Abstract
We report epidemiological and clinical investigations on ten pediatric SARS-CoV-2 infection cases confirmed by real-time reverse transcription PCR assay of SARS-CoV-2 RNA. Symptoms in these cases were nonspecific and no children required respiratory support or intensive care. Chest X-rays lacked definite signs of pneumonia, a defining feature of the infection in adult cases. Notably, eight children persistently tested positive on rectal swabs even after nasopharyngeal testing was negative, raising the possibility of fecal–oral transmission.
Subject terms: Paediatrics, Public health
Children infected with the COVID-19 outbreak coronavirus, SARS-CoV-2, show mild symptoms but prolonged shedding of viral RNA in feces, suggesting that the fecal–oral route might play a role in virus transmission.
Main
The outbreak of SARS-CoV-2 (formerly 2019-nCoV) infection emerged in December 2019 in Wuhan, Hubei Province, China1,2. By 25 February 2020, there had been 77,780 confirmed cases including 2,666 deaths in China and over 2,459 confirmed cases in 33 other countries3.
The genome of the new virus and early epidemiological and clinical features of the infection in adults have been reported4–6. The infection is estimated to have a mean incubation period of 5.2 d and commonly causes fever, cough, myalgia and pneumonia in patients4. To date there is a paucity of information regarding SARS-CoV-2 infection in children.
We here report the epidemiological and clinical features of ten children infected with SARS-CoV-2 and tested for evidence of viral excretion through the gastrointestinal and respiratory tracts.
By 20 February 2020, a total of 745 children and 3,174 adults, most of whom had either close contact with diagnosed patients or had members of the family reporting familial outbreaks in the previous 2 weeks, were screened by nasopharyngeal swab real-time PCR with reverse transcription (RT–PCR) for SARS-CoV-2 infection. Overall, 10 children (1.3%) and 111 adults (3.5%) tested positive. The 2.7-fold difference between children and adults is statistically significant (P = 0.002). All ten pediatric patients were admitted to our hospital, a treatment center for SARS-CoV-2 infection designated by the local municipal government.
Of the ten patients, six were male and four were female, with ages ranging from 2 months to 15 years (Table 1). Four had definite contact history with a confirmed patient, seven were from families with a cluster of infection and seven had travel history to epidemic areas in Hubei Province 2 weeks before the onset of infection.
Table 1.
Epidemiological and clinical characteristics of n = 10 independent pediatric patients confirmed with SARS-CoV-2 infection
| Characteristics | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | Patient 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Age (months) | 74 | 151 | 85 | 165 | 14 | 41 | 188 | 164 | 2 | 21 |
| Sex | Male | Female | Female | Male | Male | Male | Female | Male | Female | Male |
| Exposure history | ||||||||||
| Travel to Wuhan city | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ |
| Travel to other cities of Hubei provincial | ✗ | ✓ | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✗ | ✗ |
| Contact with person with 2019-nCoV | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✗ | ✗ | ✓ | ✗ |
| Having family cluster outbreak | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗ | ✗ | ✓ | ✓ |
