The ascent of pathogens
This week's Review Article tackles the question of the evolution of human pathogens. Analysis of what we know of the origins of 10 tropical and 15 temperate infectious diseases reveals that animals were the source of the majority of important human diseases in the recent past, and that direct contact with animals through activities such as hunting remains a major route for disease acquisition. Modern developments such as industrial food production, vaccine production and blood transfusion may be making us more vulnerable than ever to new pathogens. The authors argue that people with high exposure to wild animals, such as hunters, zoo workers and wildlife veterinarians, should be regularly screened for emergent pathogens.This would provide early warning of newly introduced disease, and supply a repository of tissue samples that would assist in reconstructing the origin of later outbreaks.
Supplementary information
The online version of this article (doi:10.1038/nature05775) contains supplementary material, which is available to authorized users.
Abstract
Many of the major human infectious diseases, including some now confined to humans and absent from animals, are ‘new’ ones that arose only after the origins of agriculture. Where did they come from? Why are they overwhelmingly of Old World origins? Here we show that answers to these questions are different for tropical and temperate diseases; for instance, in the relative importance of domestic animals and wild primates as sources. We identify five intermediate stages through which a pathogen exclusively infecting animals may become transformed into a pathogen exclusively infecting humans. We propose an initiative to resolve disputed origins of major diseases, and a global early warning system to monitor pathogens infecting individuals exposed to wild animals.
Supplementary information
The online version of this article (doi:10.1038/nature05775) contains supplementary material, which is available to authorized users.
Main
Human hunter/gatherer populations currently suffer, and presumably have suffered for millions of years, from infectious diseases similar or identical to diseases of other wild primate populations. However, the most important infectious diseases of modern food-producing human populations also include diseases that could have emerged only within the past 11,000 years, following the rise of agriculture1,2. We infer this because, as discussed below, these diseases can only be sustained in large dense human populations that did not exist anywhere in the world before agriculture. What were the sources of our major infectious diseases, including these ‘new’ ones? Why do so many animal pathogens, including virulent viruses like Ebola and Marburg, periodically infect human hosts but then fail to establish themselves in human populations?
A tentative earlier formulation1 noted that major infectious diseases of temperate zones seem to have arisen overwhelmingly in the Old World (Africa, Asia and Europe), often from diseases of Old World domestic animals. Hence one goal of this article is to re-appraise that conclusion in the light of studies of the past decade. Another goal is to extend the analysis to origins of tropical diseases3. We shall show that they also arose mainly in the Old World, but for different reasons, and mostly not from diseases of domestic animals.
These results provide a framework for addressing unanswered questions about the evolution of human infectious diseases—questions not only of practical importance to physicians, and to all the rest of us as potential victims, but also of intellectual interest to historians and evolutionary biologists. Historians increasingly recognize that infectious diseases have had major effects on the course of history; for example, on the European conquest of Native Americans and Pacific Islanders, the inability of Europeans to conquer the Old World tropics for many centuries, the failure of Napoleon’s invasion of Russia, and the failure of the French attempt to complete construction of a Panama Canal4,5,6. Evolutionary biologists realize that infectious diseases, as a leading cause of human morbidity and mortality, have exerted important selective forces on our genomes2,7.
We begin by defining five stages in the evolutionary transformation of an animal pathogen into a specialized pathogen of humans, and by considering why so many pathogens fail to make the transition from one stage to the next. We then assemble a database of 15 temperate and 10 tropical diseases of high evolutionary and/or historical impact, and we compare their characteristics and origins. Our concluding section lays out some unresolved questions and suggests two expanded research priorities. We restrict our discussion to unicellular microbial pathogens. We exclude macroparasites (in the sense of ref. 7), as well as normally benign commensals that cause serious illness only in weakened hosts. The extensive Supplementary Information provides details and references on our 25 diseases, robustness tests of our conclusions, factors affecting transitions between disease stages, and modern practices altering the risk of emergence of new diseases.
Evolutionary stages
Box 1 delineates five intergrading stages (Fig. 1) through which a pathogen exclusively infecting animals (Stage 1) may become transformed into a pathogen exclusively infecting humans (Stage 5). Supplementary Table S1 assigns each of the 25 major diseases discussed (Supplementary Note S1) to one of these five stages.
