Abstract
Chromatin structure plays a decisive role in gene regulation through the actions of transcriptional activators, coactivators, and epigenetic machinery. These trans-acting factors contribute to gene expression through their interactions with chromatin structure. In yeast INO1 activation, transcriptional activators and coactivators have been defined through intense study but the mechanistic links within these trans-acting factors and their functional implications are not yet fully understood. In this study, we examined the crosstalk within transcriptional coactivators with regard to the implications of Snf2p acetylation during INO1 activation. Through various biochemical analysis, we demonstrated that both Snf2p and Ino80p chromatin remodelers accumulate at the INO1 promoter in the absence of Snf2p acetylation during induction. Furthermore, nucleosome density and histone acetylation patterns remained unaffected by Snf2p acetylation status. We also showed that cells experience increased sensitivity to copper toxicity when remodelers accumulate at the INO1 promoter due to the decreased CUP1 expression. Therefore, our data provide evidence for crosstalk within transcriptional co-activators during INO1 activation. In light of these findings, we propose a model in which acetylation-driven chromatin remodeler recycling allows for efficient regulation of genes that are dependent upon limited co-activators.
Introduction
Transcriptional activation is a highly-regulated process that involves the recruitment of various transcription activators and coactivators to a gene’s upstream regulatory region [1–2]. One critical step during the activation process is the participation of chromatin remodeling activities [3]. There are at least two main epigenetic mechanisms by which remodeling activities can influence gene activation. The first is the recruitment of histone modifying enzymes to regulatory regions, which can perform various modifications, such as phosphorylation or acetylation, on histones [4]. The second is the recruitment of chromatin remodelers to perform nucleosome repositioning [5]. As such, biological functions of these remodeling activities have been defined.
INO1 encodes for inositol-3-phosphate-synthase (I-3-P synthase), which converts glucose-6-phosphate (G-6-P) to inositol-3-phosphate (I-3-P). I-3-P is then dephosphorylated to form inositol by inositol monophosphatases, encoded by INM1 and INM2. This inositol, which is repressive to INO1, then leads to the synthesis of PI through the actions of PIS1, which codes for PI synthase [6–7]. As such, INO1 expression is responsible for the rate-limiting step of de novo phospholipid synthesis in yeast, and it has been studied intensively [8–11].
It has been demonstrated that the expression of INO1 is an Ino2p activator-dependent event [12–13]. Ino2p binds as an Ino2p/Ino4p heterodimer to the INO1 upstream activating sequence (UAS), and subsequently recruits coactivators, such as chromatin remodelers, Snf2p and Ino80p, as well as, histone acetylases, Esa1p and Gcn5p [8–11]. Chromatin remodelers, Snf2p and Ino80p, exhibit high occupancy levels before INO1 induction, and then dissociate from the promoter as INO1 induction commences [10]. Histone acetyltransferases (HATs), Gcn5p and Esa1p, on the other hand, exhibit the opposite pattern of recruitment in which they are present in low numbers at the INO1 promoter during repression, but greatly increase upon INO1 induction [11]. Therefore, the players that are involved in INO1 activation have been identified.
Although both Snf2p and Ino80p are key remodelers of INO1 expression [8–9], the interplay of these remodelers with other transcriptional coactivators present at the INO1 promoter has not been fully elucidated. Recently, it has been suggested that chromatin remodelers can be regulated by post-translational modifications to their domains or various subunits. These modifications can include phosphorylation of SWI/SNF or acetylation of SWI/SNF, RSC, and SWI families [14–19]. Such modifications suggest mechanistic links between HAT and chromatin remodelers and this may have an impact on the regulation of gene expression. For example, it has been shown that HAT Gcn5p is capable of acetylating chromatin remodeler Snf2p, and this can lead to the dissociation of the SWI/SNF complex from acetylated histones, and reduces its association with promoters in vivo [17]. The implications of this acetylation and subsequent dissociation, however, have yet to be explored with regard to transcriptional regulation and other possible mechanisms.
In this study, we have performed biochemical analyses to examine the implications of Snf2p acetylation during INO1 activation. We also examined whether or not Snf2p acetylation and occupancy changes influenced other gene activities, as many coactivators are involved in regulating large numbers of genes within cells and may need highly regulated recycling patterns. Our results showed that the accumulation of unacetylatable Snf2p at the INO1 promoter may have significantly impacted other Snf2p-regulated genes’ expression.
Results
Unacetylatable Snf2p leads to the accumulation of chromatin remodelers at the INO1 promoter
Transcriptional activation is a highly regulated process that requires the recruitment of an assortment of activators and coactivators to a gene’s upstream regulatory region. Previous studies have suggested that the post-translational modification of the chromatin remodeler Snf2p can lead to its dissociation from gene promoters [17]; however, this has yet to be studied for its biological implication on gene expression. To this end, we have chosen the INO1 system to understand the implications of Snf2p acetylation at the INO1 promoter. It has been shown that both chromatin remodelers Ino80p and Snf2p depart from INO1 promoter region once transcription is commenced [8, 11, 20]. Furthermore, both Esa1p and Gcn5p HATs were recruited right before the remodelers’ departure. These results suggest that the action of departure might result from the remodelers’ acetylation. To confirm the acetylation was indeed necessary for Snf2p dissociation, chromatin immunoprecipitation (ChIP) was performed using WT cells and an unAcSnf2p mutant in which the acetylatable lysine residues of the remodeler were replaced with unacetylatable arginine residues [17]. In both strains, it has been engineered so that Snf2p contained a C-terminal double-FLAG tag. A snf2Δ strain was utilized as a negative control.
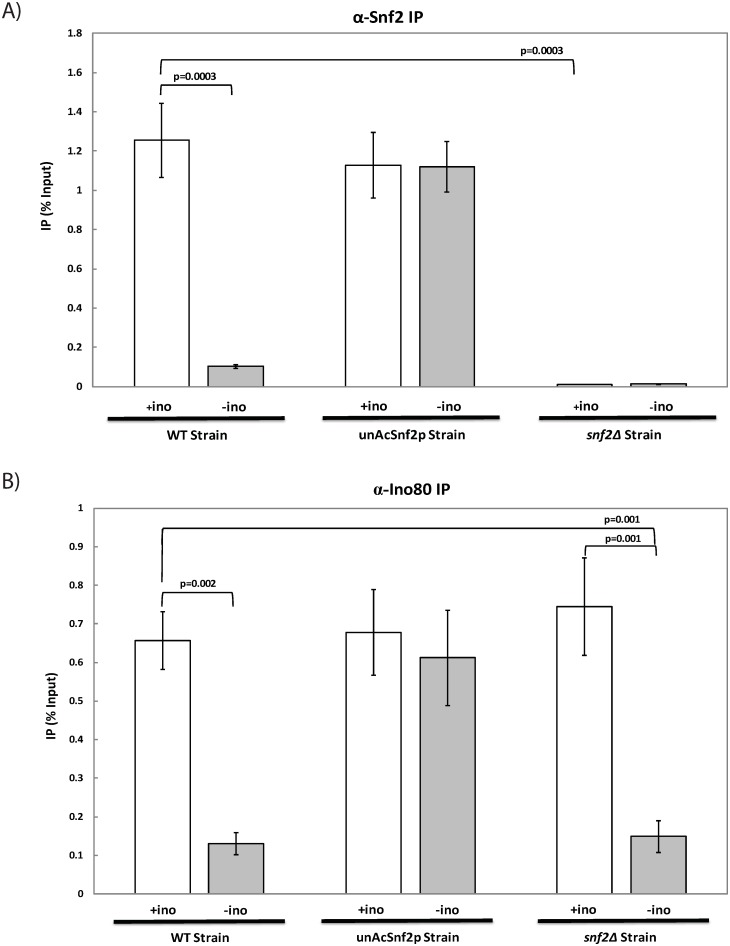
For repressing and inducing conditions, the IP signals of Snf2p-FLAG were examined at the upstream regulatory sequence (URS) of INO1 and were subsequently normalized to the INO1 input and mock. In the WT strain, the IP values demonstrated a significant difference (p<0.01), as the Snf2p-FLAG-IP were 1.26% ±0.19% under repressing conditions and 0.10% ±0.01% under inducing conditions, which demonstrated the dissociation of Snf2p from the promoter region (Fig 1A). This result is in agreement with previous findings [10]. On the other hand, IP values for the Snf2p-FLAG-IP in the unAcSnf2p mutant were 1.13% ±0.17% and 1.12% ±0.13% under repressing and inducing conditions, respectively (Fig 1A). The difference is insignificant (p = 0.996), and this observation indicated that unacetylated Snf2p does not depart from the INO1 promoter. The negative control, snf2Δ, demonstrated minimal IP values with no significant difference between repressing and inducing conditions, with 0.01% ±0.001% and 0.01% ±0.002% under repressing and inducing conditions, respectively. Therefore, our data demonstrate the accumulation of unacetylated Snf2p at the INO1 promoter upon INO1 induction in the absence of Snf2p acetylation, suggesting that Snf2p acetylation is critical to promoter dissociation.
Fig 1. Unacetylatable Snf2p leads to the accumulation of chromatin remodelers at the INO1 promoter instead of dissociating.
Real-time PCR analysis of DNA immunoprecipitated through Chromatin Immunoprecipitation (ChIP) with an antibody against (A) FLAG-tagged Snf2p (α-Snf2p) or (B) Ino80p (α-Ino80p) in WT cells, unAcSnf2p cells, and snf2Δ cells that were grown to mid-log phase (A600 = 1.0) in 10μM inositol Synthetic Complete (SC) media and were subsequently subjected to repressing or inducing conditions, 100 μM inositol (+ino; open bars) and 0 μM inositol (-ino; light grey bars) containing synthetic complete media, respectively, for 2 hours prior to collection. All experiments were performed with three repeats and all PCR reactions were done in at least triplicate. The IP for the INO1 promoter is graphed as mean ± standard deviation normalized to input and mock.
