Biodiversity is good for you
Changes in biodiversity have the potential to either increase or reduce the incidence of infectious disease in plants and animals — including humans — because they involve interactions among species. At a minimum, this requires a host and a pathogen; often many more species are involved, including additional hosts, vectors and other organisms with which these species interact. Felicia Keesing and colleagues review the evidence that reduced biodiversity affects the transmission of infectious diseases of humans, other animals and plants. Despite important questions still to be answered, they conclude that the evidence that biodiversity exerts a protective effect on infectious diseases is sufficiently strong to include biodiversity protection as a strategy to improve health.
Supplementary information
The online version of this article (doi:10.1038/nature09575) contains supplementary material, which is available to authorized users.
Subject terms: Infectious diseases, Epidemiology, Biodiversity
Abstract
Current unprecedented declines in biodiversity reduce the ability of ecological communities to provide many fundamental ecosystem services. Here we evaluate evidence that reduced biodiversity affects the transmission of infectious diseases of humans, other animals and plants. In principle, loss of biodiversity could either increase or decrease disease transmission. However, mounting evidence indicates that biodiversity loss frequently increases disease transmission. In contrast, areas of naturally high biodiversity may serve as a source pool for new pathogens. Overall, despite many remaining questions, current evidence indicates that preserving intact ecosystems and their endemic biodiversity should generally reduce the prevalence of infectious diseases.
Supplementary information
The online version of this article (doi:10.1038/nature09575) contains supplementary material, which is available to authorized users.
Main
In June 2010, a new organization, the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES)—patterned after the Intergovernmental Panel on Climate Change (IPCC)—was established to assess changes to the diversity of life on the Earth and how these changes will affect human well-being1. Human well-being would be adversely affected by biodiversity losses if ecosystems with reduced biodiversity are less able to provide the ecosystem services—such as carbon sequestration, nutrient cycling and resistance to drought—on which humans rely. In recent years, a consensus has emerged that ecosystem functions decline as biodiversity is lost2. Here we examine how biodiversity affects the transmission and emergence of infectious diseases and evaluate the evidence that reduced disease transmission is an important ecosystem service provided by high biodiversity.
Biodiversity encompasses the diversity of genes, species and ecosystems. Increases in human populations have resulted in an unprecedented and precipitous loss of biodiversity3. Current extinction rates are estimated to be at least 100–1,000 times background extinction rates and future extinction rates (over the next 50 years) are estimated to be 10 to 100 times present extinction rates3. A large proportion of species in all assessed taxa are currently threatened with extinction (12% of birds, 23% of mammals, 32% of amphibians; 31% of gymnosperms; 33% of corals4) and the best estimate of population trends of birds, mammals, amphibians, reptiles and fish indicates that since 1970 global population sizes have declined by almost 30% (ref. 5). Global and local extinction rates of some taxa, particularly microbes, have not been well characterized. For the many organisms that are symbionts of other organisms, extinction of their hosts can cause their extinction too6. Collectively, these declines and extinctions are caused by changing the Earth’s ecosystems to meet growing demands for food, fresh water, fibre, timber and fuel, and by climate change.
Changes in biodiversity have the potential to affect the risk of infectious disease exposure in plants and animals—including humans—because infectious diseases by definition involve interactions among species. At a minimum, these species include a host and a pathogen; often many more species are involved, including additional hosts, vectors and other organisms with which these species interact. Intriguingly, biodiversity may play a dual role in the emergence and transmission of infectious diseases. On the one hand, high biodiversity may provide a larger potential source of novel pathogens, but on the other hand, biodiversity can reduce further pathogen transmission for both long-established and newly emerging diseases. We first review the effects of biodiversity on the transmission of established diseases and then turn to disease emergence.
Biodiversity and pathogen transmission
Transmission of pathogens between species
Biodiversity loss might affect disease transmission through several mechanisms (Box 1). If the effect of each species on pathogen transmission were entirely idiosyncratic, one would expect that diversity declines would be equally likely to cause a decrease or an increase in disease transmission in the remaining species. However, in recent years, a consistent picture has emerged—biodiversity loss tends to increase pathogen transmission and disease incidence. This pattern occurs across ecological systems that vary in type of pathogen, host, ecosystem and transmission mode (Table 1). As an example, West Nile virus is a mosquito-transmitted virus for which several species of passerine birds act as hosts. Three recent studies detected strong correlations between low bird diversity and increased human risk or incidence of West Nile encephalitis in the United States7,8,9. Communities with low avian diversity tend to be dominated by species that amplify the virus, inducing high infection prevalence in mosquitoes and people, while communities with high avian diversity contain many species that are less competent hosts. For hantavirus pulmonary syndrome, a directly transmitted zoonotic disease, correlational and experimental studies have shown that a lower diversity of small mammals increases the prevalence of hantaviruses in their hosts, thereby increasing risk to humans (Box 2). Diversity has a similar effect for plant diseases, with species losses increasing the transmission of two fungal rust pathogens that infect perennial rye grass and other plant species10.
Table 1.