Figure 1. Illustration of the five stages through which pathogens of animals evolve to cause diseases confined to humans.
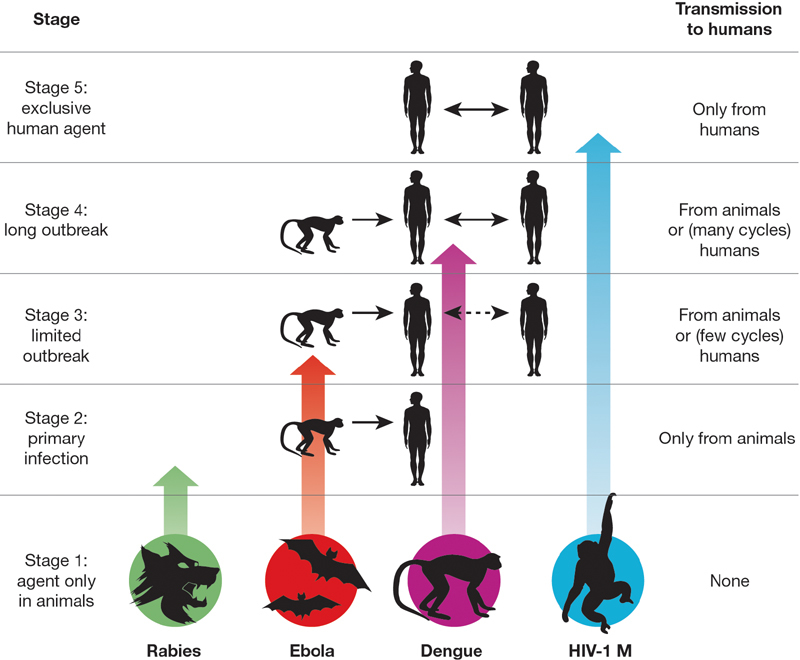
(See Box 1 for details.) The four agents depicted have reached different stages in the process, ranging from rabies (still acquired only from animals) to HIV-1 (now acquired only from humans).
A large literature discusses the conditions required for a Stage 5 epidemic to persist2,7. Briefly, if the disease infects only humans and lacks an animal or environmental reservoir, each infected human introduced into a large population of susceptible individuals must on average give rise during his/her contagious lifespan to an infection in at least one other individual. Persistence depends on factors such as the duration of a host’s infectivity; the rate of infection of new hosts; rate of development of host protective immunity; and host population density, size and structure permitting the pathogen’s regional persistence despite temporary local extinctions.
Less well understood are two of the critical transitions between stages, discussed in Box 2. One is the transition from Stage 1 to Stage 2, when a pathogen initially confined to animals first infects humans. The other is the transition from Stage 2 to Stages 3 and 4 (see also Supplementary Note S2), when a pathogen of animal origin that is nevertheless transmissible to humans evolves the ability to sustain many cycles of human-to-human transmission, rather than just a few cycles before the outbreak dies out (as seen in modern Ebola outbreaks).
Box 1: Five stages leading to endemic human diseases.
We delineate five stages in the transformation of an animal pathogen into a specialized pathogen of humans (Fig. 1). There is no inevitable progression of microbes from Stage 1 to Stage 5: at each stage many microbes remain stuck, and the agents of nearly half of the 25 important diseases we selected for analysis (Supplementary Table S1) have not reached Stage 5.
• Stage 1. A microbe that is present in animals but that has not been detected in humans under natural conditions (that is, excluding modern technologies that can inadvertently transfer microbes, such as blood transfusion, organ transplants, or hypodermic needles). Examples: most malarial plasmodia, which tend to be specific to one host species or to a closely related group of host species.
• Stage 2. A pathogen of animals that, under natural conditions, has been transmitted from animals to humans (‘primary infection’) but has not been transmitted between humans (‘secondary infection’). Examples: anthrax and tularemia bacilli, and Nipah, rabies and West Nile viruses.
• Stage 3. Animal pathogens that can undergo only a few cycles of secondary transmission between humans, so that occasional human outbreaks triggered by a primary infection soon die out. Examples: Ebola, Marburg and monkeypox viruses.
