Abstract
Cross-species transmission of viruses from wildlife animal reservoirs poses a marked threat to human and animal health1. Bats have been recognized as one of the most important reservoirs for emerging viruses and the transmission of a coronavirus that originated in bats to humans via intermediate hosts was responsible for the high-impact emerging zoonosis, severe acute respiratory syndrome (SARS)2–10. Here we provide virological, epidemiological, evolutionary and experimental evidence that a novel HKU2-related bat coronavirus, swine acute diarrhoea syndrome coronavirus (SADS-CoV), is the aetiological agent that was responsible for a large-scale outbreak of fatal disease in pigs in China that has caused the death of 24,693 piglets across four farms. Notably, the outbreak began in Guangdong province in the vicinity of the origin of the SARS pandemic. Furthermore, we identified SADS-related CoVs with 96–98% sequence identity in 9.8% (58 out of 591) of anal swabs collected from bats in Guangdong province during 2013–2016, predominantly in horseshoe bats (Rhinolophus spp.) that are known reservoirs of SARS-related CoVs. We found that there were striking similarities between the SADS and SARS outbreaks in geographical, temporal, ecological and aetiological settings. This study highlights the importance of identifying coronavirus diversity and distribution in bats to mitigate future outbreaks that could threaten livestock, public health and economic growth.
Subject terms: Viral epidemiology, Viral reservoirs
Analysis of viral samples from deceased piglets shows that a bat coronavirus was responsible for an outbreak of fatal disease in China and highlights the importance of the identification of coronavirus diversity and distribution in bats in order to mitigate future outbreaks of disease.
Main
The emergence of SARS in southern China in 2002, which was caused by a previously unknown coronavirus (SARS-CoV)11–15 and has led to more than 8,000 human infections and 774 deaths (http://www.who.int/csr/sars/en/), highlights two new frontiers in emerging infectious diseases. First, it demonstrates that coronaviruses are capable of causing fatal diseases in humans. Second, the identification of bats as the reservoir for SARS-related coronaviruses, and the fact that SARS-CoV3–10 probably originated in bats, firmly establishes that bats are an important source of highly lethal zoonotic viruses, such as Hendra, Nipah, Ebola and Marburg viruses16.
Here we report on a series of fatal swine disease outbreaks in Guangdong province, China, approximately 100 km from the location of the purported index case of SARS. Most strikingly, we found that the causative agent of this swine acute diarrhoea syndrome (SADS) is a novel HKU2-related coronavirus that is 98.48% identical in genome sequence to a bat coronavirus, which we detected in 2016 in bats in a cave in the vicinity of the index pig farm. This new virus (SADS-CoV) originated from the same genus of horseshoe bats (Rhinolophus) as SARS-CoV.
From 28 October 2016 onwards, a fatal swine disease outbreak was observed in a pig farm in Qingyuan, Guangdong province, China, very close to the location of the first known index case of SARS in 2002, who lived in Foshan (Extended Data Fig. 1a). Porcine epidemic diarrhoea virus (PEDV, a coronavirus) had caused prior outbreaks at this farm, and was detected in the intestines of deceased piglets at the start of the outbreak. However, PEDV could no longer be detected in deceased piglets after 12 January 2017, despite accelerating mortality (Fig. 1a), and extensive testing for other common swine viruses yielded no results (Extended Data Table 1). These findings suggested that this was an outbreak of a novel disease. Clinical signs are similar to those caused by other known swine enteric coronaviruses17, 18 and include severe and acute diarrhoea and acute vomiting, leading to death due to rapid weight loss in newborn piglets that are less than five days of age. Infected piglets died 2–6 days after disease onset, whereas infected sows suffered only mild diarrhoea and most sows recovered within two days. The disease caused no signs of febrile illness in piglets or sows. The mortality rate was as high as 90% in piglets that were five days or younger, whereas in piglets that were older than eight days, the mortality dropped to 5%. Subsequently, SADS-related outbreaks were found in three additional pig farms within 20–150 km of the index farm (Extended Data Fig. 1a) and, by 2 May 2017, the disease had caused the death of 24,693 piglets at these four farms (Fig. 1a). In farm A alone, 64% (4,659 out of 7,268) of all piglets that were born in February died. The outbreak has abated, and measures that were taken to control SADS included separation of sick sows and piglets from the rest of the herd. A qPCR test described below was used as the main diagnostic tool to confirm SADS-CoV infection.
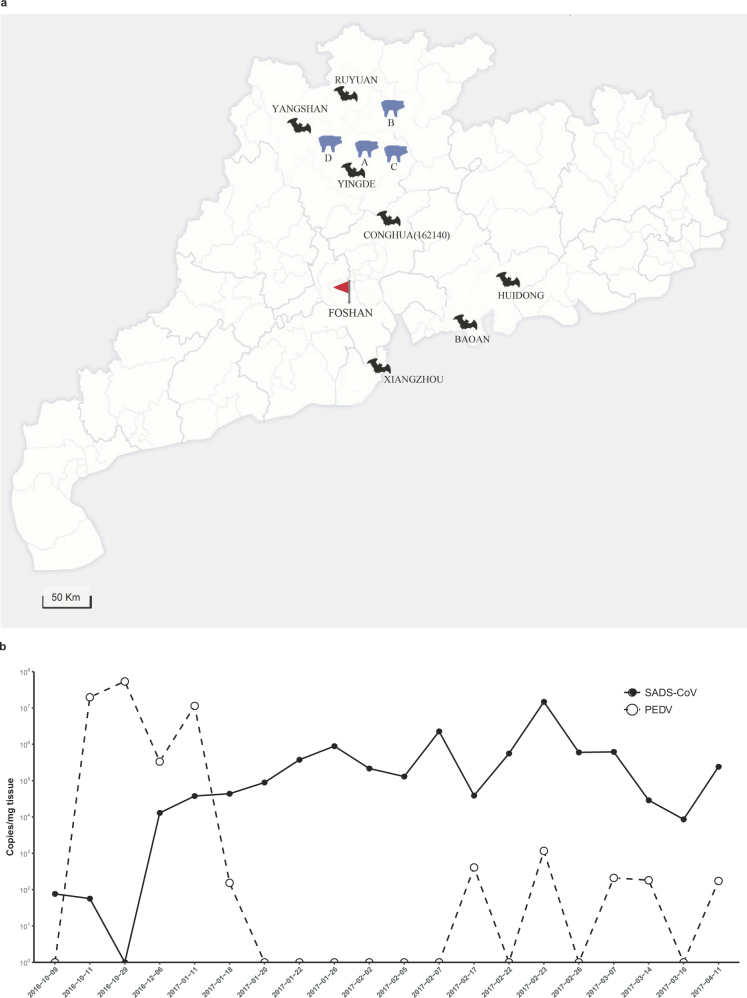
Extended Data Fig. 1. Map of outbreak locations and sampling sites in Guangdong province, China and the co-circulation of PEDV and SADS-CoV during the initial outbreak on farm A.
a, SADS-affected farms are labelled (farms A–D) with blue swine silhouettes following the temporal sequence of the outbreaks. Bat sampling sites are indicated with black bat silhouettes. The bat SADSr-CoV that is most closely related to SADS-CoV (sample 162140) originated in Conghua. The red flag marks Foshan city, the site of the SARS index case. b, Pooled intestinal samples (n = 5 or more biological independent samples) were collected at dates given on the x axis from deceased piglets and analysed by qPCR. The viral load for each piglet is shown as copy number per milligram of intestine tissue (y axis).
