Abstract
Infectious diseases have for centuries ranked with wars and famine as major challenges to human progress and survival. They remain among the leading causes of death and disability worldwide. Against a constant background of established infections, epidemics of new and old infectious diseases periodically emerge, greatly magnifying the global burden of infections. Studies of these emerging infections reveal the evolutionary properties of pathogenic microorganisms and the dynamic relationships between microorganisms, their hosts and the environment.
Main
Emerging infections (EIs) can be defined as “infections that have newly appeared in a population or have existed previously but are rapidly increasing in incidence or geographic range”1. EIs have shaped the course of human history and have caused incalculable misery and death. In 1981, a new disease — acquired immune deficiency syndrome (AIDS) — was first recognized. As a global killer, AIDS now threatens to surpass the Black Death of the fourteenth century and the 1918–1920 influenza pandemic, each of which killed at least 50 million people2,3. Of the ‘newly emerging’ and ‘re-emerging/resurging’ diseases that have followed the appearance of AIDS (Fig. 1), some have been minor curiosities, such as the 2003 cases of monkeypox imported into the United States4, whereas others, such as severe acute respiratory syndrome (SARS), which emerged in the same year5, have had a worldwide impact. The 2001 anthrax bioterrorist attack in the United States6 falls into a third category: ‘deliberately emerging’ diseases. EIs can be expected to remain a considerable challenge for the foreseeable future. Here we examine the nature and scope of emerging and re-emerging microbial threats, and consider methods for their control. We emphasize that emergence results from dynamic interactions between rapidly evolving infectious agents and changes in the environment and in host behaviour that provide such agents with favourable new ecological niches.
Figure 1. Global examples of emerging and re-emerging infectious diseases, some of which are discussed in the main text.
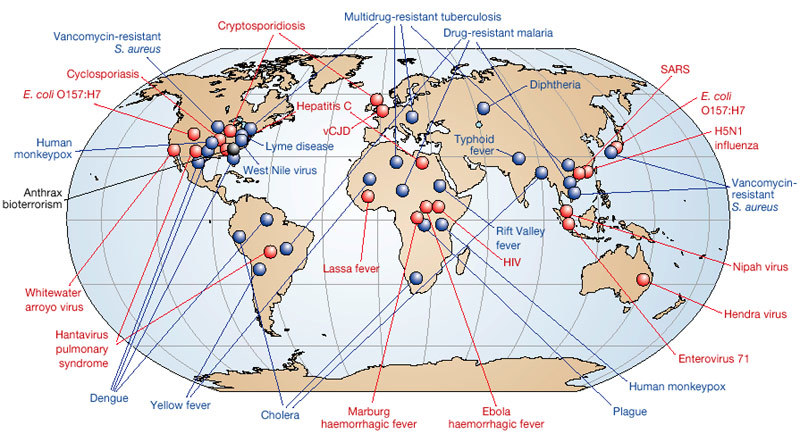
Red represents newly emerging diseases; blue, re-emerging/resurging diseases; black, a ‘deliberately emerging’ disease. Adapted, with permission, from ref. 23.
Global burden of infectious diseases
About 15 million (>25%) of 57 million annual deaths worldwide are estimated to be related directly to infectious diseases; this figure does not include the additional millions of deaths that occur as a consequence of past infections (for example, streptococcal rheumatic heart disease), or because of complications associated with chronic infections, such as liver failure and hepatocellular carcinoma in people infected with hepatitis B or C viruses7 (Fig. 2).
Figure 2. Leading causes of death worldwide.
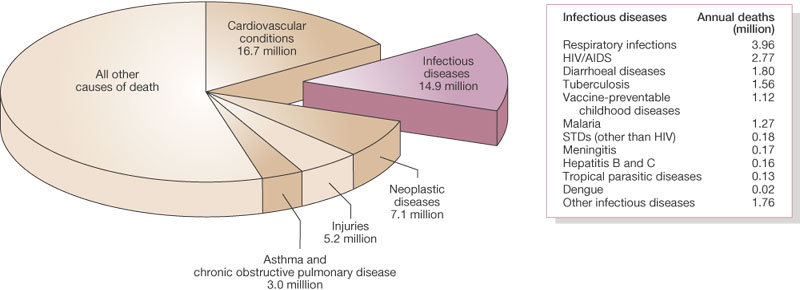
About 15 million (>25%) of 57 million annual deaths worldwide are the direct result of infectious disease. Figures published by the World Health Organization (see http://www.who.int/whr/en and ref. 7).
The burden of morbidity (ill health) and mortality associated with infectious diseases falls most heavily on people in developing countries8, and particularly on infants and children (about three million children die each year from malaria and diarrhoeal diseases alone7). In developed nations, infectious disease mortality disproportionately affects indigenous and disadvantaged minorities9.
Emerging infections in historical context
EIs have been familiar threats since ancient times. They were once identified by terms such as λoιµóς (loimos)10, and later as ‘pestilences’, ‘pestes’, ‘pests’ and ‘plagues’. Many examples can be cited in addition to the Black Death and the 1918 influenza pandemic, such as certain biblical pharaonic plagues and the unidentified Plague of Athens, which heralded the end of Greece's Golden Age11. The Age of Discovery, starting in the fifteenth century, was a particularly disastrous period with regard to the spread of infectious diseases. Importation of smallpox into Mexico caused 10–15 million deaths in 1520–1521, effectively ending Aztec civilization12,13. Other Amerindian and Pacific civilizations were destroyed by imported smallpox and measles13,14,15,16,17. Historians have referred to these events as apocalypses16 and even as genocide15.
For centuries, mankind seemed helpless against these sudden epidemics. But the establishment of the germ theory and the identification of specific microbes as the causative agents of a wide variety of infectious diseases18,19,20 led to enormous progress, notably the development of vaccines and ultimately of antimicrobials20. In fact, the era of the identification of microbes had barely begun18 when optimists at the end of the nineteenth century predicted the eradication of infectious diseases21. By the 1950s, which witnessed the widespread use of penicillin, the development of polio vaccines and the discovery of drugs for tuberculosis, complacency had set in22, and in 1967, the US Surgeon General stated that “the war against infectious diseases has been won”23.
Some experts remained sceptical, aware of recurrent lessons from history. They were less persuaded by successes than alarmed by failures such as the lack of progress against infections in the developing world and the global spread of antimicrobial resistance. Richard Krause, then the director of the US National Institute of Allergy and Infectious Diseases, warned in 1981 (ref. 24) that microbial diversity and evolutionary vigour were still dynamic forces threatening mankind. As Krause was completing his book The Restless Tide24, AIDS — one of history's most devastating pandemics — was already insidiously emerging. The emergence of AIDS led to renewed appreciation of the inevitability and consequences of the emergence of infectious diseases25,26,27,28,29,30,31. In the past 25 years, some of the factors that resulted in AIDS have also led to re-emergences of historically important diseases such as cholera, diphtheria, trench fever and plague. Many re-emergences have been catalysed by wars, loss of social cohesion, and natural disasters such as earthquakes and floods, indicating the importance not only of microbial and viral factors, but also of social and environmental determinants25,26,27,28,29,30,31.