Previously, it has been shown that both remodelers, Ino80p and Snf2p, are required at the INO1 promoter to activate INO1 transcription [10–11], and that both remodelers accumulate in the absence of the histone acetyltransferases, Esa1p and Gcn5p [11]. Since Ino80p is required to recruit Snf2p to the INO1 promoter [10], it is interesting to examine how Snf2p accumulation can affect Ino80p at the INO1 promoter in the absence of Snf2p acetylation. To this end, ChIP coupled with qPCR was performed on WT, unAcSnf2p, and snf2Δ strains.
IP signals using an antibody against Arp8p (Arp8p-IP) of INO80 were examined at the INO1 URS under repressing and inducing conditions, respectively, and were subsequently normalized to INO1 input and INO1 mock DNA. In the WT strain, the IP values for the Arp8p-IP were 0.78±0.07 under repressing conditions and 0.17% ±0.03% upon induction of INO1, suggesting a significant dissociation of the Ino80p from the promoter upon INO1 induction (p<0.01) (Fig 1B). Similarly, the Arp8p-IP were 0.74% ±0.13% with repressing conditions and 0.15% ±0.04% under inducing conditions in the snf2Δ strain as Ino80p arrives at the INO1 prior to and independent of Snf2p [10]. In the unAcSnf2p mutant strain, on the other hand, the IP values for the Arp8p-IP in the repressed condition were statistically similar to the induced condition, 0.67% ±0.11% under repressing conditions and 0.61% ±0.12% under inducing conditions (Fig 1B). These results suggest the absence of Ino80p dissociation from the INO1 promoter when Snf2p cannot be acetylated. Therefore, both Snf2p and Ino80p accumulate at the INO1 promoter in the absence of Snf2p acetylation.
The acetylation of Snf2p is not required for INO1 activity
Since it was observed that Snf2p acetylation is required for Snf2p dissociation from the INO1 promoter, it was subsequently instructed to determine the implication of acetylation on INO1 expression through growth analysis. Since INO1 plays a rate-limiting role in the de novo synthesis of inositol in cells, cells that have impeded INO1 expression are expected to demonstrate a growth deficiency in media lacking inositol, whereas cells with fully functioning INO1 gene activity can successfully synthesize inositol that is absent from the environment and can thus still thrive in inositol depleted media.
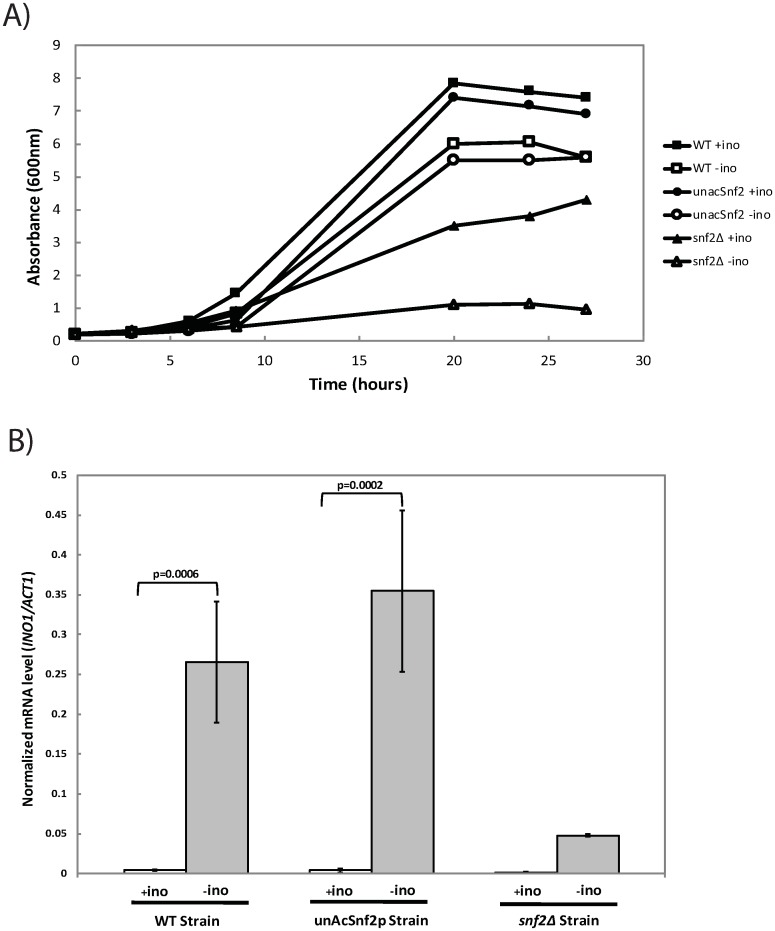
In media containing 100 μM inositol, repressing conditions, the WT type strain thrived and reached an average peak optical density A600nm of approximately 7.9±0.07. In inositol depleted media, inducing conditions, the WT cells were still able to survive, but only reached an average peak A600nm of approximately 6.0±0.07 (Fig 2A). The generation times are 274.29 min/generation and 292.68 min/generation for the WT under repressing and inducing conditions, respectively. The unAcSnf2p strain demonstrated a growth pattern similar to the WT in both the INO1 repressing and inducing conditions (p = 0.3 and p = 0.14, respectively), with an average peak A600nm around 7.5±0.14 in the presence of inositol and around 5.6±0.00 in the absence of inositol. Here, the generation times are 279.07 and 301.36 min/generation for the unAcSnf2p strain under repressing and inducing conditions, respectively. The snf2Δ strain, however, demonstrated a significantly different growth pattern compared to the unAcSnf2p strain (p<0.01 for repressing and inducing conditions), as the average peak A600nm in 100μM inositol was around 5.0±0.07, but in 0μM inositol it was only around 1.3±0.07. Taken together, these results demonstrated that the unAcSnf2p strain exhibited growth patterns very similar to the WT strain rather than the snf2Δ strain, suggesting that Snf2p acetylation is not required for cell growth in the absence of inositol. This implies that INO1 expression is not influenced by the unacetylatable Snf2p.
Fig 2. The INO1 induction is indepedent of Snf2p acetylation.
(A) Growth of yeast cells in the presence or absence of 100 μM myo-inositol. WT cell: (■) SC + inositol, (▯) SC—inositol. Mutant UnAcSnf2p cell: (●) SC + inositol, (○) SC—inositol. Mutant snf2Δ cell: (▲) SC + inositol, (Δ) SC—inositol. Cells were grown in SC media and were subsequently washed and diluted to an A600nm of 0.2 in repressing or inducing conditions, 100μM inositol (+ino) and 0μM inositol (-ino) synthetic complete media, respectively. (B) INO1 mRNA was examined by qRT-PCR analysis. Cells were grown in SC media to mid-log phase (A600 = 1.0) and were subsequently washed and resuspended in repressing (+ino; open bars) or inducing (-ino; light grey bars) conditions, respectively. Cells were grown for 2 hours prior to collection. The expression ratio for INO1 mRNA/ACT11 mRNA is graphed as mean standard ± deviation. All experiments were repeated at least three times, and in each experiment PCR reactions were done in triplicate.
To further identify that INO1 expression is independent of the Snf2p acetylation, qRT-PCR analysis was employed. A significant difference in INO1 mRNA levels between repressing and inducing conditions, 0.005±0.001 and 0.266±0.076, under repressing and inducing conditions, respectively; p<0.01, was observed in the WT strain, as well as in the unAcSnf2p strain, 0.004±0.003 and 0.355±0.101, under repressing and inducing conditions, respectively; p<0.01. On the other hand, no significant differences of INO1 expression were observed in the snf2Δ strain (Fig 2B). This demonstrated that the INO1 expression patterns in the unacetylatable strain were comparable to those observed in the WT strain rather than the deletion strain, as both strains demonstrated significant upregulation of INO1 upon induction. Taken together with the growth analyses, the mRNA analyses confirmed that the acetylation of Snf2p was not critical for INO1 activation.