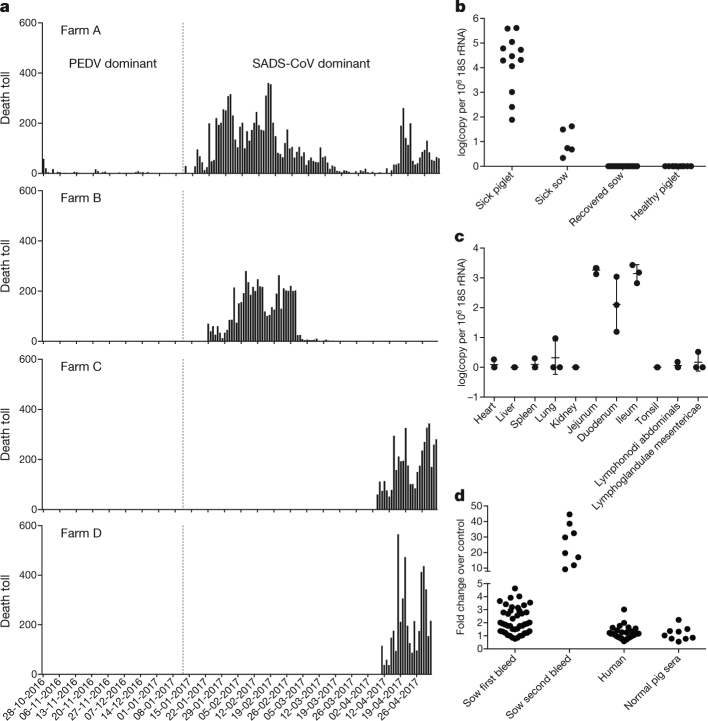
Fig. 1. Detection of SADS-CoV infection in pigs in Guangdong, China.
a, Records of daily death toll on the four farms from 28 October 2016 to 2 May 2017. b, Detection of SADS-CoV by qPCR. The y axis shows the log(copy number per 106 copies of 18S rRNA). n = 12 sick piglets, 5 sick sows, 16 recovered sows and 10 healthy piglets. c, Tissue distribution of SADS-CoV in diseased pigs. n = 3. Data are mean ± s.d.; dots represent individual values. d, Detection of SADS-CoV antibodies. n = 46 sows from whom serum was first taken in the first three weeks of the outbreak (First bleed), n = 8 sows from whom serum was taken again (Second bleed) at more than one month after the onset of the outbreak, n = 8 sera from healthy pig controls, n = 35 human sera from pig farmers.
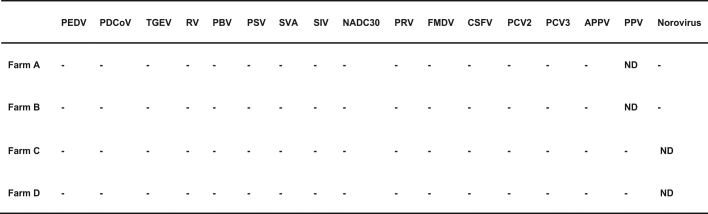
Extended Data Table 1.
List of all known swine viruses tested by PCR at the beginning of the of SADS outbreak investigation on the four farms
Faeces, intestine or faecal swabs collected from January to April 2017 were tested. Sampling type and number of samples per farm were as follows. Farm A: 1 fecal sample, 20 intestinal sample and 6 faecal swabs; farm B: 1 faecal sample and 15 intestinal samples; farm C: 2 intestinal sample and 1 faecal swab; farm D: 5 faecal sample and 1 faecal swab. The dash indicates a negative PCR result. ND, not determined. APPV, atypical porcine pestivirus; CSFV, classical swine fever virus; FMDV, foot and mouth disease virus; NADC30, porcine reproductive and respiratory syndrome virus, strain NADC30; PBV, porcine picobirnavirus; PCV2, porcine circovirus 2; PCV3, porcine circovirus 3; PDCoV, porcine deltacoronavirus; PEDV, porcine epidemic diarrhoea virus; PPV, porcine parvovirus; PRV, porcine pseudorabies virus; PSV, porcine sapelovirus; RV, porcine rotavirus; SIV, swine influenza virus; SVA, porcine senecavirus A; TGEV, porcine transmissible gastroenteritis virus.
A sample collected from the small intestine of a diseased piglet was analysed by metagenomics analysis using next-generation sequencing (NGS) to identify potential aetiological agents. Of the 15,256,565 total reads obtained, 4,225 matched sequences of the bat CoV HKU2, which was first detected in Chinese horseshoe bats in Hong Kong and Guangdong province, China19. By de novo assembly and targeted PCR, we obtained a 27,173-bp CoV genome that shared 95% sequence identity to HKU2-CoV (GenBank accession number NC_009988). Thirty-three full genome sequences of SADS-CoV were subsequently obtained (8 from farm A, 5 from farm B, 11 from farm C and 9 from farm D) that were 99.9% identical to each other (Supplementary Table 1).
Using qPCR targeting the nucleocapsid gene (Supplementary Table 2), we detected SADS-CoV in acutely sick piglets and sows, but not in recovered or healthy pigs on the four farms, nor in nearby farms that showed no evidence of SADS. The virus replicated to higher titres in piglets than in sows (Fig. 1b). SADS-CoV displayed tissue tropism of the small intestine (Fig. 1c), as observed for other swine enteric coronaviruses20. Retrospective PCR analysis revealed that SADS-CoV was present on farm A during the PEDV epidemic, where the first strongly positive SADS-CoV sample was detected on 6 December 2016. From mid-January onwards, SADS-CoV was the dominant viral agent detected in diseased animals (Extended Data Fig. 1b). It is possible that the presence of PEDV early in the SADS-CoV outbreak may have somehow facilitated or enhanced spillover and amplification of SADS. However the fact that the vast majority of piglet mortality occurred after PEDV infection had become undetectable suggests that SADS-CoV itself causes a lethal infection in pigs that was responsible for these large-scale outbreaks, and that PEDV does not directly contribute to its severity in individual pigs. This was supported by the absence of PEDV and other known swine diarrhoea viruses during the peak and later phases of the SADS outbreaks in the four farms (Extended Data Table 1).
We rapidly developed an antibody assay based on the S1 domain of the spike (S) protein using a luciferase immunoprecipitation system21. Because SADS occurs acutely and has a rapid onset in piglets, serological investigation was conducted only in sows. Among 46 recovered sows tested, 12 were seropositive for SADS-CoV within three weeks of infection (Fig. 1d). To investigate possible zoonotic transmission, serum samples from 35 farm workers who had close contact with sick pigs were also analysed using the same luciferase immunoprecipitation system approach and none were positive for SADS-CoV.
