Many patients experiencing sudden loss of lung tissue somehow undergo full recovery; here this recovery is traced to a discrete population of lung stem cells that are not only essential for lung regeneration but can be cloned and then transplanted to other mice to contribute new lung tissue.
Supplementary information
The online version of this article (doi:10.1038/nature13903) contains supplementary material, which is available to authorized users.
Subject terms: Adult stem cells
Stem cells for lung regeneration
The extent to which patients can recover after massive loss of lung tissue has been unclear. However, clinical experience has shown that large-scale lung regeneration can occur in children and adults following catastrophic lung damage, and previous studies in mice have linked a subset of cells from distal airway to a regeneration process observed after H1N1 influenza virus mediated injury. Two papers published in this issue show that when the epithelial cells lining the interior of the lung are damaged, a rare stem cell population is induced to proliferate and migrate to the damaged site where they differentiate into several cell types. Frank McKeon and colleagues describe a rare subset of mouse distal airway cells that proliferate after exposure to influenza. These cells can contribute to lung regeneration after transplantation and maintain their intrinsic lineage commitment in cell culture, suggesting that such cell subsets may have potential in stem-cell-based therapies. Harold Chapman and colleagues used lineage tracing to identify a population of quiescent cells in the mouse distal lung that are activated after bleomycin or influenza-mediated injury. These cells express cytokeratin 5 and repair the epithelium through a Notch signalling pathway, although persistent Notch signalling in this context then leads to the formation of cysts. Data from patients suffering from lung fibrosis also show the presence of hyperactive Notch and similar cysts.
Supplementary information
The online version of this article (doi:10.1038/nature13903) contains supplementary material, which is available to authorized users.
Abstract
Lung diseases such as chronic obstructive pulmonary disease1 and pulmonary fibrosis2 involve the progressive and inexorable destruction of oxygen exchange surfaces and airways, and have emerged as a leading cause of death worldwide. Mitigating therapies, aside from impractical organ transplantation, remain limited and the possibility of regenerative medicine has lacked empirical support. However, it is clinically known that patients who survive sudden, massive loss of lung tissue from necrotizing pneumonia3,4 or acute respiratory distress syndrome5,6 often recover full pulmonary function within six months. Correspondingly, we recently demonstrated lung regeneration in mice following H1N1 influenza virus infection, and linked distal airway stem cells expressing Trp63 (p63) and keratin 5, called DASCp63/Krt5, to this process7. Here we show that pre-existing, intrinsically committed DASCp63/Krt5 undergo a proliferative expansion in response to influenza-induced lung damage, and assemble into nascent alveoli at sites of interstitial lung inflammation. We also show that the selective ablation of DASCp63/Krt5 in vivo prevents this regeneration, leading to pre-fibrotic lesions and deficient oxygen exchange. Finally, we demonstrate that single DASCp63/Krt5-derived pedigrees differentiate to type I and type II pneumocytes as well as bronchiolar secretory cells following transplantation to infected lung and also minimize the structural consequences of endogenous stem cell loss on this process. The ability to propagate these cells in culture while maintaining their intrinsic lineage commitment suggests their potential in stem cell-based therapies for acute and chronic lung diseases.
Supplementary information
The online version of this article (doi:10.1038/nature13903) contains supplementary material, which is available to authorized users.
Main
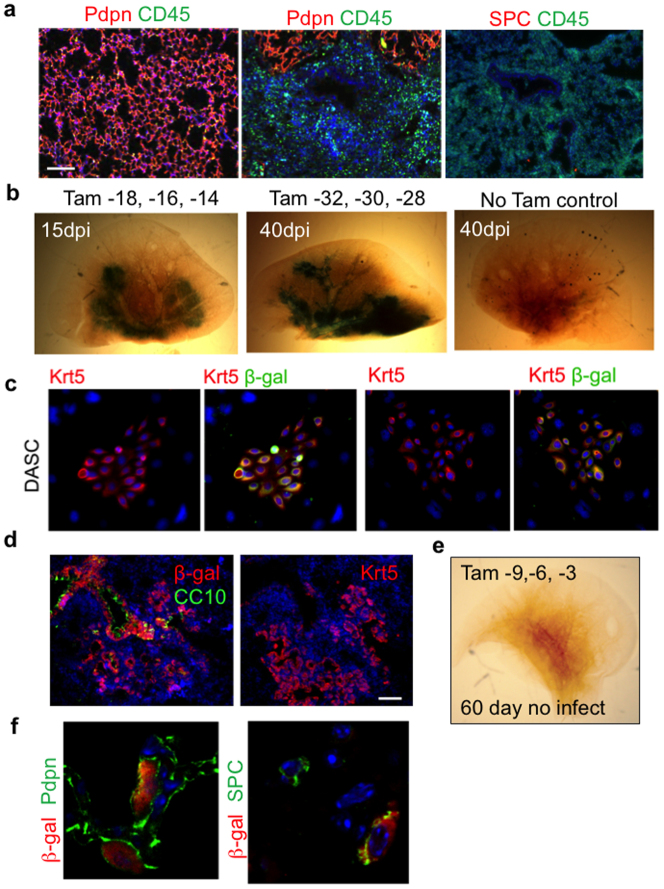
H1N1 influenza virus infection of murine lung triggers a process of leukocyte infiltration and lung damage similar to that of acute respiratory distress syndrome (ARDS)7,8,9,10 (Fig. 1a). Damaged regions are marked by densely packed, CD45+ neutrophils and macrophages11 and an absence of markers for type I (Pdpn+) and type II (SPC+) pneumocytes (Fig. 1a; Extended Data Fig. 1a). Despite the local destruction of alveoli, these same regions harbour discrete clusters of p63+Krt5+ epithelial cells proposed to be the early stages of de novo alveoli formation (Fig. 1a)7. Three dimensional reconstruction of serial sections of lungs at 15 days post-infection (dpi) reveal a broad distribution of Krt5+ cells along the axis of the bronchioles (Fig. 1b; Supplementary Video 1).
Figure 1. Lineage tracing of Krt5+ cells following viral infection.
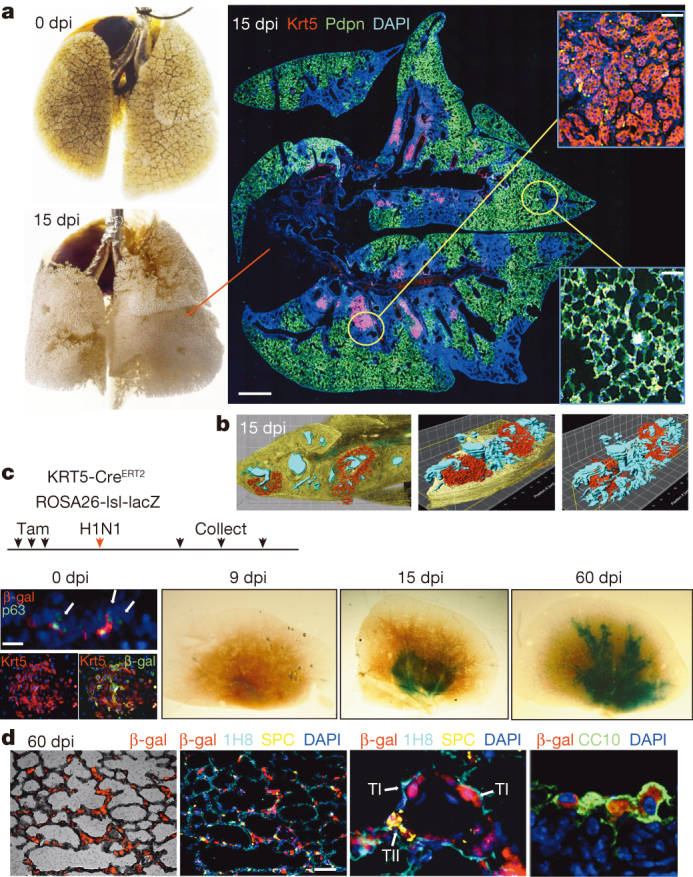
a, Left, mouse lung before and after viral infection. Right, immunofluorescence images of infected lung of anti- Krt5 (red), anti-Pdpn (green) with DNA counterstain (DAPI, blue). Scale bar, 1 mm. Insets, high magnification of indicated regions. n = 10 mice. Scale bars, 100 μm. b, Three-dimensional reconstruction of anti-Krt5 (red) from serial sections of infected lung (bronchioles, blue). Grid, 100 × 100 μm. c, Schematic of lineage tracing experiment following tamoxifen treatment to reveal lacZ expression 0, 9, 15 and 60 days post infection (n = 3 mice for each time point and control). At 0 dpi, rare clusters of p63+ cells in bronchioles express E. coli β-galactosidase (left top, arrows). Scale bar, 20 μm. Colony of Krt5+ DASC co-expressing β-galactosidase at 0 dpi (left bottom). At 9, 15 and 60 dpi, whole lungs stained by X-gal. d, Immunofluorescence in 60 dpi lung sections with indicated antibodies. TI, TII indicate type I and II pneumocytes, respectively. Scale bar, 100 μm.
