Abstract
Over the past 20 years, several coronaviruses have crossed the species barrier into humans, causing outbreaks of severe, and often fatal, respiratory illness. Since SARS-CoV was first identified in animal markets, global viromics projects have discovered thousands of coronavirus sequences in diverse animals and geographic regions. Unfortunately, there are few tools available to functionally test these viruses for their ability to infect humans, which has severely hampered efforts to predict the next zoonotic viral outbreak. Here, we developed an approach to rapidly screen lineage B betacoronaviruses, such as SARS-CoV and the recent SARS-CoV-2, for receptor usage and their ability to infect cell types from different species. We show that host protease processing during viral entry is a significant barrier for several lineage B viruses and that bypassing this barrier allows several lineage B viruses to enter human cells through an unknown receptor. We also demonstrate how different lineage B viruses can recombine to gain entry into human cells, and confirm that human ACE2 is the receptor for the recently emerging SARS-CoV-2.
Subject terms: SARS virus, Virus-host interactions, Viral transmission
This study describes the development of an approach to rapidly screen lineage B betacoronaviruses, such as SARS-CoV and the recently emerged SARS-CoV-2, for receptor usage and their ability to infect cell types from different species. Using it, they confirm human ACE2 as the receptor for SARs-CoV-2 and show that host protease processing during viral entry is a significant barrier for viral entry.
Main
Severe acute respiratory syndrome-related coronavirus (SARS-CoV) first emerged in humans in 2003 after transmitting from animals in open-air markets in China1,2. Shortly thereafter, several genetically related viruses were identified in Chinese horseshoe bats (Rhinolophus sinicus)3–7. At the same time, improvements in next-generation sequencing technology led to a boom of virus discovery, uncovering thousands of virus sequences in wild animal populations around the world. While most of these viruses have never been found in humans, many are genetically similar to known human viruses within the betacoronavirus genus. The betacoronaviruses are further divided into four lineages (that is, A–D). Lineage B, which includes SARS-CoV and the newly emerging SARS-CoV-2, has approximately 200 published virus sequences, whereas lineage C, which includes Middle East respiratory syndrome-related coronavirus (MERS-CoV), has over 500 viral sequences.
Every year, additional CoV sequences are discovered. However, there is a massive knowledge gap in the field, as very little work is performed after the viral sequences are published. Therefore, it is unknown whether these viruses have the potential to emerge in human populations.
Current methods for studying betacoronaviruses are technically demanding. Viral isolation from field samples is rarely successful and reverse genetics recovery of recombinant virus is labour intensive and expensive, as the synthesis of a single genome can cost upwards of US$15,000. These limitations are prohibitive to studying CoVs at the scale at which they are discovered.
Cell entry is an essential component of cross-species transmission, especially for the betacoronaviruses. All CoVs encode a surface glycoprotein, spike, which binds to the host-cell receptor and mediates viral entry8. For betacoronaviruses, a single region of the spike protein called the receptor-binding domain (RBD) mediates the interaction with the host-cell receptor. After binding the receptor, a nearby host protease cleaves the spike, which releases the spike fusion peptide, facilitating virus entry9–12. Known host receptors for betacoronaviruses include angiotensin-converting enzyme 2 (ACE2) for SARS-CoV and dipeptidyl peptidase-4 (DPP4) for MERS-CoV13,14.
Structural studies of coronaviruses have shown that the spike RBD is capable of folding independent of the rest of the spike protein and contains all of the structural information for host receptor binding15. Additionally, a previous study showed that replacing the RBD of the lineage B bat virus Rp3 allowed the virus to enter cells expressing human ACE216. We therefore developed a method to functionally test the RBDs from lineage B betacoronaviruses in place of the SARS-CoV spike RBD (Fig. 1). Synthesizing just the RBD of spike is much faster and cost effective than conventional pseudotyping methods that rely on the synthesis of the full spike sequence (~4 kilobases (kb)) for CoVs—a process that can take weeks and is cost prohibitive for large panels of spike sequences. The short turnaround time for our approach allowed us to test the receptor usage of all published, unique RBD sequences in lineage B, and to rapidly confirm the ACE2 receptor usage of the SARS-CoV-2 spike, which emerged in China in January 2020 as our study was ongoing.
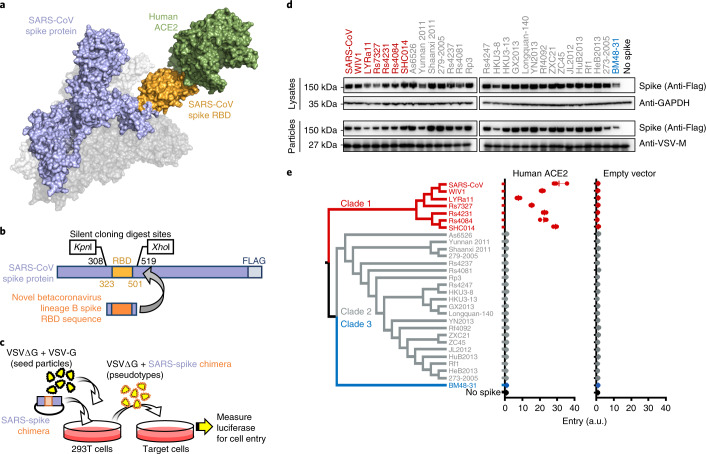
Fig. 1. Betacoronavirus lineage B entry with human ACE2 is clade specific.
a, Betacoronaviruses, including SARS-CoV, interact with the host-cell receptor via the RBD in spike (Protein Data Bank ID: 5X5B; 2AJF). b, Engineered silent mutations in SARS spike facilitated replacement of the RBD sequence. SARS spike amino acid numbers are indicated in black for the silent cloning sites and orange for the RBD. c, Outline of the experimental workflow. d, Western blot of producer cell lysates and concentrated reporter particles. The labels along the top show the origin of the RBD in the SARS-CoV spike protein. e, Cladogram of the 29 spikes tested. Cells expressing either human ACE2 or empty vector were infected with pseudotyped VSV reporter particles, and luciferase was measured and normalized to no spike as a readout for cell entry. The data are representative of three technical replicates. Vertical bars indicate mean values of all three replicates and horizontal bars indicate s.d.