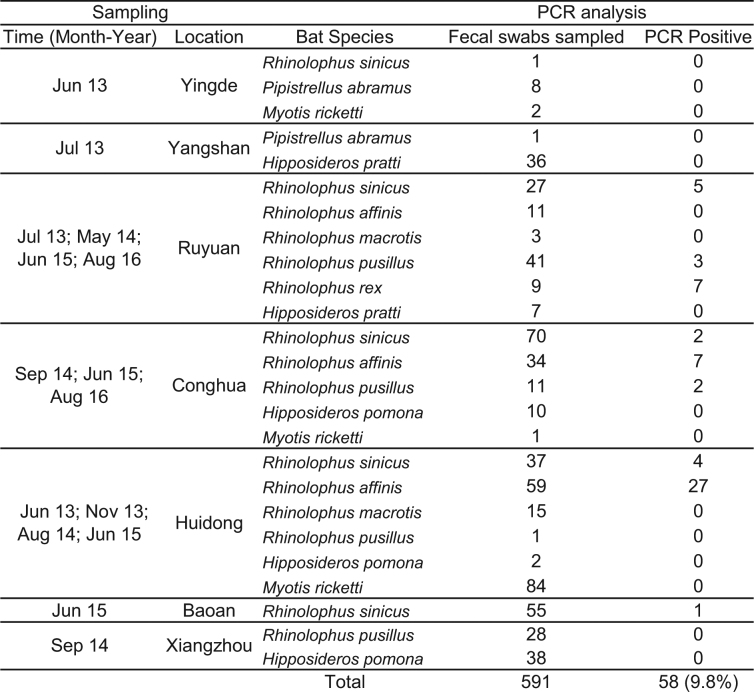
Although the overall genome identity of SADS-CoV and HKU2-CoV is 95%, the S gene sequence identity is only 86%, suggesting that the previously reported HKU2-CoV is not the direct progenitor of SADS-CoV, but that they may have originated from a common ancestor. To test this hypothesis, we developed a SADS-CoV-specific qPCR assay based on its RNA-dependent RNA polymerase (RdRp) gene (Supplementary Table 2) and screened 591 bat anal swabs collected between 2013 and 2016 from seven different locations in Guangdong province (Extended Data Fig. 1a). A total of 58 samples (9.8%) tested positive (Extended Data Table 2), all were from Rhinolophus spp. bats that are also the natural reservoir hosts of SARS-related coronaviruses3–10. Four complete genome sequences with the highest RdRp PCR-fragment sequence identity to that of SADS-CoV were determined by NGS. They are very similar in size (27.2 kb) compared to SADS-CoV (Fig. 2a) and we tentatively call them SADS-related coronaviruses (SADSr-CoV). Overall sequence identity between SADSr-CoV and SADS-CoV ranges from 96 to 98%. Most importantly, the S protein of SADS-CoV shared more than 98% sequence identity with sequences of two of the SADSr-CoVs (samples 162149 and 141388), compared to 86% with HKU2-CoV. The major sequence differences among the four SADSr-CoV genomes were found in the predicted coding regions of the S and NS7a and NS7b genes (Fig. 2a). In addition, the coding region of the S protein N-terminal (S1) domain was determined from 19 bat SADSr-CoVs to enable more detailed phylogenetic analysis.
Extended Data Table 2.
List of SADSr-CoVs detected in bats in Guangdong, China
See Extended Data Fig. 1 for sampling sites in relation to SARS and SADS outbreak locations.
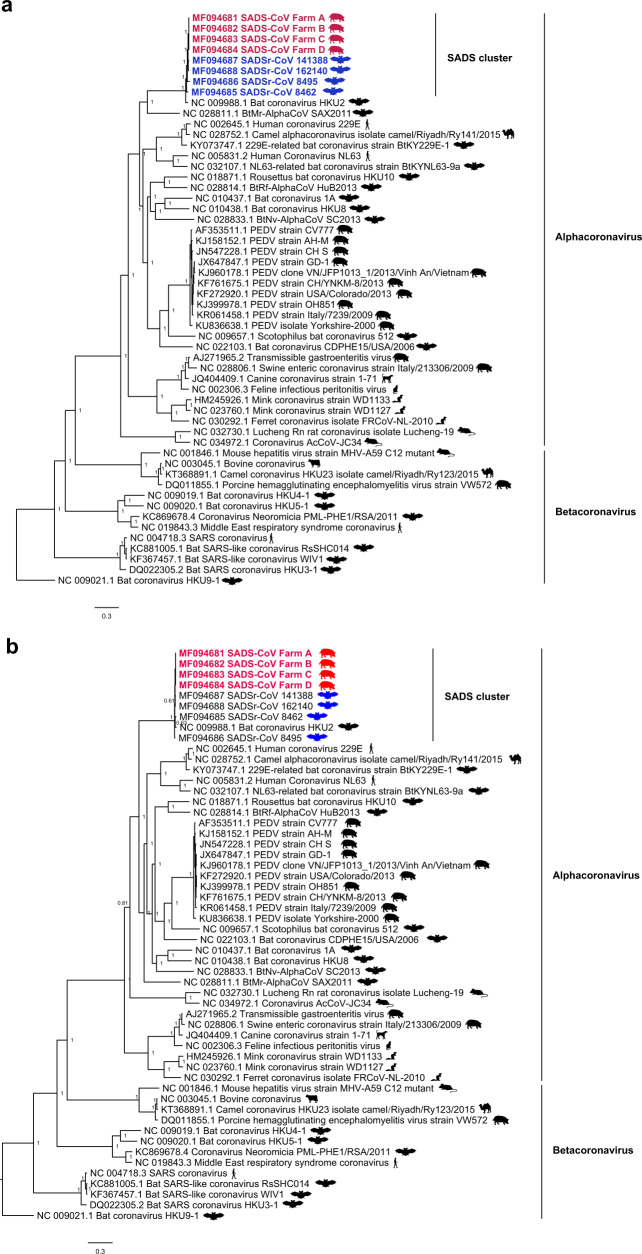
Fig. 2. Genome and phylogenetic analysis of SADS-CoV and SADSr-CoV.
a, Genome organization and comparison. Colour-coding for different genomic regions as follows. Green, non-structural polyproteins ORF1a and ORF1b; yellow, structural proteins S, E, M and N; blue, accessory proteins NS3a, NS7a and NS7b; Orange, untranslated regions. The level of sequence identity of SADSr-CoV to SADS-CoV is illustrated by different patterns of boxes: Solid colour, highly similar; Dotted fill, moderately similar; Dashed fill, least similar. b, Phylogenetic analysis of 57 S1 sequences (33 from SADS-CoV and 24 from SADSr-CoV). Different colours represent different host species as shown on the left. Scale bar, nucleotide substitutions per site.
The phylogeny of S1 and the full-length genome revealed a high genetic diversity of alphacoronaviruses among bats and strong coevolutionary relationships with their hosts (Fig. 2b and Extended Data Fig. 2), and showed that SADS-CoVs were more closely related to SADSr-CoVs from Rhinolophus affinis than from Rhinolophus sinicus, in which HKU2-CoV was found. Both phylogenetic and haplotype network analyses demonstrated that the viruses from the four farms probably originated from their reservoir hosts independently (Extended Data Fig. 3), and that a few viruses might have undergone further genetic recombination (Extended Data Fig. 4). However, molecular clock analysis of the 33 SADS-CoV genome sequences failed to establish a positive association between sequence divergence and sampling date. Therefore, we speculate that either the virus was introduced into pigs from bats multiple times, or that the virus was introduced into pigs once, but subsequent genetic recombination disturbed the molecular clock.
