Abstract
The discovery of a fourth immunoglobulin (Ig) light (L) chain isotype in sharks has revealed the origins and natural history of all vertebrate L chains. Phylogenetic comparisons have established orthology between this new shark L chain and the unique Xenopus L chain isotype σ. More importantly, inclusion of this new L chain family in phylogenetic analyses showed that all vertebrate L chains can be categorized into four ancestral clans originating prior to the emergence of cartilaginous fish: one restricted to elasmobranchs (σ-cart/type I), one found in all cold-blooded vertebrates (σ/teleost type 2/elasmobranch type IV), one in all groups except bony fish (λ/elasmobranch type II), and one in all groups except birds (κ/elasmobranch type III/teleost type 1 and 3). All four of these primordial L chain isotypes (σ, σ-cart, λ and κ) have maintained separate V region identities since their emergence at least 450 million years ago, suggestive of an ancient physiological distinction of the L chains. We suggest that, based upon unique, discrete sizes of complementarity determining regions 1 and 2 and other features of the V region sequences, the different L chain isotypes arose to provide different functional conformations in the Ig binding site when they pair with heavy chains.
Keywords: Antibody, Evolution, Immunoglobulin light chain, Isotypes, Sharks
Introduction
The classical immunoglobulins (Ig) of the vertebrate adaptive immune system, made up of heavy (H) and light (L) chains, provide obligatory defense against all extracellular, and some intracellular pathogens [1]. The H chain is composed of Ig superfamily (Igsf) domains, one N-terminal variable (V) domain, and two to six constant (C) domains, while the L chain always consists of one V and one C Igsf domain. A disulfide bond between the CL and the IgHC1 domains covalently joins the L chain to the H chain, and the two V domains associate non-covalently to form the antigen-binding site.
Functional VL genes are generated by random somatic rearrangement of V and joining (J) DNA segments in developing B lymphocytes [the H chain domain by V, diversity (D), and J segments]. The gene segments are flanked by recombination signal sequences (RSS), recognized by the recombination-activating genes (RAG)-1 and RAG-2, which initiate and play a central role in the gene rearrangement. After B cells encounter antigen, the V genes can be further diversified by somatic hypermutation. The V region diversity is concentrated in the three loops of the domain that bind to antigen, called the complementarity-determining regions (CDR). CDR1 and CDR2 are encoded by the V gene itself, while CDR3 is encoded by the V-J or V(D)J rearrangement junction and thus it is the most diverse CDR. While Igsf domains are common in all animal groups, bona fide Ig are present only in gnathostomes (jawed vertebrates), with sharks (elasmobranchs in the cartilaginous fish group) being the oldest living group.
It has long been known that mammals have two IgL isotypes, κ and λ, but additional isotypes have been found in other vertebrate groups. Three IgL isotypes are present in the amphibian Xenopus: κ [2], λ [3], and a unique isotype σ [4]. The κ isotype was believed to be the oldest and most evolutionarily conserved, as it is present in elasmobranchs and bony fish, but both of these vertebrate groups have other isotypes that could not be definitively classified.
Long ago it was suggested that L chain isotypes arose from a single ancestral gene [1]. However, a lack of data from key phylogenetic groups as well as no identifiable distinct functions for different IgL isotypes have made assignation of L chain isotypes controversial. Three criteria have been considered: amino acid identity, gene organization, and heptamer and nonamer spacing in the RSS (either 12 or 23 base pairs) [5]. Gene organization has recently been informative for resolving relationships among teleost (i.e., bony fish) L chain isotypes [6]. However, teleosts are amenable to such analyses since they have both phylogenetically informative cluster and translocon IgL arrangements and ample genomic data exist from several fish genome projects. For IgH and TCR, C domain homology proves most informative for phylogeny. However, phylogenetic analyses of IgL have suggested different selective pressures on the V and C domains [7], with the C domains usually clustering based more on taxonomy than isotype. All of these factors have contributed to a poor understanding of vertebrate IgL relationships, preventing the establishment of a definitive record for this antigen receptor family.
Here we report a new L chain gene from cartilaginous fish that is the ortholog of the σ isotype, previously described first in Xenopus and later in bony fish. These new data enable us to revisit the questions of L chain origins and natural history, and to propose new hypotheses for isotype function. V domain homology, CDR lengths, and RSS orientation all support a reclassification of vertebrate L chains into four ancestral clades present in cartilaginous fish and maintained in other vertebrates.
Results
Identification of a fourth shark IgL chain
As mentioned, cartilaginous fishes such as sharks and rays are the oldest group of animals that shares the basic features of the mammalian immune system. Sharks and skates have three H chain isotypes and three reported L chain isotypes: type I (NS5), type II (NS3), and type III (NS4); (note, the NS nomenclature refers to L chain genes in the nurse shark before the more general nomenclature for all cartilaginous fish L chains was agreed upon) (reviewed in [8]). The type III L chain is clearly κ, and the other two have been suggested to be more similar to λ, but without strong phylogenetic support. Most comparative immunologists believed that all of the different L chain isotypes had been identified, but this was not the case.
While probing a nurse shark spleen cDNA library under low-stringency conditions with the C domain gene of shark TCRγ (manuscript in preparation), clone 61.8 (Supporting Information Fig. 1) was isolated. Its deduced protein sequence suggested a leader peptide and Igsf V and C domains, but there was no transmembrane region typical of TCR. A telltale C-terminal cysteine residue as well as BLAST analyses revealed that the cDNA was most likely an L chain gene. Individual BLAST of the C domain showed 45–51% amino acid identities to CL domains from various cartilaginous fish species but no orthology to the three previously described nurse shark isotypes. BLAST searches with the V domain revealed 40–56% amino acid identity to some VL domains from bony fishes and frog, higher than the V domain identities to all identified shark L chains.
The cloning of an IgL chain gene when screening with a TCR probe is unprecedented, but not altogether unexpected as they are both members of the rearranging antigen receptor family [9]. A region encompassing the N-terminal Igsf canonical cysteine-encoding residue gave an exact match over ten nucleotides between the two genes, which probably served as a nucleation point for a section of substantial identity running throughout the 5′ end. Similarity at the nucleotide level for the rest of the C gene is extremely low.