Biodiversity loss can increase transmission
| Disease | Mechanism | Reference |
|---|---|---|
| Amphibian limb malformation | B | 12 |
| Bacteriophage of Pseudomonas syringae | B | 52 |
| Coral diseases | A | 53 |
| Fungal disease of Daphnia | B | 54 |
| Hantavirus disease | A, B | 23, 55, 56, 57 |
| Helminthic parasite of fish | A* | 58 |
| Lyme disease | A, B | 18, 22, 59 |
| Malaria | A | 60 |
| Puccinia rust infection of ryegrass | A* | 10 |
| Schistosomiasis | B | 12 |
| Trematode diseases of snails and birds | B | 61, 62, 63 |
| West Nile fever | A*, B* | 7, 8, 9, 64 |
Disease examples are since 2005. A more complete table, including several counterexamples, is available from the corresponding author. Mechanisms for effects were reported by authors or demonstrated in the text (A = host/vector abundance; B = host/vector/parasite behaviour; see Box 1 for details). Asterisks indicate a suggested mechanism. Other studies have been reviewed elsewhere21,65.
Recent attention has focused on assessing the mechanisms by which reduced biodiversity increases pathogen transmission (Box 1). Biodiversity loss can clearly increase transmission if it reduces predation and competition on reservoir hosts, thereby increasing their density. However, controversy has centred around whether the loss of species can increase transmission in other ways11. This is because field studies like those on West Nile virus, hantaviruses and rye grass have typically not controlled for changes in host density that can result from changes in ‘species richness’ (the number of species present in a community, which is a measure of taxonomic diversity). As a consequence, it has been difficult to separate the effects of higher density from those of reduced diversity. Recent experiments confirm that increases in disease transmission can occur when species richness declines even if host density stays constant. One of the best examples comes from a study of Schistosoma mansoni, a trematode that causes schistosomiasis in humans. The parasite alternately infects snails and humans via free-living infectious stages. Host snails were placed in tanks at a constant density either alone or with one or two other species of non-host snails and then exposed to the parasite12. In single-species treatments, host snails were 30% more likely to be infected because parasites in multi-species treatments often ended up in dead-end hosts. Increased parasite–host encounter rates caused by reduced diversity are sufficient to increase disease transmission for Schistosoma.
The loss of species can increase encounter rates between pathogens and hosts, as in the Schistosoma example, when the lost species are not hosts for the pathogen. But if the lost species are indeed hosts capable of transmission, this declining diversity could also reduce the total number of hosts, thereby decreasing transmission if all else remains equal13,14. Certainly reductions in the number of hosts can reduce the number of vectors15 and also their infection prevalence16,17, but empirical examples are relatively rare, in part because the issue has been neglected, and also because all else rarely remains equal. For example, the loss of hosts can cause compensatory increases in the abundances of other hosts, such that total host abundance changes little relative to total host abundance in more diverse communities. Even when total host abundance does decline in less diverse systems, differences in host quality among species can alter simple correlations between host abundance and infection risk18.
Pathogen transmission is not always a function of host density. For example, the number of infectious bites delivered by highly mobile vectors like mosquitoes can be independent of the density of the host population14. Transmission of directly transmitted pathogens like hantaviruses can also be independent of host density if transmission involves behavioural encounters, for example, aggressive interactions between rodents, and if the frequency of these encounters does not vary much with host density14,19. In systems like these, the loss of host species can actually increase transmission if the lost hosts are suboptimal for parasite development and reproduction; this is because these suboptimal hosts absorb pathogens but are poor at transmitting them.
In sum, reducing biodiversity can increase disease transmission when the lost species are either not hosts for the pathogen or are suboptimal ones. For pathogens for which transmission is a function of host density, loss of diversity is most likely to increase transmission if the loss causes an increase in the density of competent hosts. The number and diversity of examples of pathogens for which species loss leads to increases in total transmission suggests that these conditions are frequently met (Table 1). Additional studies in other disease systems would better establish the generality of these relationships.
Box 1: Effects of biodiversity on disease transmission.
The loss of biodiversity can affect the transmission of infectious diseases65 by changing:
(1) The abundance of the host or vector. For plants, seeding experimental fields with plant species that are not hosts for fungal pathogens decreased threefold the pathogen load of species that are hosts, apparently by reducing host density through competition66. On the other hand, a greater diversity of host species can sometimes increase pathogen transmission by increasing the abundance of vectors67.
(2) The behaviour of the host, vector or parasite. In a more diverse community, one of the parasitic worms that causes schistosomiasis (which infects 200 million people worldwide) is more likely to end up in an unsuitable intermediate host. This can reduce the probability of subsequent infection of humans by 25–99% (ref. 68). For hantavirus in Utah, USA, rodent hosts on more diverse plots are more likely to come in contact with heterospecific mammals and less likely to come in contact with conspecifics, reducing the probability of transmission of the virus55. In principle, higher diversity could influence behaviours with a resulting increase in disease transmission65 or could alter the evolutionary dynamics of virulence and transmission pathways.
(3) The condition of the host or vector. In experimental rice fields in China, rice plants in genetically diverse mixtures had drier leaves because the mixture changed microclimatic conditions69. As a consequence, infection with rice blast fungus was less prevalent in diverse fields. Genetically diverse plantings can also lead to induced resistance in host plants because they are exposed to similar pathogens that are specialists on the other cultivars70.
For some disease systems (for example, Lyme disease), multiple mechanisms operate in concert, leading to a compounding effect of biodiversity loss on increased disease transmission (Table 1).
Box 2: Case study of hantavirus pulmonary syndrome.
Hantaviruses are a group of negative-stranded RNA viruses associated with murid rodents. They can cause severe morbidity and mortality in humans, with case-fatality rates near 40% (ref. 71). Infected rodents shed hantavirus in saliva, urine and faeces; transmission to humans occurs through inhalation of aerosolized excreta as well as through rodent bites72. The risk of human exposure increases as the density and infection prevalence of rodent reservoirs increase72.
