Abstract
Pathogen evolution is a potential threat to the long-term benefits provided by public health vaccination campaigns. Mathematical modeling can be a powerful tool to examine the forces responsible for the development of vaccine resistance and to predict its public health implications. We conducted a systematic review of existing literature to understand the construction and application of vaccine resistance models. We identified 26 studies that modeled the public health impact of vaccine resistance for 12 different pathogens. Most models predicted that vaccines would reduce overall disease burden in spite of evolution of vaccine resistance. Relatively few pathogens and populations for which vaccine resistance may be problematic were covered in the reviewed studies, with low- and middle-income countries particularly under-represented. We discuss the key components of model design, as well as patterns of model predictions.
Keywords: Vaccine resistance, mathematical modeling
Introduction
Biomedical public health interventions can place strong selective pressure on pathogens, potentially leading to the emergence and spread of pathogens that are resistant to antimicrobial drugs or vaccines [1]. While the evolution of resistance to antimicrobials is considered more common and well documented [2,3], and includes cases of antibiotic, antifungal, antiparasitic, and antiviral/antiretroviral resistance, pathogens can evolve in response to pressure from vaccination of populations as well [4]. Vaccine resistance has been reported with Bordetella pertussis, poliovirus, Streptococcus pneumoniae, hepatitis B, as well as veterinary vaccines [3]. For example, the spread of vaccine-resistant strains is thought to have contributed to the 1996 epidemic of pertussis in the Netherlands that occurred despite high coverage of immunization [5].
Here we define vaccine resistance as a general term that refers to reduced vaccine efficacy due to evolution of the targeted microorganism. Pathogen evolution in response to vaccination can occur through a number of mechanisms, but the best studied are escape mutation and strain replacement [7]. Both escape mutation and strain replacement involve population-level genetic diversity in pathogen susceptibility to the vaccine: escape mutation is the development of a de novo mutation in the vaccine-targeted (VT) strain that confers resistance after immunization rollout, and strain replacement is the increase in incidence of an already circulating resistant or non-vaccine targeted strain (NVT) following the decrease in incidence of the VT strain after vaccine rollout. Both strain replacement and escape mutation can affect disease transmission dynamics in a vaccinated population, and can alter disease severity as well as prevalence and incidence. Higher virulence, or degree of harm to host, is associated with higher replication and a greater chance of transmission to new hosts, but is also associated with increased mortality in the host, potentially cutting short the opportunity to transmit [8,9]. Vaccines that extend infected host lifespans reduce that fitness cost, potentially selecting for more virulent pathogens [4]. This higher virulence could occur via genotypes or strains that either allow for better evasion of the host immunity or for higher replication rates. Not all reductions in vaccine effectiveness are necessarily due to vaccine resistance: e.g. resurgence of pertussis in Sweden was shown to be explained by changes in age-specific contact patterns rather than pathogen evolution [10].
The public health value of vaccines is evaluated using information from animal models, safety trials, efficacy trials, and larger observational studies of real-world effectiveness [11]. Efficacy trials, by design, are insufficiently powered to detect population-level emergence and spread of vaccine resistance, while many observational studies of effectiveness occur over too short a time span to observe this phenomenon. Mathematical models are an important tool for understanding transmission of infectious diseases in the context of public health interventions like vaccination, because they allow researchers to simplify complicated environments into controllable model components [12]. Depending on the research question, model designs tend to be structured as either compartmental, agent-based, or statistical models. Compartmental models provide epidemic information calculated from the rates of movement between disease categories: susceptible, infected, and other disease states, if relevant, e.g. recovered or re-infected. Agent-based models also incorporate movement into, out of, and within disease states in a population, but this is determined at an individual level for each agent in a population. Statistical models use observational data with statistical methods like regression to make predictions about transmission dynamics. Compartmental models are typically preferred in scenarios where transmission systems of limited complexity allow for simpler representation of disease dynamics, while agent-based models gain realism at the expense of computational feasibility and more intensive assumptions. Investigators parameterize their models with information from epidemiological studies or theoretical values and can vary factors of interest to their questions. While no model can fully represent a disease scenario, models aim to include (and identify) the key mechanisms that drive public health outcomes. Because investigators control every aspect of their model, it is necessary to explicitly account for any phenomena at play, such as vaccine resistance, in order for it to factor into predictions.
Many of the existing model-based projections of vaccine impact were estimated without incorporating potential mechanisms for the evolution of vaccine resistance. Without accounting for vaccine resistance, these models may overestimate the positive impact of certain vaccines. We propose that our understanding of vaccines as public health instruments is incomplete without representation of the potential for vaccine resistance. While modeling studies have increased our theoretical understanding of vaccine resistance by testing the conditions under which vaccine resistance may emerge and spread, very few of them have predicted the epidemiological consequences of resistance. We conducted a systematic literature search for epidemic models that examined the public health consequences of vaccine resistance with the intention of summarizing model-based predictions for vaccine resistance, discussing key themes that arise in this diverse collection of work, and informing future modeling studies by describing the design and implementation of published vaccine resistance models.
Methods
We used the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) framework for identifying and evaluating literature related to our research questions. A record search of peer-reviewed studies published in English was conducted in PubMed in June 2018. Three search term groupings were used to capture studies that addressed resistance, vaccines, and models:
Host-pathogen interactions [mesh term] or microbial interactions/immunology* [mesh term] or immune evasion or vaccine resistan* or serotype replacement or strain replacement
and immunization [mesh term] or vaccines [mesh term] or vaccination* or vaccin* or immunization
and model*
The lead author (MCR) reviewed the resultant list of publications to determine if each met the guidelines for inclusion. Studies must have mathematically modeled the impact of immunization on pathogen evolution. We were interested in actionable modeling research with explicit consideration of public health outcomes of vaccine resistance, and therefore we excluded studies that did not extend their analyses to predict epidemiological outcomes in metrics such as vaccine effectiveness or disease incidence, and we excluded primarily analytical models, defined as those with the objective of exploring theoretical concepts and with limited or no parameterization based on existing or historical epidemics. Human and veterinary vaccination scenarios were included. During the full text review, we noted any relevant studies referenced by the authors that were not captured in the PubMed search and screened them according to the same review process. We reviewed all study titles for and included those with possible relevance to our interests. We then reviewed study abstracts and further excluded studies if they did not look at vaccine resistance or did not use a mathematical model. Finally, in reviewing the remaining full-texts, we eliminated studies that did not report outcomes in epidemiological terms or were not public health motivated. A flow diagram of the study screening process is shown in Figure 1.