We show that lineage B RBDs divide into functionally distinct clades and that several previously unappreciated viruses exhibit compatibility with an unknown receptor on human cells. We also show that these clades are capable of recombining to impart human host-cell entry phenotypes, and that, beyond the RDB–receptor interaction, host protease processing is another species barrier encountered by lineage B betacoronaviruses during cell entry.
Results
ACE2 entry is lineage B clade 1 specific
The RBD of lineage B betacoronaviruses is a single, continuous domain that contains all of the structural information necessary to interact with the host receptor (Fig. 1a,b). We introduced silent mutations in the codon-optimized coding sequence for SARS-CoV to facilitate replacing the SARS RBD with the RBD from other lineage B viruses (Fig. 1b). All lineage B sequences were downloaded from online repositories and parsed to 29 unique RBD sequences, representing all published variations of the lineage B RBD (Extended Data Fig. 1a,b). These 29 RBDs phylogenetically cluster into three clades, as previously described5, but these RBD clades were not apparent in phylogenetic analysis of other viral sequences, such as the RNA-dependent RNA polymerase (Extended Data Fig. 1c). All 29 RBDs were codon optimized, synthesized and cloned in place of the SARS RBD, generating chimeric spike expression constructs. We then produced vesicular stomatitis virus (VSV) reporter particles pseudotyped with the chimeric spikes (Fig. 1c)17. We chose VSV over lentiviruses as our pseudotype platform because lentiviral pseudotypes have failed to accurately reflect viral entry with bat coronavirus spike protein7. All constructs exhibited similar levels of expression in producer cells and incorporation into VSV pseudotypes, except the chimera with BM48-31, which displayed somewhat reduced expression compared with the wild-type SARS spike (Fig. 1d). We then infected baby hamster kidney (BHK) cells expressing the receptor for SARS-CoV or empty vector (Fig. 1e) and observed that only clade 1, which includes SARS-CoV, could enter cells transfected with human ACE2 (Fig. 1e).
Extended Data Fig. 1. Lineage B RBD panel assembly and phylogenetic analysis.
a, Coronavirus sequences were downloaded from NCBI and further parsed to 29 unique RBD variants. b, Virus isolate name, accession number, clade, host species and location of identification listed for the 29 unique lineage B RBDs used in this study. c, Cladograms for the spike RBD and coronavirus RNA-dependent RNA polymerase (nsp12). d, Overview of experimental timeline.
Protease enhances clade 2 entry
After binding the host receptor, host-cell protease cleaves spike, releasing the fusion peptide and allowing for host-cell entry18. Previous studies have shown that an absence of the host protease or incompatibility between the host protease and viral spike can block viral entry19–21. To circumvent host-cell protease incompatibility or absence, we protease treated our pseudotype panel and infected a wide variety of cell types from different host species (Fig. 2 and Extended Data Fig. 2a,b). In the absence of exogenous protease, only clade 1 infected cells from African green monkey kidney, human gastrointestinal tract, human liver and porcine kidney, in agreement with previous studies. Surprisingly, exogenous protease enhanced entry of a subset of clade 2 spike chimeras in nonhuman primate, bat and human cells (Fig. 2). Importantly, VSV-G-pseudotyped particles were able to produce luciferase signal in all cell lines tested in this study (Extended Data Fig. 2).
Fig. 2. Trypsin enhances lineage B entry in various cell lines.
a–c, African green monkey (AGM; a), human (b) or bat cells (c) were infected with VSV particles pseudotyped with lineage B chimeric spikes. Pseudotypes were either left untreated or incubated with trypsin before addition to the cells. Luciferase was measured and normalized to particles produced without spike. Data in all panels are representative of three technical replicates. Vertical bars indicate mean values of all three replicates and horizontal bars indicate s.d. GI, gastrointestinal.
Extended Data Fig. 2. Additional cell lines tested without protease.
a, Additional human and cell lines or b, bat-derived cell lines derived from other species were infected with VSV-reporter particles pseudotyped with chimeric spikes and luciferase was measured a readout for cell entry. Trypsin was not used in these infections. c, All cell lines tested in this study supported entry and reporter expression of VSV-g pseudotyped particles. For all panels shown are the data for 3 techinical replicates, horizontal bars indicate the s.d. and vertical bars indicate the mean value of all 3 replicates.
Clade 2 entry is receptor dependent
Next, we tested human variants of known betacoronavirus receptors for their ability to mediate cell entry of clade 2 and 3 spike chimeras. We also tested human aminopeptidase N (APN)—a common receptor for alphacoronaviruses (Fig. 3a). Protease treatment only enhanced entry of clade 1 RBDs on cells expressing human ACE2, but not human DPP4 or APN. No entry was observed with clade 2 or 3 spikes, regardless of receptor or protease addition. Human DPP4—a receptor for the lineage C betacoronaviruses—only mediated entry of MERS-CoV, whereas APN only mediated entry of the alphacoronavirus HCoV-229E (Fig. 2b). In the absence of receptor, no entry was observed for any of the pseudotypes, suggesting that protease-mediated entry is receptor dependent (Fig. 2b).
