Abstract
From 28 young children in the Netherlands, we isolated a paramyxovirus that was identified as a tentative new member of the Metapneumovirus genus based on virological data, sequence homology and gene constellation. Previously, avian pneumovirus was the sole member of this recently assigned genus, hence the provisional name for the newly discovered virus: human metapneumovirus. The clinical symptoms of the children from whom the virus was isolated were similar to those caused by human respiratory syncytial virus infection, ranging from upper respiratory tract disease to severe bronchiolitis and pneumonia. Serological studies showed that by the age of five years, virtually all children in the Netherlands have been exposed to human metapneumovirus and that the virus has been circulating in humans for at least 50 years.
Main
The Paramyxovirinae and Pneumovirinae subfamilies of the Paramyxoviridae family include several major pathogens of humans and animals. The Pneumovirinae are taxonomically divided in the Pneumovirus and the Metapneumovirus genera1. Human respiratory syncytial virus (hRSV), the type species of the Pneumovirus genus, is the single most important cause of lower respiratory tract infections (RTIs) during infancy and early childhood worldwide2,3. Other members of the Pneumovirus genus include bovine respiratory syncytial virus (bRSV), ovine respiratory syncytial virus (oRSV) and pneumonia virus of mice (PVM). Avian pneumovirus (APV), previously known as turkey rhinotracheitis virus (TRTV), is the etiological agent of an upper RTI of turkeys4,5 and the sole member of the recently assigned Metapneumovirus genus1. Until now, metapneumoviruses have not been associated with infections or disease in mammals. The classification of the two genera is based primarily on their gene constellation; metapneumoviruses lack non-structural proteins NS1 and NS2 and the gene order is different from that of pneumoviruses (RSV, 3′-NS1-NS2-N-P-M-SH-G-F-M2-L-5′; APV, 3′-N-P-M-F-M2-SH-G-L-5′)6,7,8.
In the past few decades, many etiological agents of respiratory tract illnesses (RTI) have been identified9,10. However, a proportion of these illnesses still cannot be attributed to known pathogens. We have now isolated a previously undiscovered paramyxovirus from nasopharyngeal aspirate samples taken from 28 epidemiologically unrelated children in the Netherlands suffering from RTI during the past 20 years. Of the patients, 27 were below the age of 5 years and 13 of these were infants between the ages of 0 and 12 months. The clinical symptoms of these children were largely similar to the RTI caused by hRSV, ranging from mild respiratory problems to severe cough, bronchiolitis and pneumonia, often accompanied by high fever, myalgia and vomiting. Some of these patients were hospitalized and needed mechanical ventilation.
The virus isolates exhibited a paramyxovirus-like morphology in negative contrast electron microscopy and were closely related both antigenically and genetically. Based on sequence homology and gene constellation, the viruses seemed to be a tentative new member of the Metapneumovirus genus that we have provisionally named human metapneumovirus (hMPV). Serological surveys showed that by the age of five years virtually all children in the Netherlands have been exposed to hMPV and that the virus has been circulating in humans for at least half a century.
Virus isolation and characterization
Within our diagnostic virology setting, we isolated 28 unidentifiable viruses from patients in the Netherlands with RTI over the past 20 years. Of the 26 patients for whom personal information was available, eight (31%) were females, 13 (50%) were between the ages of 0 and 12 months, and 12 (46 %) were between the ages of 1 and 5 years. All viruses were isolated from samples collected in the winter months. The unidentified virus isolates replicated slowly in tertiary monkey kidney (tMK) cells, very poorly in Vero cells and A549 cells, and could not be propagated in Madin Darby canine kidney (MDCK) cells or chicken embryo fibroblasts (CEF). The cytopathic effects induced by these virus isolates were virtually indistinguishable from those caused by hRSV, with characteristic syncytia formation followed by rapid internal disruption of the cells and subsequent detachment from the culture plate. The cells usually displayed cytopathic effects at days 10–14 post inoculation, slightly later than the cytopathic effects induced by hRSV-containing specimens (data not shown). After 14 days of virus propagation, the viral titers in the cultures were on average 10,000 TCID50 (50% tissue culture infectious doses) per ml. Supernatants of infected tMK cells were used for electron microscopy analysis, which revealed the presence of paramyxovirus-like pleiomorphic particles in the range of 150–600 nm, with short envelope projections in the range of 13–17 nm (Fig. 1). Similar to some other paramyxoviruses such as selected hRSV and parainfluenza virus isolates, nucleocapsids were rarely observed.
Figure 1. Electron micrograph of hMPV particles.