Nucleosome density, histone acetylation patterns and transcription machinery at INO1 promoter are not influenced by the unacetylation of Snf2p
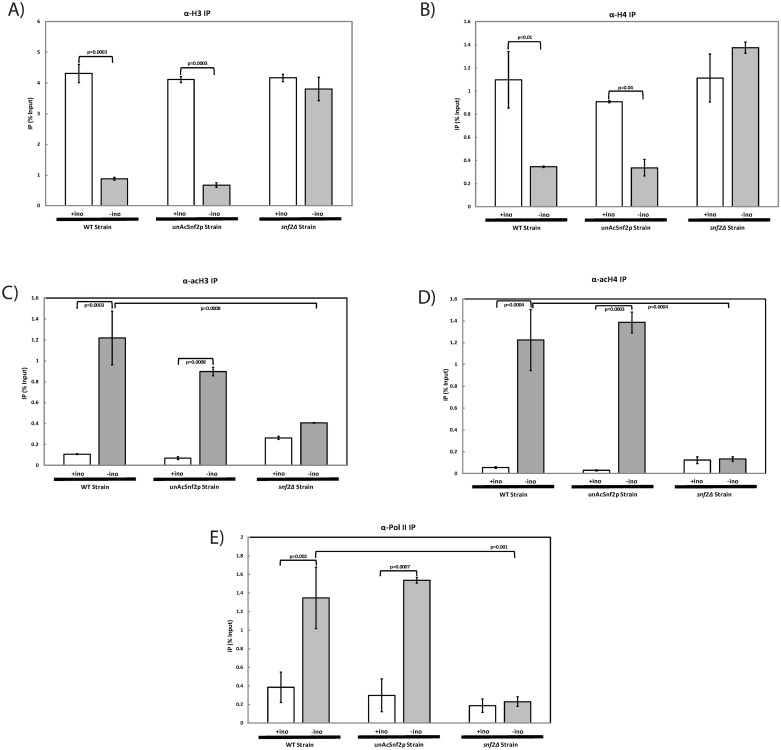
Since remodelers Snf2p and Ino80p accumulated at the INO1 promoter and INO1 expression is not influenced by the Snf2p unacetylation, it is interesting to see if remodeling activity is affected under the same condition. In order to examine nucleosome density, ChIP coupled with qPCR was performed in which antibodies targeted histones H3 and H4. In the WT strain, the H3-IP values demonstrated a significant difference in nucleosome density upon induction of INO1 (p<0.01), which is expected as chromatin remodelers are modifying the chromatin structure. Under these same conditions, the unAcSnf2p mutant demonstrated a similar nucleosome density pattern as the wild type strain, with a significant difference in H3 nucleosome density upon INO1 induction (p<0.01), which suggested that the chromatin remodeling activities at the INO1 promoter were not affected by Snf2p unacetylation (Fig 3A). Similar results were also observed with regard to histone H4 (p = 0.01; Fig 3B). These results suggest that chromatin structure is still subject to remodeling activities in the absence of Snf2p acetylation.
Fig 3. Chromatin remodeling activities and the recruitment of transcription machinery at INO1 promoter are independent of Snf2p acetylation.
Real-time PCR analysis of DNA immunoprecipitated through ChIP with an antibody against (A) Histone H3 (α-H3); (B) Histone H4 (α-H4); (C) Acetylated Histone H3 (α-acH3); (D) Acetylated Histone H4 (α-acH4); (E) RNA polymerase II (α-Pol II) at the INO1 promoter for WT cells, unAcSnf2p, and snf2Δ cells that were grown to mid-log phase (A600 = 1.0) in SC and were subsequently washed and subjected to repressing (+ino; open bars) or inducing (-ino; light grey bars) conditions, respectively, for 2 hours prior to collection. All experiments were performed with three repeats and all PCR reactions were done in at least triplicate. The IP for the INO1 promoter is graphed as mean ± standard deviation normalized to input and mock.
To evaluate histone acetylation, ChIP coupled with qPCR was performed using antibodies against acetylated H3 (acH3) and acetylated H4 (acH4). In the WT strain, the acH3-IP values were 0.11% ±0.005% and 1.22% ±0.26% under repressing and inducing conditions, respectively (Fig 3C). This demonstrated a significant increase in acetylated H3 upon induction (p<0.01). A similar pattern was also observed for the unAcSnf2p mutant in which the acH3-IP values were 0.07% ±0.01% and 0.90% ±0.04% under repressing and inducing conditions, respectively. On the contrary, the snf2Δ strain demonstrated no significant difference between repressing and inducing conditions (Fig 3C). Acetylated H4 also demonstrated a similar pattern in both the WT strain and the unAcSnf2p strain. Under WT repressing and inducing conditions, the acH4-IP values were 0.06% ±0.01% and 1.22% ±0.28%, respectively, which demonstrated a significant increase of H4 acetylation during induction conditions. In the unAcSnf2p mutant strain, a significant increase upon induction was also observed with acH4-IP values of 0.03% ±0.004% under repressing conditions and 1.39% ±0.10% under inducing conditions, whereas the snf2Δ demonstrated no significant difference (Fig 3D). These results suggest that even though chromatin remodelers are accumulated at the INO1 promoter in unAcSnf2p cells, the acetylation of histones in this region is still maintained.
We next wanted to examine RNA polymerase recruitment to the INO1 promoter through IP using an antibody directed against RNA Pol II (Pol II). In the WT strain, Pol II-IP had IP values of 0.39% ±0.16% and 1.35% ±0.33% under repressing and inducing conditions, respectively. This showed a significant increase of RNA pol II recruitment during induction (p<0.01) (Fig 3E). A similar pattern was observed in the unAcSnf2p mutant strain, with IP values of 0.30% ±0.18% and 1.54% ±0.03% under repressing and inducing conditions, respectively, whereas the snf2Δ mutant exhibited no significant difference between repressing and inducing conditions. Taken together, these results confirmed that the recruitment of the RNA polymerase II to the INO1 promoter was not dependent upon the acetylation of Snf2p. Furthermore, this validates the INO1 expression results that INO1 was expressed in unAcSnf2p mutant cells under inducing conditions (Fig 2B).
INO1 expression, nucleosome density, recruitment of remodelers, and remodeling activities are independent of Snf2p acetylation when switching from inducing conditions to repressing conditions
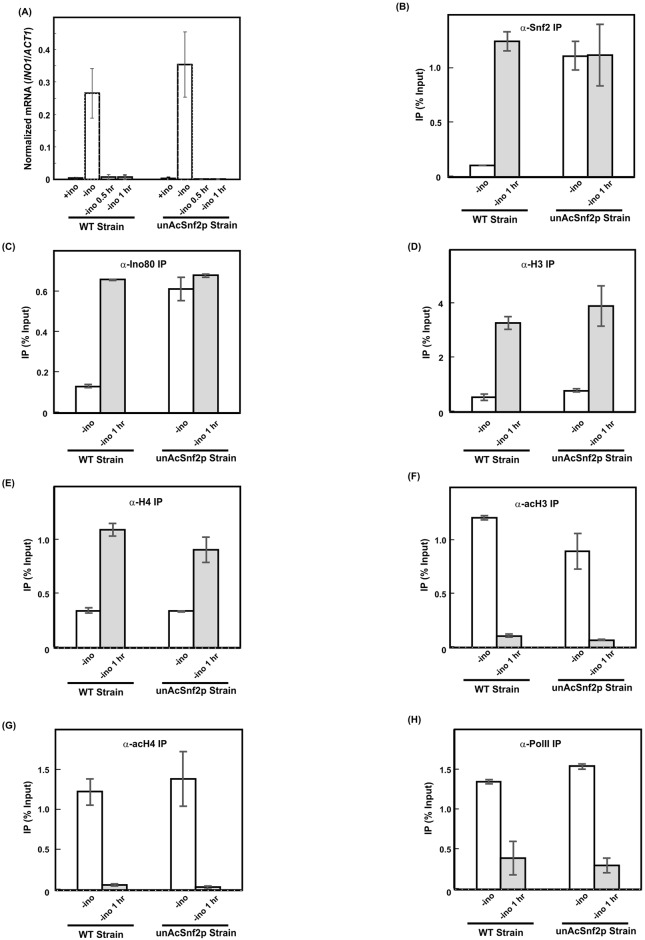
Since it was observed that the loss of Snf2p acetylation primarily affects the release of chromatin remodelers and that INO1 expression is not influenced in the absence of Snf2p acetylation, we are interested in examining the impact if INO1 is induced first, then subsequently exposed to repressing conditions. From the qRT-PCR analysis, INO1 mRNA levels were high under inducing conditions for both WT strain and the unAcSnf2p strain (Fig 4A). However, INO1 expression was downregulated for both WT and unAcSnf2p cells if the inducing conditions was employed for 30 min or 1 hr followed by the introduction of 1hr 30 min or 1 hr repressing conditions, respectively. This demonstrated that the INO1 expression patterns in the unacetylatable strain were comparable to those observed in the WT strain, as both strains demonstrated significant upregulation of INO1 upon induction and significant downregulation of INO1 upon repression. Taken together, the mRNA analyses showed that the acetylation of Snf2p was not critical for INO1 expression upon switching between INO1 induction and repression switch.
Fig 4. The INO1 induction and chromatin remodeling activities are indepedent of Snf2p acetylation when switching from inducing conditions to repressing conditions.
(A) INO1 mRNA was examined by qRT-PCR analysis. Cells were grown in SC media to mid-log phase (A600 = 1.0) and were subsequently washed and resuspended in inducing conditions for 30 min or 1hr, respectively. Subsequently, cells were switched to repressing conditions for another 1 hr 30 min or 1 hr prior to collection. The expression ratio for INO1 mRNA/ACT11 mRNA is graphed as mean standard ± deviation. All experiments were repeated at least three times, and in each experiment PCR reactions were done in triplicate. (B-H) Real-time PCR analysis of DNA immunoprecipitated through Chromatin Immunoprecipitation (ChIP) with an antibody against (B) FLAG-tagged Snf2p (α-Snf2p), (C) Ino80p (α-Ino80p), (D) Histone H3 (α-H3); (E) Histone H4 (α-H4); (F) Acetylated Histone H3 (α-acH3); (G) Acetylated Histone H4 (α-acH4); (H) RNA polymerase II (α-Pol II) at the INO1 promoter for WT cells and unAcSnf2p cells that were grown to mid-log phase (A600 = 1.0) in SC and were subsequently washed and subjected to inducing (-ino; open bars) or switching (1 hr inducing followed by 1 hr repressing conditions) (-ino 1hr; light grey bars) conditions, respectively, for total 2 hours incubation prior to collection. All experiments were performed with three repeats and all PCR reactions were done in at least twice. The IP for the INO1 promoter is graphed as mean ± standard deviation normalized to input and mock.