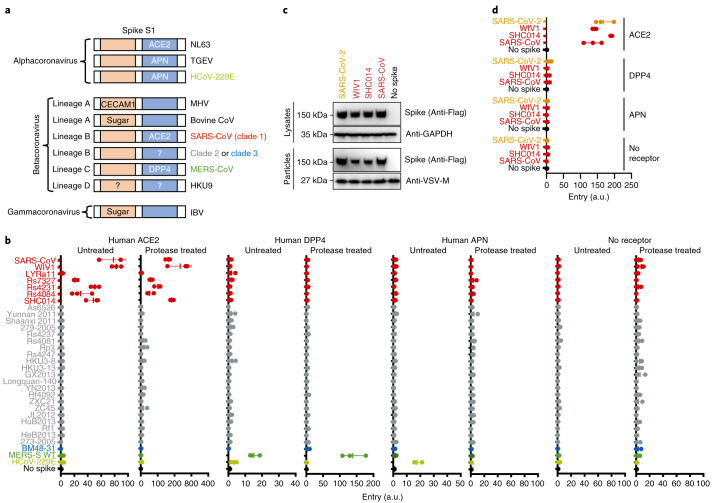
Fig. 3. Lineage B entry into cells with known CoV receptors.
a, Schematic of known coronavirus spikes and their receptors. N-terminal RBD found in spike S1 of lineage A betacoronaviruses and gammacoronaviruses is indicated in orange. C-terminal RBD found in spike S1 of alphacoronaviruses and lineage B betacoronaviruses is indicated in blue. CECAM1, carcinoembryonic antigen-related cell adhesion molecule 1; HCoV, human coronavirus; IBV, infectious bronchitis virus; MHV, mouse hepatitis virus; TGEV, transmissible gastroenteritis coronavirus. b, Pseudotyped particles were either left untreated or treated with trypsin and subsequently used to infect BHK cells expressing the indicated coronavirus receptors. WT, wild type. c, Expression and pseudotype incorporation of SARS-S–SARS-CoV-2 RBD chimeras. SARS-S, SARS-CoV spike. The top labels indicate the origin of the RBD in the SARS spike protein. d, Pseudotypes were used to infect BHK cells expressing known receptors without protease treatment. The y axis labels indicate the origin of the RBD in the SARS spike protein. Data for all panels represent three technical replicates. Vertical bars indicate mean values of all three replicates and horizontal bars indicate s.d.
Receptor usage of SARS-CoV-2
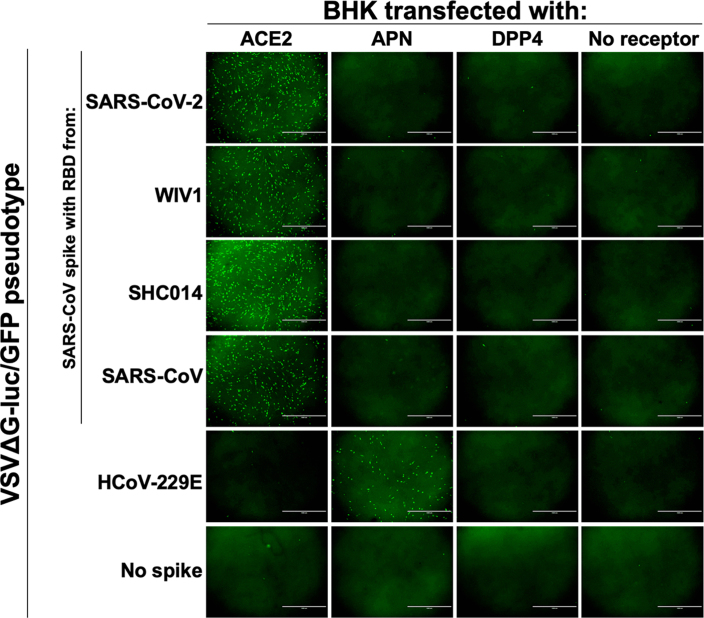
While our study was ongoing, a lineage B virus tentatively named SARS-CoV-2 was identified as the cause of a pneumonia outbreak in Hubei, China. Once the sequence was publicly available, we synthesized, cloned and tested the RBD from SARS-CoV-2 in our assay with human variants of known coronavirus receptors. The chimeric SARS–SARS-CoV-2 spike protein expressed was incorporated into particles similarly to other clade 1 chimeric spikes and was capable of entering cells expressing human ACE2, but not any of the other receptors tested (Fig. 3c,d and Extended Data Fig. 3).
Extended Data Fig. 3. 2019-nCoV uses human ACE2 to enter cells.
VSVΔG-luciferase/GFP particles were pseudotyped with the indicated spikes and used to infect BHKs transfected with known coronavirus receptors. Microscopy images were taken 20 hours post-infection. Scale bar indicates 1000 um.