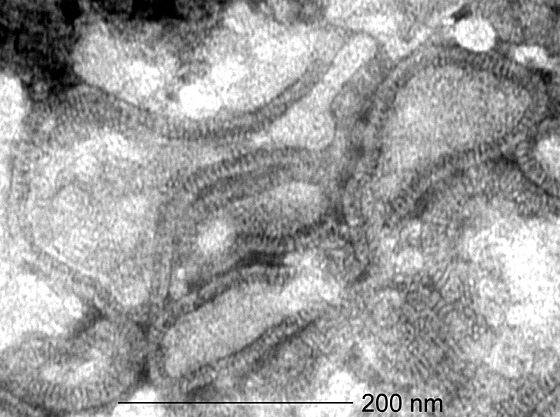
Virus concentrated from infected tMK–cell culture supernatants were visualized by negative contrast electron microscopy after PTA staining24. Magnification, ×92,000.
Consistent with the biochemical properties of enveloped viruses such as the Paramyxoviridae, standard chloroform treatment11 resulted in a 10,000-fold reduction of the TCID50 for tMK cells. Virus-infected tMK cell-culture supernatants did not display hemagglutinating activity with turkey, chicken or guinea pig erythrocytes. In tMK-cell cultures, virus replication was found to be dependent on trypsin. These combined virological data indicated that the newly identified virus is a member of the Paramyxoviridae family.
We isolated RNA from the supernatant of tMK cells infected with 15 of the unidentified virus isolates for reverse transcription (RT)-PCR analyses using primer sets specific for known paramyxoviruses (human parainfluenza virus (hPIV) types 1–4, mumps virus, measles virus, hRSV, simian virus type 5 (SV-5), Sendai virus and Newcastle disease virus (NDV)). We carried out RT-PCR assays at low stringency in order to detect potentially related viruses and used RNA isolated from homologous virus stocks as positive controls. Whereas the controls all reacted positive with the respective virus-specific primers, the newly identified virus isolates did not (data not shown), indicating the virus was not closely related to the viruses for which primer sets had been selected.
We next inoculated ferrets and guinea pigs intranasally with two of the virus isolates (without prior purification) to raise virus-specific antisera. The animals did not display any clinical symptoms. The antisera did not react in immune fluorescence assays (IFA) with cells infected with a panel of paramyxoviruses and orthomyxoviruses (hPIV types 1–4, hRSV, influenza virus type A and B). All animals seroconverted as shown in virus neutralization assays and IFA against the homologous viruses. We next tested the 28 unidentified virus isolates with the guinea pig and ferret pre- and post-infection sera. tMK cell cultures infected with each of the 28 virus isolates reacted positively in IFA with the post-infection animal sera, indicating that they were serologically related or identical.
RAP-PCR
To obtain sequence information on the unknown virus isolates, we used a random PCR amplification strategy known as RAP-PCR (refs. 12,13). To this end, we infected tMK cells with one of the virus isolates (isolate 00-1) or with hPIV-1 that served as a control. We sequenced 20 differentially displayed bands specific for the unidentified virus. When we used the deduced amino-acid sequences to search for homologies against sequences in the Genbank database using the BLAST software (http://www.ncbi.nlm.nih.gov/BLAST), 10 of 20 fragments displayed resemblance to APV/TRTV sequences. These 10 fragments were located in the genes coding for the nucleoprotein (N; fragments 1 and 2), the matrix protein (M; fragment 3), the fusion protein (F; fragments 4, 5, 6 and 7) and the polymerase protein (L; fragments 8, 9 and 10) (Fig. 2). We can exclude the possibility of contamination of the human samples with APV/TRTV as we do not have this virus in our laboratories. We next designed PCR primers to complete the sequence information for the 3′ end of the viral genome based on our RAP-PCR fragments as well as published leader and trailer sequences for the Pneumovirinae7 (Fig. 2). In total, we have generated 5.7 kilobases of sequence information of the viral genome. Analyses of the sequences of these fragments revealed the absence of NS1 and NS2 open reading frames (ORFs) at the extreme 3′ end of the viral genome (Fig. 3a) and positioning of the putative F ORF immediately adjacent to the putative M ORF (Fig. 3b). This genomic organization resembles that of the metapneumovirus APV, which is consistent with the sequence homology. It should be noted however that the genomic organization of hMPV cannot be simply deduced from that of APV, and requires further analysis.
Figure 2. Schematic representation of genomic fragments obtained from hMPV.