Extended Data Fig. 2. Bayesian phylogenetic tree of the full-length genome and the ORF1a and ORF1b sequences of SADS-CoV and related coronaviruses.
a, Bayesian phylogenetic tree of the full-length genome. b, Bayesian phylogenetic tree of the ORF1a and ORF1b sequences. Trees were constructed using MrBayes with the average standard deviation of split frequencies under 0.01. The host of each sequence is represented as a silhouette. Newly sequenced SADS-CoVs are highlighted in red, bat SADSr-CoVs are shown in blue and previously published sequences are shown in black. Scale bars, nucleotide substitutions per site.
Extended Data Fig. 3. Phylogeny and haplotype network analyses of the 33 SADS-CoV strains from the four farms.

a, Phylogenetic tree constructed using MrBayes. The GTR+GAMMA model was applied and 20 million steps were run, with the first 10% removed as burn in. Viruses from different farms are labelled with different colours. Scale bar, nucleotide substitutions per site. b, Median-joining haplotype network constructed using ProART. In this analysis, ɛ = 0 was used. The size of the circles represents the number of samples. The larger the circle, the more samples it includes.
Extended Data Fig. 4. Recombination analysis for SADS-CoV and related CoVs.
The potential genetic recombination events were detected using RDP. For each virus strain, different colours represent different sources of the genomes.
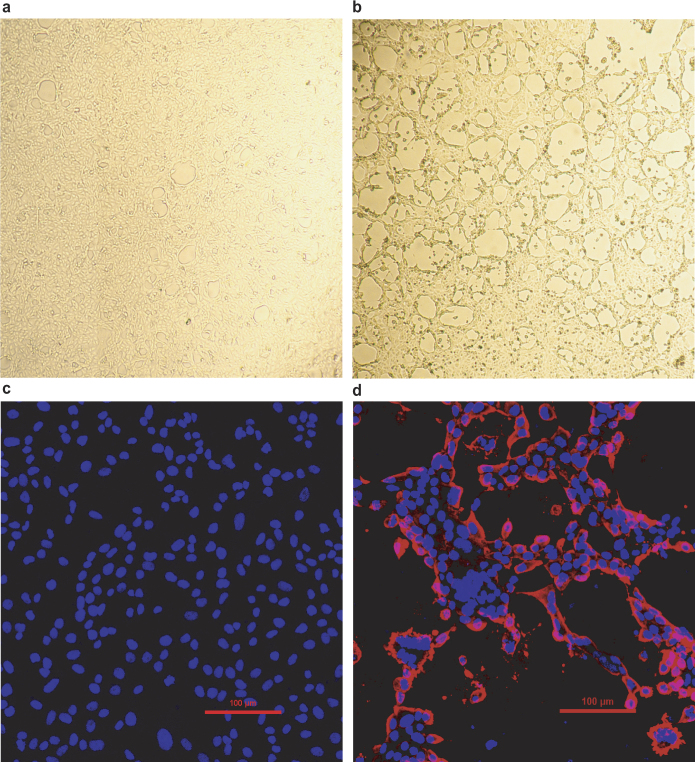
For viral isolation, we tried to culture the virus in a variety of cell lines (see Methods for details) using intestinal tissue homogenates as starting material. Cytopathogenic effects were observed in Vero cells only after five passages (Extended Data Fig. 5a, b). The identity of SADS-CoV was verified in Vero cells by immunofluorescence microscopy (Extended Data Fig. 5c, d) and by whole-genome sequencing (GenBank accession number MG557844). Similar results were obtained by other groups22, 23.
Extended Data Fig. 5. Isolation and antigenic characterization of SADS-CoV.
a, b, Vero cells are shown 20 h after infection with mock (a) or SADS-CoV (b). c, d, Mock or SADS-CoV-infected samples stained with rabbit serum raised against the recombinant SADSr-CoV N protein (red) and DAPI (blue). The experiment was conducted independently three times with similar results. Scale bars, 100 μm.
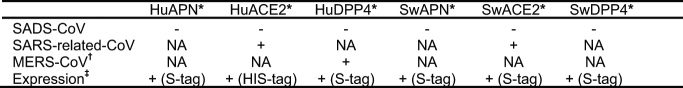
Known coronavirus host cell receptors include angiotensin-converting enzyme 2 (ACE2) for SARS-related CoV, aminopeptidase N (APN) for certain alphacoronaviruses, such as human (H)CoV-229E, and dipeptidyl peptidase 4 (DPP4) for Middle East respiratory syndrome (MERS)-CoV24–26. To investigate the receptor usage of SADS-CoV, we tested live or pseudotyped SADS-CoV infection on HeLa cells that expressed each of the three molecules. Whereas the positive control worked for SARS-related CoV and MERS-CoV pseudoviruses, we found no evidence of enhanced infection or entry for SADS-CoV, suggesting that none of these receptors functions as a receptor for virus entry for SADS-CoV (Extended Data Table 3).
Extended Data Table 3.
Test of SADS-CoV entry and infection in Hela cells expressing known coronavirus receptors
*Gene accession numbers for the genes used in this study: human APN, M22324.1; human ACE2, NM_021804; human DPP4, NM_001935.3; SwAPN (swine APN), NM_214277.1; SwACE2 (swine ACE2), NM_001116542.1; SwDPP4 (swine DPP4), NM_214257.1.
†For MERS-CoV infection, HIV-pseudovirus was used.
‡Expression of APN, ACE2 and DPP4 was confirmed by antibodies against fused tags.
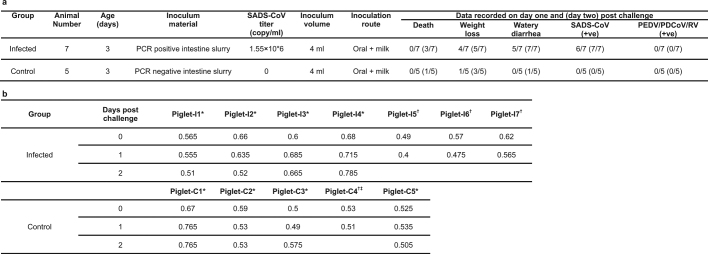
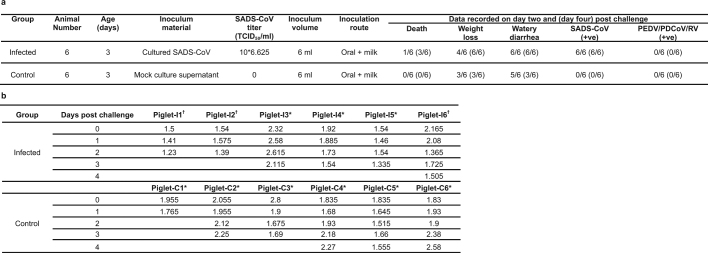
To fulfill Koch’s postulates for SADS-CoV, two different types of animal challenge experiments were conducted (see Methods for details). The first challenge experiment was conducted with specific pathogen-free piglets that were infected with a tissue homogenate of SADS-CoV-positive intestines. Two days after infection, 3 out of 7 animals died in the challenge group whereas 4 out of 5 survived in the control group. Incidentally, the one piglet that died in the control group was the only individual that did not receive colostrum due to a shortage in the supply. It is thus highly likely that lack of nursing and inability to access colostrum was responsible for the death (Extended Data Table 4). For the second challenge, healthy piglets were acquired from a farm in Guangdong that had been free of diarrheal disease for a number of weeks before the experiment, and were infected with the cultured isolate of SADS-CoV or tissue-culture medium as control. Of those inoculated with SADS-CoV, 50% (3 out of 6) died between 2 and 4 days after infection, whereas all control animals survived (Extended Data Table 5). All animals in the infected group suffered watery diarrhoea, rapid weight loss and intestinal lesions (determined after euthanasia upon experiment termination, Extended Data Tables 4, 5). Histopathological examination revealed marked villus atrophy in SADS-CoV inoculated farm piglets four days after inoculation but not in control piglets (Fig. 3a, b) and viral N protein-specific staining was observed mainly in small intestine epithelial cells of the inoculated piglets (Fig. 3c, d).
