Dear Editor,
In a recent study, we reported the metabolic alterations related to acute respiratory distress syndrome (ARDS) in patients with influenza A pneumonia (IAP) [1]. We hypothesized that other types of acute lung injury causing ARDS may share the same metabolic alterations. As proof of concept, we have analyzed the metabolomic profiles of patients with and without ARDS induced by Streptococcus pneumoniae (SPP) (ARDS = 13; no ARDS = 17) or IAP (ARDS = 12; no ARDS = 18). Some of the results of these studies have been previously reported in the form of an abstract [2]. Patients with SPP were older (p < 0.05) and presented more often with renal failure (p < 0.05) (Table-S1). Serum samples were obtained within 24 h of presentation to the emergency department, before starting mechanical ventilation, and examined by nuclear magnetic resonance spectroscopy, as previously described [1, 3]. We quantified 16 characteristic serum metabolites, and statistical significance was determined using a Bonferroni-corrected Student t test, assuming unequal variance, with p < 0.05 considered significant.
Principal component analysis (PCA) [4] (Fig. 1a) differentiated patients with and without ARDS, irrespective of the infectious pathogen, indicating that ARDS induces adaptations in similar molecular pathways regardless of the etiology. When we focused on SPP patients, PCA (Fig. 1b) provided nearly perfect discrimination between patients with and without ARDS, as we had observed previously in IAP patients [1]. Patients with ARDS showed lower serum concentrations of glucose (−41%, p < 0.01), alanine (−45%, p < 0.05), methylhistidine (−47%, p < 0.001), fatty acids (−40%, p < 0.001), citrate (−25%, p < 0.05), creatine (−40%, p < 0.05), creatinine (−20%, p < 0.05) and valine (−20%, p < 0.05), whereas acetone (100%, p < 0.05) concentration increased. These findings are in line with our previous findings in IAP patients with and without ARDS [1], and indicate impairment of normal cell energy production [5].
Fig. 1.
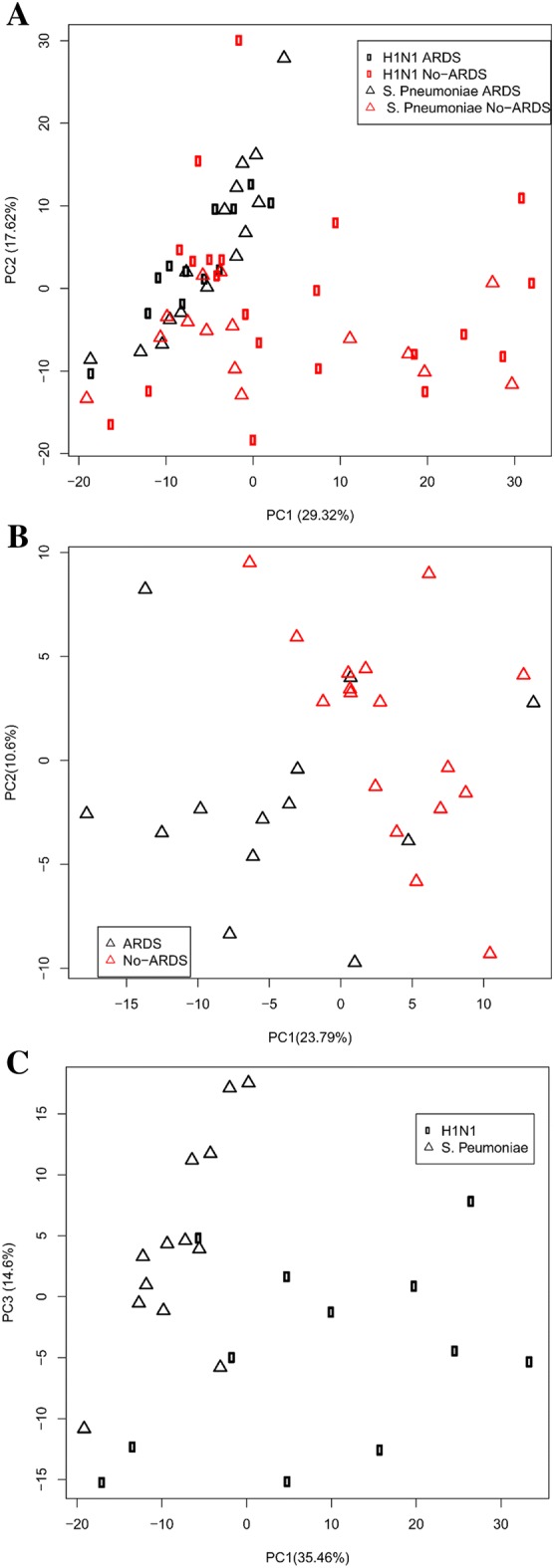
a Score plot of PCA performed on the NMR data of serum samples from patients diagnosed with S. pneumoniae pneumonia (open triangle) and H1N1 influenza virus (open square), with ARDS (black) and without ARDS (red); b score plot of PCA performed on the NMR data of serum samples from patients diagnosed with S. pneumoniae pneumonia with ARDS (black) and without ARDS (red); c score plot of PCA performed on the NMR data of serum samples from patients with ARDS induced by S. pneumoniae (open triangle) or H1N1 influenza virus (open square)
Additionally, the metabolomic fingerprints of ARDS patients with IAP or SPP were compared to identify specific metabolic biomarkers of the pathogen responsible for ARDS (Fig. 1c). Increased levels of glutamine (37%, p < 0.01), methylguanidine (88%, p < 0.001) and phenylalanine (56%, p < 0.01), previously identified as biomarkers of ARDS induced by IAP, may indicate a specific metabolic pattern of this infection. Alterations in lactate (−184%, p < 0.01) and creatine (−43%, p < 0.05) may be considered a metabolic pattern of SPP. We have also detected significant differences in common metabolic alterations, such as glucose (60%, p < 0.01), methylhistidine (29%, p < 0.01) and alanine (−79%, p < 0.05), that