Abstract
Spike (S) proteins of coronaviruses, including the coronavirus that causes severe acute respiratory syndrome (SARS), associate with cellular receptors to mediate infection of their target cells1,2. Here we identify a metallopeptidase, angiotensin-converting enzyme 2 (ACE2)3,4, isolated from SARS coronavirus (SARS-CoV)-permissive Vero E6 cells, that efficiently binds the S1 domain of the SARS-CoV S protein. We found that a soluble form of ACE2, but not of the related enzyme ACE1, blocked association of the S1 domain with Vero E6 cells. 293T cells transfected with ACE2, but not those transfected with human immunodeficiency virus-1 receptors, formed multinucleated syncytia with cells expressing S protein. Furthermore, SARS-CoV replicated efficiently on ACE2-transfected but not mock-transfected 293T cells. Finally, anti-ACE2 but not anti-ACE1 antibody blocked viral replication on Vero E6 cells. Together our data indicate that ACE2 is a functional receptor for SARS-CoV.
Supplementary information
The online version of this article (doi:10.1038/nature02145) contains supplementary material, which is available to authorized users.
Main
So far two types of coronavirus surface receptor have been identified5. The group II coronavirus mouse hepatitis virus (MHV) uses murine carcinoembryonic antigen-related cell adhesion molecules (CEACAMs), members of the immunoglobulin superfamily of receptors6. A number of group I coronaviruses, for example human coronavirus 229E, transmissible gastroenteritis virus and feline infectious peritonitis virus, require the zinc metalloprotease aminopeptidase N (APN, CD13) for entry into their target cells7,8,9. Recently, a distinct coronavirus has been identified as the aetiological agent of SARS, an acute pulmonary syndrome characterized by an atypical pneumonia that results in progressive respiratory failure and death in close to 10% of cases10,11,12,13. Analysis of the SARS-CoV genome suggests that this virus does not belong to any of the three defined coronavirus groups, and that the SARS-CoV S protein is similarly distinct14,15. Similar to the analogous human immunodeficiency virus (HIV) and influenza proteins, the S proteins of some coronaviruses—including MHV and the group III coronavirus infectious bronchitis virus—are cleaved into two subunits (S1 and S2) by a cellular protease in virus-producing cells16,17. The S proteins of other coronaviruses, including those of group I and probably SARS-CoV, are not cleaved in virus-producing cells14,15,18. Nonetheless, S1 and S2 domains of these latter S proteins can be identified through their homology with the S1 and S2 subunits of cleaved coronavirus S proteins. The S1 domains of all characterized coronaviruses mediate an initial high-affinity association with their respective receptors19,20,21.
The African green monkey kidney cell line Vero E6 permits replication of SARS-CoV10. We first investigated whether the S1 domain of the SARS-CoV S protein could bind to Vero E6 cells. Figure 1a demonstrates that a protein expressing residues 12–672 of the SARS-CoV S protein, fused to the Fc domain of human immunoglobulin-γ1 (IgG1; S1–Ig), specifically recognized a moiety present on Vero E6 cells but did not bind human kidney 293T cells. Using this same fusion protein (Fig. 1c) or a carboxy terminally tagged form of the S1 domain (Fig. 1b), a protein band of approximately 110 kDa could be immunoprecipitated from metabolically labelled Vero E6 cells lysed with 0.3% n-decyl-β-d-maltopyranoside in phosphate-buffered saline. When the immunoprecipitated protein was incubated with PNGase F, an enzyme that removes N-glycosylation, two bands were observed at approximately 80–85 and 100 kDa (Fig. 1c, lanes 3 and 4).
Figure 1. A 110 kDa protein associates with the S1 domain of SARS-CoV S protein.