In a field study in Oregon, USA, the only variable significantly linked to infection prevalence in deer mouse host populations was mammalian species diversity, with the prevalence of the hantavirus Sin Nombre virus rising from 2% to 14% as diversity declined. Deer mouse population density was not statistically associated with Sin Nombre virus infection prevalence, suggesting that high diversity reduced intraspecific encounters rather than host abundance56. A study in Utah, USA55, also found a negative correlation between small-mammal diversity and Sin Nombre virus infection prevalence in deer mice. As in Oregon, high diversity reduced infection prevalence apparently by reducing intraspecific encounters rather than by reducing host density, a result supported by experiments19.
The conclusions of these studies were supported by an experimental study of hantaviruses in small mammal communities of Panamá23. In replicated plots, small-mammal diversity was reduced by trapping and removing species that are not hosts for the virus; infection prevalence in hosts was compared on manipulated and unmanipulated plots (Box 2 figure). Experimentally reduced small-mammal diversity caused an increase in the density of host species and also in seroconversion rates and seroprevalence within hosts (Box 2 figure).
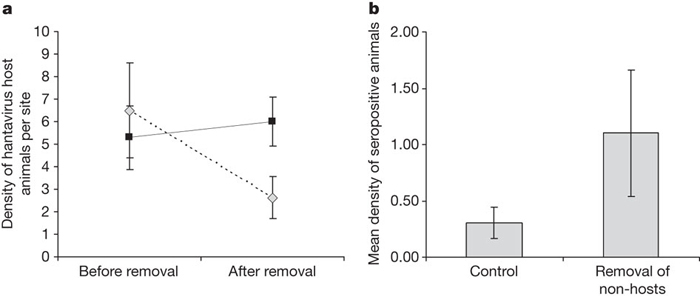
Effects of experimental removal of species.
a, Mean (± standard error) population abundance of hantavirus hosts in Panamá in field plots before and after non-host species had been removed (solid line), and in unmanipulated controls (dashed line). Hosts on control plots underwent a strong seasonal decline in abundance, whereas those on plots where non-hosts were experimentally removed did not. b, Mean (± standard error) density of seropositive (currently or previously infected) animals on plots fromwhich non-hosts had been removed and on control plots. Analysed from data provided in ref. 23.
Species diversity versus species identity
The loss of particular species in a community clearly has the potential to increase disease transmission. But does reducing diversity itself increase transmission, or is increased transmission the consequence of the removal of particular species? The answer depends on how species composition changes as richness changes20,21. For example, if those host species most responsible for amplifying the pathogen tend to persist or even thrive as biodiversity is lost, then disease risk will consistently increase as biodiversity declines. On the other hand, if amplifying species tend to disappear as biodiversity declines, then biodiversity loss will tend to reduce disease risk. These hypothetical possibilities indicate the importance of understanding both the non-random sequences by which species are lost from communities, and whether the species that tend to occur only in more species-rich communities tend to amplify or buffer pathogen transmission.
In several case studies, the species most likely to be lost from ecological communities as diversity declines are those most likely to reduce pathogen transmission. In the Lyme disease system of eastern North America, for example, the white-footed mouse is simultaneously the most abundant host species, the most competent host for the Lyme bacterium, and the highest-quality host for immature tick vectors18 (Fig. 1). As a consequence, this host species infects a high proportion of the ticks within forest communities. The white-footed mouse is also an ecologically resilient species, present in both species-rich and species-poor communities22. In contrast, Virginia opossums are poor hosts for the pathogen, kill the vast majority of ticks that attempt to feed on them, and are absent from many low-diversity forest fragments and degraded forests where mice are abundant18,22. Therefore, as biodiversity is lost, the host with a strong buffering effect—the opossum—disappears, while the host with a strong amplifying effect—the mouse—remains. The primary hosts for the pathogens that cause West Nile encephalitis, hantavirus pulmonary syndrome, and bartonellosis also appear to be resilient species that increase in abundance as biodiversity is lost7,23,24.
Figure 1. Roles of host species in the transmission of Lyme disease in the northeastern USA.
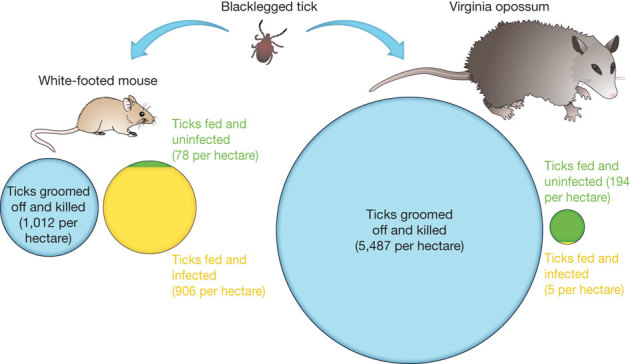
Lyme disease is transmitted to humans by the bite of an infected blacklegged tick (Ixodes scapularis). Immature ticks can acquire the infection if they feed on an infected host and can become infectious to humans if they subsequently survive to the next life stage. White-footed mice are abundant in northeastern forests and feed many ticks18. Ticks that attempt to feed on Virginia opossums are likely to be groomed off and killed. Green-and-yellow circles show the mean number of ticks per hectare fed by mice or opossums; yellow shading shows the proportion of ticks infected after feeding. Blue circles show the mean number of ticks per hectare groomed off and killed. Ticks that feed on mice are highly likely to become infected with the bacterium that causes Lyme disease, whereas those that feed on opossums are not.