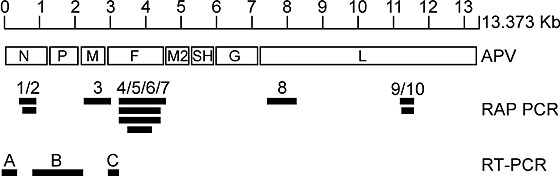
A schematic diagram of APV (3′ to 5′ end, left to right) is shown above the fragments obtained with RAP-PCR and RT-PCR on virus isolate 00-1. Fragments 1–10 were obtained using RAP-PCR. Fragment A was obtained with a primer in RAP-PCR fragments 1 and 2 and a primer designed on basis of alignment of leader and trailer sequences of APV and RSV (ref. 7). Fragment B was obtained with primers designed in RAP-PCR fragments 1 and 2 and RAP-PCR fragment 3. Fragment C was obtained with primers designed in RAP-PCR fragment 3 and RAP-PCR fragments 4–7.
Figure 3. Nucleotide and deduced amino-acid sequences for selected regions of the hMPV genome.

a and b, The nucleotide sequences for the 3′ end of the viral genome and the intergenic region between M and F ORFs. Note that the underlined sequence in a refers to the primer used for PCR amplification, and therefore does not necessarily reflect the actual hMPV leader sequence. c–f Comparison of the amino acid sequences of the putative N (panel c), P (panel d), M (panel e) and F (panel f) ORFs of hMPV, aligned with those of APV and RSV. Residues that differ between isolate 00-1 and the other viruses are shown, identical amino acids are represented by periods, gaps are represented by dashes.
The translated sequences for the N, P, M and F ORFs were aligned with the amino-acid sequences of hRSV and APV (Fig. 3c–f). For N, these alignments revealed a 20% homology with hRSV and 52% homology with APV. The amino-acid sequences for P revealed 25% homology with hRSV and 67% with APV; for M, 37% with hRSV and 87% with APV; and for F, 32% with hRSV and 80% homology with APV.
Phylogeny
As an indicator for the relationship between the newly identified virus isolates and members of the Pneumovirinae, phylogenetic trees were constructed based on the N, P, M and F ORFs of these viruses. In all four phylogenetic trees, the newly identified virus isolate was most closely related to APV (Fig. 4). From the four serotypes of APV that have been described14,15, APV serotype C, the metapneumovirus found primarily in birds of the United States, showed the closest resemblance to the newly identified virus; however, only limited sequence information for APV serotype D is available.
Figure 4. Phylogenetic analysis of ORFs of hMPV and selected pneumoviruses.
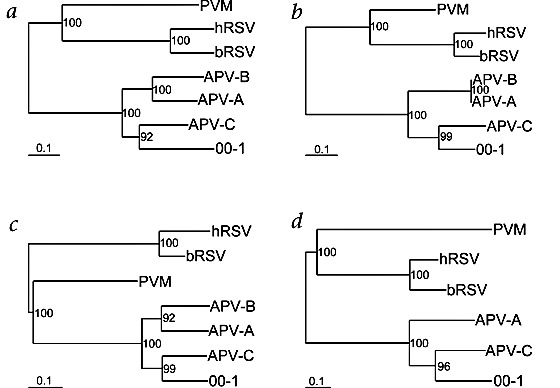
a–d, The F (panel a), N (panel b), M (panel c), and P (panel d) ORFs of hMPV isolate 00-1 were aligned with those of other members of the genus Pneumovirinae, and phylogenetic trees were generated by maximum likelihood analyses using 100 bootstraps and 3 jumbles. The scale representing the number of nucleotide changes is shown for each tree. Bootstrap values are based on the consensus trees.
Experimental infection of birds and cynomolgus macaques
Since hMPV appeared to be closely related to APV, we wished to test whether hMPV is a primary human pathogen or an avian pathogen that can also infect humans. We inoculated four juvenile turkeys, four juvenile chickens and four juvenile cynomolgus macaques with 50,000 TCID50 of hMPV on their conjunctivae and in their respiratory tracts. We collected cloaca and/or throat samples and tested them for the presence of hMPV by RT-PCR. During the three weeks follow-up, none of the birds showed clinical signs or virus replication as determined by RT-PCR using RNA isolated from the throat and cloaca swabs. In contrast, the virus replicated efficiently in the respiratory tract of all four monkeys as shown by RT-PCR of RNA isolated from the throat swabs. Whereas viral RNA was not detectable in samples taken at day 1, virus replication peaked between days 2 and 8 after inoculation. Two of the monkeys presented with mild upper respiratory tract signs that upon histological analysis proved to be associated with suppurative rhinitis (data not shown). Although the detailed pathological, virological and immunological analyses are incomplete at present, these preliminary data indicate that hMPV is a primate pathogen associated with respiratory disease.