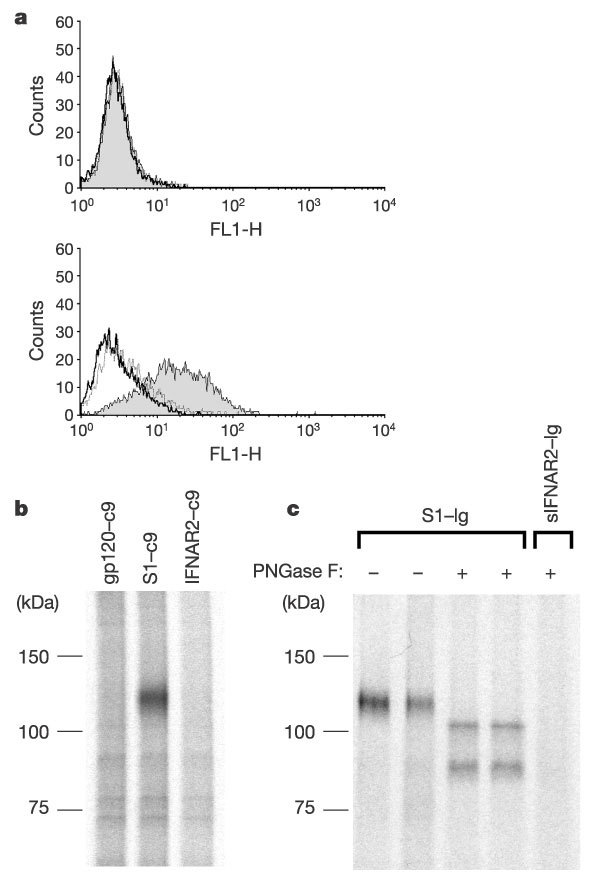
a, A fusion protein of the S1 domain (S1–Ig, shaded area), or of the first 327 residues of that domain (S1(327)–Ig, dotted line) with the Fc domain of human IgG1, or culture medium alone (thick line) was incubated with 293T (top panel) or Vero E6 (bottom panel) cells. Binding of fusion proteins to cells was measured by flow cytometry using a FITC-labelled anti-human IgG secondary antibody. b, Metabolically labelled Vero E6 cell lysates were immunoprecipitated with HIV-1 gp120, SARS-CoV S1 domain, or the ectodomain of human IFNAR2, each containing a tag (C9) at its C terminus, and an anti-tag antibody. Immunoprecipitates were analysed by SDS–PAGE. c, Labelled Vero E6 cell lysates were immunoprecipitated with S1–Ig or soluble IFNAR2–Ig. Immunoprecipitates were treated or not, as indicated, with PNGase F, and analysed by SDS–PAGE.
We then analysed the 110 kDa band by trypsin digestion and mass spectrometry. Three human proteins were identified whose sequences were consistent with the masses of tryptic fragments obtained from this band. Two of these proteins, myosin 1b and major vault protein, do not localize to the cell surface and are ubiquitously expressed, therefore we did not analyse them further. Eight independent tryptic fragments consistent with sequences comprising 17% of the amino acid sequence of human ACE2 were also identified (Supplementary Information). Because both the tissue distribution and subcellular localization of ACE2 are appropriate for a receptor for SARS-CoV3,22,23, we cloned it from complementary DNA obtained from human lung for further analysis.
293T cells transfected with plasmid expressing ACE2, but not those transfected with vector alone, were specifically recognized by S1–Ig (Fig. 2a). A soluble form of ACE2, but not that of the related enzyme ACE1, blocked the association of S1–Ig with Vero E6 cells (Fig. 2b). Furthermore, S1–Ig immunoprecipitated a soluble form of ACE2 (Fig. 2c, lane 5) that was recognized by an anti-ACE2 antibody (lane 8). When the approximately 110 kDa soluble form of ACE2 was incubated with PNGase F, an 85 kDa band was observed (Fig. 2c, lanes 5 and 6), indicating that the lower band observed in PNGase-F-treated Vero E6 immunoprecipitates is ACE2 (Fig. 1c, lanes 3 and 4). These data demonstrate a specific, high-affinity association between the S1 domain of the SARS-CoV S protein and ACE2.