Clade determinants for ACE2 usage
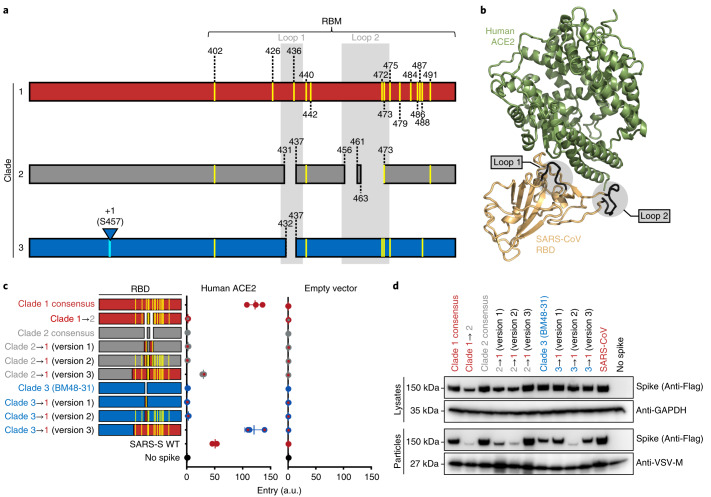
Consensus sequences of the three lineage B clades showed several key differences between these groups. Only clade 1 RBDs contain all 14 residues that have been shown through crystallography to interact with human ACE2 (Fig. 4a and Extended Data Fig. 4). The majority of these residues are absent from clades 2 and 3, which contain additional deletions in surface-exposed loops that cluster at the interface with ACE2 (Fig. 4a,b). We generated a series of clade consensus RBD variants to determine the minimum number of mutations needed to impart ACE2 function on clade 2 and 3 RBDs (Fig. 4c). Introducing the two loop deletions from clade 1 in clade 2 resulted in reduced spike expression, impaired pseudotype incorporation and loss of cell entry (Fig. 4c,d). Restoring these loops in clades 2 and 3 from the loops found in clade 1 did not enhance entry with ACE2 (Fig. 4c; 2 → 1 and 3 → 1 (version 1)). Introducing all 14 ACE2 contact points in clade 2 or 3 also failed to restore ACE2 entry (Fig. 4c; 2 → 1 and 3 → 1 (version 2)). Only replacing all 14 contact points and the surrounding amino acids (known as the receptor-binding motif (RBM)) led to increased ACE2 entry with clade 2 and 3 RBDs (Fig. 4c; 2 → 1 (version 3) = clade 2 residues 322–400 + clade 1 residues 400–501; 3 → 1 (version 3) = clade 3 residues 322–385 + clade 1 residues 386–501).
Fig. 4. Lineage B clade-specific determinants for human ACE2 usage.
a, Schematic overview of clades 1, 2 and 3 of the betacoronavirus lineage B RBD. Shown in yellow are the 14 residues that contact ACE2. Loop deletions are shown for clades 2 and 3. b, Structure of human ACE2 and the SARS-S RBD (Protein Data Bank ID: 2AJF), with the loops highlighted in grey. c, VSV pseudotypes were used to infect BHKs transfected with either human ACE2 or empty vector. The data are representative of three technical replicates. Vertical bars indicate mean values of all three replicates and horizontal bars indicate s.d. d, Western blot of producer cell lysates and concentrated pseudotyped particles. The top labels show the source of the RBD in the spike protein.
Extended Data Fig. 4. Lineage B panel RBD sequence features.
a, Amino acid sequences corresponding to SARS-spike residues 317 through 500 were aligned with ClustalW. Contact points between SARS-spike and human ACE2 are indicated with an (*). Clade 2 sequences are shown as compared to clade 2 As6526, with identical residues indicated with a (.) and sites that vary between clade 2 viruses highlighted in purple. Loop deletions are highlighted in orange. b, Amino acid alignment of 2019-nCoV RBD and consensus RBD sequences for clade 1 and 2 and BM48-31 (clade 3). Loop deletions are highlighted in orange.
Full spike and RBD chimeras are comparable
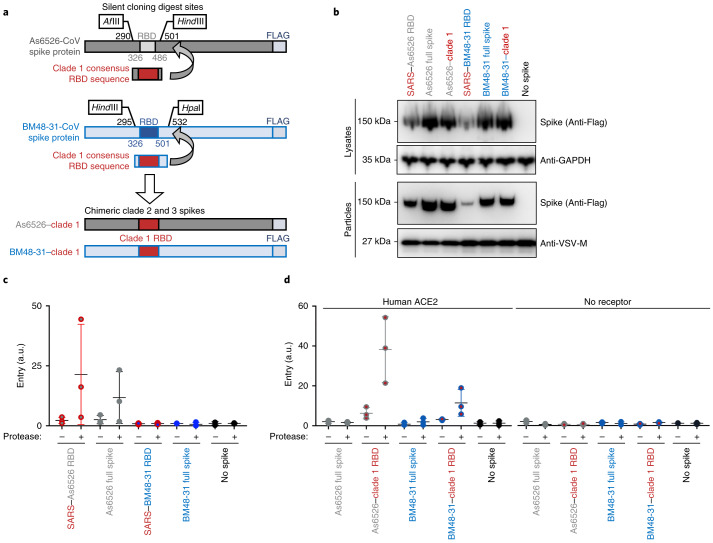
Next, we synthesized full-length clade 2 and 3 spikes to compare with our RBD chimeras. We selected the clade 2 spike As6526 because it consistently gave strong entry signal in human cells following protease treatment (Fig. 2b), as well as BM48-31, the only clade 3 spike in our panel. As we did for SARS-CoV spike, clade 2 and 3 spikes were codon optimized and FLAG tagged, and silent mutations were introduced to facilitate replacing their RBD with the consensus RBD from clade 1 (Fig. 5a). All chimeric constructs expressed similarly, with the exception of the SARS–BM48-31 RBD chimera, which exhibited reduced expression and incorporation (Figs. 1d and 5b). Protease treatment enhanced entry of both the As6526 clade 2 RBD chimera and full-length spike into Huh cells (Fig. 5c). Protease treatment had no effect on either the BM48-31 clade 3 chimera or full-length spike (Fig. 5c). Taken together, these findings show that SARS lineage B RBD chimeras reflect the entry phenotype of full-length lineage B spikes.
