Abstract
The 1918 influenza pandemic killed 20–40 million people worldwide1, and is seen as a worst-case scenario for pandemic planning. Like other pandemic influenza strains, the 1918 A/H1N1 strain spread extremely rapidly. A measure of transmissibility and of the stringency of control measures required to stop an epidemic is the reproductive number, which is the number of secondary cases produced by each primary case2. Here we obtained an estimate of the reproductive number for 1918 influenza by fitting a deterministic SEIR (susceptible-exposed-infectious-recovered) model to pneumonia and influenza death epidemic curves from 45 US cities: the median value is less than three. The estimated proportion of the population with A/H1N1 immunity before September 1918 implies a median basic reproductive number of less than four. These results strongly suggest that the reproductive number for 1918 pandemic influenza is not large relative to many other infectious diseases2. In theory, a similar novel influenza subtype could be controlled. But because influenza is frequently transmitted before a specific diagnosis is possible and there is a dearth of global antiviral and vaccine stores, aggressive transmission reducing measures will probably be required.
Supplementary information
The online version of this article (doi:10.1038/nature03063) contains supplementary material, which is available to authorized users.
Main
The emergence of a new pandemic influenza subtype is expected3. Pandemics have occurred regularly throughout this century (1918, 1957 and 1968)4, and opportunities for the generation of new subtypes5 have persisted and probably expanded3. Pandemic influenza spreads rapidly, has high attack rates and kills millions of people worldwide1,4,6. The 1918 pandemic was particularly destructive. The case fatality proportion (CFP) was ten times higher than in all other influenza pandemics and was unusually high in young adults4,7.
Understanding the great speed with which influenza is transmitted is crucial to effective preparation for pandemics. The doubling time for an epidemic curve (about three days in the 1918 pandemic) is a function of the reproductive number (R) and the serial interval (ν), which is the average time between a primary and secondary case. The magnitude of R determines the intensity of measures required to halt transmission. The components of ν, that is, the duration of the latent and infectious periods, determine how and when these measures must be applied. Estimates of R for pandemic influenza vary widely, ranging from 1.68 to 20 (refs 8–12), and are thus of limited value for pandemic preparation. Furthermore, R depends on both the infectious agent and the host population; for example, estimates for measles vary between rural and urban populations2. To our knowledge, there are no estimates of the range of R values that might be expected if a strain similar to 1918 pandemic influenza emerges.
We estimated R by fitting a deterministic SEIR model with homogeneous mixing to the excess pneumonia and influenza (P&I) death curves for 45 cities (Fig. 1). We assumed distributions of infectiousness consistent with previous studies8 and with viral shedding data4, giving mean latent and infectious periods of 1.9 days and 4.1 days, respectively. Infections in the SEIR model were transformed to P&I mortality by assuming a 2% CFP7,13. Time to death was based on influenza autopsy reports14,15,16, with a mean survival time of two weeks (see Supplementary Information). Excess deaths were defined as the deaths above the median for 1910–16 (described previously17, see Fig. 1 and Supplementary Information). The choice of baseline does not substantially affect the results; the 1918 influenza CFP was so much higher than for previous epidemics that any baseline is a small proportion of the 1918 P&I death curve (see Supplementary Information).
Figure 1. Graph of the logarithm of excess P&I deaths in 1918 for the ten most populous cities in the US.
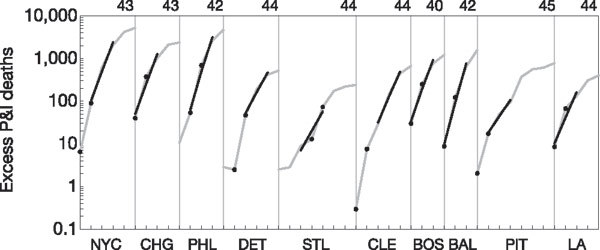
Curves from each city are separated by vertical bars for clarity, with the week number listed above each peak. Curves are shown from left to right, in order of decreasing population size: New York City (NYC), Chicago (CHG), Philadelphia (PHL), Detroit (DET), St Louis (STL), Cleveland (CLE), Boston (BOS), Baltimore (BAL), Pittsburgh (PIT) and Los Angeles (LA). Raw data are shown as grey lines. Black lines indicate model fits for the initial R estimates. Black dots indicate the weeks used for the extreme R estimates.