| Signs and symptoms at admission | ||||||||||
| Fever (maximum, duration) | 39.1 °C, 23 h | 39.2 °C, 5 d | 38.4 °C, 3 d | ✗ | 37.8 °C, 3 h | ✗ | 38.5 °C, 1 h | 38.5 °C, 1 d | ✗ | 38.0 °C, 30 min |
| Cough | ✓ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ |
| Sore throat | ✗ | ✓ | ✓ | ✗ | ✗ | ✗ | ✗ | ✓ | ✓ | ✗ |
| Rhinorrhea | ✗ | ✓ | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ |
| Diarrhea | ✓ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ |
| More than one sign or symptom | ✓ | ✓ | ✓ | ✗ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ |
| Chest X-ray findings | ||||||||||
| Only lung markings increased | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗ | ✓ | ✗ | ✗* |
| Chest computed tomography scan | ||||||||||
| Mottling and ground-glass opacity | ✗ | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗* |
| Blood routine | ||||||||||
| Leukocytes (× 109 l−1; normal range 5–12) | 12 | 4.6 | 7.9 | 4.7 | 7.8 | 8.8 | 4.0 | 7.8 | 10.8 | NA |
| Neutrophils (× 109 l−1; normal range 2.0–7.2) | 9.24 | 3.21 | 6.48 | 1.97 | 1.08 | 2.76 | 1.99 | 5.1 | 1.23 | NA |
| Lymphocytes (× 109 l−1; normal range 1.55–4.80) | 1.35 | 0.89 | 0.65 | 2.17 | 6.19 | 4.73 | 1.66 | 1.75 | 9.05 | NA |
| Platelets (× 109 l−1; normal range 140–440) | 229 | 173 | 301 | 141 | 363 | 286 | 291 | 240 | 342 | NA |
| Hemoglobin (g l−1; normal range 105–145) | 115 | 124 | 116 | 145 | 115 | 135 | 137 | 163 | 102 | NA |
| Coagulation function | ||||||||||
| APTT (s; normal range 28–45) | 43.1 | 46.9 | 42.1 | 45.8 | 40.5 | 35.3 | NA | NA | 41.5 | NA |
| PT (s; normal range 11–15) | 13.5 | 14.4 | 13.1 | 15.3 | 12.1 | 13.3 | NA | NA | 13.6 | NA |
| D-dimer (μg l−1; normal range <0.5) | 0.41 | 0.28 | 0.33 | 0.26 | 0.32 | 0.24 | 0.23 | 0.5 | 0.84 | NA |
| Blood biochemistry | ||||||||||
| Albumin (g l−1; normal range 40.0–55.0) | 44.6 | 47.9 | 39.8 | 44.5 | 45.1 | 46 | 48.3 | 48.2 | 39.5 | NA |
| ALT (U l−1; normal range 9–50) | 20 | 6 | 11 | 11 | 31 | 21 | 14 | 17 | 172 | NA |
| AST (U l−1; normal range 5–60) | 33 | 17 | 23 | 16 | 62 | 36 | 20 | 22 | 127 | NA |
| Total bilirubin (μmol l−1; normal range 2.0–17.0) | 3.5 | 5.9 | 2.6 | 8.7 | 2.8 | 3.3 | 3.8 | 6.6 | 4.9 | NA |
| BUN (mmol l−1; normal range 2.1–7.1) | 3.5 | 2.4 | 2.5 | 3.61 | 3.2 | 4.6 | 4.8 | 5.9 | 4.6 | NA |
| Scr (μmol l−1; normal range 18–62) | 29 | 49 | 29 | 55 | 22 | 29 | 50 | 59 | 15 | NA |
| CK (U l−1; normal range 45–390) | 117 | 64 | 71 | 64 | 278 | 75 | 113 | 103 | 112 | NA |
| LDH (U l−1; normal range 159–322) | 217 | 165 | 217 | 138 | 379 | 252 | 218 | 203 | 367 | NA |
| Glucose (mmol l−1; normal range 4.1–5.9) | 4.32 | 5.56 | 5.7 | 4.2 | 4.4 | 6.68 | 4.94 | 4.69 | 6.94 | NA |
| Infection-related biomarkers | ||||||||||
| Procalcitonin (ng ml−1; normal range <0.1) | 0.103 | 0.125 | 0.121 | <0.1 | <0.1 | <0.1 | <0.1 | 0.203 | 0.11 | NA |
| ESR (mm h−1; normal range 0–15) | 25 | 4 | 6 | 2 | 33 | 2 | 20 | 7 | 2 | NA |
| Serum ferritin (ng ml−1; normal range 22–322) | 84.6 | 63.6 | 58.2 | 178.4 | 9.5 | 47.4 | 73.9 | 304.4 | 919 | NA |
| CRP (mg l−1; normal range 0.0–6.0) | 2.43 | 0.31 | 16.13 | 0.3 | 1.14 | 0.3 | 11.86 | 22.05 | 1.04 | NA |
✓, with; ✗, without; *, recovered phase; APTT, activated partial thromboplastin time; PT, prothrombin time; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; Scr, serum creatinine; CK, creatine kinase; LDH, lactate dehydrogenase; ESR, erythrocyte sedimentation rate; CRP, C-reactive protein; NA, not available.