Newly emerging and newly recognized infections
The classification of EIs as ‘newly emerging’, ‘re-emerging/resurging’ or ‘deliberately emerging’ is useful because the underlying causes of emergence and the optimal prevention or control responses frequently differ between the groups. Newly emerging infections are those that have not previously been recognized in man. Many diverse factors contribute to their emergences (see Box 1); these include microbial genetic mutation and viral genetic recombination or reassortment, changes in populations of reservoir hosts or intermediate insect vectors, microbial switching from animal to human hosts, human behavioural changes (notably human movement and urbanization), and environmental factors. These numerous microbial, host and environmental factors interact to create opportunities for infectious agents to evolve into new ecological niches, reach and adapt to new hosts, and spread more easily between them.
The AIDS model
Any discussion of recent EIs must begin with the human immunodeficiency virus (HIV) that causes AIDS. HIV has so far infected more than 60 million people worldwide33. Before jumping to humans an estimated 60–70 years ago34, perhaps as a consequence of the consumption of ‘bush meat’ from non-human primates, HIV-1 and HIV-2 had ample opportunity to evolve in hosts that were genetically similar to man (the chimpanzee, Pan troglodytes, and the sooty mangabey, Cercocebus atys). But HIV/AIDS might never have emerged had it not been for disruptions in the economic and social infrastructure in post-colonial sub-Saharan Africa. Increased travel, the movement of rural populations to large cities, urban poverty and a weakening of family structure all promoted sexual practices, such as promiscuity and prostitution, that facilitate HIV transmission34,35,36,37. Such complex interactions between infectious agents, hosts and the environment are not unique to the epidemiology of HIV/AIDS. The examples cited below further illustrate how changes in population density, human movements and the environment interact to create ecological niches that facilitate microbial or viral adaptation.
Dead-end transmission of zoonotic and vector-borne diseases
Some infectious agents that have adapted to non-human hosts can jump to humans but, unlike HIV, are not generally transmitted from person to person, achieving only ‘dead end’ transmission. Infections in animals that are transmitted to humans (zoonoses), and those transmitted from one vertebrate to another by an arthropod vector (vector-borne diseases), have repeatedly been identified as ranking among the most important EIs25,26. Examples include the arenavirus haemorrhagic fevers (Argentine, Bolivian, Venezuelan and Lassa haemorrhagic fevers) and hantavirus pulmonary syndrome (HPS). Viruses in these groups have co-evolved with specific rodent species whose contact with humans has increased as a result of modern environmental and human behavioural factors. Farming, keeping domestic pets, hunting and camping, deforestation and other types of habitat destruction all create new opportunities for such infectious agents to invade human hosts25,26,27,28,29,30,31. The first epidemic of HPS, detected in the southwestern region of the United States in 1993 (ref. 38), resulted from population booms of the deer mouse Peromyscus maniculatis, in turn caused by climate-related and recurrent proliferation of rodent food sources. Increased rodent populations and eventual shortages of food drove expanded deer mouse populations into homes, exposing people to virus-containing droppings. The 1998–1999 Malaysian Nipah virus epidemic39 further illustrates the influence of human behaviours and environmental perturbations on newly emerging human infections. Pigs crammed together in pens located in or near orchards attracted fruit bats whose normal habitats had been destroyed by deforestation and whose droppings contained the then-unknown paramyxovirus. Virus aerosolization caused infection of pigs, with overcrowding leading to explosive transmission rates and ultimately to infections in pig handlers.
Variant Creutzfeldt–Jakob disease (vCJD) is another example of a zoonotic disease emerging in humans. vCJD is caused by the human-adapted form of the prion associated with the emerging epizootic (large-scale animal outbreak) of bovine spongiform encephalopathy (BSE)40, commonly known as mad cow disease. The ongoing BSE epizootic/vCJD epidemic, primarily affecting Great Britain, probably resulted from the now-abandoned practice of supplementing cattle feed with the pulverized meat and bones of previously slaughtered cattle. BSE itself is suspected to have emerged because of even earlier use of cattle feed containing the agent of sheep scrapie, a prion disease recognized by farmers more than 250 years ago41. Alarmingly, the new BSE prion has become uncharacteristically promiscuous: unlike most known prions, it readily infects multiple species in addition to humans. This suggests the possibility of further emerging diseases associated with prions with currently unknown transmissibility to humans40. The recent reports of variant strains of the BSE prion42 suggest that the BSE agent could be a more serious threat than other animal prions.
Environmentally persistent organisms
Infectious agents indirectly transmitted to or between humans by way of human-modified environments account for other emerging zoonoses, as well as certain non-zoonotic diseases, which are discussed below. For example, legionnaires' disease, first identified in 1976, is caused by Legionella pneumophila, whose emergence as a human pathogen might not have occurred were it not for the environmental niche provided by air-conditioning systems26. Campylobacter jejuni and Shiga-toxin-producing Escherichia coli (E. coli O157:H7 and other agents of haemolytic–uraemic syndrome) infect agricultural animals, gaining access to humans through food, milk, water or direct animal contact. Other enteric pathogens, such as the vibrios causing classical cholera (re-emerging; see below) and serogroup O139 cholera, and the zoonotic protozoa Cryptosporidium parvum and Cyclospora cayetanensis26, seem to have come from environmental or animal organisms that have adapted to human-to-human ‘faecal–oral’ transmission through water.
Old microbes cause new diseases
Some EIs come from microorganisms that once caused familiar diseases, but which now cause new or previously uncommon diseases. Streptococcus pyogenes caused a fatal pandemic of scarlet and puerperal fevers between 1830 and 1900 (ref. 44). Scarlet fever, then the leading cause of death in children, is now rare, but has been largely supplemented by other streptococcal complications such as streptococcal toxic shock syndrome, necrotizing fasciitis and re-emergent rheumatic fever45. When new microbes are discovered, their emergences as disease-causing pathogens may be delayed. For example, in 1883, Robert Koch was unable to show that the newly discovered Koch–Weeks bacillus caused serious disease. More than a century later, a fatal EI dubbed Brazilian purpuric fever was linked to virulent clonal variants of Haemophilus influenzae biogroup aegyptius (the Koch–Weeks bacillus)46. Although the bases of emergences of new and more severe diseases caused by S. pyogenes and H. influenzae biogroup aegyptius are not fully known, in both cases complex microbial genetic events are suspected. The distinctive clonal variants associated with severe H. influenzae biogroup aegyptius disease have been shown by PCR (polymerase chain reaction)-based subtractive genome hybridization to contain not only a unique plasmid, but also unique chromosomal regions, some of which are encoded by bacteriophages47. This research has narrowed the search for virulence determinants to unique proteins, some of which may have been acquired from other organisms by horizontal gene transfer. Streptococcus pyogenes has been studied more extensively, but the basis of severe disease emergence seems to be more complex than for H. influenzae biogroup aegyptius. Many factors associated with streptococcal virulence have been identified in strains bearing the M1 surface protein as well as in other M protein strains, among them bacteriophage-encoded superantigen toxins and a protein known as sic (streptococcal inhibitor of complement), which seems to be strongly selected by human host mucosal factors. Several lines of evidence suggest that changes in streptococcal virulence reflect genetic changes associated with phage integration, large-scale chromosomal rearrangements and possibly the shuffling of virulence cassettes (clusters of genes responsible for pathogenicity), followed by rapid human spread and immune selection48,49.