Whether an organism’s host competence and its resilience to factors that reduce biodiversity are causally related is an unresolved but critical issue. Traits that make a host resilient to biodiversity loss may also make them susceptible to pathogen infection and transmission. Such a relationship would explain the frequency with which the link between diversity loss and disease transmission has been observed in nature (Table 1). For plants, species that are fast-growing and nutrient-rich with relatively high metabolic rates—characteristics of ‘weedy’ species—can be more competent hosts for arthropod vectors and plant pathogens than those with less weedy traits25. Plants with these weedy traits are also more likely to become more abundant when plant diversity declines26. Consequently, the very species that have traits permitting persistence in degraded and species-poor ecosystems are also more likely to carry high pathogen and vector burdens. A similar pattern may occur in vertebrates—resilience in the face of disturbances that cause biodiversity loss, such as habitat destruction and fragmentation, is facilitated by life-history features such as high reproductive output and intrinsic rates of increase27. Vertebrates with these features tend to invest minimally in some aspects of adaptive immunity28,29,30; we hypothesize that this may make them more competent hosts for pathogens and vectors. Understanding the interrelationships among pathogen transmission, biodiversity loss and interspecific differences in immune function is an important area for future research. Such studies would illuminate how frequently resilient species are also those that increase pathogen transmission, and might provide general rules about the impact of biodiversity loss on disease transmission.
Diversity within individual hosts
Could changes in biodiversity within the bodies of organisms also alter pathogen transmission? Recent improvements in the ability of researchers to detect unculturable microbial species have allowed documentation of the tremendous diversity of microbes upon and within plants and animals. In human bodies, for example, 90% of all cells are microbial31. A number of studies have begun to show links between diseases and the diversity of an organism’s ‘microbiome’.
Changes in the composition of microbiomes are frequently associated with infection and disease. For example, corals suffering from white plague disease have microbial communities distinctly different from those in healthy corals32. In humans, bacterial vaginosis results from changes in the composition of the vaginal microbial community33, and this in turn increases the risk of HIV infection34. Although changes in microbial species composition associated with infection are well-documented, few studies have investigated the effects of changes in diversity itself. In a recent investigation, patients with recurrent episodes of infection caused by the bacterium Clostridium difficile had significantly lower diversity of intestinal microbes than did control patients35. Correlational studies such as these, though intriguing, make it difficult to determine whether changes in microbial communities are the cause or the consequence of infections. But some experimental studies clearly demonstrate that increasing microbial biodiversity can protect against infection. For example, children with a history of ear infections given a mixture of five strains of Streptococcus were less likely to develop subsequent infections compared to a control group36. Similarly, reducing microbial diversity within a host can increase transmission. When mice with persistent infections of C. difficile were treated with antibiotics that reduced the diversity of intestinal microbes, they began shedding C. difficile spores at high rates37.
In some of these examples, a rich microbial community appears to regulate the abundance of endemic microbial species that can become pathogenic when overly abundant35. In other cases, high microbial species diversity can prevent colonization by invasive pathogenic species. For example, the more diverse the microbiome surrounding the roots of wheat plants, the more protected the plants were against invasion by the pathogenic bacterium Pseudomonas aeruginosa38. Similarly, piglets raised in natural environments supporting a high diversity of microbes were more resistant to invasion by pathogenic gut microbes than those raised in more sterile environments39.
The effects of microbial diversity within and upon host bodies show intriguing similarities to the effects of macroscopic species diversity on disease transmission in aquatic and terrestrial ecosystems. Further exploration of these similarities, and particularly the specific mechanisms operating within hosts, is a critical research frontier because changes in microbial diversity might accompany biodiversity loss in their hosts.
Biodiversity and pathogen emergence
For pathogens already established within ecological communities, we have shown that biodiversity loss frequently increases the rate of transmission. But what role, if any, does biodiversity have in the processes by which new pathogens emerge? Between 1940 and 2004, over 300 emerging disease events were identified in humans around the world40. Concomitantly, other emerging infectious diseases also appeared in wildlife, domesticated animals, and crop and wild plants. Emerging infectious diseases include those in which the pathogen has evolved into a new strain within the same host species, for example, through the evolution of drug resistance (methicillin-resistant Staphylococcus aureus or MRSA) or switched to new host species (for example, human immunodeficiency virus or HIV, severe acute respiratory syndrome or SARS). In some cases, the switch to new host species is accompanied by a change in geographic range (for example, West Nile virus in the Americas).
For pathogens that establish in new species, the emergence process involves multiple steps, including the initial invasion into the new host (‘spillover’), the production of transmission stages within the new host, and the establishment of the pathogen in the host population as a whole41,42. The effect of biodiversity may vary for each of these steps. For the initial invasion, biodiversity may act as a source pool. This hypothesis is supported by surveys of emerging diseases of humans: most are zoonotic—jumping to humans from other vertebrate animals43. In one recent analysis, the probability of emergence of pathogens from wildlife to humans was positively correlated with mammalian wildlife species richness when data were corrected for reporting bias40. Other environmental and socioeconomic factors that bring humans into closer contact with potentially new pathogens (for example, forest clearing for agriculture, wildlife hunting) may also contribute to this pattern. Indeed, almost half of the zoonotic diseases that have emerged in humans since 1940 resulted from changes in land use, from changes in agricultural or other food production practices, or from wildlife hunting (Fig. 2). These human activities increase rates of contact between humans and animals, which may be a critical factor underlying spillover.
Figure 2. Drivers and locations of emergence events for zoonotic infectious diseases in humans from 1940–2005.