• Stage 4. A disease that exists in animals, and that has a natural (sylvatic) cycle of infecting humans by primary transmission from the animal host, but that also undergoes long sequences of secondary transmission between humans without the involvement of animal hosts. We arbitrarily divide Stage 4 into three substages distinguished by the relative importance of primary and secondary transmission: Stage 4a. Sylvatic cycle much more important than direct human-to-human spread. Examples: Chagas’ disease and (more frequent secondary transmission approaching Stage 4b) yellow fever. Stage 4b. Both sylvatic and direct transmission are important. Example: dengue fever in forested areas of West Africa and Southeast Asia. Stage 4c. The greatest spread is between humans. Examples: influenza A, cholera, typhus and West African sleeping sickness.
• Stage 5. A pathogen exclusive to humans. Examples: the agents causing falciparum malaria, measles, mumps, rubella, smallpox and syphilis. In principle, these pathogens could have become confined to humans in either of two ways: an ancestral pathogen already present in the common ancestor of chimpanzees and humans could have co-speciated long ago, when the chimpanzee and human lineages diverged around five million years ago; or else an animal pathogen could have colonized humans more recently and evolved into a specialized human pathogen. Co-speciation accounts well for the distribution of simian foamy viruses of non-human primates, which are lacking and presumably lost in humans: each virus is restricted to one primate species, but related viruses occur in related primate species19. While both interpretations are still debated for falciparum malaria, the latter interpretation of recent origins is widely preferred for most other human Stage 5 diseases of Supplementary Table S1.
Box 2: Transitions between stages.
Transition from Stage 1 to Stage 2. Most animal pathogens are not transmitted to humans, that is, they do not even pass from Stage 1 to Stage 2. This problem of cross-species infection has been discussed previously20,21,22,23. Briefly, the probability-per-unit-time (p) of infection of an individual of a new (that is, new recipient) host species increases with the abundance of the existing (that is, existing donor) host, with the fraction of the existing host population infected, with the frequency of ‘encounters’ (opportunities for transmission, including indirect ‘encounters’ via vectors) between an individual of the existing host and of the new host, and with the probability of transmission per encounter. p decreases with increasing phylogenetic distance between the existing host and new host. p also varies among microbes (for example, trypanosomes and flaviviruses infect a wide taxonomic range of hosts, while plasmodia and simian foamy viruses infect only a narrow range), and this variation is related to a microbe’s characteristics, such as its ability to generate genetic variability, or its ability to overcome host molecular barriers of potential new hosts (such as humoral and cellular defenses or lack of cell membrane receptors essential for microbe entry into host cells).
These considerations illuminate different reasons why a given animal host species may or may not become a source of many infections in humans. For instance, despite chimpanzees’ very low abundance and infrequent encounters with humans, they have donated to us numerous zoonoses (diseases that still mainly afflict animals) and one or two established human diseases (AIDS and possibly hepatitis B) because of their close phylogenetic relationship to humans. Despite their large phylogenetic distance from humans, many of our zoonoses and probably two of our established diseases (plague and typhus) have been acquired from rodents, because of their high abundance and frequent encounters with humans in dwellings. Similarly, about half of our established temperate diseases have been acquired from domestic livestock, because of high local abundance and very frequent contact. Conversely, elephants and bats are not known to have donated directly to us any established diseases and rarely donate zoonoses, because they are heavily penalized on two or three counts: large phylogenetic distance, infrequent encounters with humans, and (in the case of elephants) low abundance. One might object that Nipah, severe acute respiratory syndrome (SARS) and rabies viruses do infect humans from bats, but these apparent exceptions actually support our conclusion. While bats may indeed be the primary reservoir for Nipah and SARS, human infections by these viruses are acquired mainly from intermediate animal hosts that frequently encounter humans (respectively, domestic pigs, and wild animals sold for food). The rare cases of rabies transmission directly to humans from bats arise because rabies changes a bat’s behaviour so that it does encounter and bite humans, which a healthy bat (other than a vampire bat) would never do.