Upon admission, seven had fever but none had a temperature above 39 °C (Table 1). Other symptom presentations included coughing (five children), sore throat (four children), nasal congestion and rhinorrhea (two children) and diarrhea (three children). One child was completely asymptomatic (patient 4). None of the patients had other symptoms commonly seen in adult patients such as lethargy, dyspnea, muscle ache, headache, nausea and vomiting and disorientation. In fact, none of them sought medical care; they were all identified and diagnosed because of their exposure history.
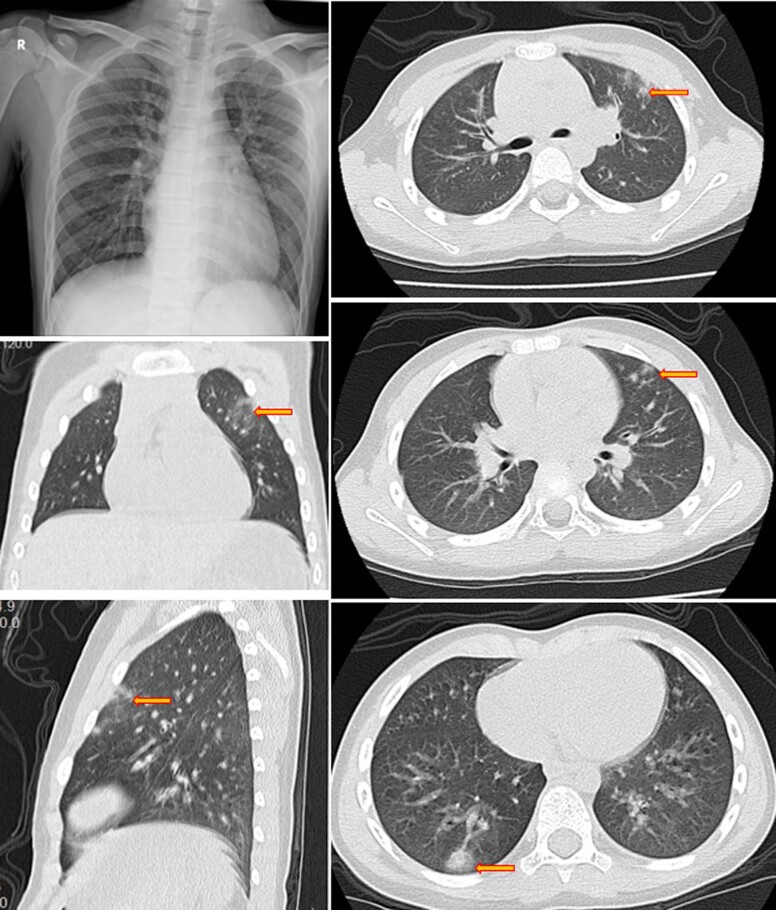
Chest X-rays of these patients were either normal or showed only coarse lung markings without unilateral or bilateral pneumonia. Chest computed tomography (CT) scans showed isolated or multiple patchy ground-glass opacities in five patients but were within normal ranges in the other five. These changes were mainly seen in the outer lung fields and few patients had subpleural bands or strips. There was no pleural effusion, enlarged lymph nodes or other changes that are typically seen in the critically ill adult patients4,6 (Supplementary Fig. 1).