Figure 1.
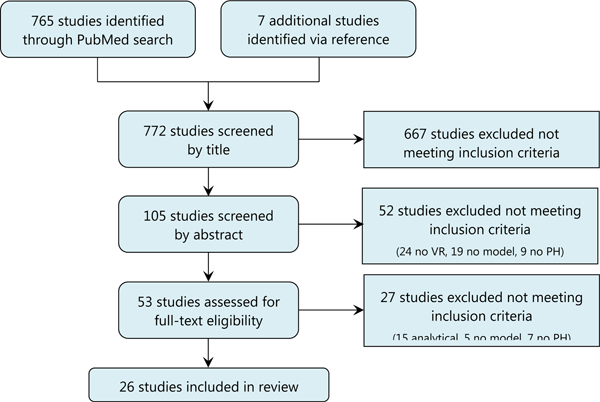
PRISMA flow diagram of study screening process
Study exclusion rationale:
“no VR” = study did not address vaccine resistance
“no model” = study did not use a mathematical model
“no PH” = study did not model public health outcomes
“analytical” = study was mainly theoretical/conceptual
After selecting the relevant literature, we looked to synthesize three main take-aways from the reviewed studies. First, we outline the state of the field of vaccine resistance modeling by describing the design and components of the reviewed models. For this, we used the descriptions of model parameters provided in each text to develop categories for the common modeling components among the studies and identify additional factors included in the studies. Second, we reviewed the key findings of each study in order to provide a general summary of model predictions for vaccine resistance. This component of the review is predominantly narrative, rather than quantitative, as heterogeneity across studies in the modeled disease and populations precludes meta-analysis.. Last, we describe the factors that were identified as important in influencing the development of vaccine resistance in different studies, and discuss themes arising from in the reviewed literature.
Results
We identified 765 studies in PubMed, with an additional seven titles retrieved from the reference lists of the PubMed studies during our full-text reviews. We excluded 667 studies with titles that fell unambiguously outside of the inclusion criteria and an additional 52 based on abstract review. A further 27 studies were removed during the full text review, most commonly due to the model being analytical (vs. epidemiologically-driven), the model not reporting outcomes in terms of public health impact (seven studies), or not using mathematical modeling (five studies). A total of 26 studies were included in the final analysis (Figure 1).
Model types
A summary of the model design and pathogen studied in each study is presented in Table 1, and a visual representation of key study characteristics is presented in Figure 2. The 26 studies explored twelve different pathogens: Streptococcus pneumoniae was most common at eleven studies, followed by two studies each for avian influenza, human influenza A, Neisseria meningitidis, rotavirus, and HIV, and one each for hepatitis B virus, Plasmodium falciparum, enterovirus A, Mycobacterium tuberculosis, and human papillomavirus. Five of the 26 studies were agent-based models [13–17], two studies were statistical models [18,19], and the remaining 19 had a compartmental structure. Twenty-three studies modeled strain replacement as the primary mechanism of resistance and three studies addressed the likelihood and impact of de novo adaptive mutations [20–22].
Table 1.
Model characteristics and parameters
| Additional Components | |||||
|---|---|---|---|---|---|
| Study Name | Pathogen | Structure | Pathogen-level | Intra-host level | Inter-host level |
| Wilson et al, 2000 | Hepatitis B | Compartmental | Co-infection Mutation rate | Age Sex | Contact rate |
| Gandon et al, 2001 | Plasmodium falciparum | Compartmental | Super-infection Virulence Natural immunity | Vector characteristics | |
| Zhang et al, 2004 | Streptococcus pneumoniae | Compartmental | Co-infection Super-infection Natural immunity | Contact rate | |
| Elbasha and Galvani, 2005 | Human papillomavirus | Compartmental | Natural immunity Co-infection | Contact rate | |
| Temime et al, 2008 | Streptococcus pneumoniae | Compartmental | Co-infection Mutation rate Invasiveness | Age | Contact rate |
| Cohen et al, 2008 | Mycobacterium tuberculosis | Compartmental | Co-infection Super-infection | ||
| Iwami, Takeuchi et al, 2009 | Avian influenza | Compartmental | Geographic area | ||
| Iwami, Suzuki et al, 2009 | Avian influenza | Compartmental | |||
| Melegaro et al, 2010 | Streptococcus pneumoniae | Compartmental | Co-infection Invasiveness | Age | Contact rate |
| Van Effelterre et al, 2010 | Streptococcus pneumoniae | Compartmental | Co-infection | Antibiotic use | |
| Invasiveness | Age | ||||
| Choi et al, 2011 | Streptococcus pneumoniae | Compartmental | Co-infection Invasiveness | Age | |
| Pitzer et al, 2011* | Rotavirus | Compartmental | Natural immunity | Age Geographic area | |
| Choi et al, 2012 | Streptococcus pneumoniae | Agent-based | Co-infection Natural immunity Invasiveness | Age | |
| Fryer and McLean, 2011 | HIV | Compartmental | Mutation rate | Host genetic factors | Contact rate |
| Alexander and Kobes, 2011 | Influenza A | Agent-based | Natural immunity | Age | Contact network |
| Flasche et al, 2013 | Streptococcus pneumoniae | Agent-based | Co-infection Natural immunity Invasiveness | Age | Contact rate |
| Bottomley et al, 2013 | Streptococcus pneumoniae | Compartmental | Co-infection Super-infection Natural immunity | Contact rate | |
| Link-Gelles et al, 2013 | Streptococcus pneumoniae | Statistical | Age Race | ||
| Nurhonen et al, 2013 | Streptococcus pneumoniae | Agent-based | Co-infection Natural immunity Invasiveness | Age Neighborhood | Contact network |
| Vickers et al, 2013 | Neisseria meningitidis | Compartmental | Natural immunity | Age | Contact rate |
| Nurhonen and Auranen, 2014 | Streptococcus pneumoniae | Statistical | Invasiveness | Age | |
| Pitzer et al, 2015* | Rotavirus | Compartmental | Re-infection Natural immunity | Age Geographic area | |
| Hogea et al, 2016 | Neisseria meningitidis | Compartmental | Co-infection Natural immunity | Age | Contact rate |
| Takahashi et al, 2016 | Enterovirus A | Compartmental | Natural immunity | ||
| Worby et al, 2017 | Influenza A | Compartmental | Natural immunity | Age | Contact rate |
| Herbeck et al, 2018 | HIV | Agent-based | Anti-retroviral use Host genetic factors Condom use Age Sex | Contact network | |
See results section for baseline model components common to all studies.