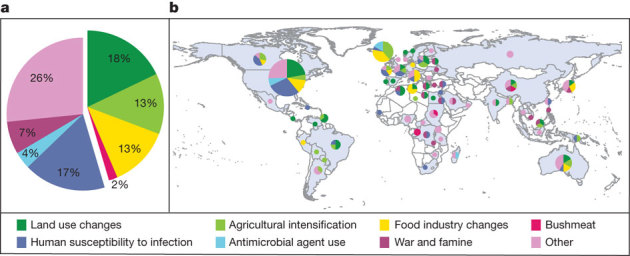
a, Worldwide percentage of emergence events caused by each driver; b, Countries in which the emergence events took place, and the drivers of emergence. The size of the circle represents the number of emergence events: for scale, the number of events in the United States was 59. Globally, almost half of these diseases resulted from changes in land use, changes in agricultural and other food production practices, or through wildlife hunting, which suggests that contact rates between humans and other animals are an important underlying cause of zoonotic disease emergence. ‘Other’ includes international travel and commerce, changes in human demographics and behaviour, changes in the medical industry, climate and weather, breakdown of public health measures, and unspecified causes. Analysed from data in ref. 40.
Once spillover of the pathogen into a new host has occurred, high densities of that host species may facilitate pathogen establishment and transmission within the new host41. For example, Nipah virus spilled over from wild fruit bats to domestic pigs in Malaysia; high densities of pigs in local farms appear to have facilitated establishment of pig-to-pig transmission, and the pathogen then spilled over from pigs to humans44. Such high densities of domesticated species are almost always associated with low biodiversity.
In contrast to emergence through host-switching, 20% of emergence events between 1940 and 2004 arose through the evolution of drug resistance40. For these cases, biodiversity of microbial communities within hosts may have a protective effect; human use of antibiotics is thought to select for resistant microbes by eliminating the great diversity of non-resistant microbial strains and species that suppress resistant strains in the absence of antibiotics. Investigations using recent advances in microbial detection support this idea45,46. Thus, reduced microbial diversity may be an important underlying cause of the emergence of drug-resistant pathogens; this too requires further investigation.
Managing pathogens by managing biodiversity
The addition of particular species—for example, natural enemies or competitors—can reduce the impacts of established pathogens. For example, experimental addition of a naturally occurring bacterium, Janthinobacterium lividum, to the skin of the endangered frog Rana mucosa eliminated frog mortality from experimental infection with chytridiomycosis, which is devastating amphibian populations worldwide47. For corals, application of phages isolated from natural communities can control the spread of bacterial infections48. The growing interest in ‘probiotics’ for humans and harvested species provides another example of this approach49.
More broadly, biodiversity itself seems to protect organisms, including humans, from transmission of infectious diseases in many cases (Table 1). Preserving biodiversity in these cases, and perhaps generally, may reduce the incidence of established pathogens. To preserve high diversity in nature, conservation scientists have developed robust methods that reflect the key principle that larger areas sustain larger numbers of species50. Methods of conserving microbial diversity within and upon bodies or in the environment are less well developed, but avoiding the overuse of antimicrobial compounds is essential. Critically, future research on the relationship between biodiversity and disease must avoid conflating the effects of biogeographic patterns of biodiversity (for example, higher diversity in lower latitudes) with those of anthropogenic reductions in extant biodiversity, because policy and management options can far more readily affect the latter than the former.
For emerging diseases, the observation that a more diverse microbiome within a host suppresses strains that are resistant to antimicrobial compounds suggests that avoiding the over-use of these compounds in medicine and agriculture can prevent the emergence of resistant strains. For pathogens that emerge by switching host species, three management approaches are warranted. First, potential emergence ‘hotspots’ could be predictable on the basis of land-use change and underlying biodiversity patterns; these areas should be targeted for surveillance of endemic wildlife pathogens that have the potential to jump host species40,51. Second, preserving and protecting intact habitats in these hotspots provides a simple, direct way of reducing human–animal contact and reduces the likelihood of emergence of new pathogens, although methods for achieving reduced contact are not always straightforward51. And third, to reduce the probability that pathogens become established and transmissible within a new host population once spillover occurs, the husbandry of high-density monocultures of domestic animals, particularly in areas at high risk of spillover, should be subject both to more intensive surveillance and to measures that reduce contact between wildlife and livestock. Managing potential emergence hotspots by attempting to eliminate them is likely to backfire because the species most resilient to habitat destruction and degradation may be those that amplify pathogen transmission.
Despite many recent advances in our understanding of biodiversity and disease, much remains to be learned. First, we must increase the number of disease systems for which we understand the effects of biodiversity loss on disease transmission across a range of spatial and temporal scales. We must also focus on how to implement specific policies informed by this science. Future research, for example, should monitor changes in epidemiology in regions in which conservation measures are imposed compared to reference sites. A major challenge will be to untangle the complex ways in which other global anthropogenic trends—such as climate change, biotic exchange, nutrient pollution, armed conflict and economic collapse—interact with biodiversity loss to influence disease dynamics, and which of these trends have the greatest impacts on human well-being. Despite remaining questions, connections between biodiversity and disease are now sufficiently clear to increase the urgency of local, regional, and global efforts to preserve natural ecosystems and the biodiversity they contain.
Supplementary information
This file contains Supplementary Table 1 and additional references. (PDF 146 kb)
Acknowledgements
We acknowledge the support of the joint NSF-NIH Ecology of Infectious Disease programme and the EPA Biodiversity and Human Health programme. M. Gillespie provided help in the preparation of the manuscript.