Transition from Stage 2 to Stage 3 or 4. Although some Stage 2 and 3 pathogens, such as the anthrax and Marburg agents, are virulent and feared, they claim few victims at present. Yet if they made the transition to Stage 4 or 5, their global impact would be devastating. Why do animal pathogens that have survived the initial jump across species lines into a human host (Stages 1 to 2) usually reach a dead end there, and not evolve past Stages 3 and 4 into major diseases confined to humans (Stage 5)? Barriers between Stages 2 and 3 (consider the rabies virus) include differences between human and animal behaviour affecting transmission (for example, animals often bite humans but humans rarely bite other humans); a pathogen’s need to evolve adaptations to the new human host and possibly also to a new vector; and obstacles to a pathogen’s spread between human tissues (for example, BSE is restricted to the central nervous system and lymphoid tissue). Barriers between Stages 3 and 4 (consider Ebola virus) include those related to human population size and to transmission efficiency between humans. The emergence of novel pathogens is now being facilitated by modern developments exposing more potential human victims and/or making transmission between humans more efficient than before24,25,26,27. These developments include blood transfusion (hepatitis C), the commercial bushmeat trade (retroviruses), industrial food production (bovine spongiform encephalitis, BSE), international travel (cholera), intravenous drug use (HIV), vaccine production (simian virus 40, SV40), and susceptible pools of elderly, antibiotic-treated, immunosuppressed patients (see Supplementary Note S2 for details).
Database and conclusions
Database
Supplementary Table S1 lists 10 characteristics for each of 25 important ‘temperate’ (15) and ‘tropical’ (10) diseases (see Supplementary Note S3 for details of this distinction). Our aim was to select well-defined diseases causing the highest mortality and/or morbidity and hence of the highest historical and evolutionary significance (see Supplementary Note S1 for details of our selection criteria). Of the 25 diseases, we selected 17 because they are the ones assessed by ref. 8 as imposing the heaviest world burdens today (they have the highest disability-adjusted life years (DALY) scores). Of the 17 diseases, 8 are temperate (hepatitis B, influenza A, measles, pertussis, rotavirus A, syphilis, tetanus and tuberculosis), and 9 are tropical (acquired immune deficiency syndrome (AIDS), Chagas’ disease, cholera, dengue haemorrhagic fever, East and West African sleeping sicknesses, falciparum and vivax malarias, and visceral leishmaniasis). We selected eight others (temperate diphtheria, mumps, plague, rubella, smallpox, typhoid and typhus, plus tropical yellow fever) because they imposed heavy burdens in the past, although modern medicine and public health have either eradicated them (smallpox) or reduced their burden. Except for AIDS, dengue fever, and cholera, which have spread and attained global impact in modern times, most of these 25 diseases have been important for more than two centuries.
Are our conclusions robust to variations in these selection criteria? For about a dozen diseases with the highest modern or historical burdens (for example, AIDS, malaria, plague, smallpox), there can be little doubt that they must be included, but one could debate some of the next choices. Hence we drew up three alternative sets of diseases sharing a first list of 16 indisputable major diseases but differing in the next choices, and we performed all 10 analyses described below on all three sets. It turned out that, with one minor exception, the three sets yielded qualitatively the same conclusions for all 10 analyses, although differing in their levels of statistical significance (see Supplementary Note S4). Thus, our conclusions do seem to be robust.
Temperate/tropical differences
Comparisons of these temperate and tropical diseases yield the following conclusions:
• A higher proportion of the diseases is transmitted by insect vectors in the tropics (8/10) than in the temperate zones (2/15) (P < 0.005, χ2-test, degrees of freedom, d.f. = 1). This difference may be partly related to the seasonal cessations or declines of temperate insect activity.
• A higher proportion (P = 0.009) of the diseases conveys long-lasting immunity (11/15) in the temperate zones than in the tropics (2/10).
• Animal reservoirs are more frequent (P < 0.005) in the tropics (8/10) than in the temperate zones (3/15). The difference is in the reverse direction (P = 0.1, NS, not significant) for environmental reservoirs (1/10 versus 6/15), but those environmental reservoirs that do exist are generally not of major significance except for soil bearing tetanus spores.
• Most of the temperate diseases (12/15) are acute rather than slow, chronic, or latent: the patient either dies or recovers within one to several weeks. Fewer (P = 0.01) of the tropical diseases are acute: 3/10 last for one or two weeks, 3/10 last for weeks to months or years, and 4/10 last for many months to decades.