Extended Data Figure 1. Lineage tracing of Krt5+ cells.
a, Left, immunofluorescence images of sections of 15 dpi lung with staining patterns of antibodies to pan-leukocyte marker CD45 and the type I pneumocyte marker Pdpn with DNA counterstained with DAPI. Right, immunofluorescence images of pan-leukocyte marker CD45 and the type II pneumocyte marker SPC. Scale bar, 150 μm. b, X-gal staining (blue) to reveal lacZ-dependent β-galactosidase activity in whole lungs after 15 and 40 days post infection following long time gaps between induction of lacZ labelling by tamoxifen and influenza infection-induced lung damage. The similarity of this long gap labelling and the short gap labelling presented in Fig. 1 argues against prolonged actions of tamoxifen in these lineage-labelling protocols. Tamoxifen is given at indicated times before infection and no-tamoxifen control is included. c, Immunofluorescence images of colonies of DASCs derived from tamoxifen-treated ROSA26-lsl-lacZ; Krt5-CreERT2 mice stained with antibodies to keratin 5 (Krt5) or Krt5 and E. coli-specific β-galactosidase. d, Histological section of lung at 15 dpi stained with E. coli-specific β-galactosidase antibody and markers of secretory cells (CC10+) and expanded stem cells (Krt5+). Scale bar, 50 μm. e, Whole-mount image of X-gal developed, uninfected lung from ROSA26-lsl-lacZ; Krt5-CreERT2 which received tamoxifen treatments at −69, −66 and −63 days before dissection. f, Histological section of 60 dpi lung stained with E. coli-specific β-galactosidase antibody and markers of type I pneumocytes (Pdpn+) and type II pneumocytes (SPC+).
To decipher the origin of these p63+Krt5+ cells appearing in response to lung damage, we performed genetic lineage-tracing of Krt5+ cells starting before infection through the cycle of lung damage and resolution. Mice expressing a tamoxifen-dependent lacZ gene under the control of the Krt5 promoter12 (Tg (KRT5-CreERT2) ROSA26-lsl-lacZ)13 were treated with tamoxifen before intratracheal delivery of H1N1 influenza virus (Fig. 1c; Extended Data Fig. 1b). At 0 dpi, lacZ activity was not detectable in whole-mount lung and yet Krt5+ cells were evident in distal lung as clusters of peribronchiolar Krt5+p63+ cells were observed in one of three to four consecutive sections of lung. Approximately 50% of these cells expressed Escherichia coli-specific β-galactosidase (7 clusters of p63+ cells were observed in 25 slides with a total of 39 p63+ cells, of which 19 were β-galactosidase positive; Fig. 1c). No labelling of other cell types in the lung was observed. In addition, colonies of distal airway stem cells (DASC) with long-term self-renewal (passage 4) were generated7,14,15 from three 0 dpi mice and stained with antibodies to Krt5 and β-galactosidase. All 256 colonies stained with anti-Krt5 antibodies and 132 co-expressed β-galactosidase (Fig. 1c; Extended data Fig. 1c). At 9 dpi, lacZ activity was subtle and restricted to the airways (Fig. 1c), consistent with our previous observations that at 9 dpi, p63+Krt5+ cells had accumulated within mouse bronchioles with no evidence of migration to interstitial regions7. By 15 dpi, however, the lacZ signal was significantly more robust and included broader regions of interstitial lung (Fig. 1c; Extended Data Fig. 1d). At 60 dpi, the lacZ signal was distributed along the conducting airways and the surrounding interstitial regions, suggesting a progressive process (Fig. 1c). Importantly, no lacZ activity was detected in the lungs of tamoxifen-treated mice in the absence of infection, indicating that the robust signal we observed was in response to lung damage (Extended data Fig. 1e). Histological analysis of the lacZ-positive regions of lung from infected mice revealed broad interstitial areas of staining corresponding to alveoli and that 72 ± 7% of 1,051 lineage-labelled cells expressed type I (1H8+ and Pdpn+) or type II (SPC+) pneumocyte markers with the remainder being secretory cells in the bronchioles (n = 3 mice; Fig. 1d; Extended Data Fig. 1d, f).
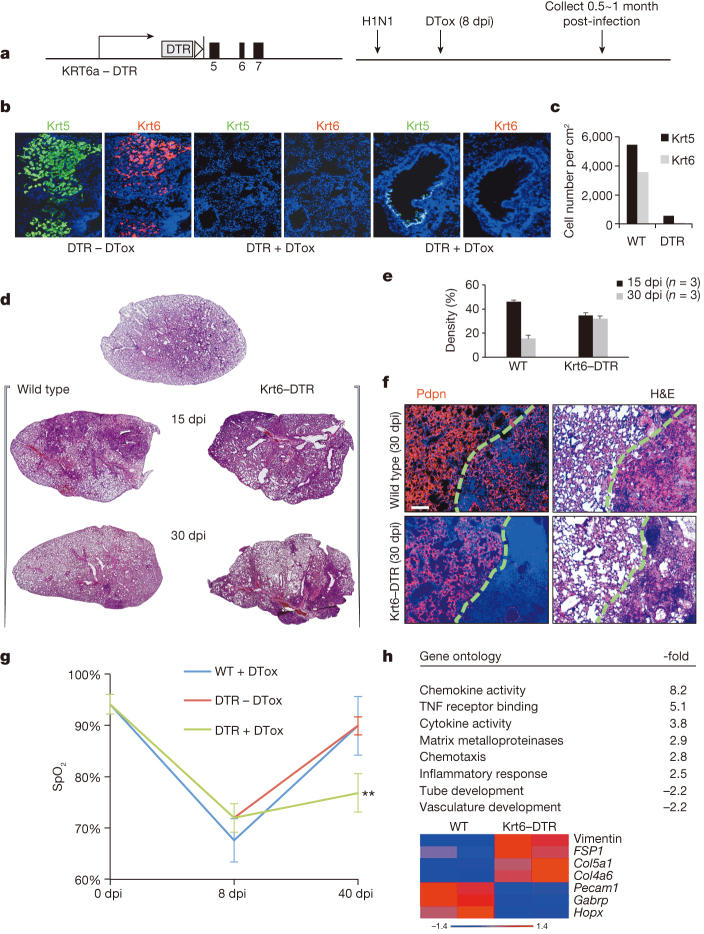
To generate a mouse model in which DASCp63/Krt5 could be conditionally ablated, we engineered the human diphtheria toxin receptor (DTR)16 into the Krt6a (Krt6) locus (Krt6–DTR; Fig. 2a; Extended Data Fig. 2a, b) as the Krt6 gene becomes activated specifically in DASCs at approximately 8–10 dpi7, rather than the p63 or Krt5 gene that are expressed in stem cells of many stratified epithelia17. Consistently, DASCp63/Krt5 from the Krt6a–DTR mice were found to co-express DTR and Krt6a in vivo and in vitro (Extended Data Fig. 2c, d). To test this ablation model in vivo, we infected the Krt6–DTR mice with influenza virus and injected diphtheria toxin at 8 dpi (Fig. 2a). By 15 dpi, exposure to toxin resulted in a rapid loss of interstitial clusters of Krt5+Krt6+ cells (Fig. 2b, c), indicating that we had generated a highly efficient ablation model. Compared to wild-type controls, Krt6–DTR mice lost 90% of Krt5+ cells and over 99% of Krt6+ cells following diphtheria toxin treatment (Fig. 2c).
Figure 2. Conditional ablation of activated DASCp63/Krt5.
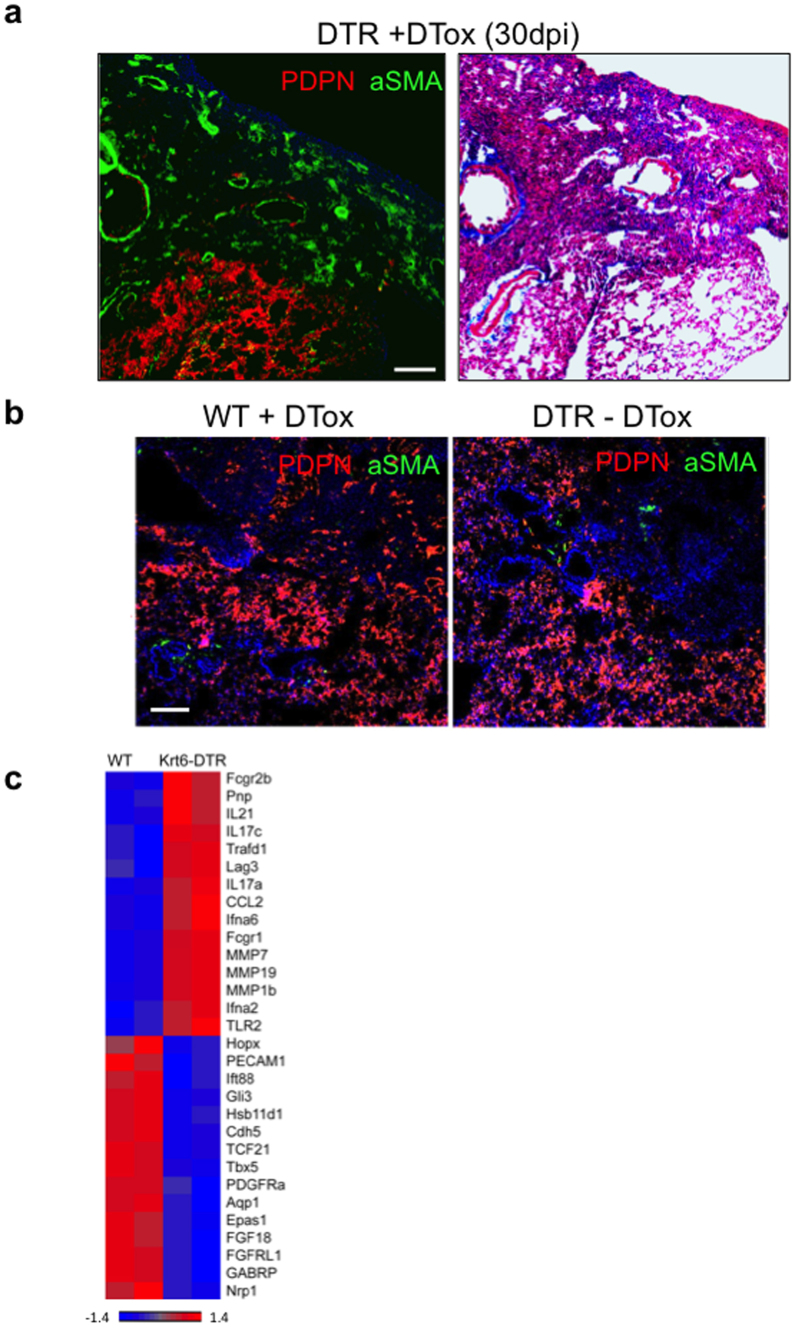
a, Modified Krt6a locus driving the human diphtheria toxin receptor and experimental scheme. DTox, diphtheria toxin. b, Immunofluorescence images of distal lung at 15 dpi with indicated diphtheria toxin (+/−DTox) condition. c, Quantification of Krt5+ and Krt6+ cells in DTox-treated mice. n = 2 mice per group, 6 sections covering the whole lung for each mouse. d, Lung sections from indicated mice following influenza virus infection and treatment with DTox. e, Morphometric analysis of interstitial densities in sections of lung (n = 3 mice per condition and time). Error bars, s.e.m. f, Anti-Pdpn (red) immunofluorescence of lung densities to reveal type I pneumocytes counterstained with DAPI (blue). Scale bar, 100 μm. H&E, haematoxylin and eosin. g, Peripheral capillary oxygen saturation (SpO2) values obtained by pulse oximetry (WT, n = 3, DTR ± DTox, n = 4) at indicated times. Error bars, s.e.m. **P < 0.01 for –DTox versus +DTox. h, Gene ontology classes and fold-change DASCp63/Krt5-ablation versus normal mouse lung at 30 dpi. Below, heat map of differentially expressed (P < 0.05) genes involved in pre-fibrosis, alveolar structure and vasculature.