Next, we wanted to examine the recruitment patterns of Snf2p and Ino80p. For inducing conditions, the IP signal of Snf2p-FLAG of WT strain was very low (Fig 4B). Upon the introduction of the repressing conditions after 1 hr induction, the IP values demonstrated a significant increase from 0.10%±0.002% to 1.26% ±0.09% (p = 0.006). On the other hand, the IP value for the Snf2p-FLAG-IP in the unAcSnf2p mutant was 1.12% ±0.14% under inducing conditions (Fig 4B). Upon switching the inducing conditions to repressing conditions, the IP value was 1.13% ±0.28%. The difference is insignificant, and this observation indicated that unacetylated Snf2p does not depart from the INO1 promoter under inducing conditions. Upon switching the conditions to repressing conditions, large amounts of unacetylated Snf2p were at the INO1 promoter. Since Ino80p also accumulated at INO1 promoter when Snf2p was unacetylated (Fig 1B), we also examined the presence of Ino80p during the switch process. In the WT strain, the IP values for the Arp8p-IP were 0.13%±0.01% under inducing conditions and 0.66% ±0.003% upon switching to repression of INO1, suggesting a significant dissociation of the Ino80p from the promoter upon INO1 induction and accumulation of Ino80p upon repression (p = 0004) (Fig 4C). In the unAcSnf2p mutant strain, on the other hand, the IP value for the Arp8p-IP in the inducing conditions was 0.61% ±0.06% (Fig 4C). Upon switching the inducing conditions to repressing conditions, the IP value was 0.68% ±0.01%. The difference is insignificant. Our results demonstrated that both Snf2p and Ino80p accumulate at the INO1 promoter in the absence of Snf2p acetylation even if we introduced inducing conditions followed by the repressing conditions.
For nucleosome density, the H3-IP value of WT strain demonstrated very low nucleosome density which is 0.52%±0.12% upon induction of INO1 (Fig 4D). Upon switching the inducing conditions to repressing conditions, the H3-IP value increased to 3.29%±0.25% (p = 0.01). Under these same conditions, the unAcSnf2p mutant demonstrated a similar nucleosome density pattern (from 0.79%±0.06% to 3.91%±0.74%; p = 0.05) as the wild type strain, which suggested that the chromatin remodeling activities at the INO1 promoter were not affected by Snf2p unacetylation. Similar results were also observed with regard to histone H4 (Fig 4E). Therefore, chromatin structure is subject to remodeling activities and is independent of Snf2p acetylation. Furthermore, we found that the INO1 chromatin structure is independent of the initial conditions. When the conditions were switched from induction to repression, the chromatin structures of both WT and unAcSnf2p strains were similar to the chromatin structure at the repressing conditions (Figs 3A, 3B, 4D and 4E).
For the histone acetylation, the acH3-IP value of the WT strain was 1.22% ±0.02% under inducing conditions (Fig 4F). Upon switching the conditions from induction to repression, the acH3-IP value was 0.11% ±0.01% (p = 0.0005). This demonstrated a significant decrease in acetylated H3 when the condition was switched to repression. A similar pattern was also observed for the unAcSnf2p mutant in which the acH3-IP values were 0.90% ±0.17% and 0.07% ±0.01% under inducing and switching to repressing conditions, respectively (p = 0.04) (Fig 4F). Acetylated H4 also demonstrated a similar pattern in both the WT strain and the unAcSnf2p strain. Under WT inducing and switching to repressing conditions, the acH4-IP values were 1.22% ±0.17% and 0.06% ±0.01% (p = 0.02), respectively, which demonstrated a significant decrease of H4 acetylation when switched to repressing conditions. In the unAcSnf2p mutant strain, a significant decrease upon switching to repressing conditions was also observed with acH4-IP values of 1.39% ±0.34% under inducing conditions and 0.03% ±0.016% when switched to repressing conditions (p = 0.05) (Fig 4G). These results suggest that even though chromatin remodelers are accumulated at the INO1 promoter in the unAcSnf2p strain, histone acetylation in this region is similar to WT strain.
We next examined RNA polymerase recruitment to the INO1 promoter. In the WT strain, Pol II-IP had IP values of 1.35% ±0.02% and 0.39% ±0.21% under inducing and switching from inducing to repressing conditions, respectively (p = 0.05) (Fig 4H). A similar pattern was observed in the unAcSnf2p mutant strain, with IP values of 1.54% ±0.03% and 0.30% ±0.10% under inducing conditions and switching to repressing conditions, respectively (p = 0.007). These results confirmed that the recruitment of the RNA polymerase II to the INO1 promoter was also independent of the acetylation of Snf2p. Taken together, our results showed that the recruitment of chromatin remodelers, chromatin structure, remodeling activities and the transcription machinery are all similar between WT strain and unAcSnf2p strain when switched from inducing to repressing conditions, suggesting that they all are independent of Snf2p acetylation during the transition period.
Increased copper sensitivity when Ino80p and Snf2p accumulate at the INO1 promoter
We have shown that INO1 expression and the recruitment of chromatin remodelers are not influenced in the absence of Snf2p acetylation. Furthermore, we also observed the accumulation of both remodelers at the INO1 promoter. However, those chromatin remodelers might be required for the expression of other genes within yeast cells. Therefore, the accumulation of Ino80p and Snf2p at the INO1 promoter might impact their availability for other genes.
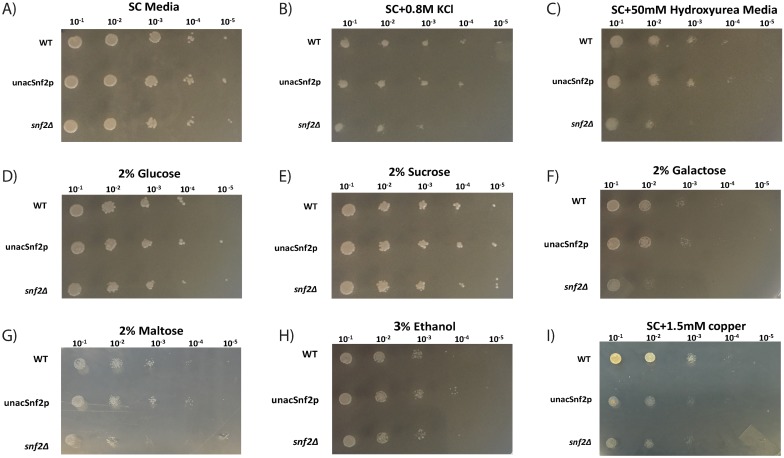
To demonstrate whether this limited availability of remodelers in unAcSnf2p cells can influence their regulated pathways, plate sensitivity assays were performed targeting osmoregulation [21], DNA damage [22], carbon source usage [23], and copper toxicity [20]. WT, unAcSnf2p, and snf2Δ strains were first plated on SC media to demonstrate cell viability. All strains demonstrated viable growth at the five dilutions used (Fig 5A; stock cell concentration 1.6x109 cells per ml diluted 10−1 through 10−5). These same strains were then analyzed under high osmolarity conditions. Osmoregulation is a common stress pathway that is highly regulated in yeast and can easily be tested for dysregulation via plate sensitivity assays, in which yeast growth is screened in the presence of high salt concentrations [24]. In the presence of high salt (0.8 M KCl), the WT strain was still visible at the greatest dilution (Fig 5B). The unAcSnf2p strain demonstrated growth comparable to the WT at most dilutions, but was not observed at the greatest dilution of 10−5. Lastly, the snf2Δ exhibited significantly diminished growth compared to the other strains, as it had very weak growth at the 10−3 dilution and no growth at the 10−4 or 10−5 dilutions. As the unAcSnf2p strain demonstrated growth more comparable to the WT versus the deletion strain, the sensitivity of the cells in the presence of osmotic stress did not appear to be significantly altered in the absence of Snf2p acetylation.
Fig 5. Increased copper sensitivity when Snf2p is unacetylatable.
WT, unAcSnf2p, and snf2Δ cellular growth in (A) SC media, (B) SC with 0.8M KCl, (C) SC with 50mM Hydroxyurea (D) 2% Glucose, (E) 2% Sucrose, (F) 2% Galactose, (G) 2% Maltose, (H) 3% Ethanol, and (I) 1.5mM copper. All cells were grown overnight, and then diluted to a starting optical density of 2.0 at 600nm absorbance as a stock solution (approximately 1.6x109 cells per ml). Five serial dilutions were plated for each strain (from left to right 10−1, 10−2, 10−3, 10−4, and 10−5). Experiments were repeated in triplicate.
Similar results were observed with regard to DNA damage sensitivity in which all strains were plated in the presence of 50 mM hydroxyurea. In the presence of hydroxyurea, the WT strain and the unAcSnf2p strain both demonstrated similar growth with each being observed through the 10−4 dilution, whereas the snf2Δ strain was unable to viably grow beyond the 10−2 dilution (Fig 5C). As with the osmotic results, the DNA damage sensitivity plates demonstrated that the lack of Snf2p acetylation did not hinder DNA damage repair.
Another highly regulated process in yeast cells, which is known to be associated with SWI/SNF family remodelers, is the utilization of various carbon sources in the form of fermentable substrates, such as glucose, sucrose, galactose, and maltose, as well as non-fermentable carbon sources, such as ethanol [23–25, 26]. In snf2Δ strains, for example, this regulatory mechanism is known to be disrupted so cells grow slower in sucrose media, and fail to grow in galactose [25]. To determine if the availability of Snf2p affects the utilization of alternate carbon sources, synthetic complete plates with 2% glucose, 2% sucrose, 2% galactose, 2% maltose, or 3% ethanol were utilized. We observed that the unAcSnf2p strain survived all conditions, including the fermentable carbon sources, such as 2% glucose, 2% sucrose, 2% galactose, and 2% maltose, as well as the non-fermentable carbon source of 3% ethanol (Fig 5D, 5E, 5F, 5G and 5H). In fact, on each plate, the unAcSnf2p mutant grew comparable to the WT. These results suggest that the accumulation of both Ino80p and Snf2p at the INO1 promoter does not affect the utilization of alternate carbon sources.