• A somewhat higher proportion of the diseases (P = 0.08, NS) belongs to Stage 5 (strictly confined to humans) in the temperate zones (10/15 or 11/15) than in the tropics (3/10). The paucity of Stage 2 and Stage 3 diseases (a total of only 5 such diseases) on our list of 25 major human diseases is noteworthy, because some Stage 2 and Stage 3 pathogens (such as anthrax and Ebola) are notoriously virulent, and because theoretical reasons are often advanced (but also denied) as to why Stage 5 microbes with long histories of adaptation to humans should tend to evolve low morbidity and mortality and not cause major diseases. We discuss explanations for this outcome in Supplementary Note S5.
Most (10/15) of the temperate diseases, but none of the tropical diseases (P < 0.005), are so-called ‘crowd epidemic diseases’ (asterisked in Supplementary Table S1), defined as ones occurring locally as a brief epidemic and capable of persisting regionally only in large human populations. This difference is an immediate consequence of the differences enumerated in the preceding five paragraphs. If a disease is acute, efficiently transmitted, and quickly leaves its victim either dead or else recovering and immune to re-infection, the epidemic soon exhausts the local pool of susceptible potential victims. If in addition the disease is confined to humans and lacks significant animal and environmental reservoirs, depletion of the local pool of potential victims in a small, sparse human population results in local termination of the epidemic. If, however, the human population is large and dense, the disease can persist by spreading to infect people in adjacent areas, and then returning to the original area in a later year, when births and growth have regenerated a new crop of previously unexposed non-immune potential victims. Empirical epidemiological studies of disease persistence or disappearance in isolated human populations of various sizes have yielded estimates of the population required to sustain a crowd disease: at least several hundred thousand people in the cases of measles, rubella and pertussis2,7. But human populations of that size did not exist anywhere in the world until the steep rise in human numbers that began around 11,000 years ago with the development of agriculture1,9. Hence the crowd epidemic diseases of the temperate zones must have evolved since then.
Of course, this does not mean that human hunter/gatherer communities lacked infectious diseases. Instead, like the sparse populations of our primate relatives, they suffered from infectious diseases with characteristics permitting them to persist in small populations, unlike crowd epidemic diseases. Those characteristics include: occurrence in animal reservoirs as well as in humans (such as yellow fever); incomplete and/or non-lasting immunity, enabling recovered patients to remain in the pool of potential victims (such as malaria); and a slow or chronic course, enabling individual patients to continue to infect new victims over years, rather than for just a week or two (such as Chagas’ disease).
Pathogen origins
(See details for each disease in Supplementary Note S10). Current information suggests that 8 of the 15 temperate diseases probably or possibly reached humans from domestic animals (diphtheria, influenza A, measles, mumps, pertussis, rotavirus, smallpox, tuberculosis); three more probably reached us from apes (hepatitis B) or rodents (plague, typhus); and the other four (rubella, syphilis, tetanus, typhoid) came from still-unknown sources (see Supplementary Note S6). Thus, the rise of agriculture starting 11,000 years ago played multiple roles in the evolution of animal pathogens into human pathogens1,4,10. Those roles included both generation of the large human populations necessary for the evolution and persistence of human crowd diseases, and generation of large populations of domestic animals, with which farmers came into much closer and more frequent contact than hunter/gatherers had with wild animals. Moreover, as illustrated by influenza A, these domestic animal herds served as efficient conduits for pathogen transfers from wild animals to humans, and in the process may have evolved specialized crowd diseases of their own.
It is interesting that fewer tropical than temperate pathogens originated from domestic animals: not more than three of the ten tropical diseases of Supplementary Table S1, and possibly none (see Supplementary Note S7). Why do temperate and tropical human diseases differ so markedly in their animal origins? Many (4/10) tropical diseases (AIDS, dengue fever, vivax malaria, yellow fever) but only 1/15 temperate diseases (hepatitis B) have wild non-human primate origins (P = 0.04). This is because although non-human primates are the animals most closely related to humans and hence pose the weakest species barriers to pathogen transfer, the vast majority of primate species is tropical rather than temperate. Conversely, few tropical but many temperate diseases arose from domestic animals, and this is because domestic animals live mainly in the temperate zones, and their concentration there was formerly even more lop-sided (see Supplementary Note S8).