Complete blood count, urine and stool analyses, coagulation function, blood biochemistry and infection biomarkers were tested upon admission (Table 1). Almost all test results were normal in the patients except for patient 9. Few cases had leukopenia, leukocytosis, lymphopenia or elevated transaminase, which in contrast are frequently seen in adult patients. Viral testing for influenza-A virus (H1N1, H3N2, H7N9), influenza B virus, respiratory syncytial virus, parainfluenza virus, adenovirus, SARS-CoV and MERS-CoV were negative in all patients.
Out of nine patients (the tenth patient was transferred from another hospital that did not conduct cytokine tests upon admission), seven showed elevated interleukin (IL)-17F and five of these patients had concurrent elevation of IL-22 (Supplementary Table 1). Five patients had elevated IL-6.
All patients received antiviral therapy with α-interferon oral spray initiated from admission (8,000 U, two sprays, three times a day). Patient 1 was the very first pediatric case of SARS-CoV-2 infection diagnosed in Guangzhou and also treated with azithromycin 10 mg kg−1 per day for 5 d and IVIG 300 mg kg−1 per day for 3 d. No patient required respiratory support or intensive care unit care.
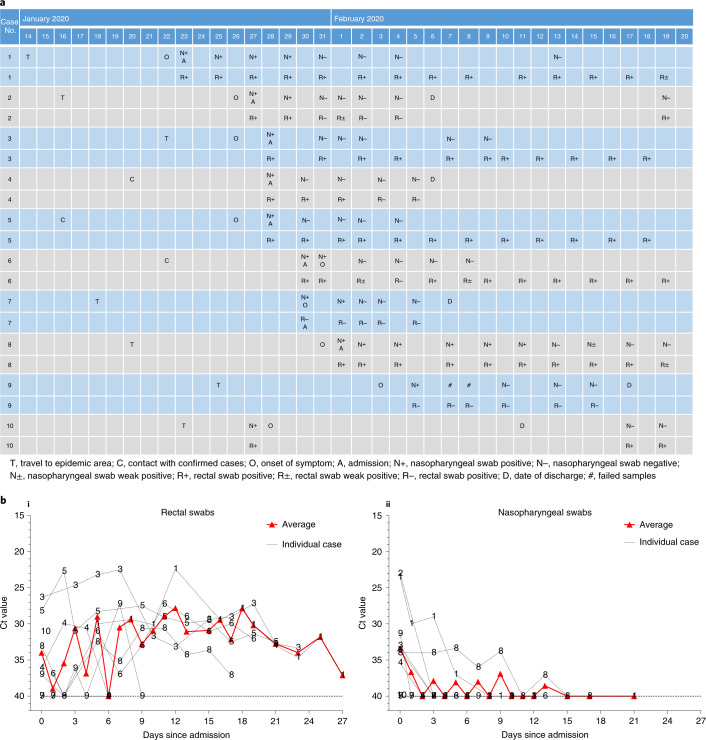
We followed the pattern of viral excretion from respiratory and gastrointestinal tracts in all ten patients by a chronological series of nasopharyngeal and rectal swab samples using real-time RT–PCR (Fig. 1a). Patient 4 was asymptomatic but tested positive on multiple occasions. Patient 6 was asymptomatic on the day his nasopharyngeal swab tested positive and then developed nasal congestion and rhinorrhea the following day. The remaining eight patients had positive tests soon after the onset of symptoms. In addition, eight of ten patients also had real-time RT–PCR-positive rectal swabs, suggesting potential fecal viral excretion. Moreover, eight of ten patients (patients 1–6, 8 and 10) demonstrated persistently positive real-time RT–PCR tests of rectal swabs after their nasopharyngeal testing had become negative.
Fig. 1. Chronology of major epidemiological events and molecular testing results of n = 10 independent pediatric patients confirmed with SARS-CoV-2 infection.
a, Dates of exposure, illness onset and sampling and real-time RT–PCR results of nasopharyngeal swabs and rectal swabs. The total number of patients was n = 10 and real-time RT–PCR was assayed only once for one type of sample at one time point from one independent patient. Colors in the figure represent individual patients. b, Chronological changes in Ct values of Orf1ab and N genes using real-time RT–PCR after hospital admission. The Ct values of Orf1ab and N genes on real-time RT–PCR detected in rectal swabs obtained from n = 10 independent patients (i) and Ct values in nasopharyngeal swabs from n = 10 independent cases (ii). The Ct value is supposed to be inversely related to viral RNA copy numbers and a value of 40 means the virus is molecularly undetectable.
