp. Cell Res. 2007;313:2920–2936. doi: 10.1016/j.yexcr.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cipriani G., Rapizzi E., Vannacci A., Rizzato R., Moroni F., Chiarugi A. Nuclear poly(ADP-ribose) polymerase-1 rapidly triggers mitochondrial dysfunction. J. Biol. Chem. 2005;280:17227–17234. doi: 10.1074/jbc.M414526200. [DOI] [PubMed] [Google Scholar]
- 37.Putics A., Gorbalenya A.E., Ziebuhr J. Identification of protease and ADP-ribose 1″-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 2006;87:651–656. doi: 10.1099/vir.0.81596-0. [DOI] [PubMed] [Google Scholar]
- 38.Bick M.J., Carroll J.W., Gao G., Goff S.P., Rice C.M., MacDonald M.R. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 2003;77:11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacDonald M.R., Machlin E.S., Albin O.R., Levy D.E. Zinc finger antiviral protein acts synergistically with an interferon-induced factor for maximal activity against alphaviruses. J. Virol. 2007;81:13509–13518. doi: 10.1128/JVI.00402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerns J.A., Emerman M., Malik H.S. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008;4:150–158. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otto H., Reche P.A., Bazan F., Dittmar K., Haag F., Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Virág L. The expanding universe of poly(ADP-ribosyl)ation. Cell. Mol. Life Sci. 2005;62:719–720. doi: 10.1007/s00018-004-4502-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kustatscher G., Hothorn M., Pugieux C., Scheffzek K., Ladurner A.G. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 44.Minaga T., Kun E. Probable helical conformation of poly(ADP-ribose). The effect of cations on spectral properties. J. Biol. Chem. 1983;258:5726–5730. [PubMed] [Google Scholar]
- 45.Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 46.Peränen J., Rikkonen M., Hyvönen M., Kääriäinen L. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- 47.Uchida T., Win K.M., Suzuki K., Komatsu K., Iida F., Shikata T., et al. Serial transmission of a putative causative virus of enterically transmitted non-A, non-B hepatitis to Macaca fascicularis and Macaca mulatta. Jpn. J. Exp. Med. 1990;60:13–21. [PubMed] [Google Scholar]
- 48.Panzeter P.L., Realini C.A., Althaus F.R. Noncovalent interactions of poly(adenosine diphosphate ribose) with histones. Biochemistry. 1992;31:1379–1385. doi: 10.1021/bi00120a014. [DOI] [PubMed] [Google Scholar]
- 49.Malanga M., Pleschke J.M., Kleczkowska H.E., Althaus F.R. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J. Biol. Chem. 1998;273:11839–11843. doi: 10.1074/jbc.273.19.11839. [DOI] [PubMed] [Google Scholar]
- 50.Magden J., Takeda N., Li T., Auvinen P., Ahola T., Miyamura T., et al. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J. Virol. 2001;75:6249–6255. doi: 10.1128/JVI.75.14.6249-6255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laakkonen P., Hyvönen M., Peränen J., Kääriäinen L. Expression of Semliki Forest virus nsP1-specific methyltransferase in insect cells and in Escherichia coli. J. Virol. 1994;68:7418–7425. doi: 10.1128/jvi.68.11.7418-7425.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]