Figure 2. A high-affinity association between ACE2 and the S1 domain.
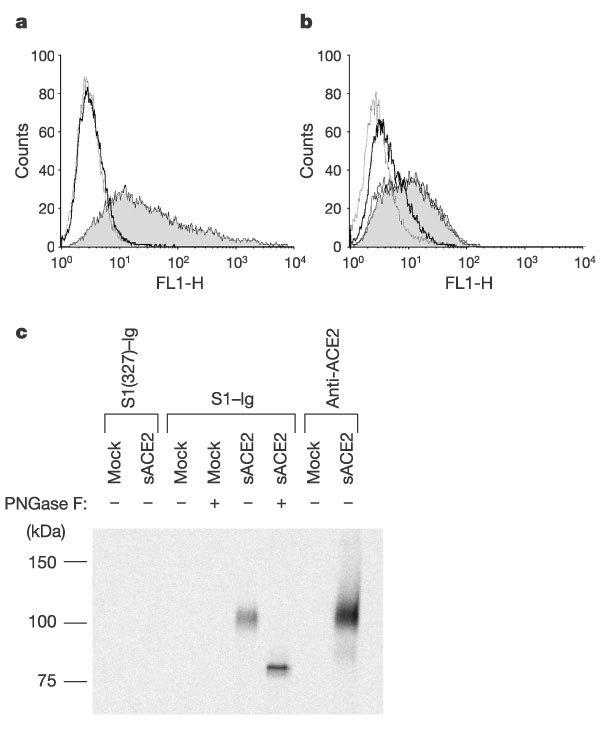
a, 293T cells transfected with plasmid encoding ACE2 (shaded area and dotted line), or with vector alone (thick solid line), were analysed by flow cytometry with S1–Ig (shaded area and thick solid line) or S1(327)–Ig (dotted line). b, Vero E6 cells were incubated with S1–Ig (shaded area, thin solid line and thick solid line), or with medium alone (dotted line), in the presence of soluble ACE1 (thin solid line) or ACE2 (thick solid line), or without either protein (shaded area and dotted line). c, Supernatants of radiolabelled 293T cells transfected with plasmid encoding soluble ACE2 or with vector alone (mock) were immunoprecipitated with S1(327)–Ig, S1–Ig, or an anti-ACE2 antibody. Immunoprecipitates were treated or not, as indicated, with PNGase F, and analysed by SDS–PAGE.
Proteins that mediate fusion between viral and cellular membranes can in some cases also do so between cells that express the viral fusion protein and those that express the viral receptor. For instance, cells expressing the HIV-1 envelope glycoprotein gp160 can form multinucleated syncytia with cells expressing the HIV-1 receptors CD4 and CCR5 (ref. 24). Because syncytia have been observed in Vero E6 cultures infected with SARS-CoV, in SARS-CoV-infected primates, and in SARS patients10,12,13, we investigated whether 293T cells expressing the SARS-CoV S protein could fuse with 293T cells expressing ACE2. Figure 3a shows that these cells formed syncytia efficiently. As expected, 293T cells transfected with CD4 and CCR5 formed many syncytia with cells expressing HIV-1 gp160 (top left), but not with cells expressing SARS-CoV S protein (bottom left). In contrast, 293T cells expressing ACE2 did not form syncytia with cells expressing gp160 (top right), but formed many large syncytia with cells expressing S protein (bottom right). 293T cells expressing S protein also efficiently formed syncytia with ACE2- but not mock-transfected HeLa cells (not shown). Syncytia formation between S protein- and ACE2-expressing cells was blocked by more than 95% by affinity-purified antibody raised against ACE2, but not by control antibody (Fig. 3b). These data demonstrate that the ability of the SARS-CoV S protein to mediate cell–cell fusion is dependent on the presence of ACE2.