Orthology to L chains from other ectothermic vertebrates
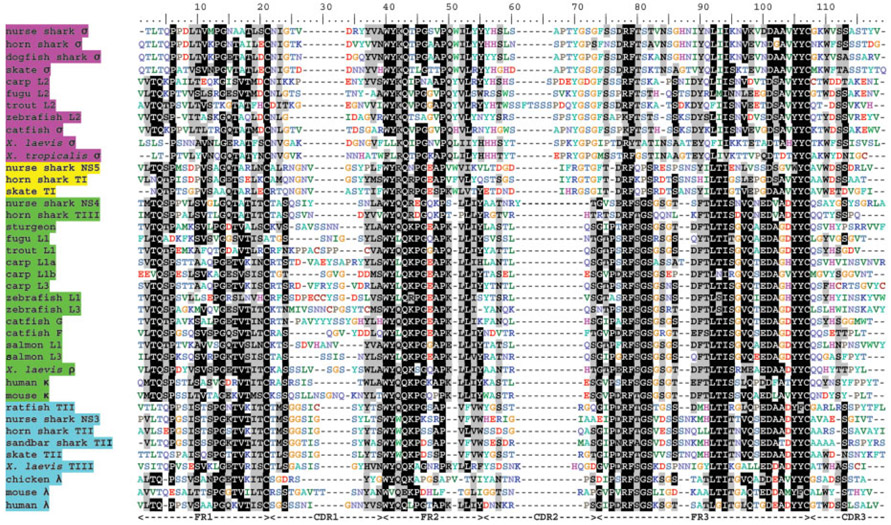
When the predicted V domain of clone 61.8 is aligned with V domains of other vertebrate L chains, unexpectedly the highest similarity is seen with the L2 isotype of teleost fish and IgLσ from the amphibian Xenopus laevis (Fig. 1). We retain the σ designation for the new shark L chain [4].
Figure 1.
V domain alignment. Hyphens denote gaps. Common names of organisms and original IgL nomenclature are highlighted based on our proposed four isotype scheme; σ = pink, σ-cart = yellow, κ = green, and λ = blue (consistent throughout rest of figures). Accession numbers of sequences used are zebrafish NITR (NM001005577), skate I (L25568), horn shark I (X15315), nurse shark NS5 (AAV34678.1), chicken λ (M24403), human λ (AAA59013), mouse λ (AA053422), X. laevis TIII (L76575), ratfish II (L25549), skate II (L25566), horn shark II (L25560), sandbar shark II (M81314), X. laevis ρ (XELIGLVAA), horn shark III (L25561), nurse shark NS4 (GSU15144), mouse κ (MUSIGKACN), human κ (S46371), catfish F (U25705), salmon L3 (AF406956), Fugu L1 (AB126061), carp L1b (AB073332), zebrafish L3 (AB246193), catfish G (L25533), sturgeon (CAB44624), trout L1 (X65260), salmon L1 (AF273012), zebrafish L1 (AF246185), carp L1a (AB073328), carp L3 (AB073335), trout L2 (AAB41310), carp L2 (AB091113), Fugu L2 (DQ471453), zebrafish L2 (AF246183), catfish σ (CK403931), X. laevis σ (S78544), X. tropicalis σ (scaffold 289), dogfish σ (CX662707), skate σ (CV222129), nurse shark σ (EF114759) and horn shark σ (EF114760). Nurse shark NS3 sequence is from the dissertation of Andrew Greenberg at the University of Miami. β-Strand predictions are shown at bottom as solid lines.
We then found additional examples of IgLσ from other cold-blooded vertebrates. Nurse shark σC was used to isolate an ortholog from a horn shark spleen cDNA library. Screenings in silico resulted in undescribed IgLσ orthologs from two other elasmobranchs, the dogfish shark and a batoid, the little skate. Xenopus tropicalis expressed sequence tags (EST) as well as genomic scaffolds identified the isotype in a second Xenopus species, separated by 50 million years from X. laevis. Additionally, an IgL expressed sequence tag was recently found [6] from the bony fish channel catfish with high homology to σ and teleost L2 that was clearly not of the described catfish isotypes F or G, which are both in the κ class [10, 11]. No σ sequences were detected in any of the eu- or metatherian mammal databases; thus this isotype arose early in vertebrate evolutionary history and has been maintained in all cold-blooded vertebrates examined.
The alignment of all V domains showed a striking conservation of long CDR2 size in σ V regions (relative to other isotypes), in species separated by up to 450 million years (Fig. 1). Furthermore, a consensus sequence of PxYGxGFS (amino acids 68–75) is also well conserved over very long phylogenetic distances. Conversely, the σ CDR1 is relatively short compared to other L chains. No such signature sequence/feature is seen in the C domain alignment (Supporting Information Fig. 2).
Phylogenetic analyses
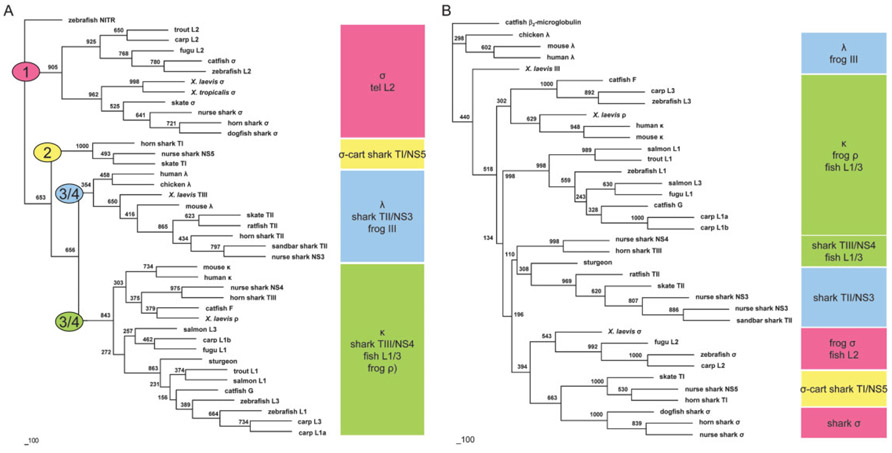
An analysis based upon VL domains of diverse species and isotypes was performed (Fig. 2A). The sequences group into four distinct clades, regardless of the number of gap columns trimmed from the alignment or the dendrogram-generating algorithms used. As expected, based on the previous BLAST and sequence alignments, σ from elasmobranchs groups with σ from amphibians, L2 from bony fish, and the catfish σ annotated here. Shark type I has no known orthologs outside of elasmobranchs, but is clearly more closely related to σ and we rechristen it σ-cart; previous suggestions that this isotype was most similar to λ are highly unlikely [9]. However, consistent with previous predictions [12], cartilaginous fish type II now clearly clusters with the tetrapod λ isotypes, including the Xenopus and chicken orthologs.