Patients 2, 4, 7 and 10 were discharged home after two consecutive negative real-time RT–PCR tests (separated by at least 24 h) on rectal swabs. Their nasopharyngeal and rectal swabs were repeated weekly after discharge. Patients 4 and 7 remained negative during follow-up but patient 2 had a positive rectal swab again 13 d after discharge. Interestingly, the mother of patient 2, who was hospitalized for SARS-CoV-2 infection (COVID-19) and discharged from a different hospital during the same time period, also had a positive rectal swab test on the same day as her child. A similar phenomenon was also observed in patient 10. He was hospitalized in the Eighth People’s Hospital of Guangzhou between 27 January and 11 February and discharged after two consecutive negative results for both nasopharyngeal and rectal swabs obtained 24 h apart. He was however re-admitted to our hospital because his rectal swab test became positive again on 17 February, although his nasopharyngeal swab test remained negative. As of 20 Feb, all remaining patients (patients 1, 3, 5, 6, 8 and 9) were still testing positive for rectal swabs and continued to be under hospital isolation and observation, although they had all recovered from their illness and had become asymptomatic.
As suggested in a recent study on a similar topic7, we used the cycle threshold (Ct) values of the serial rectal and nasopharyngeal swab tests to approximately indicate viral load (inversely related to Ct value) in these patients to show its change over time. Viral RNA measurements suggest that viral shedding from the digestive system might be greater and last longer than that from the respiratory tract (Fig. 1b).
Clinical features of SARS-CoV-2 infection in adults have been reported elswhere4,8,9. However, few pediatric cases are published and their clinical features have yet to be documented. Compared to adult patients, the ten pediatric patients reported here had clinically milder symptoms and showed fewer alterations in radiological and laboratory testing parameters. For example, none of the ten patients showed clear clinical signs or chest X-ray findings consistent with pneumonia, a typical feature seen in the initial adult patients4,6. Mild and atypical presentations of the infection in children may make it difficult to detect. Indeed, all the patients reported here were found through screening of suspected cases.
We also observed positive real-time RT–PCR results in rectal swabs in eight out of ten pediatric patients, which remained detectable well after nasopharyngeal swabs turned negative, suggesting that the gastrointestinal tract may shed virus and fecal–oral transmission may be possible. Indeed, fecal–oral transmission does exist with other respiratory viruses10. These findings also suggest that rectal swab-testing may be more useful than nasopharyngeal swab-testing in judging the effectiveness of treatment and determining the timing of termination of quarantine12. However, we do not have evidence of replication-competent virus in fecal swabs, which is required to confirm the potential for fecal–oral transmission.
Methods
Study design and participants
In this single-center prospective observational study, between 22 January 2020 and 20 February 2020, 745 ‘highly suspected’ children were screened by real-time RT–PCR using nasopharyngeal swabs to detect people with SARS-CoV-2 infection. Ten children tested positive and were admitted to Guangzhou Women and Children’s Medical Center. They were all recruited to this study. A child who came in contact with a patient confirmed with SARS-CoV-2 infection within the previous 14 d or who was a member of a familial outbreak of the virus was considered ‘highly suspected’. All ‘highly suspected’ children were placed under home quarantine until a SARS-CoV-2 diagnosis was ruled out by real-time RT–PCR.
Data on demographics, contact/exposure history, clinical presentations on admission, chest X-ray and laboratory tests were collected. Laboratory data collected on each patient included complete blood count, urine and stool analysis, blood biochemistry, coagulation function, biomarkers of infection, viral testing through nasopharyngeal and rectal swabs, as well as immunological testing (six items) and a cytokine profile (14 cytokines). Examination of viral excretion from the respiratory and gastrointestinal tract was conducted by serial real-time RT–PCR of nasopharyngeal and rectal swab samples.