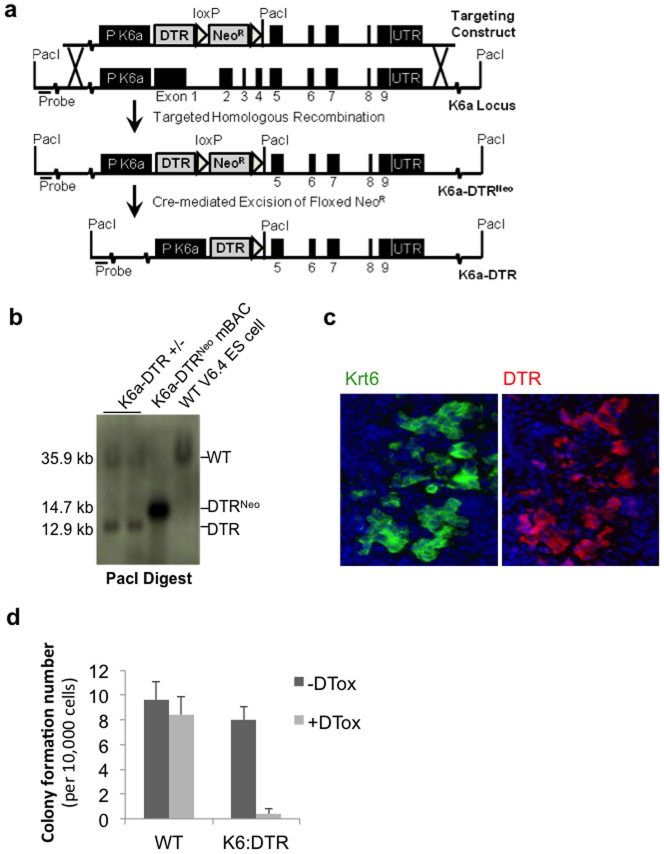
Extended Data Figure 2. Conditional DASCp63/Krt5 ablation mouse model.
a, Schematic of Krt6a locus, the targeting vector constructed to introduce the human diphtheria toxin receptor (DTR). b, The structure of the modified Krt6a locus in embryonic stem cells screened by Southern blot. c, Co-expression of Krt6 and DTR in Krt5+ pods in 15 dpi lung. d, Histogram showing resistance of wild-type, 12 dpi DASCp63/Krt5 to diphtheria toxin (DTox) and the sensitivity of DASCp63/Krt5/DTR to diphtheria toxin. n = 3 mice per group. Error bars, s.e.m.
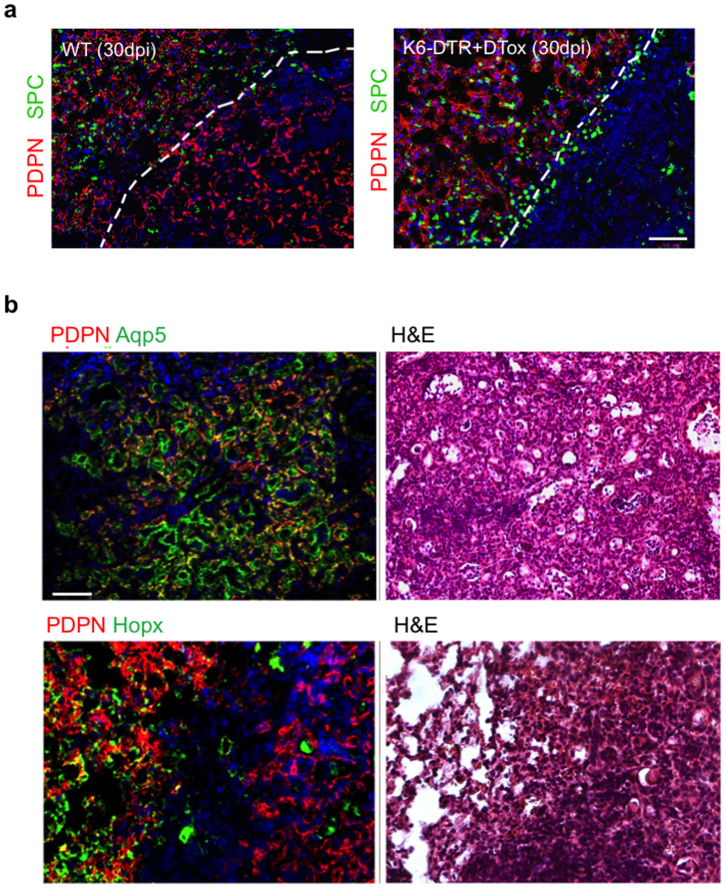
We next evaluated the effect of ablating DASCp63/Krt5 on the process of lung regeneration itself. Focal infiltrates of leukocytes appearing as haematoxylin-eosin staining densities form at about 10–15 dpi in wild-type mice and are typically resolved over the next 15–45 days7 (Fig. 2d, e). However, in diphtheria-toxin-treated Krt6–DTR mice, these densities fail to resolve over time (Fig. 2d, e). The persistent damage in DASCp63/Krt5-ablated lungs is also evident by whole-lung genome expression analyses (Extended Data Fig. 3a, false discovery rate (FDR) q < 0.001). Histological comparisons of the persistent densities at 30 dpi revealed the possible basis for this difference. Although wild-type lungs at 30 dpi still display some lung densities, nearly all of these densities were negative for the CD45 leukocyte marker and instead possess unusual networks of Pdpn+ type I pneumocytes (Fig. 2f; Extended Data Fig. 3b). These networks of type I pneumocytes lacked markers of type II pneumocytes (Extended Data Fig. 4). Interestingly, these networks stain positive for other type I pneumocyte markers (Aqp5) but not all (for example, Hopx; Extended Data Fig. 4), suggesting the possibility that the type I pneumocytes in these networks are undergoing a maturation process. Remarkably, similar alveoli-like structures formed by type I pneumocytes lacking type II pneumocytes in anatomical analyses of mice recovering from infection by the NWS influenza A virus were reported nearly 40 years ago18. In contrast to the type I pneumocytes networks in persistent densities of wild-type mice, DASCp63/Krt5-ablated mice showed no such alveolar networks at 30 dpi (Fig. 2f) and instead maintained persistent infiltration of leukocytes evidenced by anti-CD45 staining (Extended Data Fig. 3b).