Extended Data Table 4.
Experimental infection of SPF piglets using intestine tissue homogenate
Experimental details can be found in the Methods. a, Animals were recorded every day for signs of disease, including weight loss, diarrhoea and death. PCR on DNA from faecal swabs was carried out to monitor the presence of SADS-CoV or other pig viruses. b, Daily body weight record of all piglets. Weights are in kg.
*Euthanized on the indicated day for further analysis.
†Animal died during the experiment.
‡The only animal that did not receive colostrum in this experiment due to shortage in supply.
Extended Data Table 5.
Experimental animal infection of farm piglets using cultured SADS-CoV
Experimental details can be found in the Methods. a, Animals were recorded every day for signs of disease, including weight loss, diarrhoea and death. PCR on DNA from faecal swabs was carried out to monitor the presence of SADS-CoV or other pig viruses. b, Daily body weight record of all piglets. Weights are in kg.
*Euthanized on the indicated day for further analysis.
†Animal died during the experiment.
Fig. 3. Immunohistopathology of SADS-CoV infected tissues.
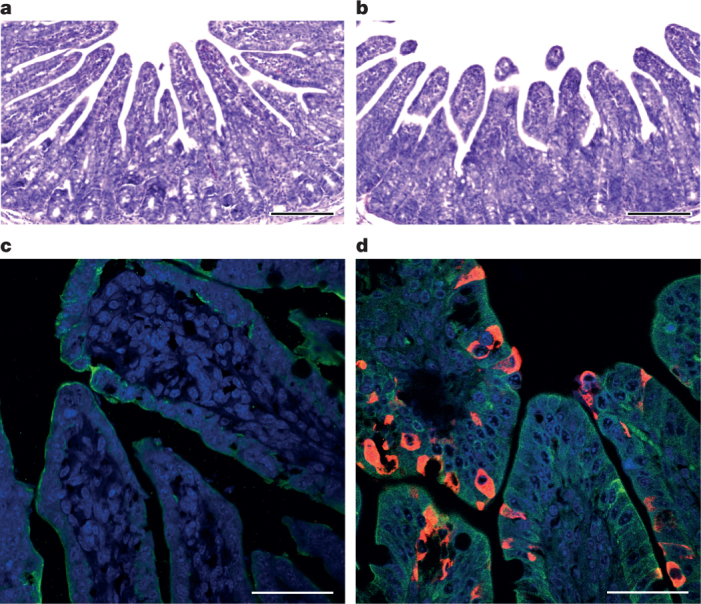
a–d, Sections of jejunum tissue from control (a, c) and infected (b, d) farm piglets four days after inoculation were stained with haematoxylin and eosin (a, b) or rabbit anti-SADSr-CoV N serum (red), DAPI (blue) and mouse antibodies against epithelial cell markers cytokeratin 8, 18 and 19 (green) in (c, d). SADS-CoV N protein is evident in epithelial cells and deeper in the tissue of infected piglets, which exhibit villus shortening. Scale bars, 200 μm (a, b) and 50 μm (c, d). The experiment was conducted three times independently with similar results.
The current study highlights the value of proactive viral discovery in wildlife, and targeted surveillance in response to an emerging infectious disease event, as well as the disproportionate importance of bats as reservoirs of viruses that threaten veterinary and public health1. It also demonstrates that by using modern technological platforms, such as NGS, luciferase immunoprecipitation system serology and phylogenetic analysis, key experiments that traditionally rely on the isolation of live virus can be performed rapidly before virus isolation.
Methods
Sample collection
Bats were captured and sampled in their natural habitat in Guangdong province (Extended Data Fig. 1) as described previously4. Faecal swab samples were collected in viral transport medium (VTM) composed of Hank’s balanced salt solution at pH 7.4 containing BSA (1%), amphotericin (15 μg ml−1), penicillin G (100 units ml−1) and streptomycin (50 μg ml−1). Stool samples from sick pigs were collected in VTM. When appropriate and feasible, intestinal samples were also taken from deceased animals. Samples were aliquoted and stored at –80 °C until use. Blood samples were collected from recovered sows and workers on the farms who had close contact with sick pigs. Serum was separated by centrifugation at 3,000g for 15 min within 24 h of collection and preserved at 4 °C. Human serum collection was approved by the Medical Ethics Committee of the Wuhan School of Public Health, Wuhan University and Hummingbird IRB. Human, pigs and bats were sampled without gender or age preference unless indicated (for example, piglets or sows). No statistical methods were used to predetermine sample size.
Virus isolation
The following cells were used for virus isolation in this study: Vero (cultured in DMEM and 10% FBS); Rhinolophus sinicus primary or immortalized cells generated in our laboratory (all cultured in DMEM/F12 and 15% FBS): kidney primary cells (RsKi9409), lung primary cells (RsLu4323), lung immortalized cells (RsLuT), brain immortalized cells (RsBrT) and heart immortalized cells (RsHeT); and swine cell lines: two intestinal porcine enterocytes cell lines, IPEC (RPMI1640 and 10% FBS) and SIEC (DMEM and 10% FBS), three kidney cell lines PK15, LLC-PK1 (DMEM and 10% FBS for both) and IBRS (MEM and 10% FBS), and one pig testes cell line, ST (DMEM and 10% FBS). All cell lines were tested free of mycoplasma contamination, species were confirmed and authenticated by microscopic morphologic evaluation. None of the cell lines was on the list of commonly misidentified cell lines (by the ICLAC).
Cultured cell monolayers were maintained in their respective medium. PCR-positive pig faecal samples or the supernatant from homogenized pig intestine (in 200 μl VTM) were spun at 8,000g for 15 min, filtered and diluted 1:2 with DMEM supplemented with 16 μg ml−1 trypsin before addition to the cells. After incubation at 37 °C for 1 h, the inoculum was removed and replaced with fresh culture medium containing antibiotics (below) and 16 μg ml−1 trypsin. The cells were incubated at 37 °C and observed daily for cytopathic effect (CPE). Four blind passages (three-day interval between every passage) were performed for each sample. After each passage, both the culture supernatant and cell pellet were examined for the presence of virus by RT–PCR using the SADS-CoV primers listed in Supplementary Table 2. Penicillin (100 units ml−1) and streptomycin (15 μg ml−1) were included in all tissue culture media.