PowerPoint slides
Author Contributions
F.K. and R.S.O. conceived the review. F.K., L.K.B., P.D., A.D., C.D.H., R.D.H., P.H., A.J., K.E.J., C.E.M., S.S.M. and R.S.O. wrote and edited the text. T.B. prepared Fig. 2.
Competing interests
The authors declare no competing financial interests.
References
- 1.Marris, E. New UN science body to monitor biosphere. Nature 10.1038/news.2010.297 (2010)
- 2.Naeem S, Bunker D, Hector A, Loreau M, Perrings C. Biodiversity, Ecosystem Functioning, and Human Wellbeing: an Ecological and Economic Perspective. 2009. [Google Scholar]
- 3.Mace GM, Masundire H, Baillie JEM. Ecosystems and Human Well-Being: Current State and Trends: Findings of the Condition and Trends Working Group. 2005. [Google Scholar]
- 4.IUCN. IUCN Red List of Threatened Species Version 2010.2. 〈http://www.iucnredlist.org〉 (downloaded on, 29 June 2010)
- 5.Loh J, et al. 2010 and Beyond: Rising to the Biodiversity Challenge. 2008. [Google Scholar]
- 6.Dobson AP, et al. Homage to Linnaeus: How many parasites? How many hosts? Proc. Natl Acad. Sci. USA. 2008;105:11482–11489. doi: 10.1073/pnas.0803232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allan BF, et al. Ecological correlates of risk and incidence of West Nile virus in the United States. Oecologia. 2009;155:699–708. doi: 10.1007/s00442-008-1169-9. [DOI] [PubMed] [Google Scholar]
- 8.Ezenwa VO, Godsey MS, King RJ, Guptill SC. Avian diversity and West Nile virus: testing associations between biodiversity and infectious disease risk. Proc. R. Soc. Lond. B. 2006;273:109–117. doi: 10.1098/rspb.2005.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swaddle J, Calos P. Increased avian diversity is associated with lower incidence of human West Nile infection: observation of the dilution effect. PLoS ONE. 2008;3:e2488. doi: 10.1371/journal.pone.0002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roscher C, Schumacher J, Foitzik O, Schulze E-D. Resistance to rust fungi in Lolium perenne depends on within-species variation and performance of the host species in grasslands of different plant diversity. Oecologia. 2007;153:173–183. doi: 10.1007/s00442-007-0713-3. [DOI] [PubMed] [Google Scholar]
- 11.Begon M. Infectious Disease Ecology: Effects of Ecosystems on Disease and of Disease on Ecosystems. 2008. pp. 12–29. [Google Scholar]
- 12.Johnson PTJ, Lund P, Hartson RB, Yoshino T. Community diversity reduces Schistosoma mansoni transmission and human infection risk. Proc. R. Soc. Lond. B. 2009;276:1657–1663. doi: 10.1098/rspb.2008.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudolf VH, Antonovics J. Species coexistence and pathogens with frequency-dependent transmission. Am. Nat. 2005;166:112–118. doi: 10.1086/430674. [DOI] [PubMed] [Google Scholar]
- 14.Dobson AP. Population dynamics of pathogens with multiple host species. Am. Nat. 2004;164:S64–S78. doi: 10.1086/424681. [DOI] [PubMed] [Google Scholar]
- 15.Cecère MC, Gürtler RE, Chuit R, Cohen J. Effects of chickens on the prevalence of infestation and population density of Triatoma infestans in rural houses of northwest Argentina. Med. Vet. Entomol. 1997;11:383–388. doi: 10.1111/j.1365-2915.1997.tb00426.x. [DOI] [PubMed] [Google Scholar]
- 16.Bouma M, Rowland M. Failure of passive zooprophylaxis: cattle ownership in Pakistan is associated with a higher prevalence of malaria. Trans. R. Soc. Trop. Med. Hyg. 1995;89:351–353. doi: 10.1016/0035-9203(95)90004-7. [DOI] [PubMed] [Google Scholar]
- 17.Laurenson MK, Norman RA, Gilbert L, Reid HW, Hudson PJ. Identifying disease reservoirs in complex systems: mountain hares as reservoirs of ticks and louping-ill virus, pathogens of red grouse. J. Anim. Ecol. 2003;72:177–185. doi: 10.1046/j.1365-2656.2003.00688.x. [DOI] [Google Scholar]
- 18.Keesing F, et al. Hosts as ecological traps for the vector of Lyme disease. Proc. R. Soc. Lond. B. 2009;276:3911–3919. doi: 10.1098/rspb.2009.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clay CA, Lehmer EM, St Jeor S, Dearing MD. Testing mechanisms of the dilution effect: deer mice encounter rates, Sin Nombre virus prevalence and species diversity. EcoHealth. 2009;6:250–259. doi: 10.1007/s10393-009-0240-2. [DOI] [PubMed] [Google Scholar]
- 20.Ostfeld RS, LoGiudice K. Community disassembly, biodiversity loss, and the erosion of an ecosystem service. Ecology. 2003;84:1421–1427. doi: 10.1890/02-3125. [DOI] [Google Scholar]
- 21.Johnson PTJ, Thieltges DW. Diversity, decoys and the dilution effect: how ecological communities affect disease risk. J. Exp. Biol. 2010;213:961–970. doi: 10.1242/jeb.037721. [DOI] [PubMed] [Google Scholar]
- 22.LoGiudice K, et al. Impact of host community composition on Lyme disease risk. Ecology. 2008;89:2841–2849. doi: 10.1890/07-1047.1. [DOI] [PubMed] [Google Scholar]