Recent research has demonstrated that the chromatin remodelers, Snf2p and Ino80p are also required at the CUP1 promoter [20]. Thus, to determine if Snf2p and Ino80p accumulation at the INO1 promoter is detrimental to copper resistance, all strains were plated on synthetic complete plates containing 1.5 mM. In the presence of 1.5 mM copper, WT cells demonstrated strong viability until the 10−4 dilution, whereas the unAcSnf2p strain demonstrated significant sensitivity, with very weak growth even at the 10−1 dilution, comparable to the snf2Δ strain (Fig 5I). This suggests that Snf2p and Ino80p accumulation at the INO1 promoter has dramatic influence on yeast cells’ resistance to copper.
The increased copper sensitivity resulted from the decreased CUP1 expression
To further examine the detail of increased copper sensitivity for unAcSnf2p strain, WT cells, unAcSnf2p, and snf2Δ cells were grown in the presence or absence of copper under un-induced (100 μM inositol) and induced conditions (0 μM inositol) for INO1.
In 0 μM inositol, all three strains were able to grow in the absence of copper as expected, although snf2Δ cells exhibited a significantly reduced growth pattern compared to when inositol was present. As expected, there was no significant difference between WT and unAcSnf2p growth in these circumstances when no toxic copper was present. In the presence of 1.5 mM copper and 0 μM inositol, however, the WT cells were once again able to survive, while the unAcSnf2p cells were now sensitive to the copper, comparable to snf2Δ cells (Fig 6A). The unAcSnf2p cells only reached an A600nm of 0.59 after 24 hours. This is when chromatin remodelers have dissociated from the INO1 promoter in WT cells but remain accumulated in unAcSnf2p cells. This suggests that copper toxicity defense is hindered in unAcSnf2p cells under INO1 inducing conditions when chromatin remodelers are accumulated at the INO1 promoter.
Fig 6. Snf2p acetylation is required for copper toxicity survival.
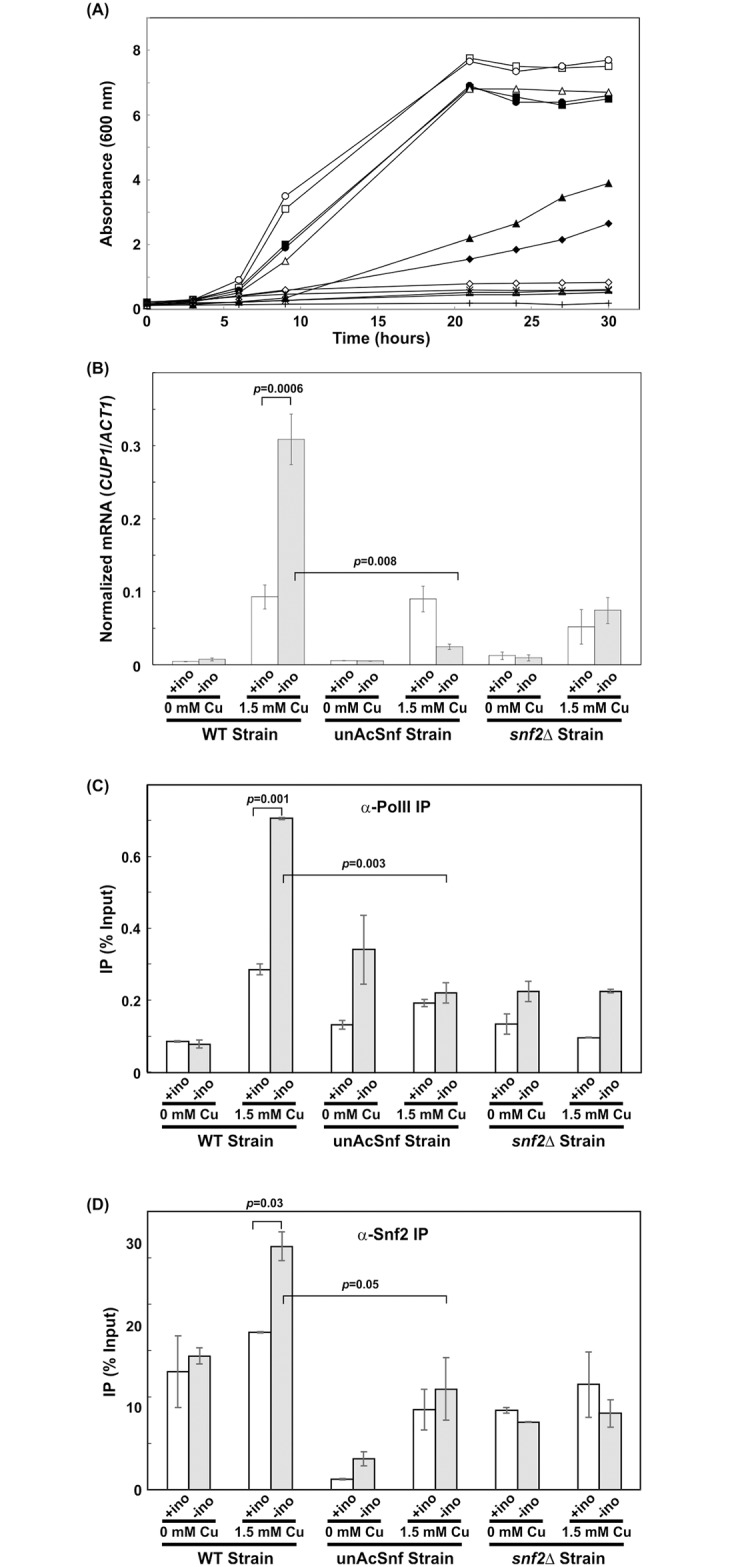
(A) Growth of yeast cells in the presence or absence of 100 μM myo-inositol with or without 1.5 mM copper. WT cell: (■) SC—inositol and 0 mM Cu2+, (▯) SC + inositol and 0 mM Cu2+, (◆) SC—inositol and 1.5 mM Cu2+, (◇) SC + inositol and 1.5 mM Cu2+. Mutant UnAcSnf2p cell: (●) SC—inositol and 0 mM Cu2+, (○) SC + inositol and 0 mM Cu2+, (*) SC—inositol and 1.5 mM Cu2+, (x) SC + inositol and 1.5 mM Cu2+. Mutant snf2Δ cell: (▲) SC—inositol and 0 mM Cu2+, (Δ) SC + inositol and 0 mM Cu2+, (+) SC—inositol and 1.5 mM Cu2+, (-) SC + inositol and 1.5 mM Cu2+. Cells were grown in SC media and were subsequently washed and diluted to an A600nm of 0.2 in repressing or inducing conditions, 100μM inositol (+ino) and 0μM inositol (-ino) synthetic complete media, respectively, with or without 1.5 mM copper. (B) INO1 mRNA was examined by qRT-PCR analysis. Cells were grown in SC media to mid-log phase (A600 = 1.0) and were subsequently washed and subjected to INO1 repressing (100 μM inositol) or inducing conditions (0 μM inositol), as well as CUP1 repressing (0 mM copper) and inducing conditions (1.5 mM copper), respectively. Cells were grown for 2 hours prior to collection. The expression ratio for INO1 mRNA/ACT11 mRNA is graphed as mean standard ± deviation. All experiments were repeated at least three times, and in each experiment PCR reactions were done in triplicate. (C-D) Real-time PCR analysis of DNA immunoprecipitated through ChIP with an antibody against (C) RNA polymerase II (α-Pol II) and (D) FLAG-tagged Snf2p (α-Snf2p) at the CUP1 TATA or UAS promoter, respectively, for WT cells, unAcSnf2p, and snf2Δ cells that were grown to mid-log phase (A600 = 1.0) in SC, and were subsequently washed and subjected to INO1 repressing (100 μM inositol) or inducing conditions (0 μM inositol), as well as CUP1 repressing (0 mM copper) and inducing conditions (1.5 mM copper), respectively. Cells were grown for 2 hours prior to collection. All experiments were performed with two repeats and all PCR reactions were done in at least triplicate. The IP for the CUP1 TATA or UAS promoter is graphed as mean ± standard deviation normalized to input and mock.
We then examined cell growth in 100 μM inositol, where cells are known to have Snf2p highly recruited and accumulated at the INO1 promoter [11]. In the absence of toxic copper, all three strains, including the snf2Δ cells, demonstrated strong growth in 100 μM inositol with no significant difference observed between the WT and unAcSnf2p cells. Unlike the 0μM inositol media, however, WT cells in 100μM inositol, when chromatin remodelers are most heavily accumulated at the INO1 promoter, were no longer able to survive in the presence of 1.5 mM copper and now demonstrated a lack of growth comparable to the unAcSnf2p cells and the snf2Δ cells (Fig 6A). As such, in the presence of 1.5mM copper which is CUP1 inducing conditions, there was a significant difference between 0 μM inositol (when remodelers have dissociated away from the INO1 promoter) and 100 μM inositol WT growth (when remodelers are most heavily accumulated at the INO1 promoter). On the other hand, the unAcSnf2p strain demonstrated growth defect in the presence of copper under both 0 μM and 100 μM inositol conditions. As such, this suggests that CUP1 expression is influenced when remodelers are accumulated at the INO1 promoter.