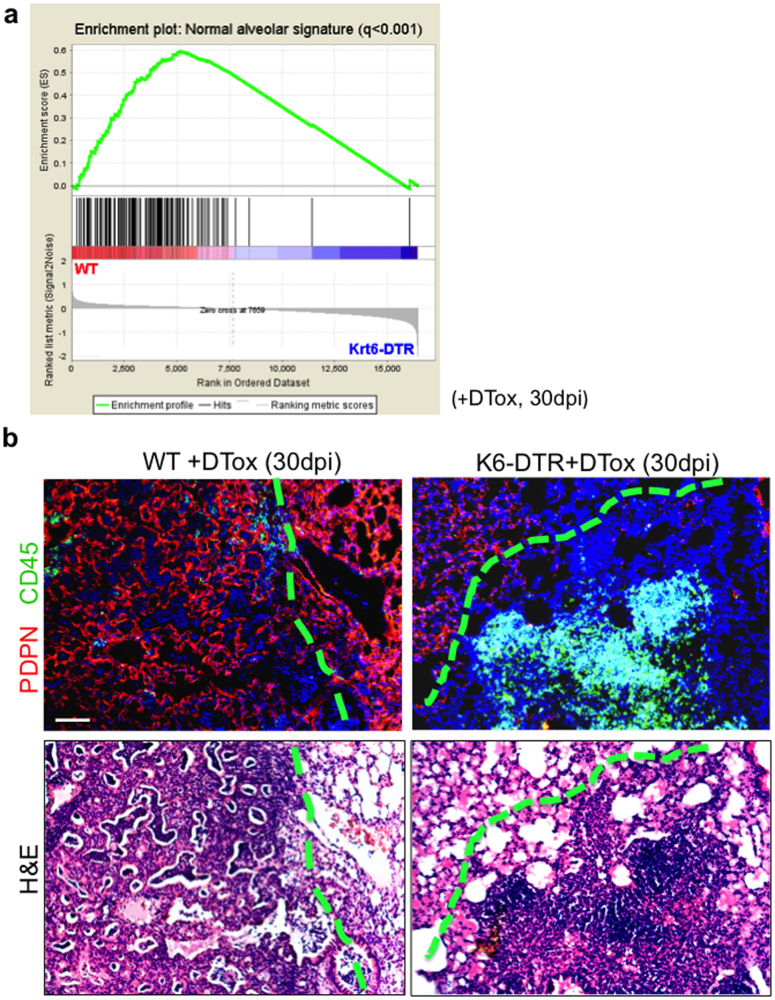
Extended Data Figure 3. Persistent damage in DASCp63/Krt5-ablated lungs.
a, Gene set enrichment analysis (GSEA) showing the overrepresentation of normal alveolar signature gene sets in WT rather than Krt6–DTR mouse lungs (whole lobes). For normal alveolar signature build up, laser capture microdissection of frozen sections was used to dissect normal alveoli region from 0 dpi lung and damaged interstitial infiltrated region from 15 dpi lung for microarray analysis. Differentially expressed genes (fold change > 5, P < 0.01) were used to develop normal alveolar gene expression signatures. b, Top panel, histological analysis of lung densities using anti-Pdpn antibodies (red) and anti-CD45 (green) to reveal type I pneumocytes and leukocyte infiltration, respectively. Left, wild-type mice showing apparently normal lung region adjacent to interstitial density having Pdpn+ network but lacking CD45+ infiltrates. Right, Krt6–DTR lung showing apparently normal region adjacent to zone of damaged interstitial lung lacking Pdpn+ network but having CD45+ infiltrates. Bottom panel, H&E staining of the same histological region. Scale bar, 100 μm.
Extended Data Figure 4. Networks of type I pneumocytes in 30 dpi mouse lung.
a, Histological analysis of lung densities using anti-Pdpn antibodies (red) and anti-SPC (green) to reveal type I and type II pneumocytes respectively. Left, wild-type mice showing apparently normal lung region adjacent to interstitial density having Pdpn+ network but lacking SPC+ cells. Right, Krt6–DTR lung showing normal region adjacent to zone of damaged interstitial lung lacking both pneumocytes. Scale bar, 100 μm. b, Top panel, histological analysis of wild-type lung densities using anti-Pdpn antibodies (red) and anti-Aqp5 (green) type I pneumocyte markers showing the interstitial density having Pdpn/Aqp5 double-positive network. Bottom panel, wild-type mice show apparently normal lung region adjacent to interstitial density having Pdpn+ network but the density lack expression of another type I pneumocyte marker, Hopx. Scale bar, 100 μm.
We next asked if the loss of DASCp63/Krt5 also affected aspects of pulmonary function in these mice. Using pulse oximetry19 to assess peripheral capillary oxygen saturation (SpO2), we found that the normal 95% SpO2 values plummeted to approximately 70% in normal and DASCp63/Krt5-ablated mice eight days following infection. However, the normal mice recovered to 90% SpO2 by 40 dpi, whereas the DASCp63/Krt5-ablated mice only reached SpO2 values approaching 75% saturation (Fig. 2g). Consistent with this apparent decline in pulmonary function, the persistent densities in the 30 dpi Krt6–DTR lung showed staining for smooth muscle actin (α-SMA; Extended Data Fig. 5), a marker of myofibroblasts known to be associated with a pre-fibrotic state of the lung20. These same interstitial regions showed weak but detectable staining with Masson’s trichrome blue, a marker of fibrosis (Extended Data Fig. 5). Correspondingly, whole-genome expression profiles of wild-type and DASCp63/Krt5-ablated lungs indicated the persistence of inflammatory gene expression and a relative decrease in gene expression linked to vasculature development in the DASCp63/Krt5-ablated lungs at 30 dpi (Fig. 2h; Extended Data Fig. 5). Moreover, the DASCp63/Krt5-ablated mice showed the presence of pre-fibrosis gene signature21,22 including vimentin, FSP1 and collagen genes (Fig. 2h). Together these data suggest that the ablation of DASCp63/Krt5 arising during acute injury results in a failure of the regenerative process with structural and functional consequences for the lung.
Extended Data Figure 5. Failure of regeneration in DASCp63/Krt5-ablated lungs.
a, Histological section through 30 dpi DASC-ablated lung (Krt6–DTR +DTox) showing normal region (Pdpn+) adjacent to interstitial density positive for α-SMA and weakly positive for Masson’s trichrome (MT) staining for fibrosis. Scale bar, 100 μm. b, Histological section through 30 dpi control lung showing normal region (Pdpn+) and interstitial density (Pdpn−) which are both negative for α-SMA. c, Expression heat map of selected, differentially expressed genes (P < 0.05) comparing wild-type mouse lungs with DASC-ablated mouse lungs at 30 dpi. Scale bar, 100 μm.
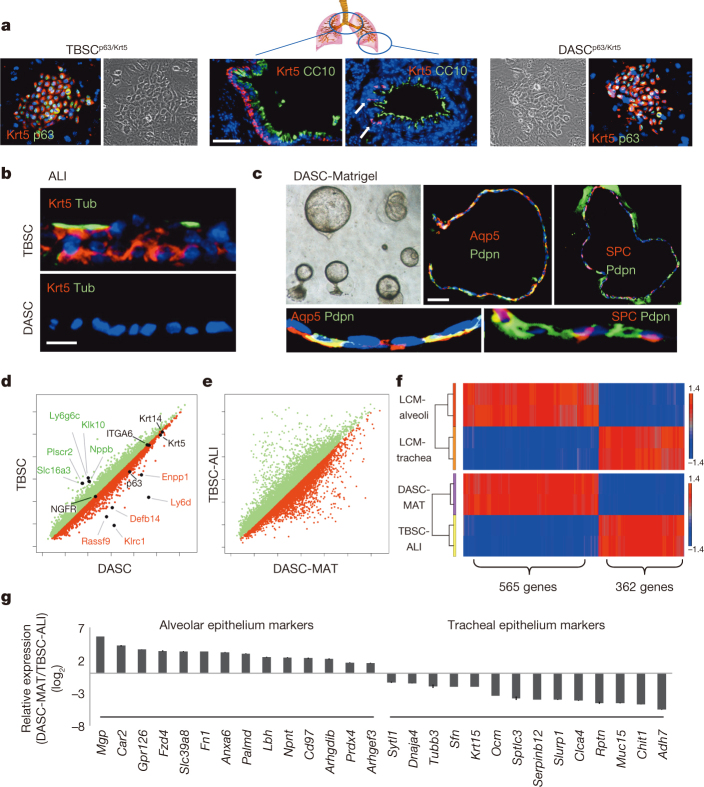
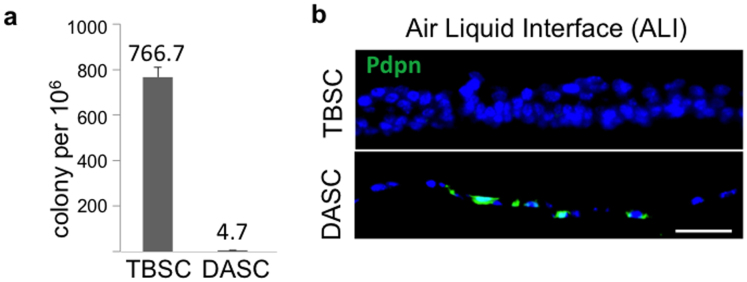
Whereas p63+Krt5+ cells are prominent features of the proximal lung, their presence in distal lung has been less clear23. This is reflected in the abundant Krt5+ cells in proximal lung (Fig. 3a) and the intermittent, peri-bronchiolar clusters seen in one of every three to four sections of distal lung (Fig. 3a, arrows). Consistently, 100-fold fewer DASCp63/Krt5 colonies arise from cell suspensions of distal lung than tracheobronchiolar stem cell (TBSCp63/Krt5) colonies from proximal lung (Extended data Fig. 6a). Regardless, both DASCp63/Krt5 and TBSCp63/Krt5 can be cloned and propagated as single-cell-derived pedigrees7 that show very different fates upon differentiation. In air–liquid interface (ALI) cultures24, TBSCs yield a stratified epithelium with Krt5+ basal cells and apical ciliated and secretory cells typical of proximal airway, while DASCs yield a monolayer of differentiated cells expressing Pdpn (Fig. 3b; Extended Data Fig. 6b). In three-dimensional Matrigel cultures, DASCs form unilaminar, alveolar-like spheres composed of cells expressing type I (Pdpn and Aqp5) and type II (SPC) pneumocyte markers (Fig. 3c). The very minor variation in gene expression between TBSCs and DASCs (less than 1% with fold change > 1.5, P < 0.05; Fig. 3d) transforms to major differences between TBSCs differentiated in air–liquid interface (TBSC-ALI) with DASCs in Matrigel (DASC-MAT), consistent with their divergent fates (Fig. 3e). To further probe the differential fates of TBSC-ALI and DASC-MAT, we used laser-capture microdissection (LCM) to generate gene expression profiles of normal tracheal and alveolar epithelium and compared with those of TBSC-ALI and DASC-MAT (Fig. 3f). Gene-set enrichment analysis revealed a strong coincidence in gene expression patterns between in vitro-differentiated TBSCs and DASCs and their in vivo counterparts (FDR q value < 0.001), including many genes not previously identified as differential markers though confirmed by publically available antibody data sets24 (Fig. 3g).