Figure 3. Syncytia formation between S-protein- and ACE2-expressing cells.
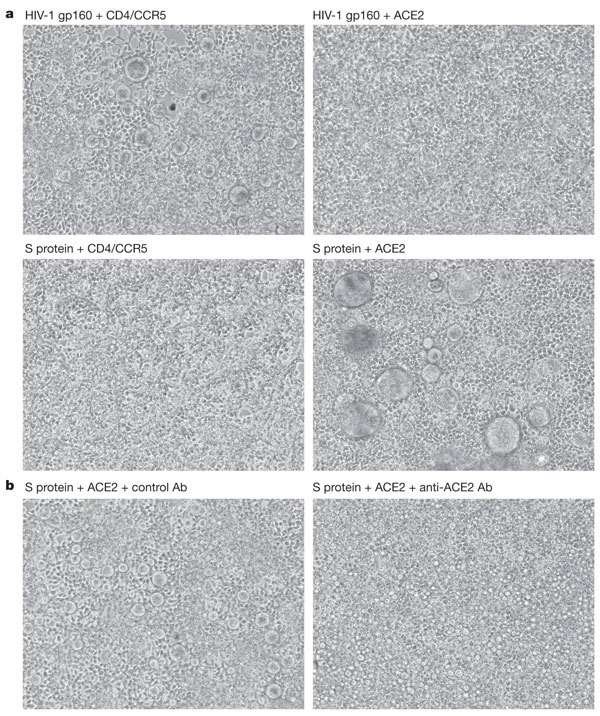
a, 293T cells transfected with plasmids encoding the HIV-1 envelope glycoprotein gp160 (top row) or SARS-CoV S protein (bottom row) were mixed at a 1:1 ratio with 293T cells transfected with plasmids encoding HIV-1 receptors CD4 and CCR5 (left column) or ACE2 (right column). b, 293T cells transfected with ACE2 plasmid were mixed at a 1:1 ratio with 293T cells transfected with plasmid encoding S protein, in the presence of 10 µg ml-1 goat polyclonal control antibody (left) or goat polyclonal anti-ACE2 antibody (right). Ab, antibody.
We then investigated the ability of ACE2 to mediate viral replication. ACE2- and mock-transfected 293T cells were incubated in the presence or absence of SARS-CoV. After 2 days in culture, numerous detached, round and floating cells were observed in cultures of infected ACE2-transfected cells (not shown), consistent with viral replication. In contrast, infected mock-transfected cells and uninfected ACE2-transfected cells were indistinguishable from uninfected control cells, and these cells displayed none of the signs of cytopathicity that were observed in infected ACE2-expressing cells. Figure 4 shows that ACE2- but not mock-transfected 293T cells supported efficient replication of SARS-CoV. We measured viral genomic RNA in cell supernatants by semi-quantitative polymerase chain reaction with reverse transcription (RT–PCR). We found that viral genome copies in ACE2-transfected cells increased by more than 100,000-fold in the first 48 h (Fig. 4a, bottom panel). In contrast, genome copies in mock-transfected cells increased tenfold in the same period (top panel), indicating some basal replication on 293T cells. In addition, virus titres in supernatants from infected ACE2- and mock-transfected cells were measured by titration onto Vero E6 cells. As shown in Fig. 4b, virus stock obtained 96 h after infection from ACE2-transfected cells efficiently replicated on Vero E6 cells, with visible cytopathic effect (CPE) up to and including the highest dilution assayed (1:6,561), whereas virus stock obtained from mock-transfected cells induced no CPE beyond a 1:27 dilution. Consistent with the basal replication observed in both infection assays, minor expression of ACE2 on 293T cells was subsequently detected by an anti-ACE2 antibody (Supplementary Information). We also investigated the ability of anti-ACE2 antibody to inhibit viral replication on Vero E6 cells. Anti-ACE1 antibody had no observable effect on virally induced cytopathicity, whereas anti-ACE2 antibody inhibited cytopathicity in a dose-dependent manner (Fig. 4c). Collectively, these data show that ACE2 contributes substantially to the efficiency of SARS-CoV replication.