may indicate greater inflammatory response and energetic requirements in SPP. The specific metabolic fingerprint was then used to develop a partial least squares model [4] for the identification of the etiological pathogens responsible for ARDS (identification success of 93.0 ± 8.3%, sensitivity 81.1 ± 9.2% and specificity 93.6 ± 7.6%). However, this must be interpreted considering that the populations being compared differed in age and prevalence of complications (Table S1). Also, differences between the two groups with regard to other variables, such as the influence of the environment, nutrition, medication or duration of illness prior to hospital presentation, may explain some of the differences found here, rather than the etiology.
In summary, the findings from our pilot study, showing that there are common metabolic alterations in ARDS regardless of its etiology, require further investigation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Funding
This research was supported by the Spanish Ministry of Economy, Industry and Competitiveness (MEIC-AEI) Grant SAF2017-84494-C2-1-R, Comunidad de Madrid grant B2017/BMD3875, Instituto de Salud Carlos III FEDER Funds from EU (FIS PI 15/1942), Programa Red Guipuzcoana de Ciencia, Tecnología e Información 2018-CIEN-000058-01 and from the Gobierno Vasco, Dpto. Industria, Innovación, Comercio y Turismo under the ELKARTEK Program (Grant no. KK-2019/bmG19). JRC received funding from the BBVA Foundation (Ayudas a Equipos de investigación científica Biomedicina 2018). CIC biomaGUNE is supported by the Maria de Maeztu Units of Excellence Program from the Spanish State Research Agency, Grant no. MDM-2017-0720.
Compliance with ethical standards
Conflicts of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jose Luis Izquierdo-Garcia and Nicolás Nin contributed equally to the manuscript.
References
- 1.Izquierdo-Garcia JL, Nin N, Jimenez-Clemente J, Horcajada JP, Arenas-Miras MDM, Gea J, Esteban A, Ruiz-Cabello J, Lorente JA. Metabolomic profile of ARDS by nuclear magnetic resonance spectroscopy in patients with H1N1 influenza virus pneumonia. Shock. 2018;50:504–510. doi: 10.1097/SHK.0000000000001099. [DOI] [PubMed] [Google Scholar]
- 2.Nin N, Izquierdo J, Cardinal P, Sanchez-Munoz I, Lopez-Cuenca S, Ruiz-Cabello J, Esteban A, Lorente J. Metabolomic analysis as a diagnostic tool for acute respiratory distress syndrome caused by viral or bacterial pneumonia in humans. Intensive Care Med. 2012;38:S1–257. doi: 10.1007/s00134-012-2683-0. [DOI] [Google Scholar]
- 3.Izquierdo-Garcia JL, Nin N, Ruiz-Cabello J, Rojas Y, de Paula M, Lopez-Cuenca S, Morales L, Martinez-Caro L, Fernandez-Segoviano P, Esteban A, Lorente JA. A metabolomic approach for diagnosis of experimental sepsis. Intensive Care Med. 2011;37:2023–2032. doi: 10.1007/s00134-011-2359-1. [DOI] [PubMed] [Google Scholar]
- 4.Lindon JC, Holmes E, Nicholson JK. Pattern recognition methods and applications in biomedical magnetic resonance. Prog Nucl Magn Reson Spectrosc. 2001;39:1–40. doi: 10.1016/S0079-6565(00)00036-4. [DOI] [Google Scholar]
- 5.Serkova NJ, Standiford TJ, Stringer KA. The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am J Respir Crit Care Med. 2011;184:647–655. doi: 10.1164/rccm.201103-0474CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