Figure 2.
Phylogenetic analysis of IgL chains. (A). Neighbor joining tree based on V domain alignment in Fig. 1. Numbers in ovals represent hypothesized order of emergence of L chain clades. (B). Neighbor joining tree based on C domain alignment in Supporting Information Fig. 2. Clusters of sequences in both trees are marked with colored blocks showing four ancestral isotypes. Bar denotes genetic distance.
As pointed out in a recent analysis of teleost L chains [6], L1 and L3 are difficult to distinguish within teleosts much less among other vertebrates; in this new tree, however, they are both clearly shown to be κ orthologs. This designation of κ genes in Fig. 2A is consistent with a previous analysis of the isotype although the relationships between Vκ genes from different species differ somewhat [13]. Thus, this phylogenetic analysis invokes a reclassification of all vertebrate L chains into four ancient groups, including the classical κ and λ isotypes.
Like in previous studies, our phylogenetic analysis of the IgLC domain is not as informative as the IgLV (Fig. 2B), as IgLC clusters more with taxonomic group than isotype [14]. Although the alignment of IgLC domains (Supporting Information Fig. 2) used to generate Fig. 2B does not show obvious hallmarks of isotype, the resulting tree is still largely consistent in topology with that made from the V domains (Fig. 2A). We believe that our analysis is more definitive simply because we were able to include many more informative groups.
Expression and CDR3 repertoire
mRNA expression in nurse shark immune tissues was monitored by northern blotting with the IgLσC probe (Supporting Information Fig. 3). Highest expression was found in spleen, followed by the epigonal organ (the mammalian bone marrow equivalent), and peripheral blood leukocytes. This expression pattern is consistent with previous studies of shark IgH and IgL expression [15]. A high-molecular-weight band (>6 kb) was seen consistently in these experiments, but has resisted all attempts at cloning. This band may represent germline transcripts from unrearranged σ genes.
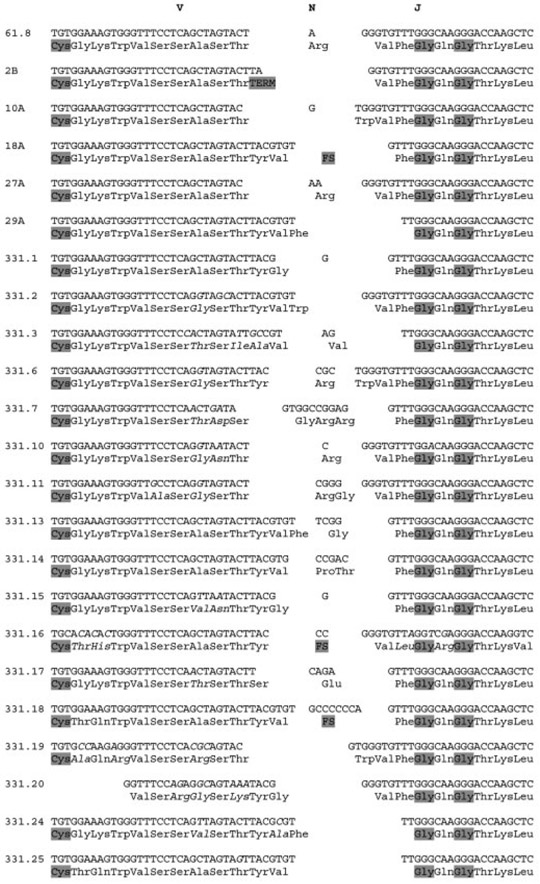
Spleen cDNA library screening and PCR provided examples of rearranged nurse shark IgLσ CDR3 (Fig. 3). Terminal deoxynucleotidyltransferase has been shown to have access to the V and J coding ends in shark L chains in a manner more consistent with H chains of other vertebrates [16], and the action of this enzyme in non-template nucleotide additions is evident in the clones from adult shark. Exonuclease activity is also suggested when the cDNA sequences in Fig. 3 are compared to the genomic sequence in S Supporting Information Fig. 4.
Figure 3.
Nurse shark IgLσ CDR3 alignment. The top six clones are from young animals, the rest from adult. Conserved cysteine of FR3 and GxG motif of FR4 are highlighted in grey, as are stop codons (TERM) and frameshifts (FS). Deviations from genomic thought due to somatic hypermutation are in italics along with any change in encoded amino acid.
Somatic hypermutation is obvious in the adult samples, at similar frequencies and often with the contiguous substitutions that have been described in other shark H and L chain isotypes [17]. It is also curious that both N region diversity and somatic hypermutation often result in arginines encoded in CDR3. This raises the possibility that Igσ could be selected to bind a particularly charged antigen moiety; paratopes with arginine have been implicated in anti-DNA-mediated autoimmunity [18]. Eleven arginine residues not encoded by V or J segments are found in just the 22 CDR3 reported here. By contrast, only four were found in 28 NS5 (σ-cart) clones. Additionally, the length of expressed σ CDR3 (mean of 8.5 amino acids with standard deviation of 0.87, calculated as exclusive number of codons between conserved cysteine of V and phenylalanine of J) seems more constrained than what was observed for nurse shark σ-cart [16].
RSS orientation supports the phylogenetic assignment
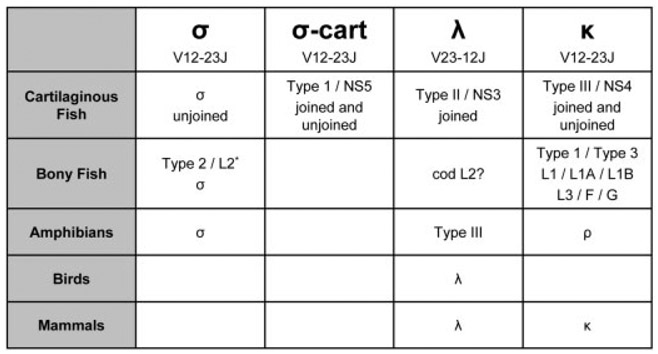
After amino acid identity, RSS orientation is the second most common characteristic used in distinguishing IgL isotypes. PCR of nurse shark genomic DNA with primers to the V and J segments of IgLσ revealed the RSS orientation of the germline gene (Supporting Information Fig. 4). Heptamer and nonamer are spaced by 12 nucleotides 3′ of the V segment and 23 nucleotides 5′ of the J segment, as is the case in mammalian κ [19, 20]. Fig. 4 summarizes the IgL nomenclature from different vertebrates including the RSS orientations that confirm the four clades proposed by the phylogenetic tree in Fig. 2a (X. tropicalis λ RSS orientation was obtained from the genome version 4.1).