Microbial agents and chronic diseases
Infectious agents that are associated with chronic diseases are one of the most challenging categories of newly emerging (or at least newly appreciated) infections. Examples include the associations of hepatitis B and C with chronic liver damage and hepatocellular carcinoma, of certain genotypes of human papillomaviruses with cancer of the uterine cervix, of Epstein–Barr virus with Burkitt's lymphoma (largely in Africa) and nasopharyngeal carcinoma (in China), of human herpesvirus 8 with Kaposi sarcoma, and of Helicobacter pylori with gastric ulcers and gastric cancer50,51,52. Some data even suggest infectious aetiologies for cardiovascular disease and diabetes mellitus53, major causes of death and disability worldwide. Other associations between infectious agents and idiopathic chronic diseases will inevitably be found.
Re-emerging and resurging infections
Re-emerging and resurging infections are those that existed in the past but are now rapidly increasing either in incidence or in geographical or human host range. Re-emergence is caused by some of the factors that cause newly emerging infectious diseases, such as microbial evolutionary vigour, zoonotic encounters and environmental encroachment. Re-emergences or at least cyclical resurgences of some diseases may also be climate-related — for example, the El Niño/Southern Oscillation (ENSO) phenomenon is associated with resurgences of cholera and malaria54.
Geographical spread of infections
The impact of both new and re-emerging infectious diseases on human populations is affected by the rate and degree to which they spread across geographical areas, depending on the movement of human hosts or of the vectors or reservoirs of infections. Travel has an important role in bringing people into contact with infectious agents55. An increase in travel-associated importations of diseases was anticipated as early as 1933, when commercial air travel was still in its infancy56. This has since been demonstrated dramatically by an international airline hub-to-hub pandemic spread of acute haemorrhagic conjunctivitis in 1981 (ref. 57), by epidemics of meningococcal meningitis associated with the Hajj, and more recently by the exportation of epidemic SARS (a newly emerging disease) from Guangdong Province, China, to Hong Kong, and from there to Beijing, Hanoi, Singapore, Toronto and elsewhere5 (Fig. 3). The persistent spread of HIV along air, trucking, drug-trafficking and troop-deployment routes is a deadly variation on this theme35,36,37.
Figure 3. Probable cases of severe acute respiratory syndrome (SARS) with onset of illness from 1 November 2002 to 31 July 2003.
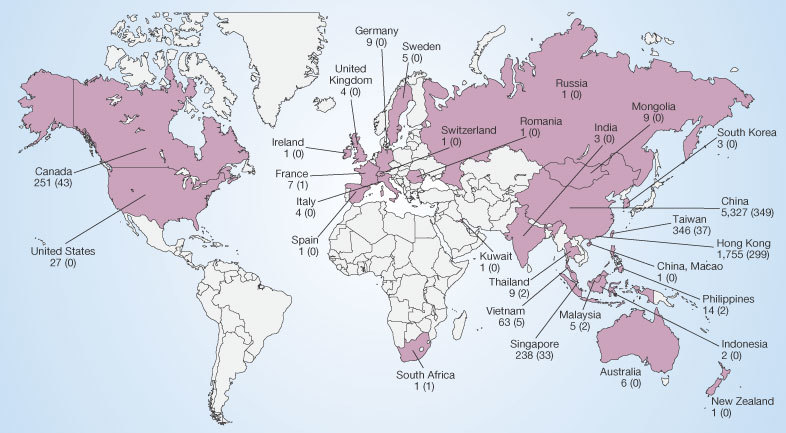
Cases are given by country. SARS-related deaths are indicated in parentheses. A total of 8,096 cases (and 774 deaths) are presented. Figures published by the World Health Organization (see http://www.who.int/csr/sars/country/en).
Malaria
Plasmodium falciparum malaria was neglected for several decades, but is now among the most important re-emerging diseases worldwide (Fig. 2). Years of effective use of dichlorodiphenyltrichloroethane (DDT) led to the abandonment of other mosquito-control programmes, but the insecticide fell into disuse because of mosquito resistance and concerns about the insecticide's potentially harmful effects on humans and wildlife. Consequently, malaria has re-emerged, and the situation has been worsened by the development of drug resistance to chloroquine and mefloquine58. Research efforts focus on the development of vaccines59 and new drugs, and on re-establishing public health measures such as the use of bed nets.
Tuberculosis
Tuberculosis is one of the most deadly re-emerging diseases (Fig. 2). The discovery of isoniazid and other drugs initially led to effective tuberculosis cures, empty sanitoria and the dismantling of public health control systems in developed nations. Consequently, by the 1980s, when tuberculosis had re-emerged in the era of HIV/AIDS, local and state health departments in the United States lacked field, laboratory and clinical staff and so had to reinvent tuberculosis-control programmes25. The remarkable re-emergence of tuberculosis was fuelled by the immune deficiencies of people with AIDS, which greatly increases the risk of latent Mycobacterium tuberculosis infections progressing to active disease, and being transmitted to others. Inadequate courses of anti-tuberculosis therapy compound the problem, leading to the emergence and spread of drug-resistant and multidrug-resistant strains60, and a need for more expensive treatment strategies such as directly observed therapy. It has been known for over a century that tuberculosis is a disease of poverty, associated with crowding and inadequate hygiene. The continuing expansion of global populations living in poverty makes tuberculosis more difficult to control.
Drug-resistant microbes
Drug resistance, another factor causing microbial and viral re-emergence, may result from mutation (for example, in the case of viruses and M. tuberculosis), or from bacterial acquisition of extraneous genes through transformation or infection with plasmids. Sequential emergences of Staphylococcus aureus that are resistant to sulpha drugs (1940s), penicillin (1950s), methicillin (1980s) and to vancomycin in 2002 (ref. 61) — a last line of antibiotic defence for some multiply drug-resistant bacteria — are troubling. Nosocomial Enterococcus faecalis became fully resistant to vancomycin by 1988, and then apparently transferred vanA resistance genes to co-infecting staphylococci61. Methicillin-resistant staphylococci are now being isolated from livestock that have been fed with growth-promoting antibiotics62, possibly contributing to resistance problems in humans. Many other important microbes have also become effective ‘resistors’, among them Streptococcus pneumoniae and Neisseria gonorrhoeae63.