Figure 3. Cloning and in vitro differentiation of DASCs and TBSCs.
a, Krt5+ cells in proximal (left) and distal (arrows, right) lung, with corresponding TBSC and DASC colonies (outside panels). Scale bar, 50 μm. b, Differentiation of TBSC and DASC in air–liquid interface cultures showing respectively stratified epithelia with ciliated (acetylated-Tub+) cells and a monolayer of differentiated cells. Scale bar, 20 μm. c, Unilaminar spheres formed by DASCs in three-dimensional Matrigel cultures and the expression of indicated markers in sections. Scale bar, 50 μm. Insets, high magnification. d, Scatter plot of gene expression of immature TBSCs and DASCs highlighting common and disparately expressed (fold change > 3, P < 0.001) genes. e, Scatter plot of gene expression of TBSC-ALI and DASC-MAT. f, Top, heat map of gene sets differentially expressed in murine alveoli and tracheal epithelium (P < 0.05). Bottom, heat map of differentially expressed genes in DASC-MAT versus TBSC-ALI (P < 0.05) informed by alveolar and tracheal data sets. g, Histogram of differentially expressed genes of DASC-MAT versus TBSC-ALI for which validating immunohistochemistry data are available (see http://www.proteinatlas.org/).
Extended Data Figure 6. Cloning and in vitro differentiation of TBSCp63/Krt5 and DASCp63/Krt5.
a, Histogram of cloning efficiency of TBSCs and DASCs on irradiated 3T3-J2 cells per 1 million tracheal or distal airway cells derived from respective tissues of adult mice. Tissues derived from 3 mice. Error bars, s.e.m. b, Immunofluorescence images of sections of TBSC and DASC air–liquid interface cultures using an antibody to the type I pneumocyte marker Pdpn (green). Sections were counterstained with DAPI (blue).
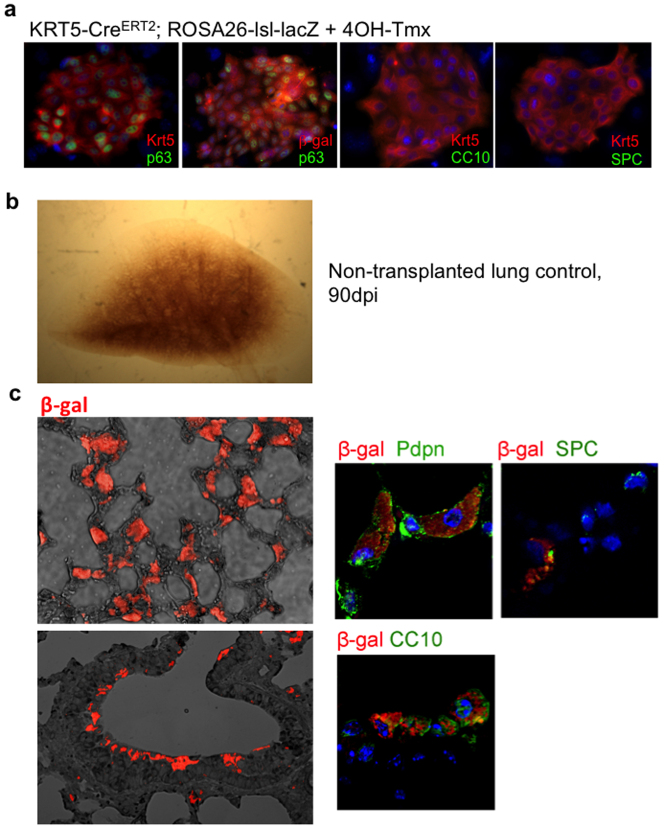
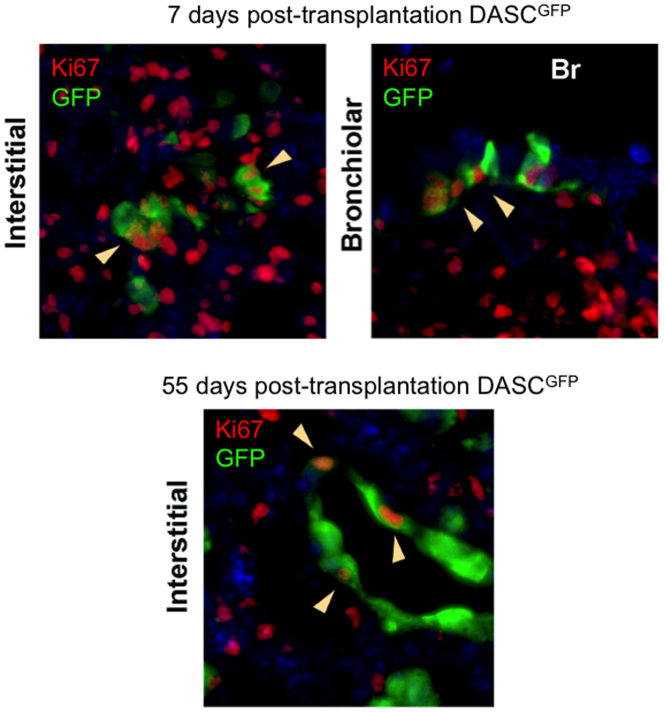
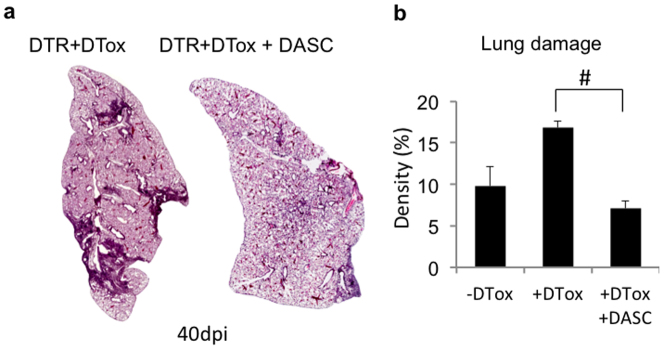
To determine if cloned lung stem cells could incorporate into damaged lung, we first generated single-cell-derived pedigrees of DASClacZ and TBSClacZ from murine lung (Fig. 4a; Extended data Fig. 7a). We delivered one million TBSClacZ or DASClacZ to syngeneic mice five days after influenza virus infection (Fig. 4a). At 40 dpi (35 days post-transplantation), DASClacZ were distributed in interstitial regions emanating from airways (Fig. 4b). At 90 dpi, DASClacZ showed a more homogenous pattern in interstitial spaces compared to 40 dpi (Fig. 4b). Significantly, mock-infected lungs, or mock-transplanted, infected lungs, showed no incorporation of DASClacZ at 35 days post-transplantation (Fig. 4b: Extended data Fig. 7b). As with the lineage-tracing experiments, we used E. coli-specific β-galactosidase antibodies to mark the transplanted cells in 90 dpi lungs and observed that at least 40% of the cells in alveolar region express pneumocytes markers (Pdpn, 1H8 and SPC) and at least 80% of the β-galactosidase-positive cells in bronchiolar region express secretory cell marker CC10 (mouse number n = 3; Fig. 4c; Extended data Fig. 7c). Gene expression analysis of the lacZ-positive regions of these lungs using laser-capture microdissection revealed a typical alveoli gene signature very different from that of immature DASCs or of damaged lung (Fig. 4d). Together these findings demonstrate that single-cell-derived pedigree lines of DASCs can readily incorporate into damaged lung during the process of lung regeneration and give rise to multiple epithelial cell types of bronchioles and alveoli. In contrast, transplanted TBSClacZ appeared confined to major airways (Fig. 4e). Parallel transplantations with green fluorescent protein (GFP)-labelled DASC (Fig. 4f) yielded similar patterns of co-labelling of lineage and type I and type II pneumocyte markers at 40 dpi seen with transplanted DASClacZ (Fig. 4f). Significantly, a fraction of these transplanted DASCGFP or their progeny continue to express the proliferation marker Ki67 even up to 60 dpi (Extended data Fig. 8), suggesting their high viability and extended contribution to the regenerative process. Lastly, morphometric analyses of diphtheria-toxin-treated, virally infected Krt6–DTR mice indicate that transplantation of DASCs results in a significant reduction of interstitial densities at 40 dpi (Extended data Fig. 9).
Figure 4. Transplantation of TBSClacZ and DASClacZ.