Figure 4.
Isotype traits and phylogenetic prevalence. RSS orientation is given under isotype, and germline-joined status in cartilaginous fish is listed under old nomenclature.
Gene organization and number
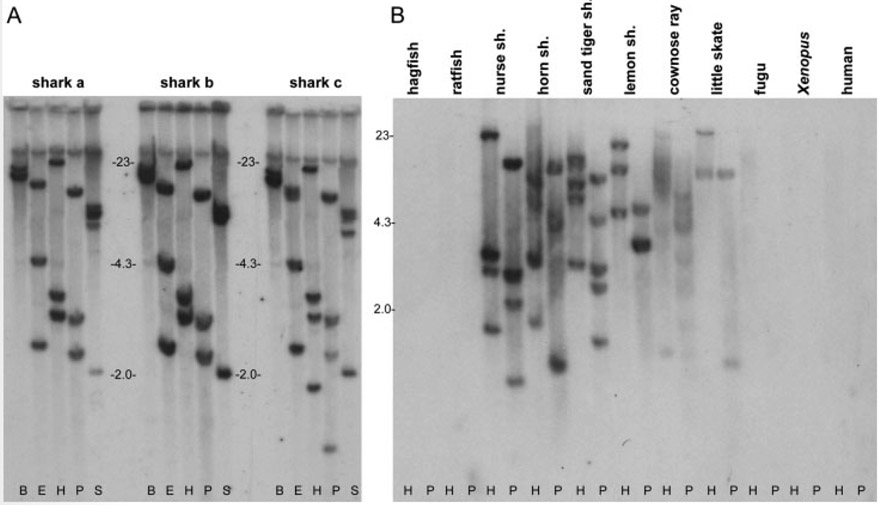
All cartilaginous fish Ig genes studied to date are in the cluster configuration, with each L chain cluster containing V, J, and C gene segments. A Southern blot of nurse shark gDNA from erythrocytes (still nucleated when mature in all non-mammalian vertebrates) was probed with IgLσC (Fig. 5A). Three bands in most digests suggest multiple loci, yet all cDNA sequence data suggest expression from just one (usually somatically mutated) locus. The similar number of bands detected with a Vσ probe (not shown), and the presence of V and J segments over a short stretch of DNA in the germline (Supporting Information Fig. 4), suggest that nurse shark σ genes are also in the cluster configuration.
Figure 5.
Genomic blotting. (A) Southern blot of genomic DNA from three nurse sharks probed with nurse shark IgLσC. Restriction endonucleases are marked at bottom: BamHI, EcoRI, HindIII, PstI, and SacI. (B) Zooblot of genomic DNA probed with nurse shark IgLσC. Markers are in kb.
Another unusual feature of shark Ig genes is the phenomenon of germline joining of V-J or V(D)J via RAG activity in germ cells [21]. In nurse shark some or all of the V-J segments are joined for each previously studied isotype. By contrast, as yet there is no evidence for a germline-joined IgLσ locus; there is no expression of an invariant rearrangement (Fig. 3) at the two developmental time points sampled.
A zooblot confirmed the presence of IgLσ in diverse chondrichthians (Fig. 5B), as suggested by the cloning in horn shark, dogfish and little skate. Under the high-stringency wash conditions used, the nurse shark probe still annealed to the batoid skate and ray, but did not recognize L2 from Fugu or Xenopus σ, or any ortholog in the holocephalian ratfish. Although the phylogenetic tree strongly supports the presence of the σ gene in Fugu and Xenopus, even orthologous immune genes rarely cross-hybridize over large phylogenetic distances. Judging by the number of hybridizing bands, two to four IgLσ loci are common in diverse elasmobranchs; for the other L chain isotypes the number of genes varies widely in different species.
The degeneracy of the genetic code provides information on the likelihood of a mutation in a particular codon to encode an amino acid replacement, sometimes referred to as “codon volatility”. For example, the arginine codon "CGA" is the least likely to mutate to another amino acid and thus has the lowest volatility score, whereas any substitution to the tryptophan and methionine codons will result in a change (thus they are most volatile). Such codon volatility in CDR of mammalian L chain isotypes has been studied by others with the hope of gaining functional insight [23].
We chose 20 IgL V sequences from a variety of isotypes and vertebrates to look for trends in codon volatility (Supporting Information Fig. 5). We confirmed the trend noted by Chang and Casali [22] of much higher CDR1 volatility vs. CDR2 in VH but not VL to be consistent in the sequences from additional VL isotypes and vertebrates that we surveyed. A more recent analysis has suggested that λ framework regions are less volatile than those of κ, allowing κ to be a more radical mutator [23]. While our analysis did not delve into conservative replacements and transition neighborhoods, we found no evidence for such a trend in codon use amongst any IgL isotypes or phylogenetic group. Our analysis implied that even VL from the same isotype and class can differ greatly in the volatility of particular frameworks and CDR. As a general rule for vertebrate L chains, we propose that the entire V domains are volatile and that we should look elsewhere for isotype functional distinction.