Opportunistic re-emerging infections
Immune deficiency associated with AIDS, and with chemotherapy for cancer, immune-mediated diseases and transplantation, has contributed to an enormous global increase in the numbers of immunosuppressed people over the past few decades (probably more than 1% of the world's population), setting the stage for the re-emergence of many opportunistic infections. HIV, which has infected more than 60 million people globally33, is the largest single cause of human immune deficiency and markedly increases vulnerability to a wide range of opportunistic pathogens, including Pneumocystis carinii, various fungi, tuberculosis, protozoa and herpesviruses64. Breakthroughs in cancer therapy and in immunosuppressive therapies used to treat immune-mediated diseases and for transplantation65,66 can also leave patients susceptible to opportunistic infections. Human organ transplantation adds a further risk of infection with undetected pathogens in donor tissues, and transplantation of animal organs introduces the risk of transmission to humans of animal microbes67.
Re-emerging zoonotic and vector-borne diseases
The emergence of zoonotic and vector-borne diseases can also be associated with human behaviours and environmental perturbation. In 2003, monkeypox — an endemic infection of African rodents — crossed the Atlantic with exported pets, which were then shipped from Texas to infect people throughout the US Midwest4. Lyme disease, caused by Borrelia burgdorferi, re-emerged in the United States as a result of suburban expansion, which brought people into increasing contact with deer, deer mice and ticks. Similarly, tick-borne encephalitis re-emerged in Russia when weekend getaways (dachas) drew city dwellers into contact with forest ticks. The simultaneous 1999 emergences of encephalitis due to West Nile virus in the United States and in Russia68,69 reflect abundances of eclectic vector mosquitoes and avian hosts in these locations. Both were probably connected to endemic sites by virus carriage in migratory birds and travellers. The remarkable geographical spread of West Nile virus in the five years since its introduction into the Western Hemisphere reflects an unfortunate confluence of viral promiscuity and ecological diversity70. Although humans are dead-end hosts for West Nile virus, the risk of infection is greatly increased by marked zoonotic viral amplification and persistence in the environment. Unlike most viruses, which tend to be fairly narrowly adapted to specific hosts, West Nile virus is known to infect more than 30 North American mosquito species, which together transmit infection to at least 150 North American bird species, many of which migrate to new and distant locations, spreading the virus to rural and urban ecosystems throughout North and Central America70.
Although West Nile virus is now a major epidemiological concern in the developed world, dengue remains the most significant and widespread flavivirus disease to have emerged globally71. A 2001–2002 epidemic in Hawaii — fortunately without fatalities — is a reminder that dengue has also re-emerged in locations once considered to be dengue-free. Usually transmitted by Aedes aegypti mosquitoes, dengue has recently been transmitted by Aedes albopictus — a vector switch of potential significance with respect to dengue re-emergence71. In the Americas, including many US southern states, A. albopictus has been spreading into areas where A. aegypti mosquitoes are not found, and persisting for longer seasonal periods, putting tens of millions more people at risk of dengue infection. Dengue re-emergence is further complicated by disturbing increases in a serious and formerly rare form of the disease, dengue haemorrhagic fever (dengue shock syndrome being its highly fatal form). These severe complications are thought to result from the evolution of dengue viruses to escape high population immunity, seen in increased viral virulence and human immunopathogenesis due to antibody-dependent enhancement of viral infection72.
Cholera is also of interest, not only as an important cause of mortality, but also because of the complexity of factors that determine its re-emergence. Both virulent and avirulent strains of these zoonotic bacteria are maintained in the environment and are rapidly evolving in association with phyto- and zooplankton, algae and crustaceans. Such environmental strains seem to act as reservoirs for human virulence genes (for example, genes for the phage-encoded cholera toxin and the toxin-coregulated pilus (TCP) factor associated with attachment), and to undergo gene transfer events that lead to new strains containing further virulence gene combinations. These result in periodic cholera emergences that cause epidemics and pandemics73. Thus, although the disease we know as cholera has appeared to be clinically and epidemiologically stable at least since the third pandemic (in the 1840s), modern evidence suggests that such apparent stability masks aggressive bacterial evolution in complex natural environments.
Influenza A
Influenza A viruses, which are endemic gastrointestinal viruses of wild waterfowl, have evolved elaborate mechanisms to jump species into domestic fowl, farm animals and humans. Periodic gene segment reassortments between human and animal viruses produce important antigenic changes, referred to as ‘shifts’. These can lead to deadly pandemics, as occurred in 1888, 1918, 1957 and 1968 (refs 74, 75). In intervening years, shifted viruses undergo continual but less dramatic antigenic changes called ‘drifts’, which allow them partially to escape human immunity raised by previously circulating influenza viruses. Influenza drift is an evolutionary success story for the virus. Influenza A has a seemingly inexhaustible repertoire of mutational possibilities at several critical epitopes surrounding the viral haemagglutinin site that attaches to human cells. It remains something of a mystery how zoonotic influenza viruses mix with each other and with human strains to acquire the additional properties of human virulence and human-to-human transmissibility.
Before 1997, mild cases of human disease associated with avian influenza viruses were occasionally reported76. These events have become more frequent, sometimes resulting in severe cases of disease and death. Avian influenza has recently made dead-end jumps to humans — for example, the 1997 Hong Kong outbreak of the newly emergent H5N1 influenza, the 2003 H7N7 epidemic in the Netherlands, the 2003–2004 H5N1 and H7N3 epizootics in Asia and elsewhere, and occasional cases of H9N2 disease (Fig. 4). Meanwhile, back-switches of human H3N2 viruses have emerged in pigs, from which both doubly mixed (pig–human) and triply mixed (pig–human–avian) viruses74,75 have been isolated. Such enzootic/zoonotic mixing is suspected to have occurred in the influenza pandemic of 1918–1920, which was caused by an H1N1 virus with an avian-like receptor-binding site77. The predicted virulence genes of this virus are now being sought from 85-year-old pathology specimens and from frozen corpses78. The implications of interspecies genetic mixing for future influenza pandemics are troubling. Although much remains speculative about how influenza viruses emerge and spread, it seems clear that the process is driven by prolific and complex viral evolution (genetic reassortment and mutational ‘drift’), interspecies mixing and adaptation, and ecological factors that bring humans into contact with animals and each other. By whatever means new influenza virus pandemic strains emerge, they eventually reach a critical threshold of human transmission beyond which epidemic and pandemic spread follows mathematically predictable patterns.
Figure 4. Documented human infections with avian influenza viruses, 1997–2004.
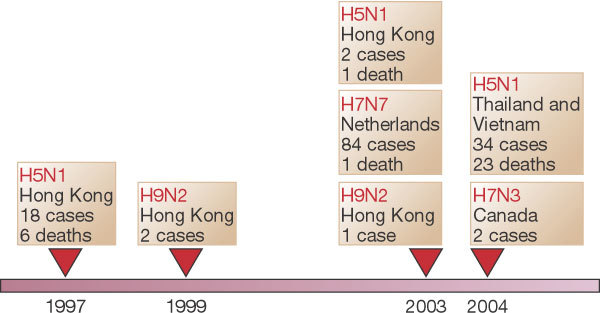
Sporadic cases of mild human illness associated with avian influenza viruses were reported before 1997. See http://www.who.int/csr/disease/influenza/en and ref. 76.