We estimate that R for 1918 pandemic influenza was approximately 2–3 (Fig. 2). The median estimated R for 45 cities was 2 (interquartile range 1.7, 2.3), based on the first three weeks of each epidemic curve with greater than one excess P&I death per 100,000 population. To address the possibility that these initial R estimates were biased downward—the model ignores the possibility of substantial slowing of exponential growth due to depletion of susceptible hosts, heterogeneous mixing and/or transmission-reducing interventions—we also fit, for each city, the two adjacent weeks of data with the greatest exponential growth rate. The median estimated R (2.7) increased and the interquartile range (2.3, 3.4) widened. The maximum initial (6.3) and extreme (6.5) R estimates were similar. The extreme R values serve as an upper bound; R estimates based on fits of other regions of the epidemic curves tended to be lower than the extreme R values (see Supplementary Information). These results strongly support the conclusion that the estimated R for 1918 pandemic influenza was not large relative to that for many other infectious diseases2.
Figure 2. Histogram of initial and extreme estimated R values for 45 cities during the 1918 influenza pandemic.
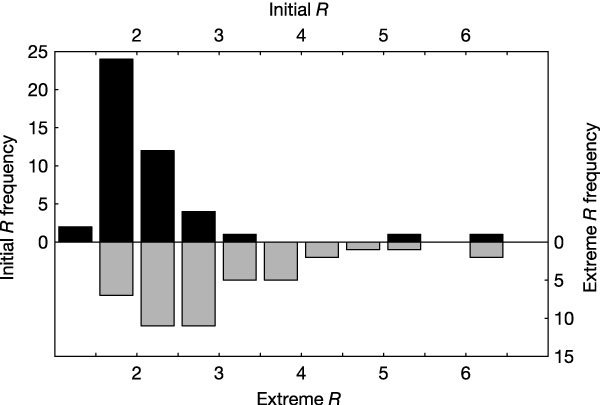
Dark bars show initial R estimates, grey bars show extreme R estimates.
The initial R estimate was not significantly correlated with factors that included latitude, longitude, population size18, population density19,20, age distribution21 or sex distribution21. These results are consistent with contemporary findings by Pearl, who sought correlates of an “epidemic index”21. The extreme R estimates were weakly positively correlated with population density in 1910 (Spearman correlation ρ = 0.5, P = 0.001) and 1920 (ρ = 0.4, P = 0.011), but only the former association remains statistically significant after the Bonferroni correction for multiple comparisons.
Our R estimates were based on excess P&I mortality data, which are surrogates for influenza case data. We assessed the potential bias of using mortality data in several ways. Sensitivity analyses indicated that the assumptions governing the case-to-death transformation in our model (the CFP and the time-to-death distribution) did not affect the magnitude of R estimates (see Supplementary Information). Furthermore, calculations described in the Supplementary Information indicate that mortality data will quickly (within two weeks of the start of the epidemic) reach exponential growth at a rate determined by R, even if transmission and mortality are concentrated in separate groups. Thus, mortality-based R estimates should not be biased. We also estimated R using symptomatic influenza reports from three of the 45 cities. They are consistent with these theoretical results, and are comparable to mortality-based estimates (see Supplementary Information).