RNA extraction, S1 gene amplification and qPCR
Whenever commercial kits were used, the manufacturer’s instructions were followed without modification. RNA was extracted from 200 μl of swab samples (bat), faeces or homogenized intestine (pig) with the High Pure Viral RNA Kit (Roche). RNA was eluted in 50 μl of elution buffer and used as the template for RT–PCR. Reverse transcription was performed using the SuperScript III kit (Thermo Fisher Scientific).
To amplify S1 genes from bat samples, nested PCR was performed with primers designed based on HKU2-CoV (GenBank accession number NC_009988.1)19 (Supplementary Table 2). The 25-μl first-round PCR mixture contained 2.5 μl 10× PCR reaction buffer, 5 pmol of each primer, 50 mM MgCl2, 0.5 mM dNTP, 0.1 μl Platinum Taq Enzyme (Thermo Fisher Scientific) and 1 μl cDNA. The 50-μl second-round PCR mixture was identical to the first-round PCR mixture except for the primers. Amplification of both rounds was performed as follows: 94 °C for 5 min followed by 60 cycles at 94 °C for 30 s, 50 °C for 40 s, 72 °C for 2.5 min, and a final extension at 72 °C for 10 min. PCR products were gel-purified and sequenced.
For qPCR analysis, primers based on SADS-CoV RdRp and N genes were used (Supplementary Table 2). RNA extracted from above was reverse-transcribed using PrimeScript RT Master Mix (Takara). The 10 μl qPCR reaction mix contained 5 μl 2× SYBR premix Ex TaqII (Takara), 0.4 μM of each primer and 1 μl cDNA. Amplification was performed as follows: 95 °C for 30 s followed by 40 cycles at 95 °C for 5 s, 60 °C for 30 s, and a melting curve step.
Luciferase immunoprecipitation system assay
The SADS-CoV S1 gene was codon-optimized for eukaryotic expression, synthesized (GenScript) and cloned in frame with the Renilla luciferase gene (Rluc) and a Flag tag in the pREN2 vector21. pREN2-S1 plasmids were transfected into Cos-1 cells using Lipofectamine 2000 (Thermo Fisher Scientific). At 48 h post-transfection, cells were collected, lysed and a luciferase assay was performed to determine Rluc expression for both the empty vector (pREN2) and the pREN2-S1 construct. For testing of unknown pig or human serum samples, 1 μl of serum was incubated with 10 million units of Rluc alone (vector) or Rluc-S1, respectively, together with 3.5 μl of a 30% protein A/G UltraLink resin suspension (Pierce, Thermo Fisher Scientific). After extensive washing to remove unbounded luciferase-tagged antigens, the captured luciferase amount was determined using the commercial luciferase substrate kit (Promega). The ratio of Rluc-S1:Rluc (vector) was used to determine the specific S1 reactivity of pig and human sera. Commercial Flag antibody (Thermo Fisher Scientific) was used as the positive control, and various pig sera (from uninfected animals in China or Singapore; or pigs infected with PEDV, TGEV or Nipah virus) were used as a negative control.
Protein expression and antibody production
The N gene from SADSr-CoV 3755 (GenBank accession number MF094702), which shares a 98% amino acid sequence identity to the SADS-CoV N protein, was inserted into pET-28a+ (Novagen) for prokaryotic expression. Transformed Escherichia coli were grown at 37 °C for 12–18 h in medium containing 1 mM IPTG. Bacteria were collected by centrifugation and resuspended in 30 ml of 5 mM imidazole and lysed by sonication. The lysate, from which N protein expression was confirmed with an anti-His-tag antibody, was applied to Ni2+ resin (Thermo Fisher Scientific). The purified N protein, at a concentration of 400 μg ml−1, was used to immunize rabbits for antibody production following published methods27. After immunization and two boosts, rabbits were euthanized and sera were collected. Rabbit anti-N protein serum was used 1:10,000 for subsequent western blots.
Amplification, cloning and expression of human and swine genes
Construction of expression clones for human ACE2 in pcDNA3.1 has been described previously5, 28. Human DPP4 was amplified from human cell lines. Human APN (also known as ANPEP) was commercially synthesized. Swine APN (also known as ANPEP), DPP4 and ACE2 were amplified from piglet intestine. Full-length gene fragments were amplified using specific primers (provided upon request). Human ACE2 was cloned into pCDNA3.1 fused with a His tag. Human APN and DPP4, swine APN, DPP4 and ACE2 were cloned into pCAGGS fused with an S tag. Purified plasmids were transfected into HeLa cells. After 24 h, expression human or swine genes in HeLa cells was confirmed by immunofluorescence assay using mouse anti-His tag or mouse anti-S tag monoclonal antibodies (produced in house) followed by Cy3-labelled goat anti-mouse/rabbit IgG (Proteintech Group).
Pseudovirus preparation
The codon-humanized S genes of SADS-CoV or MERS-CoV cloned into pcDNA3.1 were used for pseudovirus construction as described previously5, 28. In brief, 15 μg of each pHIV-Luc plasmid (pNL4.3.Luc.R-E-Luc) and the S-protein-expressing plasmid (or empty vector control) were co-transfected into 4 × 106 HEK293T cells using Lipofectamine 3000 (Thermo Fisher Scientific). After 4 h, the medium was replaced with fresh medium. Supernatants were collected 48 h after transfection and clarified by centrifugation at 3,000g, then passed through a 0.45-μm filter (Millipore). The filtered supernatants were stored at −80 °C in aliquots until use. To evaluate the incorporation of S proteins into the core of HIV virions, pseudoviruses in supernatant (20 ml) were concentrated by ultracentrifugation through a 20% sucrose cushion (5 ml) at 80,000g for 90 min using a SW41 rotor (Beckman). Pelleted pseudoviruses were dissolved in 50 μl phosphate-buffered saline (PBS) and examined by electron microscopy.
Pseudovirus infection
HeLa cells transiently expressing APN, ACE2 or DPP4 were prepared using Lipofectamine 2000 (Thermo Fisher Scientific). Pseudoviruses prepared above were added to HeLa cells overexpressing APN, ACE2 or DPP4 24 h after transfection. The unabsorbed viruses were removed and replaced with fresh medium at 3 h after infection. The infection was monitored by measuring the luciferase activity conferred by the reporter gene carried by the pseudovirus, using the Luciferase Assay System (Promega) as follows: cells were lysed 48 h after infection, and 20 μl of the lysates was taken for determining luciferase activity after the addition of 50 μl of luciferase substrate.