These models also factored in seasonality, which does not fall directly into any of the three parameter categories
Figure 2.

Tree maps of the proportion of studies (#) by pathogen, type of resistance, and region studied, as well as model structure
Model components
All of the reviewed models included a set of core parameters, primarily to set the proportions of the population in various health states (e.g. susceptible, immune, vaccinated), as well as the likelihood and rate of moving between these health states. Vaccine-specific parameters were also essential, including vaccination coverage, strain-specific efficacy, and sometimes programmatic specifics like target age group and boosters.
Beyond these core model inputs, many studies included further components and parameters. We categorized these into three main classes: 1) pathogen-level; 2) intra-host; and 3) inter-host. Pathogen-level parameters were defined as those particular to the type of pathogen studied (e.g. natural immunity was classed as a pathogen-specific parameter because while it occurs in the host, the presence and magnitude of a host immune response depends on which pathogen they are exposed to). Intra-host parameters were defined as those particular to individuals in a population (e.g. individual age or distribution of ages in the population). Inter-host parameters were defined as those pertaining to interaction between disease hosts (e.g. contact rate). There is some conceptual overlap between these parameters, (e.g. genetic factors that cause some individuals to have a more effective immune response to HIV were considered an intra-host parameter, but are a facet of natural immunity which was attributed to the pathogen-level factors).
Of the pathogen-level parameters, natural immunity was most frequently included in the reviewed models (14 studies). The studies that did not include immunity were generally of pathogens for which variation in innate and adaptive immune responses have negligible impacts on infection, re-infection, or recovery (e.g., HIV, hepatitis B, and avian influenza) [2,17,20,22–24]. Inclusion of natural immunity involved designation of the duration of natural immunity following recovery from a first infection or after birth for maternal antibody-mediated immunity [4,13–16,25–33]. For multi-strain models, investigators often specified if infection with one strain incurred any protection against re-infection, co-infection, or super-infection by other strains [2,4,13–16,20,21,25–28,33–36]. Other pathogen-level parameters were mutation rate [20–22], and virulence [4]. Nine studies included an invasiveness term to address when an individual is colonized by or is “carrying” a pathogen without having symptoms or disease [14–16,19,21,27,34–36]. For example, Nurhonen et al. (2013) calculate invasiveness as a ratio of observed invasive pneumococcal disease (IPD) incidence to the incidence of carriage episodes for each subtype of S. pneumoniae studied.
The intra-host parameters most often included were adjustments for demographic factors. 16 studies made some adjustments for age, usually specifying risk of infection or contact rate by age group [13–21,27,28,31,32,34–36]. Two studies also accounted for sex [17,20] and Link-Gelles et al (2013) included race as parameters related to differential transmission or infection. Geographic location was a component in four models [16,23,28,29]. Other host-level considerations allowed for host genetic factors [17,22], or antiretroviral/antibiotic use [17,36] to impact the risk of infection.
Inter-host considerations were made in 15 studies, and were typically represented by a contact rate [14,15,20–22,25–27,31–35], or a contact network [13,16,17], all of which varied in their complexity. One study, of theoretical malaria vaccines, also included mosquito vector-related parameters necessary to describe the transmission dynamics of interest [4].
Model predictions
A summary of the design and epidemiological predictions of each model is given in Table 2. Seven studies found that the impact of vaccine resistance on overall vaccine effectiveness would be negligible [18,21,26,28–31]. 17 studies predicted overall positive vaccine impacts despite some moderate resistance [2,4,13,16,17,19,20,22,24,26,27,31–36]. Four studies found vaccine benefits were effectively canceled out due to vaccine resistance, resulting in no net change in outcomes of interest [4,22,23,25]. In five studies, vaccines could cause harm to the overall population either by increasing prevalence compared to pre-vaccination through strain replacement or by changing the average virulence of the pathogen in unvaccinated hosts under certain conditions [2,4,22,24,32,35]. [Note: Studies that are cited multiple times in the above summaries offered multiple predictions based on their experimental questions and had varied results.] To help explore what factors might be behind whether a study predicted vaccine resistance, Table 3 presents potential explanations for the variation and summarizes the evidence in support of these hypotheses.
Table 2.
Summary of models of epidemiological impact of vaccine resistance
| Paper Name | Summary | Epidemic Setting | Pathogen, vaccine | Predicted Public Health Impact |
|---|---|---|---|---|
| Wilson et al, 2000 | Predictions for the emergence of HBV vaccine escape mutants and conditions of strain replacement in a high prevalence setting | The Gambia | Hepatitis B, HBsAg vaccine | An escape mutant is predicted to emerge and replace the wild-type VT strain, though overall HBV prevalence is expected to decrease. |
| Gandon et al, 2001 | Model of strain replacement following four types of immunization programs that either block infection, transmission, toxin production, or parasite density | High-transmission setting | Plasmodium falciparum, theoretical vaccines | Malaria mortality in unvaccinated hosts may increase due to virulence evolution when vaccines reduce disease severity but do not prevent transmission. |
| Zhang et al, 2004 | Model of strain replacement after vaccination for direct or indirect mechanisms of strain competition | General epidemic | Streptococcus pneumoniae, theoretical monovalent vaccine | Vaccine reduces overall carriage of both VT and NVT strains, with higher strain replacement occurring at higher degrees of inter-strain competition and shorter duration of natural immunity |
| Elbasha and Galvani, 2005 | Exploratory modeling of HPV strain dynamics and their impact on vaccination. Infection with one strain either reduces, increases, or has no effect on susceptibility to infection with a second strain. | General epidemic | Human papilloma virus, theoretical monovalent or universal HPV vaccines | Vaccination can indirectly reduce prevalence of an NVT strain if infection with the VT strain increases susceptibility to second infection. If the VT strain doesn't change or decreases susceptibility to re-infection, strain replacement may occur. |