Guangzhou Women and Children’s Medical Center, located in Guangzhou, Guangdong Province, China, is a specialized tertiary pediatric hospital caring for children under 18 years of age. To effectively manage patients with SARS-CoV-2 infection, the Municipal Government has put in place a centralized care policy and this hospital is a designated treatment center for children with SARS-CoV-2 infection in Guangdong Province, of which Guangzhou is the capital city. All highly suspected and confirmed pediatric cases in Guangzhou were admitted to this hospital after the SARS-CoV-2 infection was classified as a reportable disease in the Infectious Disease Law and Health and Quarantine Law in China on 20 January 2020. Diagnosis of SARS-CoV-2 infection was made using real-time RT–PCR in the virology laboratory of our hospital and then further confirmed by either Guangzhou Center for Disease Control and Prevention (CDC) or Guangdong Provincial CDC. Nasopharyngeal and rectal swabs were collected upon admission and then every 1–3 d throughout the hospitalization period. SARS-CoV-2 was tested through real-time RT–PCR of 2019-nCoVRNA as previously reported6. Two target genes, including open reading frame1ab (ORF1ab) and nucleocapsid protein (N), were simultaneously amplified and tested during the real-time RT–PCR assay.
Target 1 (ORF1ab):
Forward primer CCCTGTGGGTTTTACACTTAA;
Reverse primer ACGATTGTGCATCAGCTGA;
The probe 5′-VIC-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3′
Target 2 (N):
Forward primer GGGGAACTTCTCCTGCTAGAAT;
Reverse primer CAGACATTTTGCTCTCAAGCTG;
The probe 5′-FAM- TTGCTGCTGCTTGACAGATT-TAMRA-3′.
A Ct value <37 was defined as positive and >40 was defined as negative. These diagnostic criteria were based on the recommendation by the National CDC (China) (http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html). A medium load, defined as a Ct value of 37 to 40, required confirmation by at least two repeats.
In addition, other potential respiratory viruses were also tested using real-time RT–PCR approved by the China Food and Drug Administration. These included influenza-A virus (H1N1, H3N2, H7N9), influenza B virus, respiratory syncytial virus, parainfluenza virus, adenovirus, SARS coronavirus (SARS-CoV) and MERS coronavirus (MERS-CoV).
Data collection
Microsoft Excel (MS Excel 2013, v.15.0) was used for data collection of the epidemiological and clinical information. Epidemiological data were obtained through standard epidemiological questionnaire and interviews5. Clinical information, including clinical presentations and management, were obtained by reviewing clinical charts, nursing records and results of laboratory testing, chest X-rays (Canon SM-50HF-B-D-C mobile DR: tube voltage 65 kV, tube current 125 mA, exposure time 0.04 s) and CT scans (Toshiba’s Aquilion 64-slice CT: tube voltage 120 kV, tube current 50–100 mA, tube rotation time 0.5 s r−1, slice thickness 1.0 mm, slice interval 1.0 mm) for all children with laboratory-confirmed 2019-nCOV infection. Clinical outcome data were recorded up to the date of drafting this manuscript on 20 February 2020. The accuracy of clinical data was confirmed by two pediatricians caring for the patients.
To assess whether SARS-CoV-2 infection could induce an intense immune response that would result in cytokine storms, we collected peripheral blood on admission and tested six immunological indexes and 14 cytokines in these patients. Plasma cytokines (interferon-γ, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12p70, IL-17A, IL-17F, IL-22, tumor necrosis factor-α and tumor necrosis factor-β) were measured by using Human Cytokine Standard 27-Plex Assays panel (HumanTh1/Th2/Th1718-plex, Tianjin Quantobio) and FACSCanto II Flow Cytometry system (Becton Dickinson) for all patients according to the manufacturer’s instructions. The plasma samples from four healthy adults were used as controls for cross comparison.