Figure 4. Efficient replication of SARS-CoV in the presence of ACE2.
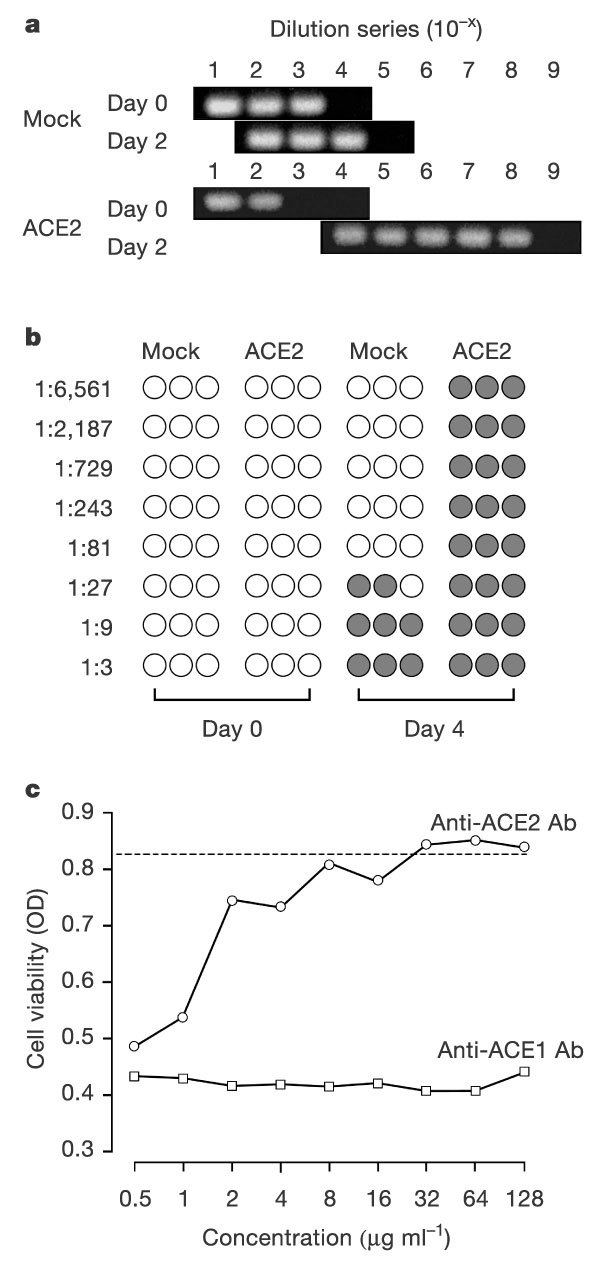
a, Mock- or ACE2-transfected 293T cells were infected with SARS-CoV for 1 h, and cell supernatants sampled 0 and 48 h after washing. Viral RNA was measured by RT–PCR at varying dilutions of supernatant. The endpoint of dilution is shown for each group. b, Supernatants (collected at days 0 and 4) of infected mock- and ACE2-transfected 293T cells were incubated at the indicated dilutions in triplicate with Vero E6 cells plated on 96-well microtitre plates. Cytopathic effect, indicated by a shaded circle, was monitored for each well 3 days after Vero E6 cell infection. c, Duplicate samples of Vero E6 cells were incubated with affinity-purified goat anti-ACE1 or anti-ACE2 antibody, at the indicated concentrations, before infection with SARS-CoV. Cells were washed and cell viability, shown as the average optical density (OD) at 490 nm wavelength, was assayed using CellTiter 96. The dotted line indicates viability of uninfected cells. No toxicity was observed in uninfected cells treated with the maximum concentration of either antibody.