| Cohen et al, 2008 | Model to predict strain competition under various TB vaccination conditions | General epidemic | Mycobacterium tuberculosis, theoretical vaccines | Degree of strain replacement depends on ability of competing strains to prevent super-infection or co-infection, as well as Ro of strains and vaccine coverage. Some scenarios predicted equilibrium TB burdens higher than pre-vaccination levels, though most combinations saw some overall reduction in TB. |
| Temime et al, 2008 | Study of scenarios where a vaccine-targeted S. pneumoniae strain escapes immune pressure by capsular switch | Europe | Streptococcus pneumoniae, PCV7 | Minimal predicted impact of vaccine escape and strain replacement on vaccine effectiveness- few excess cases of IPD due to switched pneumococcal strains. |
| Iwami, Takeuchi et al, 2009 | Model of the spread of avian flu vaccine resistance from one theoretical chicken population with a vaccination program to another without, when the two populations are linked by illegal bird trade. | General epidemic | Avian influenza, theoretical veterinary vaccine | The vaccinated poultry population will see complete replacement of the VT strain with the NVT strain. The un-vaccinated population will see a gradual rise of NVT infected birds due to the import of birds from the vaccinated population, which then co-exists with the VT strain. |
| Iwami, Suzuki et al, 2009 | Compartmental model of vaccination in a domestic poultry population with two influenza strains. | General epidemic | Avian influenza, theoretical veterinary vaccine | Strain replacement is possible following vaccination, and the degree of replacement depends on the virulence of the NVT strain, vaccine coverage, and amount of partial efficacy of the vaccine on the NVT strain |
| Van Effelterre et al, 2010 | Compartmental model to study the joint effects of antibiotic use and vaccination on the incidence of IPD caused by the Streptococcus pneumonia strain 19A | United States | Streptococcus pneumoniae, PCV7 | Antibiotic use contributed more than PCV7 vaccination did to the emergence of antibiotic non-susceptible 19A, but vaccination against 19A more impactful on disease than reducing antibiotic use |
| Melegaro et al, 2010 | Model of the impact of PCV7 on incidence of invasive disease, with herd immunity effects as well as serotype replacement. | England and Wales | Streptococcus pneumoniae, PCV7 | While incidence of IPD due to NVT strains is predicted to increase after vaccination with PCV7, overall IPD will decrease substantially unless there is cross-immunity between NVT and VT strains. |
| Fryer and McLean, 2011 | Model to understand the impact of escape mutation from a cytotoxic lymphocyte vaccine that doesn't prevent infection but reduces disease progression in infected individuals. | Sub-Saharan Africa | HIV, theoretical CTL vaccine | Rate of escape mutation in vaccinees determines the degree to which a theoretical vaccine that reduces disease severity (and therefore infectiousness) reduces overall HIV morbidity and prevalence. |
| Alexander and Kobes, 2011 | Age-based network model of influenza strain competition in a partially vaccinated population, where vaccines are rolled out either before or during the epidemic | General epidemic | Influenza A, theoretical vaccine | Strain replacement by the NVT strain peaked at intermediate vaccine coverage levels. Vaccination prior to the epidemic was most optimal, with strain replacement minimized at high coverage levels. |
| Choi et al, 2011 | Modeling newly available surveillance data of PCV7 to understand why serotype replacement was higher than original estimates | England and Wales | Streptococcus pneumoniae, PCV7 | PCV7 likely to reduce overall burden of IPD, despite moderate strain replacement. |
| Pitzer et al, 2011 | Multi-strain model of rotavirus in developed countries to quantify vaccine impact under different assumptions about natural immunity. | Developed countries | Rotavirus, RotaTeq and Rotarix | Vaccination with either available vaccine is expected to substantially reduce rotavirus disease, though some increase in NVT strains is predicted. Dynamics of heterotypic and homotypic immunity were important to the coexistence of strains pre- and post-vaccination. |
| Choi et al, 2012 | Agent-based model of IPD incidence following replacement of PCV7 with PCV13, compared to ceasing vaccination completely. | England and Wales | Streptococcus pneumoniae, PCV7 and PCV13 | Replacement of PCV7 with PCV13 likely to reduce overall burden of IPD. The more colonization with PCV13 strains prevented co-infection with NVTs, the greater the serotype replacement. |
| Bottomley et al, 2013 | Deterministic, compartmental model to predict Streptococcus pneumoniae carriage in the Gambia following vaccination with PCV13 or hypothetical vaccines with additional serotype coverage. | The Gambia | Streptococcus pneumoniae, PCV13 and four hypothetical PCV13+ vaccines | PCV13 predicted to greatly reduce VT strains, but NVT strains predicted to rise for an overall slight reduction in carriage. Hypothetical vaccines with additional serotype coverage predicted to follow similar patterns, with remaining NVT serotypes nearly replacing VT serotypes in prevalence |
| Link-Gelles et al, 2013 | Poisson regression model of IPD burden following a switch from PCV7 to PCV13 in the United States | United States | Streptococcus pneumoniae, PCV7 and PCV13 | Overall levels of IPD are predicted to decrease despite moderate levels of serotype replacement. Sensitivity analyses using no serotype replacement or high serotype replacement provided slightly higher or lower levels of reduction in IPD, respectively |
| Nurhonen et al, 2013 | Agent-based network model of Streptococcus pneumoniae carriage and IPD in Finland, factoring in indirect vaccine effects of herd immunity and serotype replacement. | Finland | Streptococcus pneumoniae, PCV7, PCV10, and PCV13 | Serotype replacement by NVT strains predicted to result in an overall slight reduction in total pneumococcal carriage. Moderate overall reductions in IPD predicted for PCV13, with PCV10 and PCV7 producing less of a reduction. |
| Vickers et al, 2013 | Dynamic, multi-strain model to compare monovalent and quadrivalent vaccines for the control of invasive meningococcal disease. | Canada | Neisseria meningitidis, monovalent C and quadrivalent ACWY conjugate vaccines | The monovalent vaccine is predicted reduce overall carriage of N. meningitidis, but with considerable strain replacement. The quadrivalent vaccine will cause minimal replacement, especially for multiple doses. |
| Flasche, et al, 2013 | Comparing two pneumococcal vaccine types, one targeting the two most prevalent strains and the other immunizing equally against all strains, to model the development of strain co-existence or replacement | United Kingdom | Streptococcus pneumoniae, theoretical bivalent or universal vaccine | Strain replacement was predicted to occur for the bivalent vaccine, such that overall pneumococcal carriage decreased slightly. The universal vaccine was generally more effective at reducing overall carriage, but its effectiveness was sensitive to increases in infectiousness of circulating strains. |