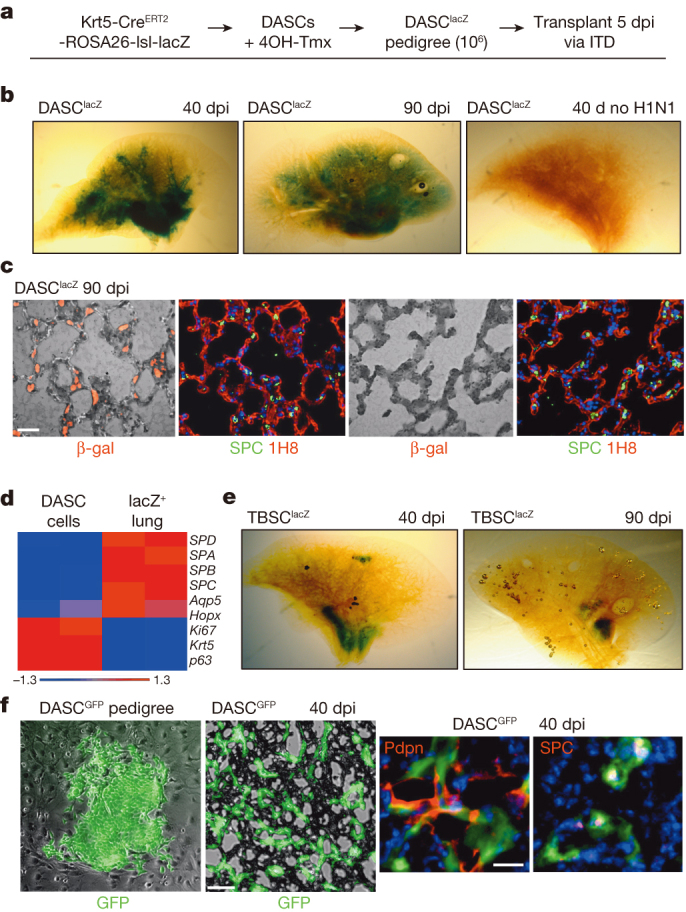
a, Schematic of pedigree generation and transplantation. 4OH-Tmx, 4-hydroxy-tamoxifen; ITD, intratracheal delivery. b, β-galactosidase activity in whole lung following DASClacZ transplantation. c, Comparison between β-galactosidase-positive (left panels) and -negative (right panels) regions of transplanted lung and markers of type I (1H8) and type II (SPC) cells. Scale bar, 50 μm. d, Heat map of selected, differentially expressed genes (P < 0.05) comparing immature DASClacZ before transplantation with laser-capture microdissected of lacZ-positive cells from transplanted lungs at 90 dpi. e, β-galactosidase activity in whole lung following TBSClacZ transplantation. f, From left, DASCGFP colony in culture; middle, cryosection of lung following DASCGFP transplantation. Scale bar, 50 μm. Right, immunofluorescence of anti-GFP, anti-Pdpn and anti-SPC in 40 dpi transplanted lung. Scale bar, 20 μm.
Extended Data Figure 7. Transplantation of DASClacZ.
a, Immunofluorescence characterization of DASCs isolated from Krt5-CreERT2;ROSA26-lsl-lacZ mice following Cre activation with 4OH-tamoxifen. From left, colony stained with antibodies to p63 (green) and Krt5 (red), p63 (green) and E. coli β-galactosidase (red), Krt5 (red) and CC10 (green), and Krt5 (red) and SPC (green). b, Whole mount image of lung 90 days after infection without stem cell transplantation. c, Left, bright field/immunofluorescence image of section of lung at 90 dpi following transplantation of DASClacZ stained with antibodies to β-galactosidase (red). Right panels, immunofluorescence images of co-staining of transplanted DASClacZ with antibodies to Pdpn, SPC, or CC10 at high magnification.
Extended Data Figure 8. Persistent proliferation of transplanted DASC.
Co-staining of antibodies to GFP (green) with the cell proliferation marker Ki67 (red) in sections of lung transplanted with DASCGFP at 12 dpi lung (7 days post transplantation) and 60 dpi lung (55 days post transplantation). Top left, immunofluorescence image of lung following transplantation of DASCGFP (7 days post-transplantation;12 dpi) stained with anti-GFP (green) and the cell cycle marker Ki67 (red, in nucleus). Top right, bronchiole co-stained with antibodies to GFP and Ki67 from 7 days post-transplantation lung. Bottom, staining of interstitial lung transplanted 55 days prior with DASCGFP with antibodies to GFP and Ki67. Arrows indicate cells co-expressing GFP and Ki67.
Extended Data Figure 9. Stem cell transplantation reduces interstitial densities in DASCp63/Krt5-ablated lungs.
a, Histological sections through entire lobe of Krt6–DTR mice with (left) and without (right) diphtheria toxin treatment forty days post-influenza infection. b, Histogram of morphometric quantification of lung densities following 40 day influenza virus infection of Krt6–DTR mice without diphtheria toxin (−DTox, mouse number n = 3), with diphtheria toxin (+DTox, n = 4), or with diphtheria toxin and transplanted DASCs (+DTox+DASC, n = 4). Error bars indicate s.e.m. and # indicates P value = 0.029 by Wilcoxon rank-sum test.
In the present work, we highlight the remarkable regenerative capacity of the lung following large-scale, acute lung damage7 and the function of a very discrete, pre-existing population of lung stem cells in this process. In addition, we demonstrate that upon transplantation, single-cell-derived DASCp63/Krt5 pedigrees contribute multiple epithelial lineages, including bronchiolar secretory cells as well as alveolar type I and type II pneumocytes, to regenerating distal lung. Thus DASCp63/Krt5 act in an emergent, conditional manner that is generally distinct from that of type II pneumocytes25,26,27,28,29, progenitor cells of limited self-renewal capacity that participate in highly focal, homeostatic lung repair. Our findings provide a mechanistic framework for the still emerging concept of lung regeneration30 and underscore potential therapeutic strategies exploiting this process.
Methods
Influenza virus infection
All mouse experiments were conducted under IACUC guidelines and approved protocols. Influenza A (H1N1) mouse-adapted PR/8/34 (VR-95, ATCC, USA) was used for all viral infections. The virus stock was amplified by V. Chow (Department of Microbiology, National University of Singapore) in chicken eggs. Virus dilutions were made in DMEM medium containing 1 μg ml−1 TPCK trypsin (Sigma-Aldrich, USA) on ice, aliquoted and stored at −80 °C. The viral titre is measured by plaque assay on Madin Darby canine kidney cells (MDCK, ATCC, USA). Virus was further diluted to final concentration in PBS on ice and use freshly. The infection of mice by H1N1 Influenza virus was performed in an Animal Biosafety Level 2 (ABSL-2) facility. Adult mice (>6 weeks old) were anaesthetized with intraperitoneal injection of ketamine (150 mg per kg body weight) + xylazine (10 mg per kg body weight). The anaesthetized mouse was rested on a stand with its front teeth hung over a suture. This causes the mouse airway to be relaxed and accessible. Using a flat forceps the tongue of the animal was drawn out of its mouth so that the anatomy can be easily visualized. Intratracheal delivery was performed by pipetting 50 μl virus directly into the larynx/trachea. Sterile PBS was administered to the control animals. The tongue of mouse was held throughout the procedure so that the virus was aspirated into the lungs.
Tissue histology
At appropriate time points, mice were euthanized by CO2 asphyxiation followed by exsanguination, and the diaphragm was carefully cut open without touching the lungs. A small incision was made in the proximal region of the trachea and lung was inflated with 4% formaldehyde using a 30G needle. The inflated lungs were dissected and fixed with 4% formaldehyde before whole mount imaging, paraffin section or cryosection. For whole mount imaging, lungs were dehydrated in graded ethanol series and sunk in BABB (benzyl alcohol/benzyl benzoate 1:2 ratio) at 4 °C overnight31. For paraffin section, lungs were processed in an automatic tissue processer (Leica Microsystems, Germany) and embedded into paraffin blocks. The blocks were cut using microtome (Leica Microsystems, Germany) to 5∼7 μm thickness at distinct planes. The sections were placed on poly-lysine coated glass slides and stored at room temperature until further use. For cryosection, lungs were embedded within Tissue-Tek O.C.T compound, solidified on dry ice and cut using a cryotome (Leica Microsystems, Germany) of 10 μm thickness.
Haematoxylin and eosin (H&E) staining was performed using standard procedures. To analyse lung damage level, interstitial densities were assessed by H&E staining backed up by type I (anti-Pdpn) and type II (anti-SPC) pneumocytes staining. A minimum of 8 axial lung interval sections (typically 400 mm2) covering >2 mm tissue depth were cut, each was stained, scored for densities, and quantified for percentage of total lung area by Zeiss AxioVision (Carl Zeiss, Germany) morphometric software. In addition, random histological sections are scored based on the general pathological morphology by blinded expert to confirm the conclusion. Stitching scanning of H&E slides were performed in histopathology lab of IMCB, A-STAR, Singapore. Masson Trichrome staining of lung fibrosis was performed using the Trichrome Staining Kit (Sigma-Aldrich, USA). The kit involves sequential staining of the sections with Biebrich Scarlet-Acid Fuchsin, PTA/PMA and Aniline Blue. After staining, sections were dehydrated and mounted using Vectamount and visualized under a light microscope (Imager Z1, Carl Zeiss, Germany).