We show here that ACE2 can be immunoprecipitated from Vero E6 cells by the S1 domain of the SARS-CoV S protein, that it mediates fusion with S-protein-expressing cells, and that it promotes viral replication in a cell line otherwise inefficient for SARS-CoV infection. Moreover, syncytia formation and viral replication were specifically inhibited by an anti-ACE2 antibody. These data demonstrate that ACE2 is a functional receptor for SARS-CoV. Real-time PCR analysis of 72 human tissues has demonstrated efficient expression of ACE2 messenger RNA in the bronchus and lung parenchyma as well as in the heart, kidney and gastrointestinal tract23. Lung and kidney are also the primary sites of expression of murine ACE2 (ref. 22). This tissue distribution is consistent with the pathology of SARS, which is characterized by an acute infection of the lungs10,11,12. SARS-CoV has also been found in kidney tissue of infected patients10, and it replicates efficiently on the primate kidney cell lines FRhK-4 and Vero E6 (refs 10, 12). Active viral replication in the small and large intestine has been observed25. Detection of virus in the faeces11 may reflect expression of ACE2 message in these tissues23. The physiological role of ACE2 in most of these tissues has not been defined, although ACE2 is thought to be an essential regulator of cardiac function26. Candidate substrates, some of which regulate vasopermeability, have been identified27. It is unclear whether SARS-CoV interferes with ACE2 enzymatic activity in a manner that contributes to the pathogenesis of SARS.
CD13, the receptor for a number of coronaviruses, is, like ACE2, a zinc metalloprotease; however, apart from this commonality the two receptors are widely divergent. This may suggest that some property of this class of proteases contributes to viral replication. However, mutations in the catalytic site of CD13 do not interfere with tissue-culture replication of transmissible gastroenteritis virus, which uses this enzyme as a receptor7,28. Similarly, our preliminary studies showed that syncytia formation mediated by SARS-CoV S protein remained efficient when two active-site histidines of ACE2 were modified to asparagine (Supplementary Information).
A number of antibodies, peptides and small compounds have been shown to bind to ACE2 (refs 29, 30). It is possible that some of these may be useful in the treatment of SARS, either by blocking the S-protein-binding site, or by inducing a conformation in ACE2 that is unfavourable to binding or fusion. Alternatively, a soluble form of the receptor itself may slow viral replication in an infected individual. Identification of ACE2 as a SARS-CoV receptor will facilitate description of the receptor-binding domain of the S protein, presumably the most effective target epitope of an S1-protein-based subunit vaccine. Also, it is likely that a cell line approved for vaccine production and made permissive for viral replication by ACE2 expression will be the most efficient large-scale producer of a whole-killed or attenuated virus for use as a vaccine. A mouse transgenic for human ACE2 may be useful as an animal model of SARS. Finally, study of the interaction between the SARS-CoV S protein and ACE2 of other animals may provide insights into the origins of the virus. Thus, if SARS returns as a threat to human health, these studies may contribute to its control.
Methods
Immunoprecipitation and identification of ACE2
Vero E6 cells were metabolically labelled for 48 h with [35S]cysteine and [35S]methionine, and lysed in 1.5 ml per 100-mm dish of 0.3% n-decyl-β-d-maltopyranoside (DDM, Anatrace) in phosphate-buffered saline (PBS) containing protease-inhibitor cocktails (Sigma and Roche) and 0.2 mM phenylmethylsulphonyl fluoride (Sigma). After removal of cell debris by centrifugation, lysate was incubated for 1 h at room temperature with 2.5 µg purified S1–Ig or human interferon-α receptor 2 (IFNAR2)–Ig fusion constructs and protein A Sepharose. Alternatively, C-terminally C9-tagged forms of the S1 domain (S1–C9) or of control proteins (HIV-1 gp120–C9 and IFNAR2–C9) were incubated with the antibody 1D4 (National Cell Culture Center) together with protein A Sepharose. Precipitates were washed twice in 0.3% DDM/PBS and once in PBS alone. Bound proteins were eluted in reducing Laemmli sample buffer at 55 °C or in non-reducing buffer at 37 °C for 10 min. Proteins were separated by SDS–polyacrylamide gel electrophoresis (PAGE) on 8% Tris-glycine gels (Invitrogen). Using this approach approximately 5 × 107 unlabelled Vero E6 cells were used to generate a distinct band of 110 kDa that could be readily visualized by Coomassie staining. This band was excised from the gel and incubated with trypsin, and the masses of tryptic fragments were determined by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Masses were compared with possible tryptic fragments of proteins in the GenBank database using Sequest software.