Fig. 5. Comparison of chimeric and full-length clade 2 and 3 spikes.
a, Design of full-length spike sequences from As6526 (clade 2) and BM48-31 (clade 3). b, Western blot of producer cell lysates and concentrated pseudotype particles. c, Pseudotypes were left untreated or treated with trypsin and subsequently used to infect Huh-7.5 cells. d, Pseudotypes with the indicated spike constructs were left untreated or treated with trypsin and subsequently used to infect BHK cells expressing human ACE2. The data are representative of three technical replicates. Vertical bars indicate s.d. and horizontal bars indicate mean values of all three replicates.
Finally, we tested whether receptor binding and protease processing are coupled. We replaced the RBD of full-length clade 2 and 3 spike with the consensus clade 1 RBD and tested pseudotypes on cells expressing ACE2. The clade 1 consensus RBD efficiently facilitated entry of both As6526 and BM48-31 spike only following protease treatment. These findings show that even though BHK cells expressing human ACE2 cells support full-length clade 1 spike entry, just having the RBD from a clade 1 virus was insufficient to mediate efficient entry. As seen in our previous experiments, protease treatment did not enhance pseudotype entry in the absence of host receptor (Fig. 5).
Discussion
Despite significant advances in next-generation sequencing technologies, which have facilitated the discovery of thousands of animal-derived viruses, tools for downstream functional assessment of these sequences are lacking. To gain traction on this ever-growing problem, we took a reductionist approach to coronavirus entry and developed a scalable, biosafety level 2-compatible method for testing only the minimal region of the virus essential for interacting with the host receptor (Fig. 1 and Extended Data Fig. 1a). Because most of these viruses have never been isolated, we resorted to synthetic biology and molecular engineering to reduce the burden of gene synthesis to just a small fragment. Thus, the cost and synthesis production time for testing several spikes for entry in our system is dramatically reduced (Extended Data Fig. 1d). In theory, this approach to functional viromics should be applicable to a wide variety of virus–host proteins and interactions.
Coronavirus entry is a multi-step process involving several distinct domains in spike that mediate virus attachment to the cell surface, receptor engagement, protease processing and membrane fusion8. While the RBD–receptor interaction is the most studied in this process, recent studies have highlighted the major role host protease processing plays as a species barrier19,22–24. Lineage C coronaviruses include MERS-CoV as well as distantly related viruses such as HKU4, HKU5 and PDF-2180 (refs. 25,26). Studies have shown that HKU4 can bind human DPP4 but requires the addition of exogenous trypsin to facilitate cell entry, and that HKU5 and PDF-2180 spikes can enter human cells through an unidentified receptor with protease treatment19,23. Analogous to these lineage C CoV studies, we observed protease-enhanced entry of lineage B CoVs (Figs. 2, 3 and 5). While it has been shown that host proteases cleave spike, allowing for downstream membrane fusion, other evidence suggests that protease may act on the receptor as well to activate it27. The addition of protease during the course of SARS-CoV infection facilitated entry in cells with low expression of ACE2 (normally insufficient to support virus entry)27. Indeed, we saw evidence of residual trypsin activity on the cells after infection in our studies, as the cell monolayer was loose compared with the untreated condition. Similarly, Menachery et al.23 also observed cell rounding during their trypsin infections. Therefore, further studies are needed to assess where trypsin is enhancing entry of coronaviruses: at the level of spike, the receptor, or both.
In the absence of exogenous protease, only clade 1 RBDs entered nonhuman primate, human and porcine cell lines (Fig. 2a,b and Extended Data Fig. 2a,b). These findings are in agreement with previous studies that have either isolated virus (WIV1) or rescued recombinant chimeric viruses (SHC014, Rs4231 and Rs7237)5,7,28. However, with trypsin, a subset of genetically similar clade 2 RBDs gained entry in these cells, suggesting that their barrier is at the level of protease processing (Fig. 2a,b). The other spikes from clade 2 and 3 did not enter the cells we tested, regardless of protease addition, suggesting an absent or incompatible receptor. Surprisingly, the protease-dependent entry phenotype was consistent in the reverse spike chimeras in which we replaced the RBD in clade 2 or 3 spike with a clade 1 RBD (Fig. 5d), suggesting that either the protease site between S1 and S2 is not compatible with the chimeric spike backbone or the protease is not expressed in these cells (Extended Data Fig. 5). Because clade 1 spikes enter cells expressing human ACE2 without the addition of protease, but clade 2–clade 1 chimeras require protease, our data show that the spike protease cleavage site may be adapted to the protease environment of the receptor-bound RBD (Extended Data Fig. 5).
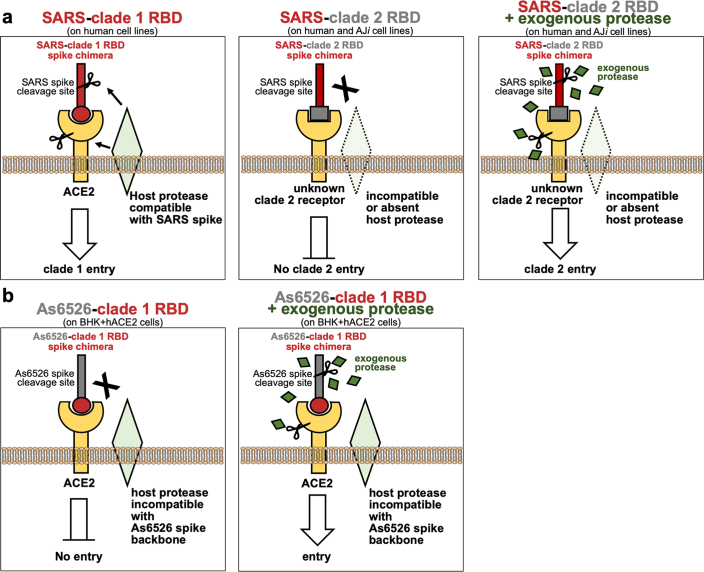
Extended Data Fig. 5. Model of lineage B entry.