To further examine whether CUP1 expression is influenced by the accumulation of unacetylatable Snf2p at INO1 promoter, CUP1 mRNA were analyzed. In the absence of copper, CUP1 is not expressed in each of the strains and inositol conditions (Fig 6B). In the presence of 1.5 mM copper, however, we observed a significant difference in CUP1 mRNA levels between INO1 repressing and inducing conditions in the WT cells. The WT CUP1 mRNA levels are 0.093±0.016 and 0.309±0.035, under INO1 repressing and inducing conditions, respectively. On the other hand, we observed no significant difference in CUP1 mRNA levels for the unAcSnf2p strain under INO1 repressing and inducing conditions. Similarly, we also found no significant difference in CUP1 mRNA levels for the snf2Δ strain under INO1 repressing and inducing conditions (Fig 6B). To rule out the possibility that it is not the stability of CUP1 mRNA but instead transcription that is adversely affected in the unAcSnf2p mutant, we also examined RNA polymerase II recruitment at the TATA region of CUP1 promoter [20]. We found that Pol II-IP values showed a similar pattern as the mRNA levels (Fig 6C). The only significant change was the increase of WT Pol II-IP values from repressing CUP1 with 0.08% ±0.01% to inducing CUP1 with 0.71%±0.003% when INO1 was in inducing conditions (p = 0.0004). All other conditions showed no significant difference. Therefore, these observations indicate that the lack of CUP1 expression in unAcSnf2p strain under both INO1 and CUP1 inducing conditions is due to the absence of RNA polymerase II instead of the CUP1 mRNA instability.
Previously, we have shown that Snf2p is present at the CUP1 promoter and it is responsible for recruiting chromatin remodeling activity to the promoter for CUP1 induction under induced conditions [20]. Here, we observed the decreased CUP1 expression in unAcSnf2p strain under INO1 inducing conditions, suggesting that the unacetylated Snf2p is not recruited to CUP1 promoter for induction. Therefore, it was instructed to examine whether the accumulation of Snf2p at unAcSnf2p strain’s INO1 promoter resulted in the lack of Snf2p at the CUP1 promoter for CUP1 induction.
For ChIP analysis, the Snf2-FLAG IP values were examined at the CUP1 upstream activation sequences (UAS) [20]. Under INO1 repressing conditions (100 μM inositol), the WT strain’s Snf2-FLAG IP values demonstrated an insignificant difference (p = 0.39), as the Snf2p-FLAG-IP were 0.26%±0.08% under CUP1 repressing conditions and 0.34% ±0.002% under CUP1 inducing conditions, which demonstrated the accumulation of Snf2p at the INO1 promoter but not the CUP1 promoter region (Fig 6D). Under the same INO1 repressing conditions, the Snf2p-FLAG-IP values in the unAcSnf2p mutant were 0.02% ±0.001% and 0.17% ±0.04% under CUP1 repressing and inducing conditions, respectively (Fig 6D). The difference is insignificant (p = 0.08), and this observation indicated that unacetylated Snf2p does not accumulate at the CUP1 promoter. The negative control, snf2Δ, demonstrated Snf2p-FLAG-IP values with no significant difference between repressing and inducing conditions (p = 0.92), with 0.17% ±0.01% and 0.23% ±0.07% under CUP1 repressing and inducing conditions, respectively. These results suggested that Snf2p accumulates at the INO1 promoter not at the CUP1 promoter under INO1 repressing conditions. Therefore, CUP1 is not expressed even in the CUP1 inducing conditions. This is in agreement with the results from growth experiments and RNA analysis (Fig 6A and 6B).
Under INO1 inducing conditions (0 μM inositol), the WT strain’s Snf2p-IP values were 0.29%±0.02% under CUP1 repressing conditions and 0.52% ±0.03% under CUP1 inducing conditions. This showed a significant difference (p = 0.02) and indicated the accumulation of Snf2p at the CUP1 promoter region (Fig 6D). Under the same INO1 inducing conditions, the Snf2p-IP values of the unAcSnf2p mutant were 0.07% ±0.02% and 0.22% ±0.07% under CUP1 repressing and inducing conditions, respectively (Fig 6D). The difference is insignificant (p = 0.16), and this observation indicated that unacetylated Snf2p does not accumulate at the CUP1 promoter. Again, snf2Δ strain’s IP values showed no significant difference between repressing and inducing conditions (p = 0.59), with 0.15% ±0.001% and 0.17% ±0.03% under CUP1 repressing and inducing conditions, respectively. These results suggested that Snf2p departs from the WT strain’s INO1 promoter and it accumulates at the CUP1 promoter when both INO1 and CUP1 are induced. On the other hand, Snf2p accumulates at the unAcSnf2p strain’s INO1 promoter not at the CUP1 promoter when both INO1 and CUP1 are induced. Therefore, Snf2p acetylation is important for its dissociation from the INO1 promoter and to be recruited to the CUP1 promoter when both are induced. Taken together with the growth analyses, the mRNA analyses and the RNA polymerase II IP analysis, the Snf2p-IP analysis confirmed that the accumulation of unacetylated Snf2p at the INO1 promoter was detrimental to CUP1 induction and proper copper protection.
Discussion
Transcriptional activation involves a series of activators and coactivators interactions with the upstream regulatory region of the genes, as well as interactions with each other and with transcription machinery during the process. Although recent research shows that some histone acetylases (e.g., Gcn5p) are capable of acetylating chromatin remodelers (e.g., SWI/SNF, RSC, or ISWI) [16–17, 27–28], the implications of this post-translational modification in the process of transcriptional activation has yet to be thoroughly examined.
Through ChIP and qPCR, we demonstrated that Snf2p acetylation is required for Snf2p dissociation from the INO1 promoter upon induction (Fig 1A). Furthermore, a lack of Snf2p acetylation resulted in an accumulation of Ino80p at the INO1 promoter (Fig 1B). Our previous findings had demonstrated that not only does Ino80p arrive at the INO1 promoter before Snf2p, but that this binding of Ino80p is necessary in order to subsequently recruit Snf2p to the promoter [10]. Since the INO80 complex binds to the promoter first and then SWI/SNF binds, it is likely that the removal of the Snf2p from the region is necessary for INO80 to be free to dissociate. Therefore, it is obvious that the majority of Ino80p remains trapped at the INO1 promoter region when unacetylatable Snf2p accumulates at INO1 promoter.
Previously, we observed that the chromatin remodeler, Snf2p, accumulated at the INO1 promoter in the absence of Gcn5p [11]. Since Snf2p is known to be acetylated by Gcn5p [17], Snf2p remains unacetylated in the absence of Gcn5p. As a result, this is similar to the unacetylatable Snf2p mutant where Ino80p accumulated at the INO1 promoter under inducing conditions (Fig 1B). Therefore, with the use of an unacetylatable Snf2p mutant, we now provide direct evidence that the dissociation was due to the acetylation of Snf2p, with a lack of acetylation resulting in an accumulation of the chromatin remodelers (Fig 1). Furthermore, Ino80p failed to dissociate from the INO1 promoter upon induction, and instead remained occupying the region, perhaps trapped by the accumulated unacetylated Snf2p, which is part of a much bulkier SWI/SNF complex.
Based on the finding that Snf2p acetylation was required for dissociation from the INO1 promoter, it was necessary to next explore the possible implications of this dissociation with regard to transcriptional activation and coactivator activities. Regarding transcriptional activation, growth survival and mRNA analyses confirmed that although the Snf2p acetylation was necessary for dissociation from the promoter, it was not necessary for INO1 gene expression, as the unAcSnf2p strain was able to survive in the absence of inositol and expressed normal INO1 mRNA levels under this inducing condition (Fig 2). This observation is in agreement with our previous finding that both Gcn5p and Esa1p are recruited to INO1 promoter after the start of INO1 induction [11]. Furthermore, our findings support the INO1 induction model where remodelers are required to be acetylated for dissociating from the promoter after the induction [8, 10–11].
INO1 activation is a highly regulated process that is heavily dependent upon the recruitment and dissociation of various coactivators, such as the chromatin remodelers, Snf2p and Ino80p, in order to modify the nucleosome structure at the promoter region and promote appropriate polymerase activity [8, 10–11]. Since we demonstrated that Snf2p acetylation was responsible for the dissociation of Snf2p from the INO1 promoter and the accumulation of both Snf2p and Ino80p was not detrimental to INO1 expression, it is reasonable to predict that chromatin remodeling activities and the recruitment of RNA polymerase II of unAcSnf2p strain should be the same as WT strain under inducing conditions. Indeed, we did observe that the nucleosome density and histone acetylation patterns are very similar for both WT and unAcSnf2p strains (Fig 3). Overall, our results suggested that acetylation plays a role beyond transcriptional activation and may instead have additional significance, possibly in the recycling of remodelers to mobilize elsewhere as proposed previously [11]. As a result, it is likely to affect the various actions that Ino80p/Snf2p would require to perform upon dissociation from the promoter. This is because both Ino80p and Snf2p are critical yeast chromatin remodelers responsible for regulating more than just the INO1 gene. Although hydroxyurea-induced DNA damage repair, osmoregulation, and carbon source utilization were not significantly affected by the accumulation of Snf2p at the INO1 promoter (Fig 5), protection from copper toxicity was noticeably impeded, as demonstrated by sensitivity plate assays (Fig 5I) and growth analyses (Fig 6A). These results suggested that Snf2p must depart from the INO1 promoter in order to be readily available at the necessary levels for the induction of CUP1, which also requires both Ino80p and Snf2p before it is able to protect cells from copper via metallothionine production [20]. This hypothesis was further supported by the observations that unAcSnf2p cells were shown to lack CUP1 induction under conditions where Snf2p was accumulated at the INO1 promoter (Fig 6B). Furthermore, we also observed that Snf2p were not recruited to CUP1 promoter and this resulted in the CUP1 repression (Fig 6D). Therefore, our findings demonstrated that Snf2p should be recycled for other purpose. Perhaps the recycle of remodelers is an economical strategy in the cells to conserve the cell metabolism.