The dynamics and determinants of such epidemic development have been studied since the nineteenth century for several infectious diseases. For influenza, both historical and prospective epidemics have been described or predicted using deterministic and stochastic mathematical models, often with surprising accuracy when compared with actual epidemic data. More complicated mathematical models that describe how diseases spread by means other than person-to-person aerosol transmission have generally been less successful in describing and predicting epidemics, but have nonetheless been helpful in planning public health responses to epidemics caused by HIV79, vCJD80 and other diseases.
Mathematical modelling is also used to determine the impact of emerging epidemics. For example, it has been difficult to estimate overall influenza mortality because fatal infections are often neither diagnosed nor accurately recorded in hospital records and death certificates, especially in the elderly. Recent epidemiological attempts to obtain improved influenza mortality estimates from seasonal excess mortality data81 have indicated that influenza mortality may be greater than was previously suspected, because influenza deaths are frequently coded under seemingly unrelated categories such as cardiovascular diseases. The same approaches also show that other influenza-like deaths may actually be due to other agents, such as respiratory syncytial virus (RSV), a common childhood virus that in the past decade has emerged as a major cause of adult mortality81.
Deliberately emerging infections
Deliberately emerging microbes are those that have been developed by man, usually for nefarious use. The term ‘deliberately emerging’ refers to both naturally occurring microbial agents such as anthrax6, and to bioengineered microorganisms such as those created by the insertion of genetic virulence factors that produce or exacerbate disease. Deliberately emerging microbes include microorganisms or toxins produced in a form that would cause maximal harm because of ease of dissemination, enhanced infectivity or heightened pathogenicity82.
Bioterrorism and biowarfare
As concepts, bioterrorism and biowarfare are probably not new. The alleged catapulting of plague-ridden corpses over enemy walls in the 1346 siege of Caffa (the modern Crimean port of Feodosia, Ukraine) and the dispatch of smallpox-impregnated blankets to Indians by British officers in the Seven Years War (1754–1763) have frequently been cited as examples of bioterrorism or biowarfare83,84.
Two modern attacks have been well documented. In 1984, an Oregon religious cult spiked restaurant salad bars with Salmonellae in an attempt to sway a local election85. A 2001 anthrax attack6, in which a terrorist mailed anthrax-spore-filled letters to prominent figures, including two US senators, resulted in illness in at least 18 people and the death of five of these individuals. Public alarm was elevated by the knowledge that Bacillus anthracis is a common and easily obtainable enzootic and soil organism found in laboratories worldwide, and that scientific technology had increased its lethality: the spores had been weaponized by being concentrated, finely milled and packed with a dispersal agent to increase their capacity to disseminate82. The United States, the United Kingdom, the Soviet Union and other nations once had sophisticated offensive bioweapons programmes that included the production of weaponized anthrax spores82. Soviet scientists continued to produce large quantities of organisms adapted for biowarfare and bioterror — among them the agents of smallpox, plague, tularaemia and Marburg virus — for several years after their signing of the Bioweapons and Toxins Treaty Convention in 1972, which forbade such activities82. By 1987, the Soviet programme was annually producing 5,000 tonnes of weaponized anthrax spores, packing them into warheads and other delivery devices82.
Before the 2001 anthrax attacks6, the US scientific community had for several years been bolstering its biodefence research capacity. The anthrax attacks greatly accelerated this expansion as part of a national defence plan, which includes efforts to provide a knowledge base for the development of effective countermeasures against agents of bioterror, such as diagnostics, therapeutics and vaccines, and to translate this knowledge into the production and delivery of such measures86. Bioterror agents have been grouped into three categories of risk87. The six category A agents (anthrax, smallpox, plague, tularaemia, viral haemorrhagic fevers and clostridial botulinum toxin) are given top priority because they are highly lethal and readily deployed as weapons. Category B and C agents include food-borne and water-borne organisms that incapacitate but usually do not kill.
Meeting the challenge of emerging infections
Infectious diseases will continue to emerge and re-emerge, leading to unpredictable epidemics and difficult challenges to public health and to microbiology and allied sciences.
Surveillance and response, the key elements in controlling EIs, be they naturally occurring or deliberately engineered, depend on rapid clinical diagnosis and detection and containment in populations and in the environment. Globally, such efforts are coordinated by the World Health Organization, which recently led a multifaceted effort to successfully contain the global SARS outbreak of 2002–2003 (ref. 88). In the United States, such efforts are led by the US Centers for Disease Control and Prevention (CDC)89, which along with state and local health departments and other agencies have been making significant strides in national surveillance–response capacity. The enormous influx of US government-funded research resources (largely through the National Institutes of Health) and public health resources (mainly through the CDC, and state and local public health agencies) in response to the increased threat of a bioterrorist attack86 will fortify the response capabilities related to all EIs.
However, it is clear that surveillance and other activities that traditionally fall within the domain of public health are not in themselves sufficient to adequately address the problem of EIs. Of critical importance are basic, translational and applied research efforts to develop advanced countermeasures such as surveillance tools, diagnostic tests, vaccines and therapeutics86. Genomics, proteomics and advances in nanotechnology90 are increasingly being exploited in diagnostic, therapeutic and microbial research applications, and in rational drug and vaccine design. Direct and computational structural determination91, prediction of protein–protein interactions between microorganisms and drugs, and sophisticated bioinformatics techniques support research in all of the above areas. These technologies have led to numerous advances in real-world utility against EIs, most notably in the development of more than 20 antiretroviral drugs that can effectively suppress HIV replication. Where they are available and properly used in HIV-infected individuals, these medications have dramatically reduced HIV morbidity and mortality92.
Gene- and protein-based microarrays can be used to detect pathogen signals, to monitor resistance to anti-infective agents, to characterize host gene responses to recent infections, and to facilitate the development of new drugs and vaccines93. Basic and applied research together have provided promising new vaccine platforms, such as recombinant proteins, immunogenic peptides, naked DNA vaccines, viral vectors of extraneous genes encoding immunogenic proteins (including chimaeras), replicons and pseudovirions94. Many novel vaccine candidates are now being developed against EIs such as HIV, Ebola virus, West Nile virus, dengue, the SARS coronavirus, tuberculosis and malaria. Of particular note are novel tuberculosis vaccines that recently entered clinical trials — the first time in more than 60 years that new approaches to vaccination for tuberculosis have been assessed in humans95. Chimaeric flavivirus vaccines for West Nile virus, dengue and Japanese encephalitis virus are effective in animal models and are in various stages of clinical testing95.
Our growing understanding of the human immune system is also helping to accelerate vaccine development. This is especially true in the case of innate immune responses, which are evolutionarily older, less specific and faster-acting than the adaptive responses that have been the traditional targets of vaccines96. As we learn more about innate immunity and its relationship with the adaptive immune system, opportunities to create more effective vaccine adjuvants will emerge. For example, synthetic DNA sequences that contain repeated CpG motifs mimic the stimulatory activity that bacterial DNA fragments exert on the innate immune system. These sequences show promise as vaccine adjuvants that accelerate and augment immune responses97. We can anticipate more progress of this kind as we continue to delineate the complex interactions between innate and adaptive immune responses.