| Nurhonen and Auranen, 2014 | Statistical model using age-specific serotype distributions to predict impact of available PCV vaccines on disease and find optimal vaccine serotype compositions. | Finland | Streptococcus pneumoniae, PCV10 and PCV13 | Vaccination can result in an overall substantial reduction in IPD in the target age group of children under the age of 5 despite serotype replacement, but the indirect effects of vaccination on the older population will be less marked. |
| Pitzer et al, 2015 | Models for fluctuations in strain composition following rotavirus vaccination in Belgium | Belgium | Rotavirus, RotaTeq and Rotarix | The number of hospitalizations due to rotavirus expected to remain low despite some strain replacement after rollout |
| Hogea et al, 2016 | Exploratory analysis of potential serogroup replacement following introduction of a vaccine targeted at serogroup B N. meningitidis in two populations | England, Wales, and Czech Republic | Neisseria meningitidis, MenB vaccine | Addition of the MenB vaccine is predicted to induce serogroup replacement by NVTs, though the extent of replacement depends on inter-strain competition and serogroup prevalence prior to vaccine introduction |
| Takahashi et al, 2016 | Multi-strain model of hand, foot, and mouth disease control by theoretical vaccines. | China | Enterovirus A (hand, foot, and mouth disease), theoretical vaccines | Some serotype replacement is predicted, but overall burden of HFMD is expected to be reduced substantially by monovalent and bivalent vaccines. |
| Worby et al, 2017 | Comparison of monovalent and bivalent flu vaccine impact on flu burden under different degrees of cross-immunity between co-circulating strains. | United States | Influenza, monovalent or bivalent A/H3N2 vaccines | Higher cross-immunity leads to greater strain replacement for the monovalent vaccine, though both the monovalent and bivalent vaccine can reduce disease |
| Herbeck et al, 2018 | Agent-based network models of HIV evolution in response to a partially effective vaccine in two epidemic settings | South Africa (heterosexual) and United States (men who have sex with men) | HIV, theoretical RV144-like vaccine | The public health impact of a partially effective HIV vaccine is expected to decrease with time as a result of replacement by vaccine-resistant strains in both epidemics. Overall vaccine efficacy drops with increasing coverage levels, and prevalence decreases less markedly with resistance. |
VT = vaccine type strain, NVT = non-vaccine type strain, PCV = pneumococcal conjugate vaccine, IPD = invasive pneumococcal disease
Table 3.
Summary of evidence for potential hypotheses to explain vaccine resistance predictions in the reviewed studies
| Does vaccine resistance vary by: | |
|---|---|
| …pathogen? | There was considerable variation in findings within studies of the same pathogen, though only S.
pneumoniae was examined in more than two studies. |
| …region? | Geographic location did not appear to influence whether studies predicted vaccine resistance; studies in the same region (e.g. European countries) had varied predictions. |
| …vaccine characteristics? | Studies of theoretical vaccines that do not yet exist predicted a wider range of vaccine resistance outcomes compared to studies of well-documented extant vaccines which more consistently predicted mild to moderate vaccine resistance (see results paragraph nine for further details). |
| …model type? | Agent-based or statistical models did not predict outcomes markedly different from the majority of studies, which used compartmental models. |
| …model parameters? | Within studies that conducted sensitivity analyses, model prediction proved to be influenced by parameter choice, though cross-immunity was the only factor that consistently influenced predictions of vaccine resistance in studies that included it (see results paragraph eleven for further details). |
Models of existing vaccines more often found overall beneficial vaccine effects, compared to models of conceptual vaccines (or vaccines in early pre-trial stages of development), which found a wider range of vaccine resistance predictions. Vaccines yet to be developed and tested will have less information available to investigators, and so models may be more exploratory. With the exception of the hand-foot-and-mouth-disease vaccine study by Takahashi et al (2016), the studies of primarily theoretical vaccines predicted a wide range of potential public health outcomes that depended on intentionally varied parameters [2,4,22]. In studies of existing vaccines, however, there was considerable agreement in predictions despite disparate pathogens and research questions. Overall, the studies of vaccines that have been in use, have trial data, or have existing homologs predicted positive health outcomes despite vaccine resistance. The exceptions to this would be the studies of established pneumococcal conjugate vaccines by Melegaro et al (2010), which varied a parameter of interest, and Bottomley et al (2013), which studied carriage of S. pneumoniae and not disease.
Naturally, scenarios where vaccination may worsen public health outcomes are of great concern. These observations did not stand alone as the key findings of any study; they tended to be minor analyses to demonstrate what could happen under certain extreme parameter values or assumptions. The two models of vaccines that modify disease severity increased the lifespan of vaccinated infected individuals, favoring an increase in pathogen virulence either because this increased the number of chances to transmit [22], or reduced the fitness cost of high virulence (host death) and selecting for overall more harmful pathogens [4]. Iwami et al (2009) predicted that conditions of high vaccine coverage combined with particularly ineffective vaccines for avian flu in a poultry population could increase prevalence of avian influenza to higher levels than pre-vaccination through emergence of a non-vaccine type (NVT) strain. For their findings of higher post-vaccination prevalence, Worby et al (2017), Cohen et al (2008), and Melegaro et al (2010) required high levels of cross-immunity between co-circulating strains, as well as greater infectiousness of the NVT strain for Worby et al (2017) and Cohen et al (2008), and introduction of the NVT strain after vaccine type (VT) epidemic peak for Worby et al (2017). Some of these negative outcomes are due to cross-immunity (discussed below). As with most of the reviewed model predictions, these six studies predicting poorer outcomes in the presence of vaccine programs are illuminating for their insight into the range of potential evolutionary outcomes, rather than concrete predictions of what vaccines will do in a population.