A final noteworthy point about animal-derived human pathogens is that virtually all arose from pathogens of other warm-blooded vertebrates, primarily mammals plus in two cases (influenza A and ultimately falciparum malaria) birds. This comes as no surprise, considering the species barrier to pathogen transfer posed by phylogenetic distance (Box 2). An expression of this barrier is that primates constitute only 0.5% of all vertebrate species but have contributed about 20% of our major human diseases. Expressed in another way, the number of major human diseases contributed, divided by the number of animal species in the taxonomic group contributing those diseases, is approximately 0.2 for apes, 0.017 for non-human primates other than apes, 0.003 for mammals other than primates, 0.00006 for vertebrates other than mammals, and either 0 or else 0.000003 (if cholera really came from aquatic invertebrates) for animals other than vertebrates (see Supplementary Note S9).
Geographic origins
To an overwhelming degree, the 25 major human pathogens analysed here originated in the Old World. That proved to be of great historical importance, because it facilitated the European conquest of the New World (the Americas). Far more Native Americans resisting European colonists died of newly introduced Old World diseases than of sword and bullet wounds. Those invisible agents of New World conquest were Old World microbes to which Europeans had both some acquired immunity based on individual exposure and some genetic resistance based on population exposure over time, but to which previously unexposed Native American populations had no immunity or resistance1,4,5,6. In contrast, no comparably devastating diseases awaited Europeans in the New World, which proved to be a relatively healthy environment for Europeans until yellow fever and malaria of Old World origins arrived11.
Why was pathogen exchange between Old and New Worlds so unequal? Of the 25 major human diseases analysed, Chagas’ disease is the only one that clearly originated in the New World. For two others, syphilis and tuberculosis, the debate is unresolved: it remains uncertain in which hemisphere syphilis originated, and whether tuberculosis originated independently in both hemispheres or was brought to the Americas by Europeans. Nothing is known about the geographic origins of rotavirus, rubella, tetanus and typhus. For all of the other 18 major pathogens, Old World origins are certain or probable.
Our preceding discussion of the animal origins of human pathogens may help explain this asymmetry. More temperate diseases arose in the Old World than New World because far more animals that could furnish ancestral pathogens were domesticated in the Old World. Of the world’s 14 major species of domestic mammalian livestock, 13, including the five most abundant species with which we come into closest contact (cow, sheep, goat, pig and horse), originated in the Old World1. The sole livestock species domesticated in the New World was the llama, but it is not known to have infected us with any pathogens1,2—perhaps because its traditional geographic range was confined to the Andes, it was not milked or ridden or hitched to ploughs, and it was not cuddled or kept indoors (as are some calves, lambs and piglets). Among the reasons why far more tropical diseases (nine versus one) arose in the Old World than the New World are that the genetic distance between humans and New World monkeys is almost double that between humans and Old World monkeys, and is many times that between humans and Old World apes; and that much more evolutionary time was available for transfers from animals to humans in the Old World (about 5 million years) than in the New World (about 14,000 years).
Outlook and future research directions
Many research directions on infectious disease origins merit more effort. We conclude by calling attention to two such directions: clarifying the origins of existing major diseases, and surveillance for early detection of new potentially major diseases.
Origins of established diseases
This review illustrates big gaps in our understanding of the origins of even the established major infectious diseases. Almost all the studies that we have reviewed were based on specimens collected opportunistically from domestic animals and a few easily sampled wild animal species, rather than on systematic surveys for particular classes of agents over the spectrum of domestic and wild animals. A case in point is our ignorance even about smallpox virus, the virus that has had perhaps the greatest impact on human history in the past 4,000 years. Despite some knowledge of poxviruses infecting our domestic mammals, we know little about poxvirus diversity among African rodents, from which those poxviruses of domestic mammals are thought to have evolved. We do not even know whether ‘camelpox’, the closest known relative of smallpox virus, is truly confined to camels as its name implies or is instead a rodent virus with a broad host range. There could be still-unknown poxviruses more similar to smallpox virus in yet unstudied animal reservoirs, and those unknown poxviruses could be important not only as disease threats but also as reagents for drug and vaccine development.