The sequencing of the human genome, the genomes of six other animals, including the mouse, and those of microbial vectors and microbes themselves (for example, P. falciparum and its mosquito vector, Anopheles gambiae), have elevated microbiology to a whole-genome level. The ability to sequence microbial genomes in a few days98 or less, and to examine host–vector–microbe interactions at both the genome level and at the tertiary protein structural level, will help us to understand the molecular mechanisms that underlie the pathogenesis of infectious disease and host defences, including resistance and immune evasion. These advances will facilitate the development of new countermeasures. Other fertile areas of research include the use of geographical information systems99 and satellite imaging to support field study and epidemic prevention (for example, predicting HPS and Rift Valley fever epidemics in indigenous areas by satellite imagery of water and vegetation related to animal reservoir and vector prevalence).
Underlying disease emergence are evolutionary conflicts between rapidly evolving and adapting infectious agents and their slowly evolving hosts. These are fought out in the context of accelerating environmental and human behavioural alterations that provide new ecological niches into which evolving microbes can readily fit. It is essential that broadly based prevention strategies, as well as new and improved countermeasures (that is, surveillance tools, diagnostics, therapeutics and vaccines), be continually tested, refined and upgraded, requiring a strengthened relationship between public health and basic and clinical sciences. The challenge presented by the ongoing conflict between pathogenic microorganisms and man has been well summarized by a noted champion of the war on EIs, Joshua Lederberg: “The future of microbes and mankind will probably unfold as episodes of a suspense thriller that could be entitled Our Wits Versus Their Genes”29. The global scientific and public health communities must confront this reality not only with wit, but also with vision and sustained commitment to meet a perpetual challenge.
Box 1: Factors involved in the emergence of infectious diseases.
Selected factors contribute to the emergence/re-emergence of infectious diseases25,26. These factors, which frequently differ for ‘newly emerging’, ‘re-emerging/resurging’ and ‘deliberately emerging’ diseases, include genetic, biological, and social, political and economic factors.
• Microbial adaptation and change
• Human susceptibility to infection
• Climate and weather
• Changing ecosystems
• Human demographics and behaviour
• Economic development and land use
• International travel and commerce
• Technology and industry
• Breakdown of public health measures
• Poverty and social inequality
• War and famine
• Lack of political will
• Intent to harm
Acknowledgements
The authors thank R. M. Krause for helpful discussions, and J. Weddle for graphic design.
Competing interests
The authors declare that they have no competing financial interests.
References
- 1.Morse SS. Factors in the emergence of infectious diseases. Emerg. Infect. Dis. 1995;1:7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohn GC. The Wordsworth Encyclopedia of Plague and Pestilence. 1995. pp. 25–26. [Google Scholar]
- 3.Johnson NPAS, Mueller J. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 2002;76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Multistate outbreak of monkeypox — Illinois, Indiana, and Wisconsin, 2003. MMWR52, 537–540 (2003). [PubMed]
- 5.Peiris JSM, et al. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 6.Jernigan DB, et al. Investigation of bioterrorism-related anthrax, United States, 2001: epidemiologic findings. Emerg. Infect. Dis. 2002;8:1019–1028. doi: 10.3201/eid0810.020353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. The World Health Report 2004 (World Health Organization, Genève, 2004).
- 8.Guerrant RL, Blackwood BL. Threats to global health and survival: the growing crises of tropical infectious diseases — an “unfinished” agenda. Clin. Infect. Dis. 1999;28:966–986. doi: 10.1086/514765. [DOI] [PubMed] [Google Scholar]
- 9.Butler JC, et al. Emerging infectious diseases among indigenous peoples. Emerg. Infect. Dis. 2001;7(suppl.):554–555. doi: 10.3201/eid0707.017732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodges N. Λoιµoλoγια — sive, Pestis Nuperæ apud Populum Londinensem Grassantis Narratio Historica. 1672. [Google Scholar]
- 11.Morens DM, Littman RJ. Epidemiology of the Plague of Athens. Trans. Am. Philol. Assoc. 1992;122:271–304. [Google Scholar]
- 12.Crosby AW. The Columbian Exchange: Biological and Cultural Consequences of 1492. 1972. [Google Scholar]
- 13.Hopkins D. Princes and Peasants. Smallpox in History. 1983. pp. 204–233. [Google Scholar]
- 14.MacArthur N. Island Populations of the Pacific. 1968. [Google Scholar]
- 15.Bushnell OA. The Gifts of Civilization. Germs and Genocide in Hawai'i. 1993. [Google Scholar]
- 16.De Bevoise K. Agents of Apocalypse: Epidemic Disease in the Colonial Philippines. 1995. [Google Scholar]
- 17.Morens DM. Measles in Fiji, 1875: thoughts on the history of emerging infectious diseases. Pac. Health Dialog. 1998;5:119–128. [Google Scholar]
- 18.Koch R. Untersuchungen über Bacterien. V. Die Aetiologie der Milzbrand-Krankheit, begründet auf die Entwicklungsgeschichte des Bacillus Anthracis. Beiträge zur Biologie der Pflanzen. 1876;2:277–310. [Google Scholar]
- 19.Worboys M. Spreading Germs: Diseases, Theories, and Medical Practice in Britain, 1865–1900. 2000. [Google Scholar]
- 20.Porter R. The Greatest Benefit to Mankind: A Medical History of Humanity from Antiquity to the Present. 1997. [Google Scholar]
- 21.Deming WC. The extermination of infectious diseases. NY Med. J. 1894;59:710–715. [Google Scholar]
- 22.Cockburn A. The Evolution and Eradication of Infectious Diseases. 1963. [DOI] [PubMed] [Google Scholar]
- 23.Fauci AS. Infectious diseases: considerations for the 21st century. Clin. Infect. Dis. 2001;32:675–685. doi: 10.1086/319235. [DOI] [PubMed] [Google Scholar]
- 24.Krause RM. The Restless Tide: The Persistent Challenge of the Microbial World. 1981. [Google Scholar]
- 25.Committee on Emerging Microbial Threats to Health. Emerging Infections. Microbial Threats to Health in the United States (eds Lederberg, J., Shope, R. E. & Oaks, S. C.) (National Academy Press, Washington DC, 1992). [PubMed]
- 26.Committee on Emerging Microbial Threats to Health in the 21st Century. Microbial Threats to Health in the United States: Emergence, Detection and Response (eds Smolinski, M. S., Hamburg, M. A. & Lederberg, J.) (National Academy Press, Washington DC, 2003). [PubMed]
- 27.Stephens DS, et al. Emerging and re-emerging infectious diseases: a multidisciplinary perspective. Am. J. Med. Sci. 1998;315:64–75. doi: 10.1097/00000441-199802000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Desselberger U. Emerging and re-emerging infectious diseases. J. Infect. 2000;40:3–15. doi: 10.1053/jinf.1999.0624. [DOI] [PubMed] [Google Scholar]
- 29.Lederberg J. Infectious history. Science. 2000;288:287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 30.Pollard AJ, Dobson AR. Emerging infectious diseases in the 21st century. Curr. Opin. Infect. Dis. 2000;13:265–275. doi: 10.1097/00001432-200006000-00011. [DOI] [PubMed] [Google Scholar]
- 31.Feldman H, et al. Emerging and re-emerging infectious diseases. Med. Microbiol. Immunol. 2002;191:63–74. doi: 10.1007/s00430-002-0122-5. [DOI] [PubMed] [Google Scholar]
- 32.McMichael, T. Human Frontiers, Environments and Disease. Past Patterns, Uncertain Futures (Cambridge Univ. Press, Cambridge, 2001).