It is unknown how similar the recent and 1918 pandemic A/H1N1 influenza viral shedding patterns are, nor is it clear how these patterns translate into transmission probability. To better understand the dependence of R on assumptions about the serial interval and its components, we plotted R estimates based on a linearized SEIR model with an exponential growth rate corresponding to the median rate found in calculating the extreme values of R (Fig. 3, see Supplementary Information). R is less than 3 for most plausible mean ν, regardless of the fraction of ν that is spent in the latent period. To observe an R above 5, the mean ν for 1918 influenza would have had to be several times longer than that of more recent influenza strains4.
Figure 3. Graph of the relationship between the serial interval (ν) and the magnitude of R.
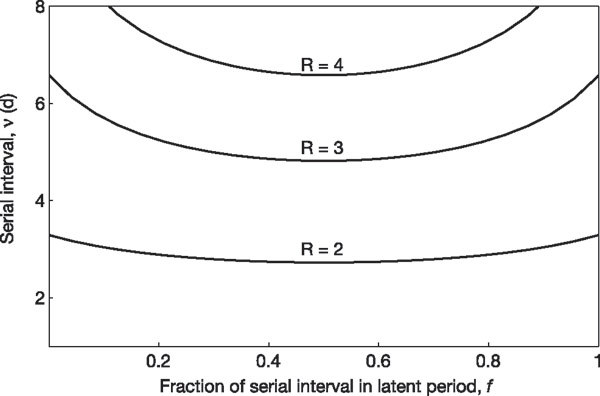
Lines indicate combinations of ν and the fraction of ν in the latent period (f) that yield constant values of R. These R estimates assume a linearized SEIR model and the median extreme exponential growth rate.
The proportion of the population susceptible at the start of the pandemic determines the relationship between R and the basic reproductive number (R0), which is the number of secondary cases generated by a primary case in a completely susceptible population2. Frost hypothesized that a 1918 pandemic-like strain spread throughout America in the spring of 1918 (ref. 22), and recent analyses support this ‘herald wave’ hypothesis23. Anecdotal evidence suggests that those who fell ill in the spring were protected from disease in the autumn pandemic24. Nevertheless, a large majority of the population was probably susceptible to the A/H1N1 pandemic strain in September 1918. In a typical epidemic transmission season, 15–25% of the population becomes infected with influenza4. The herald wave is believed to have arrived late in the 1917–18 transmission season. Using 70% as a conservative lower bound for the fraction susceptible at the start of the autumn pandemic, the medians for our initial and extreme R0 are 2.9 and 3.9.
We have presented a range of R estimates for 1918 pandemic influenza in 45 US cities. We know of one previous R estimate for 1918 pandemic influenza, obtained by fitting the entirety of a single curve of influenza deaths from cities across England and Wales10. Although the previous estimate was described by its author as having a poor fit to data10, and despite the additional assumptions implicit in that analysis, it is reassuring that our R estimates for 1918 A/H1N1 are similar. The median initial and extreme R values are slightly higher than 1957 A/H2N2 and 1968 A/H3N2 pandemic R estimates8,11 (see Supplementary Discussion), but are well below that of many other infections2. It is likely that an exceptionally high CFP in young adults, rather than an unusually high R, is what distinguishes the epidemic curve of 1918 from more recent pandemics.
For many infectious agents, explosive epidemics indicate high transmissibility. While the 1918 pandemic progressed quite rapidly, our results indicate that 1918 pandemic influenza A/H1N1 was not highly transmissible relative to other influenza subtypes8,11 and infectious agents2. A similar pandemic with an R0 of 2–4 could in principle be prevented by vaccinating or administering antiviral prophylaxis to 50–75% of the population, a figure that may require some adjustment owing to heterogeneous mixing patterns in real populations25. Unfortunately, controlling a future pandemic will not be so simple. At present, vaccine production capacity and antiviral medication stockpiles are insufficient to provide such broad coverage, even in wealthy countries3, although judicious use of available supplies may nevertheless reduce mortality during a pandemic8. Control measures based on case identification (for example, contact tracing, isolation, targeted prophylaxis and treatment) that were central to the control of severe acute respiratory syndrome (SARS), whose R was similar to that for 1918 influenza26,27,28, will only be partially successful for influenza12. For influenza, unlike SARS, substantial transmission occurs before the onset of case-defining symptoms12. This implies that measures that generally reduce contacts between persons, regardless of infection status, may be our most powerful protection against a pandemic until adequate vaccine and antiviral medicines can be produced, at which point mass-vaccination and prophylaxis may be more effective than targeted approaches.