a, SARS spike-clade 1 RBD enters cells expressing ACE2 and a host protease capable of cleaving SARS spike (left panel). While clade 2 RBDs can bind an unknown host receptor, the SARS spike backbone is incompatible with the receptor-associated protease, resulting in a lack of cleavage and entry (middle panel). The addition of exogenous protease may overcome the lack of endogenous protease cleavage of spike, resulting in receptor-dependent entry. Alternatively, the addition of exogenous protease may activate the receptor to facilitate entry. b, Replacing the RBD of As6526 spike with the clade 1 RBD allows for ACE2 interaction, but the As6526 spike backbone is incompatible with the ACE2-associated protease (left panel). Addition of exogenous protease overcomes protease incompatibility, allowing for ACE2-mediate entry (right panel).
None of the spike pseudotypes efficiently entered Rhinolophus cells, as was observed in previous studies using these cells29,30 (Fig. 2c). Surprisingly, AJi cells were selectively permissive for only clade 2 entry following protease treatment, which suggests that clade 2 RBDs interact with a receptor that is distinct from clade 1 (Fig. 2c). Unfortunately, a common problem with studies assessing viral entry in cell types from different species is accurately measuring surface protein expression of the receptor through conventional methods such as flow cytometry. We have previously shown that DPP4 from different species cross-reacts differently with the same DPP4 antibody, confounding correlations between receptor expression level and entry31.
Our results show that, despite all of them being classified as the same virus species, most lineage B betacoronaviruses do not use currently known coronavirus receptors (Figs. 1e and 3a,b). We did not observe any pseudotype entry in the presence of protease and the absence of receptor, suggesting that lineage B cell entry is still receptor dependent following protease treatment (Fig. 3b). While our study was ongoing, a lineage B betacoronavirus was identified as the etiological agent behind an outbreak of pneumonia in Wuhan, Hubei, China (SARS-CoV-2). The RBD for SARS-CoV-2 has residues and motifs found in all three clades but forms a distinct clade, so we tested it for receptor usage and observed entry of human ACE2 but not other known coronavirus receptors (Fig. 3d). Interestingly, within the backbone of SARS-CoV spike, cell entry of SARS-CoV-2 was similar to the other clade 1 spikes tested, including SARS-CoV. These findings suggest that SARS-CoV-2 is capable of using human ACE2 as efficiently, if not more, as SARS-CoV, which may help to explain the human-to-human transmissibility of this virus. Our finding that SARS-CoV-2 uses human ACE2 has now been confirmed with the full virus and a computer modelling-based approach32,33.
The RBM is a region in the carboxy-terminal half of the RBD that contains all of the residues that interface with the host receptor (Fig. 3a)15. The 14 contact points in the co-structure of the SARS RBD bound to human ACE2 are largely absent from clade 2 and 3 RBDs, which also contain deletions compared with clade 1 RBMs (Fig. 4a,b and Extended Data Fig. 4a). Simply mutating clades 2 and 3 to have these 14 contact points was insufficient to impart human ACE2 usage, and in some instances even led to protein destabilization and a loss of pseudotype incorporation of spike (Fig. 4c). This is probably because the non-contact residues in the RBM play a critical structural role for these contact points, and indeed, these non-contact residues are different between the clades (Extended Data Fig. 4).
In contrast with changing individual amino acids, our chimeric RBD constructs show that clade 2 and 3 RBDs containing the clade 1 RBM are compatible with human ACE2. Coronaviruses frequently undergo recombination, gaining large swaths of genetic material at once34,35. Taken together with our data, it is possible that recombination with a clade 1 virus will impart compatibility with human ACE2. Interestingly, the SARS-CoV-2 RBD contains most of the contact points with human ACE2 that are found in clade 1, as well as some amino acid variations that are unique to clades 2 and 3 (Extended Data Fig. 4b). Taken together with our receptor assay results, it may be possible that SARS-CoV-2 arose from recombination between clade 1 and the other clades.
As we saw with the SARS–As6526 RBD (clade 2) spike chimera, full-length As6526 spike entered cells following protease treatment, but BM48-31 (clade 3) spike did not (Figs. 2 and 5c). These data show that chimeric spikes generally reproduce the entry phenotypes of full-length spikes. Notably, the full-length As6526 spike did not enter cells as efficiently as the SARS–As6526 chimera, suggesting that other human-cell adaptations are probably needed in As6526 spike.
The capacity to predict the zoonotic potential of newly detected viruses has been severely hindered by a lack of functional data for these animal virus sequences. Here, we have developed a rapid and cost-effective platform with which to functionally test large groups of related viruses for zoonotic potential. We found that several other lineage B coronaviruses are capable of entering human cells through an unknown receptor and that lineage B spike proteins can recombine to gain entry with a known host receptor. Taken together with the latest outbreak of SARS-CoV-2 in humans, these findings underscore the importance of continued surveillance of coronaviruses at the sequence and functional levels in order to better prepare for the next emerging virus.
Methods
Cells
293T, A549, BHK, Caco-2, Huh-7.5, PK-15 and Vero cells were obtained from the American Type Culture Collection and maintained in Dulbecco’s modified Eagle’s medium (DMEM; Sigma–Aldrich) supplemented with 10% foetal bovine serum (FBS), penicillin/streptomycin and l-glutamine. The bat cell lines RhiNi/40.1, HypNi and RhiLu (from M. Müller), AJ-primary (from A. Schountz), AJi and RaKSM-2.5i (generated in our laboratory) were maintained in DMEM/F12 (Gibco) supplemented with 12% FBS, penicillin/streptomycin, non-essential amino acids, sodium pyruvate and l-glutamine. AJ-primary cells were immortalized with a lentiviral vector expressing SV40 T-antigen following the manufacturer’s instructions to generate AJi cells (abm; G203). RaKSM-2.5 primary cells have been previously described and were immortalized in this study similar to AJi cells36. Species for all of the cell lines used in this study were confirmed by cytochrome B PCR and subsequent Sanger sequencing. All of the cell lines tested negative for Mycoplasma contamination by PCR.