Immunofluorescence staining
For immunofluorescence staining, paraffin-embedded tissue slides were subjected to antigen retrieval in citrate buffer (pH 6, Sigma-Aldrich, USA) at 120 °C for 20 min with the exception of CD45 staining. Antibodies used for immunofluorescence included stem cell markers: Krt5 (1:200, EP1601Y, Thermo), Krt6 (1:100, T-18, Santa Cruz and Ab24646, Abcam), p63 (1:2, 4A4 clone, house-made); pneumocyte markers: Pdpn (1:100, M-172 and A-18, Santa Cruz), Aqp5 (1:100, G-19 and H-200, Santa Cruz), Hopx (1:100, FL-73, Santa Cruz), SPC (1:100, M-20 and FL-197, Santa Cruz), 1H8 mouse monoclonal antibodies (1:2, house-made); others: CC10 (1:100, T-18, Santa Cruz), CD45 (1:100, 30-F11, Santa Cruz), bacteria-specific β-galactosidase (1:400, A-11132, Life Technologies and Ab9361, Abcam), Ki67 (1:200, RM-9106, Thermo), alpha-SMA (1:400, 1A4, Dako), HB-EGF (1:100, Ab16783, Abcam), GFP (1:100, B-2, Santa Cruz) and acetylated alpha tubulin (1:1,000, Ab24610, Abcam). Among them, 1H8 murine monoclonal antibody was generated under IRB approval using standard methods, and validated for specific mouse type I pneumocyte staining by immunofluorescence. Alexa-conjugated secondary antibodies (1:200) were used for immunofluorescence. After staining, tissues slides underwent auto-fluorescence removal and mounting with DAPI containing mounting media (Vectashield, Vector Labs, USA). Stained slides were stored at 4 °C in the dark and images were taken using Zeiss fluorescence microscope (Observer Z1, Carl Zeiss, Germany) or Zeiss confocal microscope (LSM 510, Carl Zeiss, Germany).
Lineage tracing of Krt5+ stem cells
Krt5-CRE/ERT2 (011916-MU, MMRRC, USA) mice were crossed with Rosa26-loxP-STOP-loxP-lacZ mice (003309, Jackson Laboratory, USA) to generate mice for lineage tracing. Genotype was confirmed for each mouse using tail genomic DNA collection and PCR validation. Tamoxifen was dissolved in corn oil and freshly applied to mice at 200 mg per kg body weight through intraperitoneal injection at indicated days before influenza infection. The gap between tamoxifen and H1N1 administration was varied as indicated to control for any possible tamoxifen persistence. Sublethal doses (25 plaque forming units) of H1N1 influenza A virus were diluted in PBS and intratracheally delivered into anaesthetized mice. After infection by virus, mouse lungs were collected at various time points and subjected to standard immunofluorescence staining by two independent bacteria-specific β-galactosidase antibodies (1:400, A-11132, Life Technologies and Ab9361, Abcam) or X-gal staining. For X-gal staining, lungs were briefly fixed on ice for 30 min and subjected to X-gal (Invitrogen, USA) whole-mount staining overnight using standard protocol. After staining, lungs were washed and fixed again in 4% formaldehyde before whole-mount visualization or paraffin sectioning. For whole-mount visualization, lungs were made transparent by BABB as described before and images were taken using dissection microscope (Leica Microsystems, Germany). For paraffin section, 5∼7 μm sections were cut and immunofluorescence staining is performed following standard protocol and visualized under light microscope (Imager Z1, Carl Zeiss, Germany).
Generation of the Krt6–DTR mouse
The complementary DNA of diphtheria toxin receptor (DTR), which is also known as human heparin-binding epidermal growth factor-like growth factor (HB-EGF), and a neomycin resistance selection cassette flanked with loxPs (Floxed NeoR) with an introduced PacI restriction endonuclease site, were introduced to replace the first 4 exons of Krt6a in a modified bacterial artificial chromosome (mBAC). Retrieval and linearization of a selected section of this mBAC resulted in the targeting construct, which was electroporated into V6.4 B6.129 hybrid embryonic stem cells to be selected with G418. Single colonies screened by Southern blot analysis of PacI digests detected by a 5′ external probe (hybridizing unmodified Krt6a genomic DNA) revealed fragment size differences due to the introduced PacI site. Wild-type alleles with endogenous PacI sites returned 35.9 kb fragments, while a recombination event returned a shorter 14.7 kb. Floxed NeoR probes provided further verification for a single specific insertion. Successfully engineered embryonic stem cells were microinjected into blastocysts to generate chimaeras, which were similarly tested for germline transmission via backcrosses to C57BL/6. Progeny from crosses with FVB/N-Tg(ACTB-cre)2Mrt/J (Jackson Laboratory, USA) were screened for stable transmission of Krt6a–DTR alleles Cre-mediated excision of Floxed NeoR as indicated by a reduced 12.9 kb PacI digest fragment.
Oxygen saturation measurements
Peripheral capillary oxygen saturation (SpO2) was measured using MouseOx Plus pulse oximeter (Starr Life Sciences, USA). An S-size CollarClip sensor was applied to depilated regions of the back neck skin and mice were rested for one hour before measuring SpO2. Ten minutes before measurement, mice were anaesthetized by ketamine (150 mg per kg body weight) + xylazine (10 mg per kg body weight) intraperitoneal injection. After SpO2 readings were stable, data collection was started and SpO2 readings were recorded every second for one minute to calculate an average value.
Cloning of TBSC and DASC and in vitro differentiation
To isolate airway stem cells, trachea and lung were collected from adult mice and immersed in cold wash buffer (F12 medium, 1% Pen/Strep, 5% FBS). The trachea and two main bronchi were separated from the lungs and the lobes were cut with a sterile surgical blade into small pieces and digested with dissociation buffer (F12/DMEM, 1mg ml−1 protease, 0.005% trypsin and 10ng ml−1 DNase I) overnight with gentle rocking. The dissociated cells were washed with wash buffer, passed through a 40-μm cell strainer, counted and plated onto irradiated 3T3 feeder cells as described7,14,15. After 4 consecutive passages, single cell colonies were picked up by cloning ring and expanded. Colonies were characterized by immunofluorescence staining (E-cadherin+Krt5+p63+Pdpn−CC10−SPC−). These colonies have been passaged up to 12 months with no observable phenotypic or chromosome count changes. To compare gene expression in immature TBSCs and DASCs, whole genome expression microarrays were performed on passage 7 (P7) cells. Figure 3c is a scatterplot comparison of the TBSC and DASC whole transcriptome data. Gene names are indicted for outliers (fold change > 3, P value <0.001) typically reported as markers or well known for other biology and for which we validated expression by quantitative PCR. To activate lacZ in TBSCs and DASCs cloned from ROSA26-lsl-lacZ; Krt5-CreERT2 mice, we exposed colonies in vitro to 4-OH-tamoxifen (1 μg ml−1; Sigma-Aldrich) for 5 days at which point >70% cells express β-galactosidase and all cells were Krt5+p63+SPC−CC10−Pdpn−. GFP labelling was performed by retroviral transduction of pMX-GFP (Cell Biolab, USA) followed by manual sorting of single GFP+ DASCs to 96-well plates.
Matrigel differentiation assays were performed as previously described7. FGF10 (50 ng ml−1) was included in medium to favour distal airway differentiation. Under this condition, DASCs clustered and grew into sphere-like structures marked by unilaminar epithelia surrounding a clear lumen. TBSCs were grown on air–liquid interface (ALI) cultures for differentiation7. ALI differentiation was performed as previously described11, FGF10 was excluded and retinoid acid (50 nM) included in medium to favour proximal airway differentiation. Under ALI condition, TBSC forms a stratified epithelium structure marked by ciliated cells (acetylated-Tub+) and basal cells (Krt5+). In addition, we used laser capture microdissection (LCM, Zeiss PALM) to dissect mouse trachea epithelium, alveoli and damaged lung interstitium for microarray analysis to develop tissue specific gene expression signatures.
Orthotopic transplantation of stem cells
To perform orthotropic transplantation of stem cells into lung, adult mice were infected with 25 plaque forming units H1N1 influenza virus five days prior transplantation. Stem cell pedigrees were expanded in culture and harvested by differential trypsinization to remove feeder cells. One million cells were diluted in 50 μl DMEM/F12 medium for transplantation into each mouse. Adult mice (>6 weeks old) were anaesthetized with intraperitoneal injection of ketamine (150 mg per kg body weight) + xylazine (10 mg per kg body weight) and rested on a stand gesture. Intratracheal aspiration was performed by pipetting the virus directly into trachea via mouth. For rescue of Krt6–DTR phenotype by DASC transplantation, 3×105 cells were transplanted on 8 dpi (the same day DTox was given) and control mice received medium alone.
Laser capture microdissection (LCM)
For LCM, fresh, non-fixed or X-gal stained tissue samples were embedded in Tissue-Tek O.C.T. compound (Sakura, Japan) on dry ice. 10 µm cryosections were mounted on PEN Membrane slides (Leica Microsystems, Germany). Slides were dehydrated in 95%, 75%, 50% nuclease-free ethanol for 30 s each, stained in Arcturus HistoGene staining solution (Life Technologies, USA) for 1 min then dehydrated in 50%, 75%, 95%, 100% ethanol for 30 s each and xylene for 5 min. Slides were allowed to air dry and LCM was performed immediately using an inverted microscope and PALM Robo software (Carl Zeiss, Germany). Cut elements were catapulted onto an Adhesive Cap-500 tube (Carl Zeiss) and subsequently transferred into a PCR tube with 50 µl Arcturus PicoPure extraction buffer. RNA was isolated using the Arcturus PicoPure RNA Isolation kit (Life Technologies, USA).