Rapid application of control measures for a 1918-like influenza pandemic will be crucial. The short time between primary and secondary cases (ν) means that case numbers in a 1918-like pandemic will rise rapidly, with incident infections and the ensuing deaths doubling about every three days. Increased passenger travel relative to 1918 will facilitate the spread of a new virus across the globe. It is imperative that real-time surveillance information be shared freely, and that preventive measures be taken very early in a new pandemic. Therefore, while the relatively modest reproductive number estimated for 1918 pandemic influenza suggests the feasibility of controlling a similar future pandemic, significant planning and investment will be required to facilitate a rapid and effective response.
Supplementary information
This file includes Supplementary Data, Methods, Discussion and Figure Legends. (DOC 100 kb)
Histogram of the proportion of infected hosts assumed to be infectious between 1 and 8 days post infection in the model. (PDF 15 kb)
Histogram of the proportion of all deaths occurring on each day in the five weeks following infection. (PDF 15 kb)
Transmission rate ratio, the adult:child CFP ratio (m) and the number of serial intervals required for the exponential growth rate to reflect the magnitude of R. (PDF 213 kb)
Spreadsheet of the weekly excess pneumonia and influenza mortality data originally published in Collins, S. D., Frost, W. H., Gover, M. & Sydenstricker, E. Mortality from Influenza and Pneumonia in 50 Large Cities of the United States 1910-1929. Public Health Reports 45, 2277-2328 (1930). (XLS 23 kb)
Acknowledgements
We thank J. Wallinga and D. Olson for discussions and C. Raviola for data procurement. This work was supported in part by the National Defense Science and Engineering Graduate Fellowship (C.E.M.), the Medical Scientist Training Program Fellowship (C.E.M.), NIAID (J.M.R. and M.L.) and the Ellison Medical Foundation (M.L.).
Competing interests
The authors declare that they have no competing financial interests.
References
- 1.Patterson KD, Pyle GF. The geography and mortality of the 1918 influenza pandemic. Bull. Hist. Med. 1991;65:4–21. [PubMed] [Google Scholar]
- 2.Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. 1991. [Google Scholar]
- 3.Webby RJ, Webster RG. Are we ready for pandemic influenza? Science. 2003;302:1519–1522. doi: 10.1126/science.1090350. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson K, Webster RG, Hay AJ. Textbook of Influenza. 1998. [Google Scholar]
- 5.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simonsen L, et al. Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J. Infect. Dis. 1998;178:53–60. doi: 10.1086/515616. [DOI] [PubMed] [Google Scholar]
- 7.Frost WH. Statistics of influenza morbidity with special reference to certain factors in case incidence and case fatality. Public Health Rep. 1920;35:584–597. doi: 10.2307/4575511. [DOI] [Google Scholar]
- 8.Longini IM, Jr, Halloran ME, Nizam A, Yang Y. Containing pandemic influenza with antiviral agents. Am. J. Epidemiol. 2004;159:623–633. doi: 10.1093/aje/kwh092. [DOI] [PubMed] [Google Scholar]
- 9.Gog JR, Rimmelzwaan GF, Osterhaus AD, Grenfell BT. Population dynamics of rapid fixation in cytotoxic T lymphocyte escape mutants of influenza A. Proc. Natl Acad. Sci. USA. 2003;100:11143–11147. doi: 10.1073/pnas.1830296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spicer CC, Lawrence CJ. Epidemic influenza in Greater London. J. Hyg. (Lond.) 1984;93:105–112. doi: 10.1017/S0022172400060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rvachev LA, Longini IM., Jr A mathematical model for the global spread of influenza. Math. Biosci. 1985;75:3–22. doi: 10.1016/0025-5564(85)90064-1. [DOI] [Google Scholar]