Examination of known CoV receptors for SADS-CoV entry/infection
HeLa cells transiently expressing APN, ACE2 or DPP4 were prepared using Lipofectamine 2000 (Thermo Fisher Scientific) in a 96-well plate, with mock-transfected cells as controls. SADS-CoV grown in Vero cells was used to infect HeLa cells transiently expressing APN, ACE2 or DPP4. The inoculum was removed after 1 h of absorption and washed twice with PBS and supplemented with medium. SARS-related-CoV WIV167 and MERS-CoV HIV-pseudovirus were used as positive control for human/swine ACE2 or human/swine DPP4, respectively. After 24 h of infection, cells were washed with PBS and fixed with 4% formaldehyde in PBS (pH 7.4) for 20 min at room temperature. SARS-related-CoV WIV16 replication was detected using rabbit antibody against the SARS-related-CoV Rp3 N protein (made in house, 1:100) followed by Cy3-conjugated goat anti-rabbit IgG (1:50, Proteintech)7. SADS-CoV replication was monitored using rabbit antibody against the SADSr-CoV 3755 N protein (made in house, 1:50) followed by FITC-conjugated goat anti-rabbit IgG (1:50, Proteintech). Nuclei were stained with DAPI (Beyotime). Staining patterns were examined using confocal microscopy on a FV1200 microscope (Olympus). Infection of MERS-CoV HIV-pseudovirus was monitored by luciferase 48 h after infection.
High-throughput sequencing, pathogen screening and genome assembly
Tissue from the small intestine of deceased pigs was homogenized and filtered through 0.45-μm filters before nucleic acid extraction and ribosomal RNA was depleted using the NEBNext rRNA Depletion Kit (New England Biolabs). Metagenomics analysis of both RNA and DNA viruses was performed. For RNA virus screening, the sequencing library was constructed using Ion Total RNA-Seq Kit v2 (Thermo Fisher Scientific). For DNA virus screening, NEBNext Fast DNA Fragmentation & Library Prep Set for Ion Torrent (New England Biolabs) was used for library preparation. Both libraries were sequenced on an Ion S5 sequencer (Thermo Fisher Scientific). An analysis pipeline was applied to the sequencing data, which included the following analysis steps: (1) raw data quality filtering; (2) host genomic sequence filtering; (3) BLASTn search against the virus nucleotide database using BLAST; (4) BLASTx search against the virus protein database using DIAMOND v.0.9.0; (5) contig assembling and BLASTx search against the virus protein database. For whole viral genome sequencing, amplicon primers (provided upon request) were designed using the Thermo Fisher Scientific online tool with the HKU2-CoV and the SADS-CoV farm A genomes as references, and the sequencing libraries were constructed using NEBNext Ultra II DNA Library Prep Kit for Illumina and sequenced on an MiSeq sequencer. PCR and Sanger sequencing was performed to fill gaps in the genome. Genome sequences were assembled using CLC Genomic Workbench v.9.0. 5′-RACE was performed to determine the 5′-end of the genomes using SMARTer RACE 5′/3′ Kit (Takara). Genomes were annotated using Clone Manager Professional Suite 8 (Sci-Ed Software).
Phylogenetic analysis
SADS-CoV genome sequences and other representative coronavirus sequences (obtained from GenBank) were aligned using MAFFT v.7.221. Phylogenetic analyses with full-length genome, S gene and RdRp were performed using MrBayes v.3.2. Markov chain Monte Carlo was run for 20–50 million steps using the GTR+G+I model (general time reversible model of nucleotide substitution with a proportion of invariant sites and γ-distributed rates among sites). The first 10% was removed as burn-in. The association between phylogenies and phenotypes (for example, host species and farms) was assessed by BaTS beta-build2, with the trees obtained in the previous step used as input. For SADS-CoVs, a median-joining network analysis was performed using PopART v.1.7, with ɛ = 0. Phylogenetic analysis of the 33 full-length SADS-CoV genome sequences was performed using RAxML v.8.2.11, with GTRGAMMA as the nucleotide substitution model and 1,000 bootstrap replicates. The maximum likelihood tree was used to test the molecular clock using TempEst v.1.5. Potential genetic recombination events in our datasets were detected using RDP v.4.72.
Animal infection studies
Experiments were carried out strictly in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The use of animals in this study was approved by the South China Agricultural University Committee of Animal Experiments (approval number 201004152).
Two different animal challenge experiments were conducted. Pigs were used without gender preference. In the first experiment, which was conducted before the virus was isolated, we used three-day old specific pathogen-free (SPF) piglets of the same breeding line, cared for at a SPF facility, fed with colostrum (except one). These piglets were bred and reared to be free of PEDV, CSFV, SIV, PCV2 and PPV infections, and were routinely tested for viral infections using PCR. We also conducted NGS to further confirm that these were animals were free of infection of the above viruses before the animal experiment, and to demonstrate that the animals were free of SADS-CoV infection. The intestinal tissue samples from healthy and diseased animals (intestinal samples excised from euthanized piglets, then ground to make slurry for the inoculum and NGS was performed to confirm no other pig pathogens were found in the samples), were used to feed two groups of 5 (control) and 7 (infection) animals, respectively. For the second experiment, isolated SADS-CoV was used to infect healthy piglets from a farm in Guangdong, which had been free of diarrheal disease for a number of weeks. These piglets were from the same breed as those on SADS-affected farms, to eliminate potential host factor differences and to more accurately reproduce the conditions that occurred during the outbreak in the region. Both groups of piglets were cared for at a known pig disease-free facility. Again, qPCR and NGS were used to make sure that there was no other known swine diarrhoea virus present in the virus inoculum or any of the experimental animals. Two groups (6 for each group) of three-day old piglets were inoculated with SADS-CoV culture supernatant or normal cell culture medium as control. NGS and qPCR were used to confirm that there were no other known swine pathogens in the inoculum.
For both experiments, animals were recorded daily for signs of diseases, such as diarrhoea, weight loss and death. Faecal swabs were collected daily from all animals and screened for known swine diarrhoea viruses by qPCR. Weight loss was calculated as the percentage weight loss compared the original weight at day 0 with a threshold of >5%. It is important to point out that piglets when they are three days old tend to suffer from diarrhoea and weight loss when they are taken away from sows and the natural breast-feeding environment even without infection. At experimental endpoints, piglets were humanely euthanized and necropsies performed. Pictures were taken to record gross pathological changes to the intestines. Ileal, jejunal and duodenal tissues were taken from selected animals and stored at –80 °C for further analysis.
Haematoxylin and eosin and immunohistochemistry analysis
Frozen (–80 °C) small intestinal tissues including duodenum, jejunum and ileum taken from the experimentally infected pigs were pre-frozen at –20 °C for 10 min. Tissues were then embedded in optimal cutting temperature (OCT) compound and cut into 8-μm sections using the Cryotome FSE machine (Thermo Fisher Scientific). Mounted microscope slides were fixed with paraformaldehyde and stained with haematoxylin and eosin for histopathological examination.
For immunohistochemistry analysis, a rabbit antibody raised against the SADSr-CoV 3755 N protein was used for specific staining of SADS-CoV antigen. Slides were blocked by incubating with 10% goat serum (Beyotime) at 37 °C for 30 min, followed by overnight incubation at 4 °C with the rabbit anti-3755 N protein serum (1:1,000) and mouse anti-cytokeratin 8+18+19 monoclonal antibody (Abcam), diluted 1:100 in PBST buffer containing 5% goat serum. After washing, slides were then incubated for 50 min at room temperature with Cy3-conjugated goat-anti-rabbit IgG (Proteintech) and FITC-conjugated goat-anti-mouse IgG (Proteintech), diluted 1:100 in PBST buffer containing 5% goat serum. Slides were stained with DAPI (Beyotime) and observed under a fluorescence microscope (Nikon).