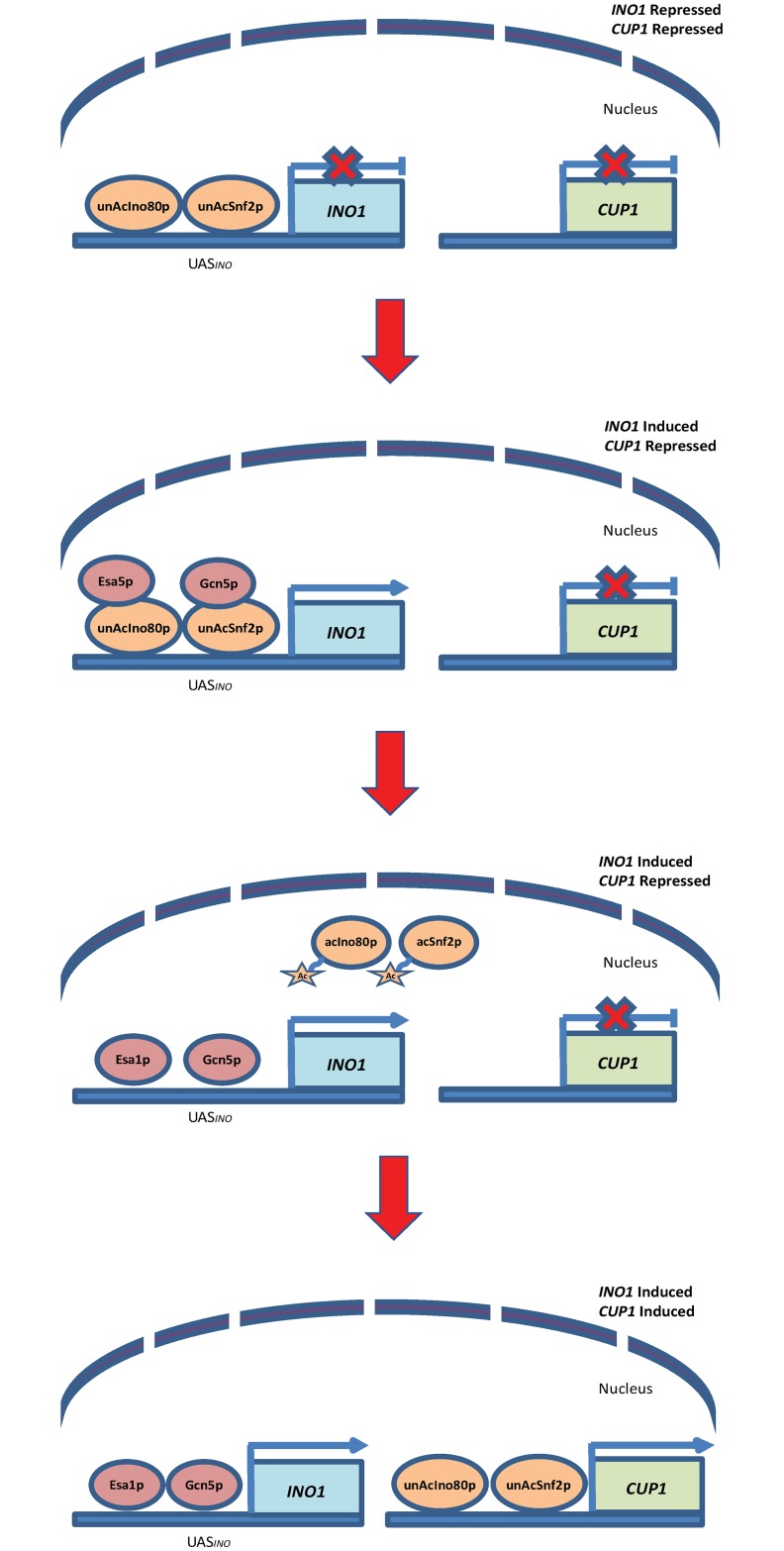
The order of events for INO1 and CUP1 induction has been examined and they demonstrate inverse patterns of remodeler recruitment [10, 20]. Under both INO1 and CUP1 inducing conditions, Snf2p is required to be recruited to the CUP1 promoter but the Snf2p departs from the INO1 promoter. Therefore, if the Snf2p is trapped at the INO1 promoter, less Snf2p will be available for CUP1 activation in the presence of copper. Here, we revealed that when chromatin remodelers are trapped at the INO1 promoter, the copper toxicity defense pathway, which is dependent upon those same remodelers for activation of CUP1, is unable to properly function and protect the cells. Our results suggest that acetylation of remodelers is necessary to allow the recycle of remodelers and a working model has been proposed according to this finding (Fig 7). In this model, the acetylation status of remodelers determines whether remodelers can bind to promoter region or not. We propose that both unacetylated Ino80p and Snf2p bind to the INO1 promoter before induction. Once INO1 is induced, histone acetylases Gcn5p and Esa1p acetylate chromatin remodelers Snf2p and Ino80p, allowing the remodelers to dissociate from the INO1 promoter. Once freed from the INO1 promoter region, the remodelers can be recruited to the CUP1 promoter, which is the gene coding for the production of a metallothionein that binds free copper to protect yeast cells from toxicity, and activate CUP1 expression. In the event that remodeler acetylation is absent, then remodelers are unable to vacate the INO1 promoter and thus fail to mobilize towards CUP1. This yields major defects in copper detoxification and protection. As such, this model describes the strategy cells adopted to use remodelers efficiently in activating gene expression, and maximizing resources.
Fig 7. Proposed model of chromatin remodeler’s recycling during gene activation.
In this model, both unacetylated Ino80p and Snf2p bind to the INO1 promoter before induction. Once INO1 is induced, histone acetylases Gcn5p and Esa1p acetylate chromatin remodelers Snf2p and Ino80p, allowing the remodelers to dissociate from the INO1 promoter. Once freed from the INO1 promoter region, the remodelers can be recruited to the CUP1 promoter and activate CUP1 expression.
Our findings have revealed insight into the mechanism of chromatin remodeler acetylation and its implications in gene expression regulation. Chromatin remodeler mobility is an active area of research, with significant advances recently being made with regard to the RSC complex, for which there are only between 200 to 2000 copies regulating the nearly 70,000 nucleosomes in need of condensing within yeast [29]. This disparity between copy numbers of nucleosomes and the remodelers that regulate them is even more pronounced with regard to Snf2p with only 100–500 copies per nucleus [30], yet no studies have elucidated the implications at this point. So even though Snf2p acetylation was not necessary for INO1 transcriptional activation, we demonstrated that it is still a significant post-translational modification in yeast cells, as our results strongly suggest a recycling role for increased mobility of remodelers may be in effect for chromatin remodeler acetylation. As Snf2p and Ino80p, known regulators of CUP1, accumulate at the INO1 promoter in the absence of Snf2p acetylation, other genes requiring Snf2p and Ino80p may also be affected, since Snf2p is required for the activation of nearly 5% of all yeast genes, yet is fairly rare within the cells at an estimated 100–500 copies per nucleus [30]. Perhaps, the inverse remodeler recruitment pattern observed with certain genes, such as INO1 compared to CUP1, is normal in the cells and may provide maximum and efficient usage of the remodelers for the transcriptional activation.
Materials and methods
Yeast strains and growth conditions
Wild Type (BY4741; MATa his3Δ200 leu2Δ0 met15Δ0 ura3Δ0 SNF2-FLAG::LEU; a gift from Dr. Workman’s lab [17]) and unAcSnf2p (MATa his3Δ200 leu2Δ0 met15Δ0 ura3Δ0 snf2-K1493R-K1497R-FLAG::LEU; a gift from Dr. Workman’s lab [17]), as well as snf2Δ (MATa his3Δ200 leu2Δ0 met15Δ0 ura3Δ0 snf2Δ) strains were used in this study. WT and unAcSnf2p cells were grown at 30°C in Synthetic Complete (SC) media without leucine (SC-leu) containing 2% glucose (wt/vol). Snf2Δ cells were grown at 30°C in SC. Upon optical density (O.D.) between 0.8–1.2, cells were centrifuged and washed twice with media lacking any inositol to completely remove inositol. Cells were then resuspended in appropriate media with 100 μM inositol for repressing conditions or without inositol for inducing conditions, and incubated for 2 hours at 30°C.
Chromatin immunoprecipitation (ChIP)
Yeast strains were grown to mid-logarithmic phase (0.8 A600nm) in 10 μM inositol synthetic complete (SC) media, washed and subjected to conditions to repress (100 μM inositol) or induce (0 μM inositol) INO1 transcription for 2 hours prior to collection. For switching INO1 expression conditions experiment, cells were subjected to the inducing conditions (0 μM inositol) for 1 hour followed by the repressing conditions (100 μM inositol). For copper experiment, cells were grown initially in 10 μM inositol SC media and subsequently washed and diluted to an A600nm of 0.2 in INO1 repressing or inducing conditions as before, that also contained either 0 mM Cu or 1.5mM Cu. The preparation of cross-linked chromatin, immunoprecipitation procedures, and antibodies used in this study are described in the S1 Table.
Real-time qPCR using Taqman probes (Applied Biosystems) was employed to analyze IP samples, along with mock and input samples. All experiments were performed with three repeats and all PCR reactions were done in at least duplicate. The IP for each sample was calculated as:
The IP for the INO1 or CUP1 promoter was then graphed as mean standard ± deviation normalized to input and mock. Input was prepared as all genomic DNA sequences from the cell lysate without any selection or immunoprecipitation. Mock, on the other hand, was prepared as a no-antibody signal background in which all ChIP steps were performed on cell lysate, except for the addition of the selective antibody. Primer and Probe sequences utilized are listed in S2 and S3 Tables.