- 23.Suzán G, et al. Experimental evidence for reduced mammalian diversity causing increased hantavirus prevalence. PLoS ONE. 2009;4:e5461. doi: 10.1371/journal.pone.0005461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosoy M, et al. Distribution, diversity, and host specificity of Bartonella in rodents from the Southeastern United States. Am. J. Trop. Med. Hyg. 1997;57:578–588. doi: 10.4269/ajtmh.1997.57.578. [DOI] [PubMed] [Google Scholar]
- 25.Cronin, J. P., Welsh, M. E., Dekkers, M. G., Abercrombie, S. T. & Mitchell, C. E. Host physiological phenotype explains pathogen reservoir potential. Ecol. Lett. 10.1111/j.1461–0248.2010.01513.x (2010) [DOI] [PubMed]
- 26.Pilgrim ES, Crawley MJ, Dolphin K. Patterns of rarity in the native British flora. Biol. Conserv. 2004;120:161–170. doi: 10.1016/j.biocon.2004.02.008. [DOI] [Google Scholar]
- 27.Cardillo M, et al. The predictability of extinction: biological and external correlates of decline in mammals. Proc. R. Soc. Lond. B. 2008;275:1441–1448. doi: 10.1098/rspb.2008.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin LB, Hasselquist D, Wikelski M. Investment in immune defense is linked to pace of life in house sparrows. Oecologia. 2006;147:565–575. doi: 10.1007/s00442-005-0314-y. [DOI] [PubMed] [Google Scholar]
- 29.Martin LB, Weil ZM, Nelson RJ. Immune defense and reproductive pace of life in Peromyscus mice. Ecology. 2007;88:2516–2528. doi: 10.1890/07-0060.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee KA, Wikelski M, Robinson WD, Robinson TR, Klasing KC. Constitutive immune defenses correlate with life-history variables in tropical birds. J. Anim. Ecol. 2008;77:356–363. doi: 10.1111/j.1365-2656.2007.01347.x. [DOI] [PubMed] [Google Scholar]
- 31.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sunagawa S, et al. Bacterial diversity and White Plague disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 2009;3:512–521. doi: 10.1038/ismej.2008.131. [DOI] [PubMed] [Google Scholar]
- 33.Holzman C, et al. Factors linked to bacterial vaginosis in nonpregnant women. Am. J. Public Health. 2001;91:1664–1670. doi: 10.2105/AJPH.91.10.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atashili J, Poolea C, Ndumbeb PM, Adimoraa AA, Smith JS. Bacterial vaginosis and HIV acquisition: a meta-analysis of published studies. AIDS. 2008;22:1493–1501. doi: 10.1097/QAD.0b013e3283021a37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang JY, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 36.Roos K, Håkansson EG, Holm S. Effect of recolonisation with “interfering” α streptococci on recurrences of acute and secretory otitis media in children: randomised placebo controlled trial. Br. Med. J. 2001;322:1–4. doi: 10.1136/bmj.322.7280.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawley TD, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matos A, Kerkhof L, Garland J. Effects of microbial community diversity on the survival of Pseudomonas aeruginosa in the wheat rhizosphere. Microb. Ecol. 2005;49:257–264. doi: 10.1007/s00248-004-0179-3. [DOI] [PubMed] [Google Scholar]
- 39.Mulder Imke E, Schmidt Bettina, Stokes Christopher R, Lewis Marie, Bailey Mick, Aminov Rustam I, Prosser James I, Gill Bhupinder P, Pluske John R, Mayer Claus-Dieter, Musk Corran C, Kelly Denise. Environmentally-acquired bacteria influence microbial diversity and natural innate immune responses at gut surfaces. BMC Biology. 2009;7(1):79. doi: 10.1186/1741-7007-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones K, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudson P, Perkins S, Cattadori I. Infectious Disease Ecology: Effects of Ecosystems on Disease and of Disease on Ecosystems. 2008. pp. 347–367. [Google Scholar]
- 42.Wolfe N, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Epstein JH, Field HE, Luby S, Pulliam JRC, Daszak P. Nipah virus: Impact, origins, and causes of emergence. Curr. Infect. Dis. Rep. 2006;8:59–65. doi: 10.1007/s11908-006-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flanagan JL, et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J. Clin. Microbiol. 2007;45:1954–1962. doi: 10.1128/JCM.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris R, et al. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 2009;3:818–824. doi: 10.1038/ismej.2009.27. [DOI] [PubMed] [Google Scholar]
- 48.Efrony R, Atad I, Rosenberg E. Phage therapy of Coral White Plague disease: properties of phage BA3. Curr. Microbiol. 2009;58:139–145. doi: 10.1007/s00284-008-9290-x. [DOI] [PubMed] [Google Scholar]
- 49.Sleator RD, Hill C. New frontiers in probiotic research. Lett. Appl. Microbiol. 2008;46:143–147. doi: 10.1111/j.1472-765X.2007.02293.x. [DOI] [PubMed] [Google Scholar]
- 50.Margules C, Sarkar S. Systematic Conservation Planning. 2007. [Google Scholar]