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
Sequence data that support the findings of this study have been deposited in GenBank with accession codes MF094681–MF094688, MF769416–MF769444, MF094697–MF094701, MF769406–MF769415 and MG557844. Raw sequencing data that support the findings of this study have been deposited in the Sequence Read Achieve (SRA) with accession codes SRR5991648, SRR5991649, SRR5991650, SRR5991651, SRR5991652, SRR5991654, SRR5991655, SRR5991656, SRR5991657, SRR5991658 and SRR5995595.
Online content
Any Methods, including any statements of data availability and Nature Research reporting summaries, along with any additional references and Source Data files, are available in the online version of the paper at 10.1038/s41586-018-0004-7.
Supplementary information
This file contains two supplementary tables. Supplementary table 1 contains a list of nucleotide variants among the 33 SADS-CoV genomes obtained from the four farms. Supplementary table 2 contains a list of PCR primers used in this study
Source data
Acknowledgements
We thank S.-B. Xiao for providing pig cell lines, P. Burbelo for providing the luciferase immunoprecipitation system vector and L. Zhu for enabling the rapid synthesis of the S gene; the WIV animal facilities; J. Min for help with the preparation of the immunohistochemistry samples; and G.-J. Zhu and A. A. Chmura for assistance with bat sampling. This work was jointly supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDPB0301) to Z.-L.S., China Natural Science Foundation (81290341 and 31621061 to Z.-L.S., 81661148058 to P.Z., 31672564 and 31472217 to J.-Y.M., 81572045, 81672001 and 81621005 to Y.-G.T.), National Key Research and Development Program of China (2015AA020108, 2016YFC1202705, AWS16J020 and AWS15J006) to Y.-G.T.; National Science and Technology Spark Program (2012GA780026) and Guangdong Province Agricultural Industry Technology System Project (2016LM1112) to J.-Y.M., State Key Laboratory of Pathogen and Biosecurity (SKLPBS1518) to Y.-G.T., Taishan Scholars program of Shandong province (ts201511056 to W.-F.S.), NRF grants NRF2012NRF-CRP001–056, NRF2016NRF-NSFC002-013 and NMRC grant CDPHRG/0006/2014 to L.-F.W., Funds for Environment Construction & Capacity Building of GDAS’ Research Platform (2016GDASPT-0215) to LBZ, United States Agency for International Development Emerging Pandemic Threats PREDICT project (AID-OAA-A-14-00102), National Institute of Allergy and Infectious Diseases of the National Institutes of Health (Award Number R01AI110964) to P.D. and Z.-L.S.
Reviewer information
Nature thanks C. Drosten, G. Palacios and L. Saif for their contribution to the peer review of this work.
Extended data figures and tables
Author contributions
L.-F.W., Z.-L.S., P.Z., Y.-G.T., and J.-Y.M. conceived the study. P.Z., W.Z., Y.Z., S.M., X.-S.Z., B.L., X.-L.Y., H.G., D.E.A., Y.L., X.L.L. and J.C. performed qPCR, serology and histology experiments and cultured the virus. H.F., Y.-W.Z., J.-M.L., G.-Q.P., X.-P.A., Z.-Q.M., T.-T.H., Y.H., Q.S., Y.-Y.W., S.-Z.X., X.-L.-L.Z., W.-F.S. and J.L. performed genome sequencing and annotations. T.L., Q.-M.X., J.-W.C., L.Z., K.-J.M., Z.-X.W., Y.-S.C., D.L., Y.S., F.C., P.-J.G. and R.H. prepared the samples and carried out animal challenge experiments. Z.-L.S., P.D., L.-B.Z., S.-Y.L. coordinated collection of bat samples. P.Z., L.-F.W., Z.-L.S. and P.D. had a major role in the preparation of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Peng Zhou, Hang Fan, Tian Lan.
Change history
4/5/2018
The Extended Data Figures and Tables section originally published with this article was missing Tables 1–5. This has now been corrected.
Contributor Information
Peter Daszak, Email: daszak@ecohealthalliance.org.
Lin-Fa Wang, Email: linfa.wang@duke-nus.edu.sg.
Zheng-Li Shi, Email: zlshi@wh.iov.cn.
Yi-Gang Tong, Email: tong.yigang@gmail.com.
Jing-Yun Ma, Email: majy2400@scau.edu.cn.
Extended data
is available for this paper at 10.1038/s41586-018-0010-9.
Supplementary information
is available for this paper at 10.1038/s41586-018-0010-9.
References
- 1.Olival KJ, et al. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan Y, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 3.Lau SK, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 5.Ge XY, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He B, et al. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J. Virol. 2014;88:7070–7082. doi: 10.1128/JVI.00631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang XL, et al. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J. Virol. 2016;90:3253–3256. doi: 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Z, et al. ORF8-related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2016;213:579–583. doi: 10.1093/infdis/jiv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, et al. Discovery and genetic analysis of novel coronaviruses in least horseshoe bats in southwestern China. Emerg. Microbes Infect. 2017;6:e14. doi: 10.1038/emi.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu B, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drosten C, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 12.Ksiazek TG, et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 13.Marra MA, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 14.Peiris JS, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rota PA, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 16.Wang L-F, Cowled C, editors. Bats and Viruses: A New Frontier of Emerging Infectious Diseases. 1st edn. Hoboken: John Wiley & Sons; 2015. [Google Scholar]
- 17.Dong N, et al. Porcine deltacoronavirus in mainland China. Emerg. Infect. Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun D, Wang X, Wei S, Chen J, Feng L. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: a mini-review. J. Vet. Med. Sci. 2016;78:355–363. doi: 10.1292/jvms.15-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau SK, et al. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, et al. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010;155:1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burbelo PD, et al. Serological diagnosis of human herpes simplex virus type 1 and 2 infections by luciferase immunoprecipitation system assay. Clin. Vaccine Immunol. 2009;16:366–371. doi: 10.1128/CVI.00350-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong L, et al. A new bat-HKU2-like coronavirus in swine, China, 2017. Emerg. Infect. Dis. 2017;23:1607–1609. doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan Y, et al. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masters, P. S. & Perlman, S. in Fields Virology Vol. 2 (eds Knipe, D. M. & Howley, P. M.) 825–858 (Lippincott Williams & Wilkins, Philadelphia, 2013).
- 26.Raj VS, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harlow E, Lane D. Antibodies: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 28.Ren W, et al. Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and SARS-like coronavirus of bat origin. J. Virol. 2008;82:1899–1907. doi: 10.1128/JVI.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains two supplementary tables. Supplementary table 1 contains a list of nucleotide variants among the 33 SADS-CoV genomes obtained from the four farms. Supplementary table 2 contains a list of PCR primers used in this study
Data Availability Statement
Sequence data that support the findings of this study have been deposited in GenBank with accession codes MF094681–MF094688, MF769416–MF769444, MF094697–MF094701, MF769406–MF769415 and MG557844. Raw sequencing data that support the findings of this study have been deposited in the Sequence Read Achieve (SRA) with accession codes SRR5991648, SRR5991649, SRR5991650, SRR5991651, SRR5991652, SRR5991654, SRR5991655, SRR5991656, SRR5991657, SRR5991658 and SRR5995595.