Plasmids
The pATX-based VSV plasmid was used to generate a dual-reporter plasmid17. First, the coding sequence for enhanced green fluorescent protein was PCR amplified and inserted into the vector via XhoI and NheI. Firefly luciferase was PCR amplified and inserted into the vector via MluI and AvrII. This plasmid was then used to generate the replication-incompetent VSV particles, as previously described.
The spike coding sequences for SARS-CoV Urbani, As6526 and BM48-31 were codon optimized for human cells, then appended with a 5′ kozak expression sequence (GCCACC) and 3′ tetra-glycine linker followed by nucleotides encoding a FLAG-tag sequence (DYKDDDDK). For SARS-CoV spike, silent mutations were introduced around codons 308 and 519 to form KpnI and XhoI digest sites. For As6526 spike, silent mutations were introduced around codons 290 and 501 to form AflII and HindIII digest sites. For BM48-31 spike, silent mutations were introduced around codons 295 and 501 to form AflII and HindIII digest sites. These engineered spike sequences were synthesized and cloned into pcDNA3.1+ (GenScript).
Spike RBDs were first codon optimized for human cells, appended with regions of the target spike backbone to facilitate In-Fusion cloning and synthesized as double-stranded DNA fragments (Integrated DNA Technologies). SARS-CoV, As6526 or BM48-31 engineered spike plasmids were digested with their corresponding restriction enzymes and gel purified. RBD inserts were resuspended in water and In-Fusion cloned into gel-purified, digested spike backbone vectors (Takara).
Human ACE2 (Q9BYF1.2), DPP4 (XM_005246371.3) or APN (NP_001141.2) were synthesized and cloned into pcDNA3.1+ (GenScript). All DNA constructs were verified by Sanger sequencing (ACGT).
Receptor transfection
BHK cells were seeded in black 96-well plates and transfected the next day with 100 ng plasmid DNA encoding human ACE2, DPP4, APN or empty vector, using polyethylenimine (Polysciences). All downstream experiments were performed 24 h post-transfection.
Pseudotype production
To produce seed particles (VSVΔG-luc/GFP + VSV-G), 293T cells were seeded in six-well plates and transfected 24 h later with 2 µg VSV-luc/GFP, 2 µg T7 polymerase, 0.5 µg VSV-N, 0.25 µg VSV-L, 1.25 µg VSV-P and 1 µg VSV-G. VSV seed particles were harvested 48 h post-transfection. Cell supernatants were collected, cleared from cell debris by centrifugation, aliquoted and stored at −80 °C.
CoV spike pseudotypes were produced as previously described31. 293T cells were seeded onto six-well plates pre-coated with poly-l-lysine (Sigma–Aldrich) and transfected the next day with 1,200 ng of empty plasmid and 400 ng of plasmid encoding coronavirus spike or green fluorescent protein (GFP) as a no-spike control. After 24 h, transfected cells were infected with VSVΔG particles pseudotyped with VSV-G, as previously described37. After 1 h of incubating at 37 °C, cells were washed three times and incubated in 2 ml DMEM supplemented with 2% FBS, penicillin/streptomycin and l-glutamine for 48 h. Supernatants were collected, centrifuged at 500g for 5 min, aliquoted and stored at −80 °C.
Luciferase-based cell entry assay
Target cells were seeded in black 96-well plates and inoculated, in triplicate, with equivalent volumes of pseudotype stocks. For the trypsin experiments, pseudotype stocks were diluted 1:1 in DMEM without FBS, trypsin was added to a final concentration of 2,500 μg ml−1 and the samples were incubated at 37 °C for 15 min. The samples were then diluted again 1:1 in cold DMEM supplemented with 2% FBS, then added to cells. Inoculated plates were centrifuged at 1,200g at 4 °C for 1 h and incubated overnight at 37 °C. Approximately 18–20 h post-infection, Bright-Glo luciferase reagent (Promega) was added to each well, 1:1, without removing the culture media, and luciferase was measured. Relative entry was calculated as the fold-entry over the negative control, by normalizing the relative light unit for spike pseudotypes to the plate relative light unit average for the no-spike control.
Western blot
Producer cells (spike-transfected 293T) were lysed in 1% sodium dodecyl sulfate, 150 mM NaCl, 50 mM Tris-HCl and 5 mM EDTA and clarified by centrifugation at 14,000g for 20 min.
Pseudotyped particles were concentrated from producer cell supernatants that were overlaid on a 10% OptiPrep cushion in PBS (Sigma–Aldrich) and centrifuged at 20,000g for 2 h at 4 °C. Lysates and concentrated particles were analysed for FLAG (Sigma–Aldrich; A8592; 1:10,000), GAPDH (Sigma–Aldrich; G8795; 1:10,000) and/or VSV-M (Kerafast; 23H12; 1:5,000) expression on 10% Bis-Tris PAGE gel (Thermo Fisher Scientific).