Discussion
Natural history of four ancient IgL clades
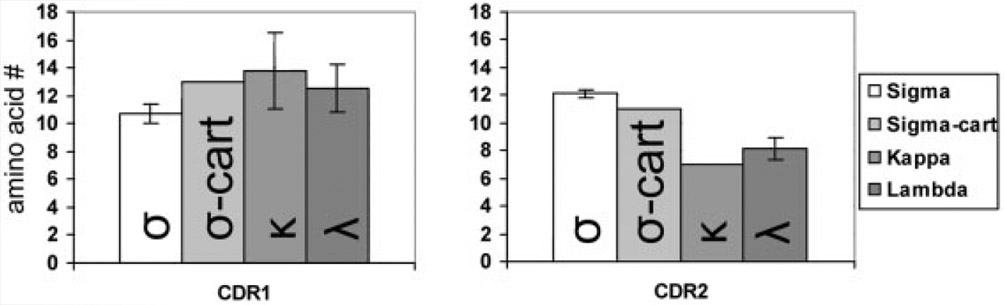
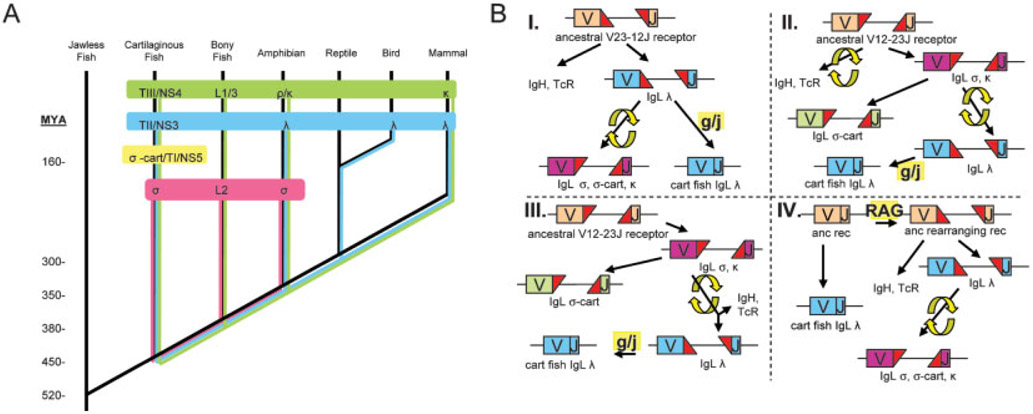
This serendipitous discovery in shark of a shark ortholog of amphibian σ has permitted a reclassification of all vertebrate L chains into four groups: σ, σ-cart, λ, and κ. As has been proposed previously [24], the L2 isotype from teleosts is also orthologous to amphibian σ, and now our work shows that the σ lineage can be traced back to the elasmobranchs. These data (and failed searches in birds and mammals) suggest that σ is the only antigen receptor chain common to, yet also restricted to, cold-blooded vertebrates. The CDR2 motif conservation, RSS orientation, and proximity in phylogenetic analyses support the close evolutionary relationship between σ and σ-cart; however, the tight clustering of the σ and σ-cart clades, as well as their distinctive CDR1 and CDR2 lengths (Fig. 6), also support separate designations for these two isotypes (Fig. 7).
Figure 6.
CDR length plot. Amino acid length of CDR1 and CDR2 of sequences in Fig. 1 are plotted by isotype. Error bars show standard deviation. Unpaired two-tailed p values for comparing σCDR1 vs. κCDR1, σCDR1 vs. λCDR1, σCDR2 vs. κCDR2, and σCDR2 vs. λCDR2 are 0.0011, 0.0046, <0.0001, and <0.0001, respectively.
Figure 7.
Summary of IgL isotype prevalence and models of IgL evolution. (A) Hypothesis of extent of IgL radiation from cartilaginous fish throughout other vertebrate groups. Colors for isotypes are magenta for κ, yellow for σ-cart, green for κ and blue for λ., and show their differential existence in different vertebrate groups since their origin in a cartilaginous fish ancestor. Approximate time of vertebrate class divergence in millions of years ago (MYA) is shown at left. (B) Four models of IgL evolution in the context of other antigen receptors where RSS swaps are presumed to be rare (evolutionarily informative) events, as shown by the two yellow arrows. Red triangles denote RSS orientation and yellow "g/j" box marks germline joining of RSS. In model IV, the yellow "RAG" box symbolizes an ancestral RAG transposon insertion event.
The 12-base-pair spacer RSS 3′ of Vκ has been thought to be an anomaly in antigen receptor genes as V segments of TCR, IgH, IgLλ, IgNAR, and NAR-TCR all have a 23-base-pair spacer at this position. However, all L chain genes, except those in the λ class, have this κ-type V12-23J arrangement (reviewed in [24]). The one exception is a cod "L2" gene (Genbank accession number GM0293808) that interestingly has higher V homology to λ, although no similar sequence has been found in other teleosts. Unfortunately, all type II/λ clusters in every cartilaginous fish species examined are V-J germline-joined, precluding RSS examination.
If the orientations of the RSS are indeed conserved indicators of isotype clades, a bigger question emerges in what they can tell us about the order of not only IgL isotype emergence but also other antigen receptors during natural history. It has been observed [25, 26] that RAG can act in the germline as a recombinase to "swap" the 12- and 23-spaced RSS between two segments in the formation of a "hybrid joint", first evidenced in shark IgH loci [27]. If such germline genomic inversion events are common, then the RSS reveal little of evolutionary relationships. But, if they are rare in the histories of these genes – as suggested by their now apparent conservation within each antigen receptor chain and isotype – they may be evolutionarily informative. If RSS swapping is exceptionally rare and RSS state is of great importance in evolutionary considerations, the most parsimonious schemes are shown in Fig. 7B.
One history (Fig. 7B, model I) has the λ isotype evolving from a common antigen receptor ancestor with a V23-12J orientation. IgL σ, σ-cart, and κ could have then evolved from λ also by hybrid joint RSS inversion. If, however, our phylogeny based on V amino acid sequence is correct (Fig. 2a), it seems more likely that λ descended from κ after an RSS swap (Fig. 7B, model II). This means that an additional RSS swap probably occurred early in the history of other antigen receptors, distinguishing them from the early IgL chains in their RSS orientations (until the λ reversion).
A similar model that only invokes one RSS swap (Fig. 7B, model III) has IgH and TCR bifurcating from the σ/κ-like IgL after the RSS flip along with λ, a model suggesting IgL (perhaps as membrane homodimers?) being ancestral to IgH and TCR. Possibly relevant to these considerations is the fact that λ is always germline-joined in cartilaginous fish, so we can add a germline/joining event in each model to yield the λ we now see in sharks. But we cannot rule out a model in which that joining of λ is not necessary. If the RAG transposon insertion event occurred in a multicopy λ-like locus, the present state of λ in cartilaginous fish could be a relic of the adaptive immune system pre-RAG (Fig. 7B, model IV). These hypotheses discount any difficulties of adding D segments for IgH, TCRβ and TCRδ and subsequent germline hybrid joint formation to get 23J orientations IgH, NAR and NAR-TCR. As genome projects in jawless and cartilaginous fish proceed, analyses of more antigen receptor loci may allow refining of these hypotheses. Or as mentioned above, if RSS swapping proves to be common, then it should not factor highly in such contemplations at all.
In addition to the clear separation of L chains into four ancestral clades, an order of evolutionary descent is suggested by the V domain tree (Fig. 2A). The bootstrap resampling technique was used to estimate statistical confidence values for each node of the tree. Randomly generated pseudosamples of the larger dataset are used to make trees by the same algorithm to test the null hypothesis (the inferred tree). The value at each node reflects the number of times out of 1000 resamples that the same bifurcation was supported by the random pseudosamples. Since all the major nodes between our proposed IgL clades were supported by at least 60% of the samples from this entire alignment (of V domains, including CDR1 and CDR2), we feel this is the most informative IgL tree generated to date.