Statistical analysis
Characteristics of the patients were described and summarized. These included epidemiological data, demographics, clinical signs and symptoms on admission, laboratory results, co-infection with other respiratory pathogens, chest radiographic findings, treatment received for 2019-nCOV and clinical outcomes. Ct values of Orf1ab and N gene using real-time RT–PCR assay were inversely related to viral RNA copy number, with Ct values of 32.04, 28.81, 25.14 and 21.54 corresponding to 5.27 × 104, 5.27 × 105, 5.27 × 106 and 5.27 × 107 copies per ml. Thus, Ct values were used to approximately evaluate the viral load detected in nasopharyngeal and rectal swabs. A chi-squared test was performed for between-group comparisons of percentages. All statistical tests were two-sided with P < 0.05 considered as statistically significant and were performed using R (v.3.1.2; R Foundation for Statistical Computing).
Ethics approval
Ethics approval was obtained from the Ethics Committee of Guangzhou Women and Children’s Medical Center and written informed consents were obtained from the parents of the included children. Data collection and analysis were also required by the National Health Commission of the People’s Republic of China to be part of a continuing public health outbreak investigation.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41591-020-0817-4.
Supplementary information
Supplementary Table 1
Acknowledgements
This work was funded by the National Key Research and Development Program of China (2019YFB1404803) and Guangzhou Regenerative Medicine Guangdong Laboratory (2020GZR110306001). We thank the CDCs of Guangzhou City and Guangdong Province for helping with confirmation of the diagnosis of the viral infection.
Extended data
Extended Data Fig. 1. An example of chest x-ray and computed tomography (CT) scan images at hospital admission (case4).
An example of chest x-ray and computed tomography (CT) scan images at hospital admission (case4).
Author contributions
J.T., H.X. and S.G. contributed to the study design and data interpretation. H.L., J.T., K.Z. and Y.X. contributed to data analysis, data interpretation and writing of the manuscript. Y.X., X.L., B.Z., C.F., Y.G., Q.G., X.S., D.Z., H.S.L. and J.S. contributed to clinical management, patient recruitment and data collection. H.Z. and B.Z. contributed to critical revision of the manuscript. All authors reviewed and approved the final version of the manuscript.
Data availability
The data in this study have been shared with the World Health Organization and will be available upon request and approval by a data access committee. The data access committee comprises four corresponding authors and there is no restriction to data access.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Alison Farrell was the primary editor on this article and managed its editorial process and peer review in collaboration with João Monteiro and the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yi Xu, Xufang Li, Bing Zhu, Huiying Liang.
Contributor Information
Huimin Xia, Email: huiminxia@hotmail.com.
Jinling Tang, Email: jltang@cuhk.edu.hk.
Kang Zhang, Email: kang.zhang@gmail.com.
Sitang Gong, Email: sitangg@126.com.
Extended data
is available for this paper at 10.1038/s41591-020-0817-4.
Supplementary information
is available for this paper at 10.1038/s41591-020-0817-4.
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E. A novel coronavirus emerging in China—key questions for impact assessment. N. Engl. J. Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 3.Coronavirus Disease (COVID-2019) Situation Report 36 (WHO, 2020).
- 4.Chen N, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, D. et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA10.1001/jama.2020.1585 (2020). [DOI] [PMC free article] [PubMed]
- 7.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holshue ML, et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan, W. et al. Clinical characteristics of 2019 novel coronavirus infection in China. N. Engl. J. Med.10.1056/NEJMoa2002032 (2020).
- 10.Zhu Z, et al. Extra-pulmonary viral shedding in H7N9 Avian Influenza patients. J. Clin. Virol. 2015;69:30–32. doi: 10.1016/j.jcv.2015.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Data Availability Statement
The data in this study have been shared with the World Health Organization and will be available upon request and approval by a data access committee. The data access committee comprises four corresponding authors and there is no restriction to data access.