Statistics and reproducibility
Each figure shows the data for three technical replicates, with the average of these replicates indicated. During the course of this study, these experiments were performed multiple times with different batches of pseudotypes and cells, and performed at different times. Thus, the results shown are representative of the biological replicates we observed. Four separate clones were recovered in parallel for the SARS–SARS-CoV-2 construct and were tested simultaneously, in triplicate. Because all four clones were identical by Sanger sequencing and behaved similarly in our entry assays, representative results with one clone are shown in Fig. 3c,d.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank A. Schountz for generously sharing primary Artibeus cells, as well as M. Muller for providing the Rhinolophus cells used in this study. We also acknowledge F. Wu, S. Zhao, B. Yu, Y.-M. Chen, W. Wang, Y. Hu, Z.-G. Song, Z.-W. Tao, J.-H. Tian, Y.-Y. Pei, M. L. Yuan, Y.-L. Zhang, F.-H. Dai, Y. Liu, Q.-M. Wang, J.-J. Zheng, L. Xu, E. C. Holmes and Y.-Z. Zhang, as well as the Wuhan Institute of Virology, Chinese Academy of Medical Sciences and Peking Union Medical College, Chinese Center for Disease Control and Prevention, and Wuhan Jinyintan Hospital for rapidly providing the genome sequence for SARS-CoV-2 and other essential outbreak information to the scientific community. This work was supported by the Intramural Research program of the National Institute of Allergy and Infectious Diseases.
Extended data
Source data
Letko full gel blots for Fig. 1d.
Luciferase values graphed in Fig. 1e.
Luciferase values graphed in Fig. 2a–c.
Unprocessed western blots for Fig. 3c.
Luciferase values graphed in Fig. 3d.
Unprocessed western blots for Fig. 4d.
Luciferase values graphed in Fig. 4c.
Unprocessed western blots for Fig. 5b.
Luciferase values graphed in Fig. 5c,d.
Luciferase values graphed in Extended Data Fig. 2a–c.
Author contributions
M.L. conceived of and designed the study, generated the RaKSM-2.5 and AJi cell lines, performed all of the experiments, analysed all of the data, assembled the figures and wrote the manuscript. A.M. generated the dual-reporter VSV system used for the pseudotype assays. V.M. secured funding and supervised the study. All authors contributed to the manuscript.
Data availability
Unprocessed gel images for Figs. 1 and 3–5 and tables of luciferase assay data for Figs. 1–5 and Extended Data Fig. 2 are included as source data. Any other data that support the findings of this study are available from the corresponding author upon request. Accession codes for all of the spike sequences used here can be found in Extended Data Fig. 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Michael Letko, Email: michael.letko@wsu.edu.
Vincent Munster, Email: vincent.munster@nih.gov.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41564-020-0688-y.
Extended data
Extended data is available for this paper at 10.1038/s41564-020-0688-y.
References
- 1.Wang M, et al. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 2005;11:1860–1865. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuiken T, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 4.Ge XY, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu B, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau SK, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menachery VD, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simmons G, Zmora P, Gierer S, Heurich A, Pohlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuyama S, et al. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertram S, et al. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011;85:13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl Acad. Sci. USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raj VS, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 16.Becker MM, et al. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl Acad. Sci. USA. 2008;105:19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzi A, et al. Vesicular stomatitis virus-based Ebola vaccines with improved cross-protective efficacy. J. Infect. Dis. 2011;204:S1066–S1074. doi: 10.1093/infdis/jir348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, et al. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl Acad. Sci. USA. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuyama S, Ujike M, Morikawa S, Tashiro M, Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl Acad. Sci. USA. 2005;102:12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kam YW, et al. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS ONE. 2009;4:e7870. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, et al. Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J. Virol. 2015;89:9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menachery VD, et al. Trypsin treatment unlocks barrier for zoonotic bat coronaviruses infection. J. Virol. 2020;94:e01774-19. doi: 10.1128/JVI.01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beniac DR, deVarennes SL, Andonov A, He R, Booth TF. Conformational reorganization of the SARS coronavirus spike following receptor binding: implications for membrane fusion. PLoS ONE. 2007;2:e1082. doi: 10.1371/journal.pone.0001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo PC, et al. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anthony SJ, et al. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. mBio. 2017;8:e00373-17. doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shulla A, et al. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adney DR, et al. Replication and shedding of MERS-CoV in upper respiratory tract of inoculated dromedary camels. Emerg. Infect. Dis. 2014;20:1999–2005. doi: 10.3201/eid2012.141280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muth D, et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8:15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann M, et al. Differential sensitivity of bat cells to infection by enveloped RNA viruses: coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS ONE. 2013;8:e72942. doi: 10.1371/journal.pone.0072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letko M, et al. Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep. 2018;24:1730–1737. doi: 10.1016/j.celrep.2018.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature10.1038/s41586-020-2012-7 (2020). [DOI] [PMC free article] [PubMed]
- 33.Wan, Y., Shang, J., Graham, R., Baric, R. S. & Li, F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol.10.1128/JVI.00127-20 (2020). [DOI] [PMC free article] [PubMed]
- 34.Liao CL, Lai MM. RNA recombination in a coronavirus: recombination between viral genomic RNA and transfected RNA fragments. J. Virol. 1992;66:6117–6124. doi: 10.1128/JVI.66.10.6117-6124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seifert, S. N. et al. Rousettus aegyptiacus bats do not support productive Nipah virus replication. J. Infect. Dis.10.1093/infdis/jiz429 (2019). [DOI] [PMC free article] [PubMed]
- 37.Takada A, et al. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl Acad. Sci. USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Unprocessed gel images for Figs. 1 and 3–5 and tables of luciferase assay data for Figs. 1–5 and Extended Data Fig. 2 are included as source data. Any other data that support the findings of this study are available from the corresponding author upon request. Accession codes for all of the spike sequences used here can be found in Extended Data Fig. 1.