From it we propose that (1) all four IgL isotypes arose in cartilaginous fish or earlier (in the placoderms?); (2) σ-cart is found only in elasmobranchs and thus is a "dead-end" isotype; (3) as mentioned, σ is found in all ectothermic vertebrates; and (4) only λ and κ are present in endothermic immune systems. Although directionality of evolution is not revealed in this unrooted tree, we use the topology of the tree to suggest the descent of the isotopes as ordered by the colored numbers in Fig. 2b. We look forward to further comparative studies of L chains in other primitive fishes, reptiles, monotremes, and marsupials to determine whether our propositions hold water.
Features of CDR1 and CDR2 fit the same four clades: A clue to functional distinctions?
Our trees (like those of others [7, 14]) suggest different selective pressures on the IgL V and C domains over evolutionary time, i.e. the V domains cluster according to isotype and the C domains group more according to taxonomy (although with the new sequences the C domains in our analysis also show isotype-specific clustering). We contend that the principal selection pressure is the distinct heterodimerization requirements of the two domains: the VL domain pairs with VH to bind antigen, while CL serves more of a structural role via associations with the IgHC1 domain. At a glance, even the alignments of the domains (Fig. 1 andSupporting Information Fig. 2) show that the four isotypes have signature features in the V, but not the C domains. The CDR1 of IgLσV is shorter than all other isotypes (chicken VLλ being the sole exception) and CDR2 is longer (extraordinarily so in trout). In fact, these two CDR display an evolutionarily conserved length for all four isotypes (Fig. 1, 6). While Vσ has the longest CDR2 but shortest CDR1, for Vκ the opposite is true. Additionally, CDR2 of Vσ has a conserved YGxG motif that is not often seen in either Vκ or Vλ CDR2.
If, as we suggest, the four L chain isotypes arose early in evolution and retained their signature features over hundreds of millions of years, there must be some functional basis for their emergence and maintenance. However, little evidence has been found for distinct functional roles of mammalian Igκ and Igλ (e.g. [28, 29]). Perhaps the difference in CDR1/2 lengths between Vσ and Vλ/κ is a useful way to begin to understand L chain function. Why are the lengths of the most diverse loops of "variable" domains so static across 400 million years of evolution? One possibility is that V regions in ancestral L chain genes evolved into isotypes that paired differently with VH, thus supporting distinct paratopes predominantly via the dissimilar CDR1 and CDR2 lengths. This could have occurred at a time when there was only a single H chain isotype, with little expansion of VH into the distinct families seen today (note, the cartilaginous fish IgM VH all belong to a single family, although there is diversity in CDR1/2 sequence between the genes [30, 31]). Studies of shark and mammalian predicted Igκ CDR conformations found similarities consistent with such a conservation of paratope support [32].
In Xenopus, Igσ was found to associate only with two of the three H chain isotypes (IgM and IgX), showing a preference of IgLσ for the two T cell-independent IgH isotypes expressed in the intestine [33]. Additionally, work in skate showed the largest disparity in the ratio of IgL isotypes (σ-cart and λ) expression in the intestine [34]. Common themes in both studies are differential use of the VL with shorter CDR1 and longer CDR2 (σ and σ-cart) compared to isotypes with the opposite trend in CDR lengths (λ and κ either in gut-associated lymphoid tissue or with IgH expressed in gut-associated lymphoid tissue. Interestingly in the monotreme platypus (representative of a more ancestral mammalian group), no Igσ has been described but Igλ V segments display exceptionally high CDR length diversity, perhaps in compensation for the loss of that diversity from Igσ [35].
Based on the preliminary data from diverse vertebrates, the existence of the four isotypes in sharks, and the IgM and IgX preference for Igσ in frog, we propose two hypotheses for how IgL isotypes evolved their distinct functions. In one, the four different isotypes provide different combinations of VL CDR1 and CDR2 length which influence the binding site topology, probably by specific support of VH CDR3; those VL with longer CDR2 such as σ associated with VH having short CDR3 [36]. This hypothesis would predict that VH genes expressed at different stages of development or in different tissues would show an L chain preference. Although crystal structures of σ and σ-cart containing Fab (with longer CDR2) do not exist, comparisons of VL CDR1 and VH CDR3 lengths in known mammalian λ- and κ-employing structures show a positive correlation (m=0.5) (Supporting Information Fig. 6). Secondly, IgL isotype C domains could be preferentially used with particular IgH isotypes. This could be regulated via IgL rearrangement and transcription through promoter and enhancer elements or through a more stochastic mechanism of prevailing best fit.
These two (VH CDR3 length and the IgH isotype) hypotheses are not mutually exclusive. For example, extant cartilaginous fish have two H chain isotypes that are expressed in lineages of B cells rather than by class switch [21, 27, 37-39]. Thus, both the V and C domains of the L chains may have coevolved with H chains to influence L chain evolution.
In summary, all vertebrate L chains can now be categorized into four clades with their origins in the cartilaginous fish or placoderms. This is supported by phylogenies based on the V domain, RSS orientations, and conserved features of the CDR1 and CDR2. The differences in sequence and structural characteristics between these isotypes could lead to the unraveling of their distinctive functions.
Materials and methods
Library and in silico screening
cDNA libraries were constructed from shark spleen RNA and screened as previously described [40]. Genomic library was made from nurse shark "yellow" in LambdaFix II (Stratagene, La Jolla, CA). Low-stringency hybridization conditions were 30% formamide washing to 2× SSC/0.1% SDS at 55°C and high-stringency hybridization conditions were 50% formamide washing to 0.2× SSC/0.1% SDS at 65°C. PCR was used to label all probes as previously described [41] and free nucleotides were removed with Quick-Spin Sephadex G-50 columns (Roche, Basel, Switzerland). Probes routinely labeled to 3×107 cpm. Primers used for the TCRγC probe were FLAJ1079 (5′-CAAACGTTCGGTCCGAAC-3′) and FLAJ1080 (5′-CAATAGACCACGATCTTCAC-3′). The IgLσC probe primers were FLAJNIK1089 (5′-CGTGACTGTGAGATCGTTGCGG-3′) and FLAJNIK1090 (5′-GACCCTTCAGTAAACCTGC-3′).