Receptor expression measurement on Vero E6 and 293T cells
Plasmids encoding the signal sequence of CD5 and a fusion of the S1 domain of the SARS-CoV S protein (residues 12–672), or the first 316 residues of that domain (12–327), with the Fc region of human IgG1 (S1–Ig and S1(327)–Ig, respectively) were transfected into 293T cells, and immunoglobulin fusion proteins were purified on protein A Sepharose beads. A total of 5 × 105 293T cells transfected with ACE2-expressing or control plasmids, or the same number of untransfected Vero E6 cells, were incubated with 15 µg ml-1 of S1–Ig or S1(327)–Ig in a volume of 100 µl. In some cases, 15 µg ml-1 of soluble forms of ACE1 or ACE2 (R&D Systems) was also included. Cells were washed in PBS with 0.5% BSA and 0.1% NaN3, incubated with FITC-labelled goat anti-human IgG Fc (Sigma), and analysed by flow cytometry.
Syncytia formation
293T cells, approximately 50% confluent on a six-well plate, were transfected by the calcium-phosphate method with plasmids encoding a codon-optimized form of the SARS-CoV S protein or of the HIV-1 envelope glycoprotein gp160 (ADA isolate). In parallel, 293T or HeLa cells were transfected with plasmids encoding ACE2 or the HIV-1 receptors CD4 and CCR5. One day after transfection, cells were trypsinized and those expressing viral-envelope proteins were mixed with cells expressing receptors at a 1:1 ratio, and plated on 12- or 24-well plates. At 24–48 h after mixing, multinucleated giant cells were observed. In some cases 10 µg ml-1 of goat anti-ACE2 antibody or of goat anti-ACE1 antibody (R&D Systems) was added at the time of cell mixing.
Infection of mock- and ACE2-transfected 293T cells
SARS-CoV (Urbani strain), provided by L. Anderson (Centers for Disease Control and Prevention), was passaged on Vero E6 cells. 293T cells transfected in 25 cm2 flasks with pcDNA3.1 alone or pcDNA3.1 expressing ACE2 were infected for 1 h with 1.4 × 103 TCID50 (50% tissue-culture infectious dose) of SARS-CoV, as measured by endpoint titration on Vero E6 cells, or left uninfected, and washed twice in culture medium. Cells were monitored for cytopathic effect for 4 days. Aliquots of cell supernatants were collected 0, 48 and 96 h after wash. RNA was recovered using a viral RNA mini-prep kit (Qiagen). Semi-quantitative RT–PCR was performed using a nested protocol as described by ref. 11. Positive control RNA was provided by C. Drosten (Bernhard Nocht Institute for Tropical Medicine, National Reference Center for Tropical Diseases). Virus titration was performed by seeding 5 × 103 Vero E6 cells per well in 96-well microtitre plates 1 day before infection. Culture supernatant from infected 293T cells was added to the first set of wells in triplicate and serially diluted. Cells were monitored for CPE 3 days after infection of Vero E6 cells. The effect of affinity-purified goat anti-ACE1 or -ACE2 antibody on SARS-CoV-induced cytopathicity was measured by reading absorbance at 492 nm of cells incubated with CellTiter 96 (Promega).