- 12.Fraser C, Riley S, Anderson RM, Ferguson NM. Factors that make an infectious disease outbreak controllable. Proc. Natl Acad. Sci. USA. 2004;101:6146–6151. doi: 10.1073/pnas.0307506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan EO. Epidemic Influenza; a Survey. 1927. [Google Scholar]
- 14.Wolbach SB. Comments on the pathology and bacteriology of fatal influenza cases, as observed at Camp Devens, Mass. Johns Hopkins Hosp. Bull. 1919;338:104–109. [Google Scholar]
- 15.MacCallum WG. Johns Hopkins Hospital Reports. 1921. [Google Scholar]
- 16.Klotz O. Studies on Epidemic Influenza. 1919. pp. 207–293. [Google Scholar]
- 17.Collins SD, Frost WH, Gover M, Sydenstricker E. Mortality from Influenza and Pneumonia in 50 Large Cities of the United States 1910–1929. Public Health Rep. 1930;45:2277–2328. doi: 10.2307/4579795. [DOI] [Google Scholar]
- 18.Bureau of the Census . Mortality Rates 1910–1920 with Population of the Federal Censuses of 1910 and 1920 and Intercensal Estimates of Population. 1923. [Google Scholar]
- 19.Bureau of the Census. Population of the 100 largest urban places: 1920. 〈http://www.census.gov/population/documentation/twps0027/tab15.txt〉 (1998).
- 20.Bureau of the Census. Population of the 100 largest urban places: 1910. 〈http://www.census.gov/population/documentation/twps0027/tab14.txt〉 (1998).
- 21.Pearl R. Influenza studies: further data on the correlation of explosiveness of outbreak of the 1918 epidemic. Public Health Rep. 1921;36:273–298. doi: 10.2307/4575894. [DOI] [Google Scholar]
- 22.Frost WH. The epidemiology of influenza. J. Am. Med. Assoc. 1919;73:313–318. doi: 10.1001/jama.1919.02610310007003. [DOI] [Google Scholar]
- 23.Olson DR, Simonsen L, Edelson PJ, Morse SS. 4th Int. Conf. on Emerging Infectious Diseases. 2004. [Google Scholar]
- 24.Stanley LL. Influenza at San Quentin Prison, California. Public Health Rep. 1919;34:996–1008. doi: 10.2307/4575142. [DOI] [Google Scholar]
- 25.Hill AN, Longini IM., Jr The critical vaccination fraction for heterogeneous epidemic models. Math. Biosci. 2003;181:85–106. doi: 10.1016/S0025-5564(02)00129-3. [DOI] [PubMed] [Google Scholar]
- 26.Riley S, et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 27.Lipsitch M, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallinga J, Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 2004;160:509–516. doi: 10.1093/aje/kwh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file includes Supplementary Data, Methods, Discussion and Figure Legends. (DOC 100 kb)
Histogram of the proportion of infected hosts assumed to be infectious between 1 and 8 days post infection in the model. (PDF 15 kb)
Histogram of the proportion of all deaths occurring on each day in the five weeks following infection. (PDF 15 kb)
Transmission rate ratio, the adult:child CFP ratio (m) and the number of serial intervals required for the exponential growth rate to reflect the magnitude of R. (PDF 213 kb)
Spreadsheet of the weekly excess pneumonia and influenza mortality data originally published in Collins, S. D., Frost, W. H., Gover, M. & Sydenstricker, E. Mortality from Influenza and Pneumonia in 50 Large Cities of the United States 1910-1929. Public Health Reports 45, 2277-2328 (1930). (XLS 23 kb)