The C and V domains of nurse shark IgLσ clone 61.8 (Supporting Information Fig. 1) was used as bait in BLAST searches of expressed sequence tags databases maintained by NCBI. IgLσ orthologs found included entries CV222129 from little skate Leucoraja erinacea, CX662707 from the dogfish Squalus acanthias, and CK403931 from channel catfish Ictalurus punctatus. Similar searches of X. tropicalis genome project scaffolds with X. laevis IgLσ [4] found an orthologous locus on scaffold 289 of version 4.1.
PCR cloning
Genomic library clones positive for both V and C domain probes were used as template in PCR reactions to amplify the intergenic V-J region. Primers FLAJNIK1092 (5′-AGGAGCTCTGGTACRCCA-3′) and FLAJ1123 (5′-CTGCAACAAAGAGCTTGGTCCC-3′) were cycled 40 times annealing at 52°C. The ~550-base-pair product was cloned into pCR2.1 (Invitrogen, Carlsbad, CA) via TA cloning and sequenced. Clones 331.x were amplified from an adult nurse shark spleen/pancreas cDNA library made in pDONR222 with Cloneminer (Invitrogen). Primers FLAJNIK1092 and FLAJNIK1114 (5′-GCTGTCCCTTGTTGCACTGTGC-3′) were employed with 30 rounds of amplification annealing at 53°C to sample CDR3 diversity.
Phylogenetic analysis
Diverse V and C domains (amino acids) from various vertebrate IgL chains were aligned with ClustalW using default parameters and then manually adjusted. V segments were trimmed before phenylalanine of g strand if no split genomic sequence was available to determine C terminus of V segment. The PHYLIP suite was used for subsequent analysis. A distance matrix was created with PROTDIST and used to draw a phylogram with NEIGHBOR. Zebrafish NITR and catfish β2-microglobulin were used as outgroups for the V and C domains, respectively, but the trees are not rooted. CDR1 length was counted as marked in Fig. 1.
Northern and Southern blotting
Total RNA was prepared for Northern blotting as described [42], and 10 μg was loaded for each lane. The nurse shark nucleotide diphosphate kinase probe used as a loading control was amplified with primers FLAJNIK1056 (5′-AACAAGGAACGAACCTTC-3′) and FLAJNIK1057 (5′-TCACTCATAGATCCAGTC-3′).
Southern blot was performed on genomic DNA from erythrocytes of three nurse sharks digested with BamHI, EcoRI, HindIII, PstI, and SacI (Roche) as described [43]. Organisms in the zooblot are Eptatretus stouti (Pacific hagfish), Hydrolagus colliei (spotted ratfish), Ginglymostoma cirratum (nurse shark), Heterodontus francisci (horned shark), Odontaspis taurus (sand tiger shark), Negaprion brevirostris (lemon shark), Rhinoptera bonasus (cownose ray), Leucoraja erinacea (little skate), Fugu rubripes (Japanese pufferfish), X. laevis (African clawed frog), and Homo sapiens (human).
Supplementary Material
Acknowledgements:
We would like to thank Bryan Buckingham for technical help with cloning and figures, and Yuko Ohta and Lynn Rumfelt for construction of libraries. Thanks to Lars Pilström and Ellen Hsu for helpful discussions, and Louis Du Pasquier and David Nemazee for critiquing the manuscript. The DNA sequences reported here were submitted to GenBank and assigned accession numbers EF114759 through EF114783. This work was supported by a fellowship (AI56963) to M.F.C. and a grant (R01RR006603) to M.F.F. from the NIH.
Abbreviations:
- Igsf:
Ig superfamily
- RSS:
recombination signal sequences
Footnotes
Supporting information for this article is available at http://www.wiley-vch.de/contents/jc_2040/2007/37263_s.pdf
References
- 1.Hill RL, Delaney R, Fellows RE and Lebovitz HE, The evolutionary origins of the immunoglobulins. Proc. Natl. Acad. Sci. USA 1966. 56: 1762–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zezza DJ, Stewart SE and Steiner LA, Genes encoding Xenopus laevis Ig L chains. Implications for the evolution of kappa and lambda chains. J. Immunol 1992. 149: 3968–3977. [PubMed] [Google Scholar]
- 3.Haire RN, Ota T, Rast JP, Litman RT, Chan FY, Zon LI and Litman GW, A third Ig light chain gene isotype in Xenopus laevis consists of six distinct VL families and is related to mammalian lambda genes. J. Immunol 1996. 157: 1544–1550. [PubMed] [Google Scholar]
- 4.Schwager J, Burckert N, Schwager M and Wilson M, Evolution of immunoglobulin light chain genes: Analysis of Xenopus IgL isotypes and their contribution to antibody diversity. EMBO J. 1991. 10: 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bengten E, Wilson M, Miller N, Clem LW, Pilstrom L and Warr GW, Immunoglobulin isotypes: Structure, function, and genetics. Curr. Top. Microbiol. Immunol 2000. 248: 189–219. [DOI] [PubMed] [Google Scholar]
- 6.Hsu E and Criscitiello MF, Diverse immunoglobulin light chain organizations in fish retain potential to revise B cell receptor specificities. J. Immunol 2006. 177: 2452–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richards MH and Nelson JL, The evolution of vertebrate antigen receptors: A phylogenetic approach. Mol. Biol. Evol 2000. 17: 146–155. [DOI] [PubMed] [Google Scholar]
- 8.Dooley H and Flajnik MF, Antibody repertoire development in cartilaginous fish. Dev. Comp. Immunol 2006. 30: 43–56. [DOI] [PubMed] [Google Scholar]
- 9.Shamblott MJ and Litman GW, Complete nucleotide sequence of primitive vertebrate immunoglobulin light chain genes. Proc. Natl. Acad. Sci. USA 1989. 86: 4684–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lobb CJ, Olson MO and Clem LW, Immunoglobulin light chain classes in a teleost fish. J. Immunol 1984. 132: 1917–1923. [PubMed] [Google Scholar]
- 11.Ghaffari SH and Lobb CJ, Structure and genomic organization of a second class of immunoglobulin light chain genes in the channel catfish. J. Immunol 1997. 159: 250–258. [PubMed] [Google Scholar]
- 12.Hohman VS, Schluter SF and Marchalonis JJ, Complete sequence of a cDNA clone specifying sandbar shark immunoglobulin light chain: Gene organization and implications for the evolution of light chains. Proc. Natl. Acad. Sci. USA 1992. 89: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sitnikova T and Nei M, Evolution of immunoglobulin kappa chain variable region genes in vertebrates. Mol. Biol. Evol 1998. 15: 50–60. [DOI] [PubMed] [Google Scholar]