Supplementary information
Supplementary Table and Figures (JPG 277 kb)
Supplementary Table and Figure Legends (DOC 20 kb)
Acknowledgements
We thank J. Coderre for assistance with RT–PCR, M. Kirk for editing, and S. H. Wang, E. Kieff, J. Sodroski, C. Gerard and N. Gerard for guidance and helpful conversations.
Competing interests
The authors declare that they have no competing financial interests.
Contributor Information
Hyeryun Choe, Email: hyeryun.choe@tch.harvard.edu.
Michael Farzan, Email: farzan@mbcrr.harvard.edu.
References
- 1.Gallagher TM, Buchmeier MJ. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes KV. SARS-associated coronavirus. N. Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- 3.Donoghue M, et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.RES.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 4.Tipnis SR, et al. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 5.Holmes KV, et al. Coronavirus receptor specificity. Adv. Exp. Med. Biol. 1993;342:261–266. doi: 10.1007/978-1-4615-2996-5_40. [DOI] [PubMed] [Google Scholar]
- 6.Dveksler GS, et al. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus-A59. J. Virol. 1993;67:1–8. doi: 10.1128/jvi.67.1.1-8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delmas B, et al. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tresnan DB, Holmes KV. Feline aminopeptidase N is a receptor for all group I coronaviruses. Adv. Exp. Med. Biol. 1998;440:69–75. doi: 10.1007/978-1-4615-5331-1_9. [DOI] [PubMed] [Google Scholar]
- 9.Yeager CL, et al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ksiazek TG, et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 11.Drosten C, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 12.Kuiken T, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier RA, et al. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rota PA, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 15.Marra MA, et al. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 16.Sturman LS, Holmes KV. Proteolytic cleavage of peplomeric glycoprotein E2 of MHV yields two 90K subunits and activates cell fusion. Adv. Exp. Med. Biol. 1984;173:25–35. doi: 10.1007/978-1-4615-9373-7_3. [DOI] [PubMed] [Google Scholar]
- 17.Jackwood MW, et al. Spike glycoprotein cleavage recognition site analysis of infectious bronchitis virus. Avian Dis. 2001;45:366–372. doi: 10.2307/1592976. [DOI] [PubMed] [Google Scholar]
- 18.Spaan W, Cavanagh D, Horzinek MC. Coronaviruses: structure and genome expression. J. Gen. Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 19.Bonavia A, Zelus BD, Wentworth DE, Talbot PJ, Holmes KV. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 2003;77:2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breslin JJ, et al. Human coronavirus 229E: receptor binding domain and neutralization by soluble receptor at 37 degrees C. J. Virol. 2003;77:4435–4438. doi: 10.1128/JVI.77.7.4435-4438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubo H, Yamada YK, Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu T, et al. Molecular cloning, mRNA expression and chromosomal localization of mouse angiotensin-converting enzyme-related carboxypeptidase (mACE2) DNA Seq. 2002;13:217–220. doi: 10.1080/1042517021000021608. [DOI] [PubMed] [Google Scholar]
- 23.Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/S0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 24.Choe H, et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/S0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 25.Leung WK, et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crackower MA, et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 27.Vickers C, et al. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- 28.Delmas B, et al. Determinants essential for the transmissible gastroenteritis virus-receptor interaction reside within a domain of aminopeptidase-N that is distinct from the enzymatic site. J. Virol. 1994;68:5216–5224. doi: 10.1128/jvi.68.8.5216-5224.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang L, et al. Novel peptide inhibitors of angiotensin-converting enzyme 2. J. Biol. Chem. 2003;278:15532–15540. doi: 10.1074/jbc.M212934200. [DOI] [PubMed] [Google Scholar]
- 30.Dales NA, et al. Substrate-based design of the first class of angiotensin-converting enzyme-related carboxypeptidase (ACE2) inhibitors. J. Am. Chem. Soc. 2002;124:11852–11853. doi: 10.1021/ja0277226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table and Figures (JPG 277 kb)
Supplementary Table and Figure Legends (DOC 20 kb)