The pessimistic forecasts of Worby et al (2017), Cohen et al (2008), and Melegaro et al (2010) highlight the importance of cross-immunity in determining how co-circulating strains interact, a theme that was common in many of the reviewed studies. Sometimes described as heterotypic immunity, cross-immunity represents a variety of inter-strain interactions relating to the ability of a strain to infect a host that was previously or is currently infected with another strain, resulting in either reinfection (subsequent infection following recovery from a previous infection), co-infection (simultaneous infection with multiple strains/types), or super-infection (infection with a second strain that replaces the first strain). We will use the term cross-immunity to refer only to pathogen-induced natural immune response, rather than a vaccine-induced immune response to multiple strains. Cross-immunity was included as an experimental parameter or set of parameters to vary in 10 studies [2,13–15,26,27,29,32,33,35]. Homotypic immunity, where previous infection with a strain reduces the chance of reinfection with the same strain, was also often considered, but was less often identified or tested as a moderator of vaccine resistance. For all of the studies that tested some measure of cross-immunity, the higher the level of cross-immunity, the greater the degree of strain replacement. This relationship between cross-immunity and vaccine resistance is thought to occur when, if cross-immunity exists between VT and NVT strains, reducing the prevalence of a VT strain by vaccination will reduce the prevalence of people naturally immune to NVT strains [37]. Elbasha and Galvani (2005) provided an interesting counter-factual to this relationship by examining the potential for previous HPV infection to increase susceptibility to reinfection by another strain, which they called synergistic, in addition to cross-immunity. They demonstrate that if cross-immunity is assumed, strain replacement will occur as expected, but if synergy is assumed, the NVT strain will actually decrease in prevalence when it loses the extra host vulnerability provided by the VT strain circulating in the population.
Discussion
We reviewed 26 studies that predicted population health impacts of vaccine resistance, covering 11 different infectious diseases. These represent a relatively small fraction of health issues for which vaccines are currently used or under development. The US Centers for Disease Control and Prevention currently recommends routine vaccination for 14 diseases, only six of which are covered in the reviewed studies, primarily more recently developed vaccines [38]. The most recent estimates from the World Health Organization report candidate vaccines for 24 different infectious diseases, five of which are addressed by models reviewed here: HIV, pneumococcal disease, tuberculosis, malaria, and rotavirus [39]. People in low and middle income countries (LMIC) stand to benefit the most from expanding vaccination [40], though just six of the 26 reviewed studies drew on data from or simulated epidemics in a LMIC. Populations for which limited data exist are naturally less-studied, and many of the unaddressed vaccines have either too few data or sufficient empirical evidence of long-term effectiveness, so the reviewed studies are by no means a comprehensive representation of the global public health impact of vaccine resistance.
The relatively small coverage of vaccine resistance in mathematical models may be a product of the recency of vaccine resistance as a concept. Published studies of vaccine resistance emerged in the 1990s and early 2000s (reviewed in Gandon and Day, 2007). It follows that there is not yet consensus in the language used to describe vaccine resistance: the term vaccine resistance itself was relatively rarely used in published literature, while specific mechanisms of resistance (e.g. strain replacement, vaccine escape) were more commonly used. Other terms such as vaccine failure, serotype replacement, or strain dynamics were also used. No NCBI Mesh Term exists at the time of writing to capture vaccine resistance or any of these related terms.
Notably, over a third of the reviewed studies concerned vaccines to prevent Streptococcus pneumoniae infection. The careful attention paid to S. pneumoniae may be due to a number of factors, not the least because it is a substantial contributor to child mortality worldwide, but also likely because heptavalent pneumococcal conjugate vaccines were introduced fairly recently in 2000, and there is sufficient epidemiological data with which to observe strain replacement [41]. The eleven S. pneumoniae studies address pneumococcal conjugate vaccines that were available or were close to public use at the time of study, primarily the heptavalent PCV7 and later PCV10 and PCV13, with the exception of Zhang et al (2004) and Flasche et al (2013), who studied purely theoretical vaccines. We see a range of predictions about vaccine resistance, but most studies describe some strain replacement in carriage and overall reductions in disease. While different from one another, these eleven studies are also complementary: each study approaches a different research question or epidemic scenario, yet together they describe the potential impact of pneumococcal conjugate vaccination on circulating S. pneumoniae and public health. Insights range from the theoretical importance of cross-immunity in determining degree of S. pneumoniae vaccine resistance, to practical guidance for selecting an optimal PCV strain composition for a European population.
Several studies used minimal epidemiological data and designed their models to illuminate a theoretical relationship. For example, the study by Gandon and coauthors (2001) used a general model of malaria epidemics to demonstrate how theoretical vaccine mechanisms could put evolutionary pressure on a pathogen and affect varied health outcomes, including increased mortality in non-vaccinated hosts. To approach an actionable response to vaccine resistance, however, models require more epidemiological data. This sensitivity to input parameters was illustrated in three studies of pneumococcal conjugate vaccination in England and Wales by the same research group. The first study, which used vaccine transmission dynamic parameters derived from the United States PCV7 vaccination program data, predicted a higher overall vaccine efficacy than England and Wales actually experienced after their PCV7 introduction [35]. Later, when preliminary vaccine surveillance data from England and Wales became available, the investigators observed that the UK epidemic indicated a much higher level of cross-immunity than was detected in the US. After refitting the same model with the more relevant local data, their predictions lined up with the empirical S. pneumoniae epidemic dynamics [34]. The updated model was adapted once more and used by the National Health Service to inform its decision to switch to the PCV13 vaccine [14]. Because England and Wales collected and used vaccine surveillance data, they were able curb the strain replacement threatening the success of their PCV program. These studies support expansion of surveillance programs to include pathogen evolution in response to vaccination.
As described in the results section, we observed that studies of more speculative vaccines tended to produce more varied results, even finding that under certain conditions vaccine resistance may overwhelm the benefits of the vaccine and cause population harm, while models of extant vaccines tended to produce more conservative estimates of vaccine resistance effects. While the motivation to do sensitivity analyses for less-studied pathogens/vaccines and limitations of data availability plausibly explain this difference, but modeling studies are not immune to publication bias as well, which may have influenced the publication of studies that reflect positive evaluations of existing vaccines or more newsworthy, dramatic results for potentially harmful vaccines. To our knowledge, tools to determine risk of bias in reviewed studies, such as the Cochrane Bias tool for clinical research [42], are not available for mathematical modeling studies, and we did not attempt to systematically evaluate the reviewed studies.
This review has several further limitations. Seven of the included studies were not identified in the PubMed search, but rather were identified in reference lists during full-text review, suggesting that, due to the diversity of terms used to describe vaccine resistance, our chosen search words may not have fully captured all of the existing mathematical models of vaccine resistance. We included both human and animal vaccines, but we identified just two animal vaccine models, limiting inference from these studies. The models were very diverse in their designs, research goals, inputs, and outcomes, making direct comparisons difficult, and limiting our ability to quantitatively summarize model predictions from included studies.