- 51.Wolfe ND, Daszak P, Kilpatrick AM, Burke DS. Bushmeat hunting, deforestation, and prediction of zoonoses emergence. Emerg. Infect. Dis. 2005;11:1822–1827. doi: 10.3201/eid1112.040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dennehy JJ, Friedenberg NA, Yang YW, Turner PE. Virus population extinction via ecological traps. Ecol. Lett. 2007;10:230–240. doi: 10.1111/j.1461-0248.2006.01013.x. [DOI] [PubMed] [Google Scholar]
- 53.Raymundo LJ, Halforda AR, Maypab AP, Kerr AM. Functionally diverse reef-fish communities ameliorate coral disease. Proc. Natl Acad. Sci. USA. 2009;106:17067–17070. doi: 10.1073/pnas.0900365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall SR, et al. Friendly competition: evidence for a dilution effect among competitors in a planktonic host–parasite system. Ecology. 2009;90:791–801. doi: 10.1890/08-0838.1. [DOI] [PubMed] [Google Scholar]
- 55.Clay C, Lehmer EM, St, Jeor S, Dearing MD. Sin Nombre virus and rodent species diversity: a test of the dilution and amplification hypotheses. PLoS ONE. 2009;4:e6467. doi: 10.1371/journal.pone.0006467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dizney LJ, Ruedas LA. Increased host species diversity and decreased prevalence of Sin Nombre virus. Emerg. Infect. Dis. 2009;15:1012–1018. doi: 10.3201/eid1507.081083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tersago K, et al. Population, environmental, and community effects on local bank vole (Myodes glareolus) Puumala virus infection in an area with low human incidence. Vector-Borne Zoonotic Dis. 2008;8:235–244. doi: 10.1089/vbz.2007.0160. [DOI] [PubMed] [Google Scholar]
- 58.Kelly DW, Paterson RA, Townsend CR, Poulin R, Tompkins DM. Has the introduction of brown trout altered disease patterns in native New Zealand fish? Freshwat. Biol. 2009;54:1805–1818. doi: 10.1111/j.1365-2427.2009.02228.x. [DOI] [Google Scholar]
- 59.Brunner J, Ostfeld RS. Multiple causes of variable tick burdens on small-mammal hosts. Ecology. 2008;89:2259–2272. doi: 10.1890/07-0665.1. [DOI] [PubMed] [Google Scholar]
- 60.Carlson JC, Dyer LA, Omlin FX, Beier JC. Diversity cascades and malaria vectors. J. Med. Entomol. 2009;46:460–464. doi: 10.1603/033.046.0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kopp K, Jokela J. Resistant invaders can convey benefits to native species. Oikos. 2007;116:295–301. doi: 10.1111/j.0030-1299.2007.15290.x. [DOI] [Google Scholar]
- 62.Thieltges DW, Bordalo MD, Caballero-Hernandez A, Prinz K, Jensen KT. Ambient fauna impairs parasite transmission in a marine parasite-host system. Parasitology. 2008;135:1111–1116. doi: 10.1017/S0031182008004526. [DOI] [PubMed] [Google Scholar]
- 63.Thieltges DW, Reise K, Prinz K, Jensen KT. Invaders interfere with native parasite-host interactions. Biol. Invasions. 2009;11:1421–1429. doi: 10.1007/s10530-008-9350-y. [DOI] [Google Scholar]
- 64.Koenig WD, Hochachka WM, Zuckerberg B, Dickinson JL. Ecological determinants of American crow mortality due to West Nile virus during its North American sweep. Oecologia. 2010;163:903–909. doi: 10.1007/s00442-010-1627-z. [DOI] [PubMed] [Google Scholar]
- 65.Keesing F, Holt RD, Ostfeld RS. Effects of species diversity on disease risk. Ecol. Lett. 2006;9:485–498. doi: 10.1111/j.1461-0248.2006.00885.x. [DOI] [PubMed] [Google Scholar]
- 66.Mitchell CE. Mitchell, C. A., Tilman, D. & Groth, J. V. Effects of grassland plant species diversity, abundance, and composition on foliar fungal disease. Ecology. 2002;83:1713–1726. doi: 10.1890/0012-9658(2002)083[1713:EOGPSD]2.0.CO;2. [DOI] [Google Scholar]
- 67.Saul A. Zooprophylaxis or zoopotentiation: the outcome of introducing animals on vector transmission is highly dependent on the mosquito mortality while searching. Malar. J. 2003;2:32–50. doi: 10.1186/1475-2875-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laracuente A, Brown RA, Jobin W. Comparison of four species of snails as potential decoys to intercept schistosome miracidia. Am. J. Trop. Med. Hyg. 1979;28:99–105. doi: 10.4269/ajtmh.1979.28.99. [DOI] [PubMed] [Google Scholar]
- 69.Zhu Y-Y, et al. Panicle blast and canopy moisture in rice cultivar mixtures. Phytopathology. 2005;95:433–438. doi: 10.1094/PHYTO-95-0433. [DOI] [PubMed] [Google Scholar]
- 70.Mundt C. Use of multiline cultivars and cultivar mixtures for disease management. Annu. Rev. Phytopathol. 2002;40:381–410. doi: 10.1146/annurev.phyto.40.011402.113723. [DOI] [PubMed] [Google Scholar]
- 71.CDC. Hantavirus pulmonary syndrome in five pediatric patients—four states, 2009. Morbidity Mortality Week. Rep.58, 1409–1412 (2009) [PubMed]
- 72.Yates TL, et al. The ecology and evolutionary history of an emergent disease: hantavirus pulmonary syndrome. Bioscience. 2002;52:989–998. doi: 10.1641/0006-3568(2002)052[0989:TEAEHO]2.0.CO;2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Table 1 and additional references. (PDF 146 kb)