- 33.Joint United Nations Programme on HIV/AIDS. AIDS Epidemic Update: December 2003 (UNAIDS, Genéve, 2003).
- 34.Sharp PM, et al. The origins of acquired immune deficiency viruses: where and when? Phil. Trans. R. Soc. Lond. B. 2001;356:867–876. doi: 10.1098/rstb.2001.0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinn TC. Population migration and the spread of types 1 and 2 human immunodeficiency viruses. Proc. Natl Acad. Sci. USA. 1994;91:2407–2014. doi: 10.1073/pnas.91.7.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daley CL, et al. The AIDS Knowledge Base. 1999. pp. 23–52. [Google Scholar]
- 37.Ronald AR. Slowing heterosexual HIV transmission. Infect. Dis. Clin. North Am. 1995;9:287–296. [PubMed] [Google Scholar]
- 38.Hjelle B, et al. A novel hantavirus associated with an outbreak of fatal respiratory disease in the southwestern United States: evolutionary relationships to known hantaviruses. J. Virol. 1994;168:592–596. doi: 10.1128/jvi.68.2.592-596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chua KB, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 40.Beisel CD, Morens DM. Variant Creutzfeldt–Jakob disease and the acquired and transmissible spongiform encephalopathies. Clin. Infect. Dis. 2004;38:697–704. doi: 10.1086/381028. [DOI] [PubMed] [Google Scholar]
- 41.Leopoldt JG. Nützliche und auf die Erfahrung gegründete Einleitung zu der Landwirthschaft, fünf Theile 348. 1759. [Google Scholar]
- 42.Casalone C, et al. Identification of a second bovine amyloidotic spongiform encephalopathy: molecular similarities with sporadic Creutzfeldt–Jakob disease. Proc. Natl Acad. Sci. USA. 2004;101:3065–3070. doi: 10.1073/pnas.0305777101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams ES. Scrapie and chronic wasting disease. Clin. Lab. Med. 2003;23:139–159. doi: 10.1016/s0272-2712(02)00040-9. [DOI] [PubMed] [Google Scholar]
- 44.Katz AR, Morens DM. Severe streptococcal infections in historical perspective. Clin. Infect. Dis. 1992;14:298–307. doi: 10.1093/clinids/14.1.298. [DOI] [PubMed] [Google Scholar]
- 45.Musser JM, Krause RM. Emerging Infections. Biomedical Research Reports. 1998. pp. 185–218. [Google Scholar]
- 46.Musser JM, Selander RK. Brazilian purpuric fever: evolutionary genetic relationships of the case clone of Haemophilus influenzae biogroup aegyptius to encapsulated strains of Haemophilus influenzae. J. Infect. Dis. 1990;161:130–133. doi: 10.1093/infdis/161.1.130. [DOI] [PubMed] [Google Scholar]
- 47.Li M-S, Farrant JL, Langford PR, Kroll JS. Identification and characterization of genomic loci unique to the Brazilian purpuric fever clonal group of H. influenzae biogroup aegyptius: functionality explored using meningococcal homology. Mol. Microbiol. 2003;47:1101–1111. doi: 10.1046/j.1365-2958.2003.03359.x. [DOI] [PubMed] [Google Scholar]
- 48.Reid SD, Hoe NP, Smoot LM, Musser JM. Group A streptococcus: allelic variation, population genetics, and host–pathogen interactions. J. Clin. Invest. 2001;107:393–399. doi: 10.1172/JCI11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beres SB, et al. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl Acad. Sci. USA. 2003;99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang Y, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 51.Parsonnet, J. (ed.) Microbes and Malignancy: Infection as a Cause of Human Cancers (Oxford Univ. Press, New York, 1999).
- 52.Sanders MK, Peura DA. Helicobacter pylori-associated diseases. Curr. Gastroenterol. Rep. 2002;4:448–454. doi: 10.1007/s11894-002-0019-x. [DOI] [PubMed] [Google Scholar]
- 53.Fredricks DN, Relman DA. Current Clinical Topics in Infectious Diseases. 1998. pp. 180–200. [PubMed] [Google Scholar]
- 54.Kovats RS, Bouma MJ, Hajat S, Worrall E, Haines A. El Niño and health. Lancet. 2003;362:1481–1489. doi: 10.1016/S0140-6736(03)14695-8. [DOI] [PubMed] [Google Scholar]
- 55.Cliff A, Haggett P, Smallman-Raynor M. Island Epidemics. 2000. pp. 165–236. [Google Scholar]
- 56.Massey A. Epidemiology in Relation to Air Travel. 1933. [Google Scholar]
- 57.Morens DM. Acute haemorrhagic conjunctivitis: dealing with a newly emerging disease. Pac. Health Dialog. 1998;5:147–153. [Google Scholar]
- 58.Wellems TE, Miller LH. Two worlds of malaria. N. Engl. J. Med. 2003;349:1496–1498. doi: 10.1056/NEJMp038127. [DOI] [PubMed] [Google Scholar]
- 59.Miller LH, Hoffman SL. Research toward vaccines against malaria. Nature Med. 1998;4(suppl.):520–524. doi: 10.1038/nm0598supp-520. [DOI] [PubMed] [Google Scholar]
- 60.Espinal MA. The global situation of MDR-TB. Tuberculosis. 2003;83:44–51. doi: 10.1016/s1472-9792(02)00058-6. [DOI] [PubMed] [Google Scholar]
- 61.Centers for Disease Control and Prevention. Staphylococcus aureus resistant to vancomycin — United States, 2002. MMWR51, 565–567 (2002).