There are many exciting directions for future research to build on the foundation laid by the studies reviewed here. Explanations for why the diphtheria, rubella, mumps, measles, and tetanus vaccines, which have been in widespread use for many decades, never developed resistance are hypothetical [3], and a model of these successful vaccines may identify correlates of vaccine effectiveness. Emphasis might be put on modeling epidemic scenarios in LMIC, where disease burdens are high and immunization programs are expanding rapidly. For vaccines that target multiple strains or subtypes of a pathogen, mathematical modeling is a valuable tool for optimizing the strain composition of these vaccines for specific populations. Similarly, targeted vaccination programs should be explored further to determine how to distribute vaccines among risk groups to minimize resistance and maximize overall benefit. For vaccines with well-understood patterns of vaccine resistance, incorporating these into cost-effectiveness analyses could help inform pragmatic program design. Finally, future vaccines are expected to offer novel designs, potentially imperfect pathogen coverage, and alternative mechanisms for protecting the public beyond traditional infection-blocking, all of which may interact with pathogens to foster resistance [43]. As these innovations develop, models of their potential for resistance evolution should follow close behind.
Conclusions
Mathematical models can illuminate the complicated relationships between pathogen characteristics, vaccine components, the emergence of vaccine resistance and the consequent impact of vaccine resistance on public health outcomes. We have seen how vaccine resistance can develop in a wide range of pathogens, with diverse implications for the health of the public. Informed vaccine development and public health policy now and in the future will depend on an improved understanding of vaccine resistance dynamics.
Acknowledgements
The authors are grateful to Sarah Safranek of the University of Washington Health Sciences Library for her guidance in developing the literature search.
Funding Source
This work was funded by the Modeling of Infectious Disease Agent Study, National Institute of General Medical Sciences Grant # R01 GM125440.
Financial Disclosures: GSG has received research grants and research support from the US National Institutes of Health, University of Washington, Bill & Melinda Gates Foundation, Gilead Sciences, Alere Technologies, Merck & Co., Janssen Pharmaceutica, Cerus Corporation, ViiV Healthcare, Bristol-Myers Squibb, Roche Molecular Systems and Abbott Molecular Diagnostics.
Footnotes
All authors attest they meet the ICMJE criteria for authorship
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors declare no conflicts of interest.
References
- [1].Levin BR, Lipsitch M, Bonhoeffer S. Population Biology, Evolution, and Infectious Disease: Convergence and Synthesis. Science 1999;283:806–9. doi: 10.1126/science.283.5403.806. [DOI] [PubMed] [Google Scholar]
- [2].Cohen T, Colijn C, Murray M. Modeling the effects of strain diversity and mechanisms of strain competition on the potential performance of new tuberculosis vaccines. Proc Natl Acad Sci 2008;105:16302–7. doi: 10.1073/pnas.0808746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kennedy DA, Read AF. Why does drug resistance readily evolve but vaccine resistance does not? Proc R Soc B Biol Sci 2017;284. doi: 10.1098/rspb.2016.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gandon S, Mackinnon MJ, Nee S, Read AF. Imperfect vaccines and the evolution of pathogen virulence. Nature 2001;414:751. doi: 10.1038/414751a. [DOI] [PubMed] [Google Scholar]
- [5].Mooi FR, van Oirschot H, Heuvelman K, van der Heide HGJ, Gaastra W, Willems RJL. Polymorphism in the Bordetella pertussis Virulence Factors P.69/Pertactin and Pertussis Toxin in The Netherlands: Temporal Trends and Evidence for Vaccine-Driven Evolution. Infect Immun 1998;66:670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Drexler JF, Grard G, Lukashev AN, Kozlovskaya LI, Böttcher S, Uslu G, et al. Robustness against serum neutralization of a poliovirus type 1 from a lethal epidemic of poliomyelitis in the Republic of Congo in 2010. Proc Natl Acad Sci U S A 2014;111:12889–94. doi: 10.1073/pnas.1323502111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Read AF, Mackinnon MJ. Pathogen evolution in a vaccinated world. Evol Health Dis 2008;2:139–152. [Google Scholar]
- [8].Alizon S, Hurford A, Mideo N, Baalen MV. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J Evol Biol 2009;22:245–59. doi: 10.1111/j.1420-9101.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- [9].André J-B, Gandon S, Koella J. Vaccination, within-host dynamics, and virulence evolution. Evolution 2006;60:13–23. doi: 10.1554/05-220.1. [DOI] [PubMed] [Google Scholar]
- [10].Rohani P, Zhong X, King AA. Contact Network Structure Explains the Changing Epidemiology of Pertussis. Science 2010;330:982–5. doi: 10.1126/science.1194134. [DOI] [PubMed] [Google Scholar]
- [11].CDC. Vaccine Testing and Approval Process | CDC; 2018. https://www.cdc.gov/vaccines/basics/test-approve.html (accessed September 17, 2018). [Google Scholar]
- [12].Huppert A, Katriel G. Mathematical modelling and prediction in infectious disease epidemiology. Clin Microbiol Infect 2013;19:999–1005. doi: 10.1111/1469-0691.12308. [DOI] [PubMed] [Google Scholar]
- [13].Alexander ME, Kobes R. Effects of vaccination and population structure on influenza epidemic spread in the presence of two circulating strains. BMC Public Health 2011;11:S8. doi: 10.1186/1471-2458-11-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Choi YH, Jit M, Flasche S, Gay N, Miller E. Mathematical Modelling Long-Term Effects of Replacing Prevnar7 with Prevnar13 on Invasive Pneumococcal Diseases in England and Wales. PLOS ONE 2012;7:e39927. doi: 10.1371/journal.pone.0039927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Flasche S, Edmunds WJ, Miller E, Goldblatt D, Robertson C, Choi YH. The impact of specific and non-specific immunity on the ecology of Streptococcus pneumoniae and the implications for vaccination. Proc R Soc B 2013;280:20131939. doi: 10.1098/rspb.2013.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nurhonen M, Cheng AC, Auranen K. Pneumococcal Transmission and Disease In Silico: A Microsimulation Model of the Indirect Effects of Vaccination. PLoS ONE 2013;8. doi: 10.1371/journal.pone.0056079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Herbeck JT, Peebles K, Edlefsen PT, Rolland M, Murphy JT, Gottlieb GS, et al. HIV population-level adaptation can rapidly diminish the impact of a partially effective vaccine. Vaccine 2018;36:514–20. doi: 10.1016/j.vaccine.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Link-Gelles R, Taylor T, Moore MR. Forecasting invasive pneumococcal disease trends after the introduction of 13-valent pneumococcal conjugate vaccine in the United States, 2010–2020. Vaccine 2013;31:2572–7. doi: 10.1016/j.vaccine.2013.03.049. [DOI] [PubMed] [Google Scholar]