Equally basic questions arise for other major pathogens. While falciparum malaria, an infection imposing one of the heaviest global burdens today, seems to have originated from a bird parasite whose descendants include both the Plasmodium falciparum infecting humans and the P. reichenowii infecting chimpanzees, malaria researchers still debate whether the bird parasite was introduced to both humans and chimpanzees12 a few thousand years ago in association with human agriculture, or instead more than five million years ago before the split of humans and chimpanzees from each other13. Although resolving this debate will not help us eradicate malaria, it is fascinating in its own right and could contribute to our broader understanding of disease emergence. In the case of rubella, a human crowd disease that must have emerged only in the past 11,000 years and for which some close relative may thus still exist among animals, no even remotely related virus is known; one or more may be lurking undiscovered somewhere. Does the recent identification of porcine rubulavirus and the Mapuera virus in bats as the closest known relatives of mumps virus mean that pigs infected humans, or that human mumps infected pigs, or that bats independently infected both humans and pigs? Is human tuberculosis descended from a ruminant mycobacterium that recently infected humans from domestic animals (a formerly prevalent view), or from an ancient human mycobacterium that has come to infect domestic and wild ruminants (a currently popular view)?
To fill these and other yawning gaps in our understanding of disease origins, we propose an ‘origins initiative’ aimed at identifying the origins of a dozen of the most important human infectious diseases: for example, AIDS, cholera, dengue fever, falciparum malaria, hepatitis B, influenza A, measles, plague, rotavirus, smallpox, tuberculosis and typhoid. Although more is already known about the origins of some of these agents (AIDS, influenza A and measles) than about others (rotavirus, smallpox and tuberculosis), more comprehensive screening is still likely to yield significant new information about even the most studied agents, as illustrated by the recent demonstration that gorillas rather than chimpanzees were probably the donor species for the O-group of human immunodeficiency virus (HIV)-114. The proposed effort would involve systematic sampling and phylogeographic analysis of related pathogens in diverse animal species: not just pigs and other species chosen for their ready availability, but a wider range of wild and domestic species whose direct contact (for example, as bushmeat) or indirect contact (for example, vector-mediated) with humans could plausibly have led to human infections. In addition to the historical and evolutionary significance of knowledge gained through such an origins initiative, it could yield other benefits such as: identifying the closest relatives of human pathogens; a better understanding of how diseases have emerged; new laboratory models for studying public health threats; and perhaps clues that could aid in predictions of future disease threats.
A global early warning system
Most major human infectious diseases have animal origins, and we continue to be bombarded by novel animal pathogens. Yet there is no ongoing systematic global effort to monitor for pathogens emerging from animals to humans. Such an effort could help us to describe the diversity of microbial agents to which our species is exposed; to characterize animal pathogens that might threaten us in the future; and perhaps to detect and control a local human emergence before it has a chance to spread globally.
In our view, monitoring should focus on people with high levels of exposure to wild animals, such as hunters, butchers of wild game, wildlife veterinarians, workers in the wildlife trade, and zoo workers. Such people regularly become infected with animal viruses, and their infections can be monitored over time and traced to other people in contact with them. One of us (N.D.W.) has been working in Cameroon to monitor microbes in people who hunt wild game, in other people in their community, and in their animal prey15. The study is now expanding to other continents and to monitor domestic animals (such as dogs) that live in close proximity to humans but are exposed to wild animals through hunting and scavenging. Monitoring of people, animals, and animal die-offs16 will serve as an early warning system for disease emergence, while also providing a unique archive of pathogens infecting humans and the animals to which we are exposed. Specimens from such highly exposed human populations could be screened specifically for agents known to be present in the animals they hunt (for example, retroviruses among hunters of non-human primates), as well as generically using broad screening tools such as viral microarrays17 and random amplification polymerase chain reaction (PCR)18. Such monitoring efforts also provide potentially invaluable repositories, which would be available for study after future outbreaks in order to reconstruct an outbreak’s origin, and as a source of relevant reagents.