- 62.Lee JH. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 2003;69:6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neu HC. The crisis in antibiotic resistance. Science. 1992;257:1064–1072. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 64.Jones JL, et al. Surveillance for AIDS-defining opportunistic illnesses, 1992–1997. MMWR. 1999;48:1–22. [PubMed] [Google Scholar]
- 65.Vento S, Cainelli F. Infections in patients with cancer undergoing chemotherapy: aetiology, prevention, and treatment. Lancet Oncol. 2003;4:595–604. doi: 10.1016/s1470-2045(03)01218-x. [DOI] [PubMed] [Google Scholar]
- 66.Singh N. Impact of current transplantation practices on the changing epidemiology of infections in transplant recipients. Lancet Infect. Dis. 2003;3:156–161. doi: 10.1016/s1473-3099(03)00546-2. [DOI] [PubMed] [Google Scholar]
- 67.Chapman LE. Xenotransplantation: public health risks — patient vs. society in an emerging field. Curr. Top. Microbiol. Immunol. 2003;278:23–45. doi: 10.1007/978-3-642-55541-1_2. [DOI] [PubMed] [Google Scholar]
- 68.Nash D, et al. The outbreak of West Nile virus infection in the New York City area in 1999. N. Engl. J. Med. 2001;344:1807–1814. doi: 10.1056/NEJM200106143442401. [DOI] [PubMed] [Google Scholar]
- 69.Platonov AE, et al. Outbreak of West Nile infection, Volgograd Region, Russia, 1999. Emerg. Infect. Dis. 2001;7:128–132. doi: 10.3201/eid0701.010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Komar N. West Nile virus: epidemiology and ecology in North America. Adv. Virus Res. 2003;61:185–234. doi: 10.1016/s0065-3527(03)61005-5. [DOI] [PubMed] [Google Scholar]
- 71.Gubler DJ. Dengue and dengue haemorrhagic fever. Clin. Microbiol. Rev. 1998;11:480–497. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morens DM. Antibody-dependent enhancement of infection and the pathogenesis of viral disease. Clin. Infect. Dis. 1994;19:500–512. doi: 10.1093/clinids/19.3.500. [DOI] [PubMed] [Google Scholar]
- 73.Faruque SM, Nair GB. Molecular ecology of toxigenic Vibrio cholerae. Microbiol. Immunol. 2002;46:59–66. doi: 10.1111/j.1348-0421.2002.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 74.Shortridge KF, Peiris JS, Guan Y. The next influenza pandemic: lessons from Hong Kong. J. Appl. Microbiol. 2003;94(suppl.):70S–79S. doi: 10.1046/j.1365-2672.94.s1.8.x. [DOI] [PubMed] [Google Scholar]
- 75.Webster RG. A molecular whodunit. Science. 2001;293:1773–1775. doi: 10.1126/science.1065206. [DOI] [PubMed] [Google Scholar]
- 76.Subbarao K, Katz J. Avian influenza viruses infecting humans. Cell Mol. Life Sci. 2000;571:1770–1784. doi: 10.1007/PL00000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stevens J, et al. Structure of the uncleaved human H1 haemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 78.Reid AH, Taubenberger JK. The origin of the 1918 pandemic influenza virus: a continuing enigma. J. Gen. Virol. 2003;84:2285–2292. doi: 10.1099/vir.0.19302-0. [DOI] [PubMed] [Google Scholar]
- 79.Anderson RM, Swinton J, Garnett GP. Potential impact of low-efficacy HIV-1 vaccines in populations with high rates of infection. Proc. R. Soc. Lond. B. 1995;261:147–161. doi: 10.1098/rspb.1995.0129. [DOI] [PubMed] [Google Scholar]
- 80.Ghani AC, Ferguson NM, Donnelly CA, Anderdson RM. Short-term projections for variant Creutzfeldt–Jakob disease onsets. Stat. Methods Med. Res. 2003;12:191–201. doi: 10.1191/0962280203sm327ra. [DOI] [PubMed] [Google Scholar]
- 81.Thompson WW, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. J. Am. Med. Assoc. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 82.Alibek K, Handelman S. Biohazard: The Chilling True Story of the Largest Covert Biological Weapons Program in the World — Told From the Inside by the Man Who Ran It. 1999. [Google Scholar]
- 83.Henschel AW. Document zur Geschichte des schwarzen Todes. Mitgetheilt und eingeleitet. Archiv für die gesammte Medicin. 1842;2:26–59. [Google Scholar]
- 84.Duffy J. Smallpox and the Indians in the American colonies. Bull. Hist. Med. 1951;25:324–341. [PubMed] [Google Scholar]
- 85.Török T, et al. A large community outbreak of salmonellosis caused by intentional contamination of restaurant salad bars. J. Am. Med. Assoc. 1997;278:389–395. doi: 10.1001/jama.1997.03550050051033. [DOI] [PubMed] [Google Scholar]
- 86.Fauci AS. Biodefence on the research agenda: the world needs new and creative ways to counter bioterrorism. Nature. 2003;421:787. doi: 10.1038/nature01480. [DOI] [PubMed] [Google Scholar]
- 87.Darling RG, Catlett CL, Huebner KD, Jarrett DG. Threats in bioterrorism: I. CDC category A agents. Emerg. Med. Clin. North Am. 2002;20:273–309. doi: 10.1016/s0733-8627(02)00005-6. [DOI] [PubMed] [Google Scholar]
- 88.World Health Organization. Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS) 〈http://www.who.int/entity/csr/sars/en/WHOconsensus.pdf〉 (2003).
- 89.Centers for Disease Control and Prevention. Preventing Emerging Infectious Diseases: A Strategy for the 21st Century (Department of Health and Human Services, Atlanta, 1998).
- 90.NIH Bioengineering Consortium. Nanoscience and Nanotechnology: Shaping Biomedical Research. Symposium Report 〈http://grants.nih.gov/grants/becon/becon_symposia.htm〉 (2002).
- 91.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 92.Dybul M, et al. Guidelines for using antiretroviral agents among HIV-infected adults and adolescents. Ann. Intern. Med. 2002;137:381–433. doi: 10.7326/0003-4819-137-5_part_2-200209031-00001. [DOI] [PubMed] [Google Scholar]
- 93.Zhu H, Bilgin M, Snyder M. Annu. Rev. Biochem. 2003;72:783–812. doi: 10.1146/annurev.biochem.72.121801.161511. [DOI] [PubMed] [Google Scholar]
- 94.Gluck R, Metcalfe IC. New technology platforms in the development of vaccines for the future. Vaccine. 2002;20:B10–B16. doi: 10.1016/s0264-410x(02)00513-3. [DOI] [PubMed] [Google Scholar]
- 95.National Institute of Allergy and Infectious Diseases. The Jordan Report: 20th Anniversary. Accelerated Development of Vaccines 〈http://www.niaid.nih.gov/dmid/vaccines/jordan20/jordan20_2002.pdf〉 (2002).
- 96.Bendelac A, Medzhitov R. Adjuvants of immunity: harnessing innate immunity to promote adaptive immunity. J. Exp. Med. 2002;195:F19–F23. doi: 10.1084/jem.20020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O'Hagan DT, Valiante NM. Recent advances in the discovery and delivery of vaccine adjuvants. Nature Rev. Drug Discov. 2003;2:727–735. doi: 10.1038/nrd1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fraser CM, Eisen JA, Nelson KE, Paulsen IT, Salzberg SL. The value of complete microbial genome sequencing (you get what you pay for) J. Bacteriol. 2002;184:6403–6405. doi: 10.1128/JB.184.23.6403-6405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cromley EK. GIS and disease. Annu. Rev. Public Health. 2003;24:7–24. doi: 10.1146/annurev.publhealth.24.012902.141019. [DOI] [PubMed] [Google Scholar]