- [19].Nurhonen M, Auranen K. Optimal Serotype Compositions for Pneumococcal Conjugate Vaccination under Serotype Replacement. PLOS Comput Biol 2014;10:e1003477. doi: 10.1371/journal.pcbi.1003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wilson JN, Nokes DJ, Carman WF. Predictions of the emergence of vaccine-resistant hepatitis B in The Gambia using a mathematical model. Epidemiol Amp Infect 2000;124:295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Temime L, Boelle P-Y, Opatowski L, Guillemot D. Impact of Capsular Switch on Invasive Pneumococcal Disease Incidence in a Vaccinated Population. PLOS ONE 2008;3:e3244. doi: 10.1371/journal.pone.0003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fryer HR, McLean AR. Modelling the Spread of HIV Immune Escape Mutants in a Vaccinated Population. PLOS Comput Biol 2011;7:e1002289. doi: 10.1371/journal.pcbi.1002289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Iwami S, Takeuchi Y, Liu X, Nakaoka S. A geographical spread of vaccine-resistance in avian influenza epidemics. J Theor Biol 2009;259:219–28. doi: 10.1016/j.jtbi.2009.03.040. [DOI] [PubMed] [Google Scholar]
- [24].Iwami S, Suzuki T, Takeuchi Y. Paradox of Vaccination: Is Vaccination Really Effective against Avian Flu Epidemics? PLoS ONE 2009;4:e4915. doi: 10.1371/journal.pone.0004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bottomley C, Roca A, Hill PC, Greenwood B, Isham V. A mathematical model of serotype replacement in pneumococcal carriage following vaccination. J R Soc Interface 2013;10:20130786. doi: 10.1098/rsif.2013.0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Elbasha EH, Galvani AP. Vaccination against multiple HPV types. Math Biosci 2005;197:88–117. doi: 10.1016/j.mbs.2005.05.004. [DOI] [PubMed] [Google Scholar]
- [27].Hogea C, Van Effelterre T, Vyse A. Exploring the population-level impact of MenB vaccination via modeling: Potential for serogroup replacement. Hum Vaccines Immunother 2015;12:451–66. doi: 10.1080/21645515.2015.1080400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pitzer VE, Bilcke J, Heylen E, Crawford FW, Callens M, Smet FD, et al. Did Large-Scale Vaccination Drive Changes in the Circulating Rotavirus Population in Belgium? Sci Rep 2015;5:18585. doi: 10.1038/srep18585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pitzer VE, Patel MM, Lopman BA, Viboud C, Parashar UD, Grenfell BT. Modeling rotavirus strain dynamics in developed countries to understand the potential impact of vaccination on genotype distributions. Proc Natl Acad Sci 2011;108:19353–8. doi: 10.1073/pnas.1110507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Takahashi S, Liao Q, Boeckel TPV, Xing W, Sun J, Hsiao VY, et al. Hand, Foot, and Mouth Disease in China: Modeling Epidemic Dynamics of Enterovirus Serotypes and Implications for Vaccination. PLOS Med 2016;13:e1001958. doi: 10.1371/journal.pmed.1001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vickers DM, Anonychuk AM, De Wals P, Demarteau N, Bauch CT. Evaluation of serogroup C and ACWY meningococcal vaccine programs: Projected impact on disease burden according to a stochastic two-strain dynamic model. Vaccine 2015;33:268–75. doi: 10.1016/j.vaccine.2013.09.034. [DOI] [PubMed] [Google Scholar]
- [32].Worby CJ, Wallinga J, Lipsitch M, Goldstein E. Population effect of influenza vaccination under co-circulation of non-vaccine variants and the case for a bivalent A/H3N2 vaccine component. Epidemics 2017;19:74–82. doi: 10.1016/j.epidem.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang Y, Auranen K, Eichner M. The Influence of Competition and Vaccination on the Coexistence of Two Pneumococcal Serotypes. Epidemiol Infect 2004;132:1073–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Choi YH, Jit M, Gay N, Andrews N, Waight PA, Melegaro A, et al. 7-Valent Pneumococcal Conjugate Vaccination in England and Wales: Is It Still Beneficial Despite High Levels of Serotype Replacement? PLOS ONE 2011;6:e26190. doi: 10.1371/journal.pone.0026190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Melegaro A, Choi YH, George R, Edmunds WJ, Miller E, Gay NJ. Dynamic models of pneumococcal carriage and the impact of the Heptavalent Pneumococcal Conjugate Vaccine on invasive pneumococcal disease. BMC Infect Dis 2010;10:90. doi: 10.1186/1471-2334-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Van Effelterre T, Moore MR, Fierens F, Whitney CG, White L, Pelton SI, et al. A dynamic model of pneumococcal infection in the United States: Implications for prevention through vaccination. Vaccine 2010;28:3650–60. doi: 10.1016/j.vaccine.2010.03.030. [DOI] [PubMed] [Google Scholar]
- [37].Lipsitch M Vaccination against colonizing bacteria with multiple serotypes. Proc Natl Acad Sci U S A 1997;94:6571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kroger A, Duchin J, Vázquez M. General Best Practice Guidelines for Immunization. 2014. https://www.cdc.gov/vaccines/hcp/acip-recs/general-recs/index.html (accessed September 20, 2018).
- [39].WHO. WHO Vaccines and diseases 2018. http://www.who.int/immunization/diseases/en/ (accessed October 24, 2018).
- [40].WHO. WHO Global Vaccine Action Plan 2011–2020. World Health Organ; 2012. http://www.who.int/immunization/global_vaccine_action_plan/GVAP_doc_2011_2020/en/ (accessed September 20, 2018). [Google Scholar]
- [41].WHO. WHO Pneumococcal disease 2011. http://www.who.int/immunization/topics/pneumococcal_disease/en/ (accessed September 20, 2018).
- [42].Higgins JP, Altman DG, Gøtzsche PC, Jüni P, Moher D, Oxman AD, et al. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. Bmj 2011;343:d5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gregorio ED, Rappuoli R. Vaccines for the future: learning from human immunology. Microb Biotechnol 2012;5:149–55. doi: 10.1111/j.1751-7915.2011.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