Supplementary information
This file contains extensive Supplementary Notes and additional references. (PDF 924 kb)
Acknowledgements
We thank L. Krain for assistance with Supplementary Note S10; M. Antolin, D. Burke, L. Fleisher, E. Holmes, L. Real, A. Rimoin, R. Weiss and M. Woolhouse for comments; and many other colleagues for providing information. This work was supported by an NIH Director’s Pioneer Award and Fogarty International Center IRSDA Award (to N.D.W.), a W. W. Smith Foundation award (to N.D.W.), and National Geographic Society awards (to J.D. and N.D.W.).
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Contributor Information
Nathan D. Wolfe, Email: nwolfe@ucla.edu
Jared Diamond, Email: jdiamond@geog.ucla.edu.
References
- 1.Diamond J. Guns, Germs, and Steel: the Fates of Human Societies. 1997. [Google Scholar]
- 2.Dobson AP, Carper ER. Infectious diseases and human population history. Bioscience. 1996;46:115–126. doi: 10.2307/1312814. [DOI] [Google Scholar]
- 3.Diamond J, Panosian C. When Disease Makes History: Epidemics and Great Historical Turning Points. 2006. pp. 17–44. [Google Scholar]
- 4.McNeill WH. Plagues and Peoples. 1976. [PubMed] [Google Scholar]
- 5.Crosby AW. Ecological Imperialism: the Biological Expansion of Europe. 1986. pp. 900–1900. [Google Scholar]
- 6.Ramenofsky A. Vectors of Death: the Archaeology of European Contact. 1987. [Google Scholar]
- 7.Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. 1991. [Google Scholar]
- 8.Lopez AD, Mathers CD, Ezzati N, Jamison DT, Murray CJL. Global Burden of Disease and Risk Factors. 2006. [PubMed] [Google Scholar]
- 9.Bellwood P. First Farmers: the Origins of Agriculture Societies. 2005. [Google Scholar]
- 10.Diamond J. Evolution, consequences, and future of plant and animal domestication. Nature. 2002;418:34–41. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 11.McNeill JR. When Disease Makes History: Epidemics and Great Historical Turning Points. 2006. pp. 81–111. [Google Scholar]
- 12.Waters AP, Higgins DG, McCutchan TF. Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proc. Natl Acad. Sci. USA. 1991;88:3140–3144. doi: 10.1073/pnas.88.8.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayala FJ, Escalante AA, Rich SM. Evolution of Plasmodium and the recent origin of the world populations of Plasmodium falciparum. Parassitologia. 1999;41:55–68. [PubMed] [Google Scholar]
- 14.Van Heuverswyn F, et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 15.Wolfe ND, et al. Naturally acquired simian retrovirus infections in central African hunters. Lancet. 2004;363:932–937. doi: 10.1016/S0140-6736(04)15787-5. [DOI] [PubMed] [Google Scholar]
- 16.Kuiken T, et al. Pathogen surveillance in animals. Science. 2005;309:1680–1681. doi: 10.1126/science.1113310. [DOI] [PubMed] [Google Scholar]
- 17.Wang D, et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones MS, et al. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 2005;79:8230–8236. doi: 10.1128/JVI.79.13.8230-8236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Switzer WM, et al. Ancient co-speciation of simian foamy viruses and primates. Nature. 2005;434:376–380. doi: 10.1038/nature03341. [DOI] [PubMed] [Google Scholar]
- 20.Taylor LH, Latham SM, Woolhouse ME. Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. B. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moya A, Holmes EC, Gonzalez-Candelas F. The population genetics and evolutionary epidemiology of RNA viruses. Nature Rev. Microbiol. 2004;2:279–288. doi: 10.1038/nrmicro863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antia R, Regoes RH, Koella JC, Bergstrom CT. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.May RM, Gupta S, McLean AR. Infectious disease dynamics: what characterizes a successful invader? Phil. Trans. R. Soc. Lond. B. 2001;356:901–910. doi: 10.1098/rstb.2001.0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morse SS. Factors in the emergence of infectious diseases. Emerg. Infect. Dis. 1995;1:7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson ME. Travel and the emergence of infectious diseases. Emerg. Infect. Dis. 1995;1:39–46. doi: 10.3201/eid0102.950201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss RA, McMichael AJ. Social and environmental risk factors in the emergence of infectious diseases. Nature Med. 2004;10:S70–S76. doi: 10.1038/nm1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains extensive Supplementary Notes and additional references. (PDF 924 kb)