- 14.Rast JP, Anderson MK, Ota T, Litman RT, Margittai M, Shamblott MJ and Litman GW, Immunoglobulin light chain class multiplicity and alternative organizational forms in early vertebrate phylogeny. Immunogenetics 1994. 40: 83–99. [DOI] [PubMed] [Google Scholar]
- 15.Rumfelt LL, Diaz M, Lohr RL, Mochon E and Flajnik MF, Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J. Immunol 2004. 173: 1129–1139. [DOI] [PubMed] [Google Scholar]
- 16.Fleurant M, Changchien L, Chen CT, Flajnik MF and Hsu E, Shark Ig light chain junctions are as diverse as in heavy chains. J. Immunol 2004. 173: 5574–5582. [DOI] [PubMed] [Google Scholar]
- 17.Lee SS, Tranchina D, Ohta Y, Flajnik MF and Hsu E, Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity 2002. 16: 571–582. [DOI] [PubMed] [Google Scholar]
- 18.Doyle CM, Han J, Weigert MG and Prak ET, Consequences of receptor editing at the lambda locus: Multireactivity and light chain secretion. Proc. Natl. Acad. Sci. USA 2006. 103: 11264–11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakano H, Huppi K, Heinrich G and Tonegawa S, Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 1979. 280: 288–294. [DOI] [PubMed] [Google Scholar]
- 20.Tonegawa S, Somatic generation of antibody diversity. Nature 1983. 302: 575–581. [DOI] [PubMed] [Google Scholar]
- 21.Lee SS, Fitch D, Flajnik MF and Hsu E, Rearrangement of immunoglobulin genes in shark germ cells. J. Exp. Med 2000. 191: 1637–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang B and Casali P, The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol. Today 1994. 15: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hershberg U and Shlomchik MJ, Differences in potential for amino acid change after mutation reveals distinct strategies for kappa and lambda light-chain variation. Proc. Natl. Acad. Sci. USA 2006. 103: 15963–15968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilstrom L, Lundqvist ML and Wermenstam NE, The immunoglobulin light chain in poikilothermic vertebrates. Immunol. Rev 1998. 166: 123–132. [DOI] [PubMed] [Google Scholar]
- 25.Lewis SM and Wu GE, The old and the restless. J. Exp. Med 2000. 191: 1631–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis SM, Hesse JE, Mizuuchi K and Gellert M, Novel strand exchanges in V(D)J recombination. Cell 1988. 55: 1099–1107. [DOI] [PubMed] [Google Scholar]
- 27.Kokubu F, Litman R, Shamblott MJ, Hinds K and Litman GW, Diverse organization of immunoglobulin VH gene loci in a primitive vertebrate. EMBO J. 1988. 7: 3413–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sverremark E, Rietz C and Fernandez C, Kappa-deficient mice are nonresponders to dextran B512: Is this unresponsiveness due to specialization of the kappa and lambda Ig repertoires? Int. Immunol 2000. 12: 431–438. [DOI] [PubMed] [Google Scholar]
- 29.Stanfield RL, Zemla A, Wilson IA and Rupp B, Antibody elbow angles are influenced by their light chain class. J. Mol. Biol 2006. 357: 1566–1574. [DOI] [PubMed] [Google Scholar]
- 30.Rumfelt LL, Lohr RL, Dooley H and Flajnik MF, Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 2004. 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen SX, Bernstein RM, Schluter SF and Marchalonis JJ, Heavychain variable regions in carcharhine sharks: Development of a comprehensive model for the evolution of VH domains among the gnathanstomes. Immunol. Cell Biol 1996. 74: 357–364. [DOI] [PubMed] [Google Scholar]
- 32.Barre S, Greenberg AS, Flajnik MF and Chothia C, Structural conservation of hypervariable regions in immunoglobulins evolution. Nat. Struct. Biol 1994. 1: 915–920. [DOI] [PubMed] [Google Scholar]
- 33.Hsu E, Lefkovits I, Flajnik M and Du PL, Light chain heterogeneity in the amphibian Xenopus. Mol. Immunol 1991. 28: 985–994. [DOI] [PubMed] [Google Scholar]
- 34.Miracle AL, Anderson MK, Litman RT, Walsh CJ, Luer CA, Rothenberg EV and Litman GW, Complex expression patterns of lymphocyte-specific genes during the development of cartilaginous fish implicate unique lymphoid tissues in generating an immune repertoire. Int. Immunol 2001. 13: 567–580. [DOI] [PubMed] [Google Scholar]
- 35.Johansson J, Salazar JN, Aveskogh M, Munday B, Miller RD, and Hellman L, High variability in complementarity-determining regions compensates for a low number of V gene families in the lambda light chain locus of the platypus. Eur. J. Immunol 2005. 35: 3008–3019. [DOI] [PubMed] [Google Scholar]
- 36.Padlan EA, Anatomy of the antibody molecule. Mol. Immunol 1994. 31: 169–217. [DOI] [PubMed] [Google Scholar]
- 37.Dooley H and Flajnik MF, Shark immunity bites back: Affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur. J. Immunol 2005. 35: 936–945. [DOI] [PubMed] [Google Scholar]
- 38.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC and Flajnik MF, A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995. 374: 168–173. [DOI] [PubMed] [Google Scholar]
- 39.Hinds KR and Litman GW, Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature 1986. 320: 546–549. [DOI] [PubMed] [Google Scholar]
- 40.Rumfelt LL, Avila D, Diaz M, Bartl S, McKinney EC and Flajnik MF, A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc. Natl. Acad. Sci. USA 2001. 98: 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohta Y, Okamura K, McKinney EC, Bartl S, Hashimoto K and Flajnik MF, Primitive synteny of vertebrate major histocompatibility complex class I and class II genes. Proc. Natl. Acad. Sci. USA 2000. 97: 4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartl S, Baish MA, Flajnik MF and Ohta Y, Identification of class I genes in cartilaginous fish, the most ancient group of vertebrates displaying an adaptive immune response. J. Immunol 1997. 159: 6097–6104. [PubMed] [Google Scholar]
- 43.Greenberg AS, Steiner L, Kasahara M and Flajnik MF, Isolation of a shark immunoglobulin light chain cDNA clone encoding a protein resembling mammalian kappa light chains: Implications for the evolution of light chains. Proc. Natl. Acad. Sci. USA 1993. 90: 10603–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.