Growth analysis
Cells were grown initially in 10 μM inositol SC media until mid-log phase and subsequently washed and diluted to an A600nm of 0.2 in repressing or inducing conditions as before. Cell growth was monitored spectrophotometrically using the A600 density until stationary phase was reached. Repeat colonies were averaged and standard deviation was calculated.
RNA preparation and qRT-PCR analysis
Cells were grown to mid-log phase (0.8 A600nm) in 10 μM inositol SC media, washed and subjected to repressing (100 μM inositol for INO1; 0 mM Cu for CUP1) or inducing conditions (0 μM inositol for INO1; 1.5 mM Cu for CUP1) for 2 hours prior to analyses. For switching INO1 expression conditions experiment, cells were subjected to the inducing conditions (0 μM inositol) for 30 min or 1 hour followed by the repressing condition (100 μM inositol) for 1 hour 30 min or 30 min, respectively. The total RNA preparation and qRT-PCR analysis was performed as described in the S1 File. The procedures of preparation and the analysis of target DNA sequence quantities are described in the S1 File as well, along with all real-time PCR primers. All experiments were performed with three repeat colonies and all PCR reactions were performed in duplicate. RNA expression levels were normalized to the constitutive ACT1 housekeeping gene using the formula 2-ΔCt in which ΔCt represents the difference between the Ct value of INO1 or CUP1 and the Ct value of ACT1. Final data was graphed as mean ± standard deviation.
Sensitivity assays
Cells were grown overnight in SC media at 300rpm, 30°C to saturation and subsequently diluted with autoclaved water to an A600nm of 2.0 (approximately 1.6 x 109 cells/ml). Five 10-fold serial dilutions of all strains were performed using autoclaved water. Each dilution was plated as a 2μl droplet onto SC plates as a control to demonstrate cell viability. Dilutions were then plated on SC plates containing 50 mM hydroxyurea, 0.8M KCl, 1.5mM copper and 3mM copper [20]. Lastly each serial dilution was plated on 2% Glucose plates, 2% Sucrose plates, 2% Galactose plates, 2% Maltose plates, and 3% Ethanol plates [23–25]. Plates were incubated for 48 hours at 30°C. All experiments were repeated in duplicate.
Copper sensitivity growth analyses
Cells were grown initially in 10 μM inositol SC media and subsequently washed and diluted to an A600nm of 0.2 in repressing or inducing conditions as before, that also contained either 0 mM Cu, 1.5mM Cu, or 3 mM Cu. Cell growth was monitored spectrophotometrically using the A600nm density until stationary phase was reached. All growth experiments were performed in duplicate; repeats were averaged and standard deviation was calculated.
Supporting information
(DOCX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We are grateful to our laboratory colleagues for technical assistance and comments on the manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
We would like to declare that CHS received funding from North Atlantic Treaty Organization (NATO) SPS G5266, National Science Foundation (NSF) MCB 0919218, Professional Staff Congress-City University of New York (PSC-CUNY) awards for this research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pacheco D, Warfield L, Brajcich M, Robbins H, Luo J, Ranish J, et al. Transcription activation domains of the yeast factors Met4 and Ino2: tandem activation domains with properties similar to the yeast Gcn4 activator. Molecular and cellular biology, 2018; 38: e00038–18. 10.1128/MCB.00038-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Molecular and cellular biology, 2000; 20: 1899–1910. 10.1128/mcb.20.6.1899-1910.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nature reviews Molecular cell biology, 2017;18: 407 10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng X, Jobin-Robitaille O, Billon P, Buisson R, Niu H, Lacoste N, et al. Phospho-dependent recruitment of the yeast NuA4 acetyltransferase complex by MRX at DNA breaks regulates RPA dynamics during resection. Proceedings of the National Academy of Sciences, 2018; 115: 10028–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahel D, Hořejší Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I., et al. Poly (ADP-ribose)–dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science, 2009; 325: 1240–1243. 10.1126/science.1177321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardocki ME, Bakewell M, Kamath D, Robinson K, Borovicka K, Lopes JM. Genomic analysis of PIS1 gene expression. Eukaryotic cell, 2005; 4: 604–614. 10.1128/EC.4.3.604-614.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaldubina A, Ju S, Vaden D, Ding D, Belmaker RH, Greenberg ML. Epi-inositol regulates expression of the yeast INO1 gene encoding inositol-1-P synthase. Molecular Psychology, 2002; 7: 174–180. [DOI] [PubMed] [Google Scholar]
- 8.Esposito M, Konarzewska P, Odeyale O, Shen C-H. Gene-wide histone acetylation at the yeast INO1 requires the transcriptional activator Ino2p. Biochemical and Biophysical Research Communications, 2009; 391: 1285–1290. 10.1016/j.bbrc.2009.12.063 [DOI] [PubMed] [Google Scholar]
- 9.Ford J, Oluwafemi O, Eskandar A, Kouba N, Shen C-H. A SWI/SNF and INO80-dependent nucleosome movement at the INO1 promoter. Biochemical and Biophysical Research Communications, 2007; 361: 974–979. 10.1016/j.bbrc.2007.07.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ford J, Odeyale O, Shen C-H. Activator-dependent recruitment of SWI/SNF and INO80 during INO1 activation. Biochemical and Biophysical Research Communications, 2008; 373: 602–606. 10.1016/j.bbrc.2008.06.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konarzewska P, Esposito M, Shen C (2012) INO1 induction requires chromatin remodelers Ino80p and Snf2p but not the histone acetylases. Biochem Biophys Res Commun 2012; 418:483–8 10.1016/j.bbrc.2012.01.044 [DOI] [PubMed] [Google Scholar]
- 12.Shetty A, Lopes JM. Derepression of INO1 transcription requires cooperation between the Ino2p-Ino4p heterodimer and Cbf1p and recruitment of the ISW2 chromatin-remodeling complex. Eukaryotic cell, 2010; 9: 1845–1855. 10.1128/EC.00144-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwank S, Ebbert R, Rautenstraub K, Schweizer E, Schuller H-J. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Research, 1995; 23: 230–237. 10.1093/nar/23.2.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartholomew B. (2014). Regulating the chromatin landscape: structural and mechanistic perspectives. Annual review of biochemistry, 2014; 83: 671–696. 10.1146/annurev-biochem-051810-093157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charles GM, Chen C, Shih SC, Collins SR, Beltrao P, Zhang X, et al. Site-specific acetylation mark on an essential chromatin-remodeling complex promotes resistance to replication stress. Proceedings of the National Academy of Sciences, 2011; 108: 10620–10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira R, Eberharter A, Bonaldi T, Chioda M, Imhof A, Becker PB. Site-specific acetylation of ISWI by GCN5. BMC molecular biology, 2007; 8:73 10.1186/1471-2199-8-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Saraf A, Florens L, Washburn M, Workman J. (2010). Gcn5 regulates the dissociation of SWI/SNF from chromatin by acetylation of Swi2/Snf2. Genes and Development, 2010; 24: 2766–2771. 10.1101/gad.1979710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muchardt C, Reyes JC, Bourachot B, Leguoy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO Journal, 1996; 15: 3394 [PMC free article] [PubMed] [Google Scholar]
- 19.Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes & Development, 1998;12:2842–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wimalarathna RN, Pan PY, Shen C. H. Co-dependent recruitment of Ino80p and Snf2p is required for yeast CUP1 activation. Biochemistry and Cell Biology, 2013;92:69–75. 10.1139/bcb-2013-0097 [DOI] [PubMed] [Google Scholar]
- 21.Davie JK, Kane CM. Genetic interactions between TFIIS and the Swi-Snf chromatin-remodeling complex. Molecular and cellular biology, 2000; 20: 5960–5973. 10.1128/mcb.20.16.5960-5973.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 1998; 95: 717–728. 10.1016/s0092-8674(00)81641-4 [DOI] [PubMed] [Google Scholar]
- 23.Schöler A, Schüller HJ. (1994). A carbon source-responsive promoter element necessary for activation of the isocitrate lyase gene ICL1 is common to genes of the gluconeogenic pathway in the yeast Saccharomyces cerevisiae. Molecular and cellular biology, 1994; 14: 3613–3622. 10.1128/mcb.14.6.3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Nadal E., Ammerer G., & Posas F. Controlling gene expression in response to stress. Nature Reviews Genetics, 2011;12: 833–845. 10.1038/nrg3055 [DOI] [PubMed] [Google Scholar]
- 25.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics, 1984;108: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abrams E, Neigeborn L, Carlson M. Molecular analysis of SNF2 and SNF5, genes required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Molecular and cellular biology, 1986; 6: 3643–3651. 10.1128/mcb.6.11.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Demark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by Gcn5 acetylation. Molecular cell, 2007; 27: 817–828. 10.1016/j.molcel.2007.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nature Structural & Molecular Biology, 2003;10: 141. [DOI] [PubMed] [Google Scholar]
- 29.Echtenkamp FJ, Gvozdenov Z, Adkins NL, Zhang Y, Lynch-Day M, Watanabe S, et al. Hsp90 and p23 molecular chaperones control chromatin architecture by maintaining the functional pool of the RSC chromatin remodeler. Molecular cell, 2016; 64: 888–899. 10.1016/j.molcel.2016.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Current opinion in genetics & development, 2000; 10: 187–192. [DOI] [PubMed] [Google Scholar]