Microarray and bioinformatics
RNAs obtained from LCM, cell colonies or whole lobe of mouse lungs were used for microarray after being amplified using WT-Ovation Pico RNA Amplification System (NuGEN, UK) and fragmented and labelled using the FL-Ovation cDNA Biotin Module V2 (NuGEN, UK). Labelled cDNA was then hybridized onto GeneChip Mouse Exon 1.0 ST Array (Affymetrix, USA) using appropriate hybridization controls and the chips were scanned and analysed as described previously7. Duplicate experiments for microarray were taken from two biological samples. To validate sample quality, probe hybridization ratios were calculated using Affymetrix Expression Console software (Affymetrix, USA). The intensity values were log2-transformed and imported into the Partek Genomics Suite 6.6 (Partek Inc., USA). Exons were summarized to genes and 1-way ANOVA was performed to identify differentially expressed genes. P values and fold-change were calculated for each analysis. Heat maps were generated using Pearson’s correlation and Ward’s method and principal component analysis was conducted using all probe sets. Gene ontology analyses were performed using the web-based GeneTrail tool32.
For the comparison between gene expression profiles of in situ alveoli and tracheal epithelium with those of in vitro differentiated TBSCs and DASCs (see Fig. 3f, g), gene set enrichment analyses were performed using web-based tools developed by the Broad Institute33,34. 605 genes highly expressed in tracheal epithelium (trachea versus alveoli, 4-fold higher, P value < 0.05) and 914 genes highly expressed in alveolar epithelium (trachea versus alveoli, 4-fold lower, P value < 0.05) were used as the queried gene sets (signature), respectively. Enrichment scores were determined after 1,000 permutations, and the permutation type was configured to the gene sets. Thereafter the whole genome expression data of DASC-MAT versus TBSC-ALI were applied to GSEA program to evaluate enrichment of signature in the fold-change ordered list. Results with normalized enrichment score > 1.4 and FDR q value < 0.001 were considered significant. The comparison between gene expression profiles of normal alveoli and damaged interstitium with those of lung tissue with or without DASC ablation (see Extended Data Fig. 3a) was performed in similar manner.
Supplementary information
Reconstruction of 3-D pattern of Krt5 staining with respect to bronchioles (blue) in portion of 15dpi infected lung from serial sections. (MP4 27357 kb)
Acknowledgements
This work was supported by grants from the Joint Council Office of the Agency for Science Technology Research Agency (ASTAR), Singapore (W.X., F.M.), Defense Advanced Research Projects Agency (DARPA, N66001-09-1-2121 to F.M.), the Johnson & Johnson ASTAR Joint Program (W.X., F.M.) and support from Connecticut Innovations (W.X., F.M.). We thank M. LaLande, B. Lane and H. Hui Ng for support, G. Wright, B. Tennent, B. Knowles and T. McLaughlin for comments on the manuscript, J. Hammer for artwork, P. Kraus for blastocyst injections, and H. Ahmad and K. L. Goh for technical assistance. We thank H. Green for advice and support.
Extended data figures and tables
PowerPoint slides
Author Contributions
Experiments were performed by W.Z., D.Z.W., S.P.G. and A.-A.L. Experimental design and conception were done by W.Z., M.V., C.P.C., W.X. and F.M.; T.Z., W.Z., X.W., S.J.L. and Y.Y. performed microarrays and computational analysis, and provided methodological advice. M.L. and W.Z. performed the serial reconstructions of infected lung.
Accession codes
Primary accessions
Gene Expression Omnibus
Competing interests
The authors declare no competing financial interests.
Footnotes
Datasets generated for this study have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database under superseries GSE60849.
Contributor Information
Wa Xian, Email: waxian@jax.org.
Frank McKeon, Email: fmckeon@jax.org.
References
- 1.Barnes PJ. Chronic obstructive pulmonary disease. N. Engl. J. Med. 2000;343:269–280. doi: 10.1056/NEJM200007273430407. [DOI] [PubMed] [Google Scholar]
- 2.Gross TJ, Hunninghake GW. Idiopathic pulmonary fibrosis. N. Engl. J. Med. 2001;345:517–525. doi: 10.1056/NEJMra003200. [DOI] [PubMed] [Google Scholar]
- 3.Kerem E, et al. Bacteremic necrotizing pneumococcal pneumonia in children. Am. J. Respir. Crit. Care Med. 1994;149:242–244. doi: 10.1164/ajrccm.149.1.8111589. [DOI] [PubMed] [Google Scholar]
- 4.Sawicki GS, Lu FL, Valim C, Cleveland RH, Colin AA. Necrotising pneumonia is an increasingly detected complication of pneumonia in children. Eur. Respir. J. 2008;31:1285–1291. doi: 10.1183/09031936.00099807. [DOI] [PubMed] [Google Scholar]
- 5.Herridge MS, et al. One-year outcomes in survivors of the acute respiratory distress syndrome. N. Engl. J. Med. 2003;348:683–693. doi: 10.1056/NEJMoa022450. [DOI] [PubMed] [Google Scholar]
- 6.Wilcox ME, et al. Radiologic outcomes at 5 years after severe ARDS. Chest. 2013;143:920–926. doi: 10.1378/chest.12-0685. [DOI] [PubMed] [Google Scholar]
- 7.Kumar PA, et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147:525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu. Rev. Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Padilla R, et al. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N. Engl. J. Med. 2009;361:680–689. doi: 10.1056/NEJMoa0904252. [DOI] [PubMed] [Google Scholar]
- 10.Matuschak GM, Lechner AJ. Acute lung injury and the acute respiratory distress syndrome: pathophysiology and treatment. Mo. Med. 2010;107:252–258. [PMC free article] [PubMed] [Google Scholar]
- 11.Short KR, Kroeze EJ, Fouchier RA, Kuiken T. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect. Dis. 2014;14:57–69. doi: 10.1016/S1473-3099(13)70286-X. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Gudjonsson JE, Woods TL, Zhang T, Johnston A, Stoll SW, Elder JT. Transgenic expression of S100A2 in hairless mouse skin enhances Cxcl13 mRNA in response to solar-simulated radiation. Arch. Dermatol. Res. 2009;301:205–217. doi: 10.1007/s00403-008-0881-y. [DOI] [PubMed] [Google Scholar]
- 13.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 14.Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/S0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 15.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl Acad. Sci. USA. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito M, et al. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nature Biotechnol. 2001;19:746–750. doi: 10.1038/90795. [DOI] [PubMed] [Google Scholar]
- 17.Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells of stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 18.Baskerville A, Thomas G, Wood M, Harris WJ. Histology and ultrastructure of metaplasia of alveolar epithelium following infection of mice and hamsters with influenza virus. Br. J. Exp. Pathol. 1974;55:130–137. [PMC free article] [PubMed] [Google Scholar]
- 19.Verhoeven D, Teijaro JR, Farber DL. Pulse-oximetry accurately predicts lung pathology and the immune response during influenza infection. Virology. 2009;390:151–156. doi: 10.1016/j.virol.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vyalov SL, Gabbiani G, Kapanci Y. Rat alveolar myofibroblasts acquire alpha-smooth muscle actin expression during bleomycin-induced pulmonary fibrosis. Am. J. Pathol. 1993;143:1754–1765. [PMC free article] [PubMed] [Google Scholar]
- 21.Tsukui T, et al. Qualitative rather than quantitative changes are hallmarks of fibroblasts in bleomycin-induced pulmonary fibrosis. Am. J. Pathol. 2013;183:758–773. doi: 10.1016/j.ajpath.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Lawson WE, et al. Characterization of fibroblast-specific protein 1 in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2005;171:899–907. doi: 10.1164/rccm.200311-1535OC. [DOI] [PubMed] [Google Scholar]
- 23.Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech. 2010;3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt D, Hubsch U, Wurzer H, Heppt W, Aufderheide M. Development of an in vitro human nasal epithelial (HNE) cell model. Toxicol. Lett. 1996;88:75–79. doi: 10.1016/0378-4274(96)03720-4. [DOI] [PubMed] [Google Scholar]
- 25.Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nature Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 26.Evans MJ, Cabral LJ, Stephens RJ, Freeman G. Transformation of alveolar type 2 cells to type 1 cells following exposure to NO2. Exp. Mol. Pathol. 1975;22:142–150. doi: 10.1016/0014-4800(75)90059-3. [DOI] [PubMed] [Google Scholar]
- 27.Sugihara H, Toda S, Miyabara S, Fujiyama C, Yonemitsu N. Reconstruction of alveolus-like structure from alveolar type II epithelial cells in three-dimensional collagen gel matrix culture. Am. J. Pathol. 1993;142:783–792. [PMC free article] [PubMed] [Google Scholar]
- 28.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xian W, McKeon F. Adult stem cells underlying lung regeneration. Cell Cycle. 2012;11:887–894. doi: 10.4161/cc.11.5.19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker K, Jährling N, Saghafi S, Dodt HU. Immunostaining, dehydration, and clearing of mouse embryos for ultramicroscopy. Cold Spring Harb. Protoc. 2013;2013:743–744. doi: 10.1101/pdb.prot076521. [DOI] [PubMed] [Google Scholar]
- 32.Keller A, et al. GeneTrailExpress: a web-based pipeline for the statistical evaluation of microarray experiments. BMC Bioinformatics. 2008;9:552. doi: 10.1186/1471-2105-9-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mootha VK, et al. PGC-1 alpha responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reconstruction of 3-D pattern of Krt5 staining with respect to bronchioles (blue) in portion of 15dpi infected lung from serial sections. (MP4 27357 kb)