Genetic heterogeneity of hMPV isolates
We next amplified small parts of the N, M, F and L ORFs (71, 143, 142 and 102 nucleotides respectively) for 9 of the 28 virus isolates by RT-PCR, then sequenced these fragments directly to minimize the possibility of errors introduced by the amplification procedure. Phylogenetic analyses revealed significant sequence variation and the existence of two potential genetic clusters of hMPV isolates obtained in the Netherlands (Fig. 5). The nucleotide sequence identity between isolates within a cluster was 90–100% for all four fragments, whereas the sequence identity for isolates between the two clusters was 81–88%. Variation was similar for the N, M, F and L fragments and did not seem to correlate with the year of virus isolation (indicated by the first two numbers in the name of the isolates). These genetic clusters might represent subgroups of the newly identified virus in analogy to other pneumoviruses such as RSV.
Figure 5. Phylogenetic analysis of 9 hMPV isolates.
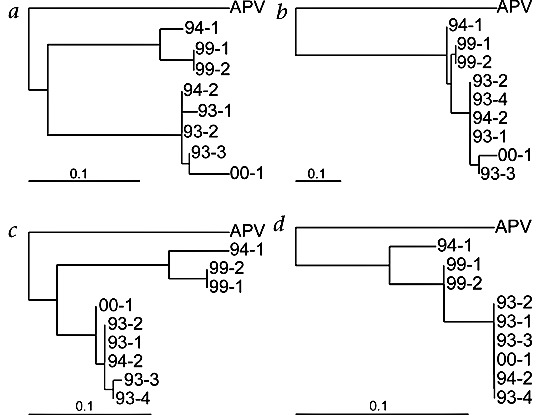
a–d, Parts of the F (panel a), N (panel b), M (panel c) and L (panel d) ORFs of the indicated hMPV isolates were amplified by PCR and sequenced directly (fragments of 142, 71, 143, and 102 nucleotides, respectively). Phylogenetic trees were generated as described in Fig. 4. The first 2 numbers in the isolate names refer to the year from which the isolate originated. We were unable to amplify the F fragment for isolate 93-4.
Seroprevalence
To study the prevalence of this virus in the human population, we tested sera from humans in different age categories by indirect IFA using tMK cells infected with one of the unidentified virus isolates. This analysis revealed that 25% of the children aged between 6 and 12 months had antibodies to the virus, and towards the age of 5 virtually all of the children had become seropositive (Table 1). In addition, 56 serum samples tested by indirect IFA were tested by virus neutralization assays. For 51 (91%) of the samples, the results of the virus neutralization assay (titer > 8) correlated with the results obtained with indirect IFA (titer > 32). Four samples that were positive in IFA (titer > 32) were negative by virus neutralization tests (titer < 8), and one serum sample that reacted negative in IFA (titer < 32) was positive in the virus neutralization test (titer > 16).
Table 1.
Seroprevalence of hMPV antibodies in humans categorized by age group
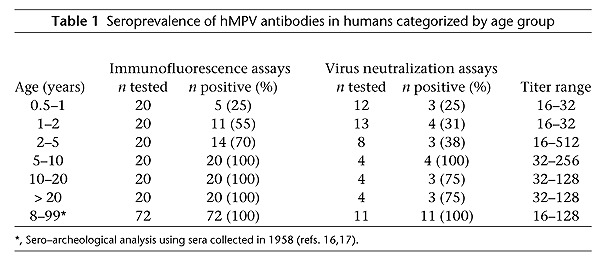
Indirect IFA conducted with 72 sera taken from humans in 1958 (aged 8–99 years)16,17 revealed a 100% seroprevalence, indicating that the virus has been circulating in the human population for more than 43 years. In addition, testing of a number of these sera in virus neutralization assays confirmed the IFA data (Table 1).
The range of antibody titers appeared higher for individuals older than 2 years as compared to children between 6 and 24 months. This observation could be related to boosting of antibody responses as a consequence of reinfection with the same or a closely related virus. However, more detailed serological and virological analyses will be required to substantiate this speculation.
Discussion
Here we describe the characterization of a new member in the Paramyxoviridae family, isolated from 28 children with RTI in the Netherlands. The children presented with clinical symptoms reminiscent of those caused by hRSV infection, ranging from mild upper respiratory tract disease to severe bronchiolitis and pneumonia. As is seen with hRSV-infected children, very young hMPV-infected children appeared to require hospitalization and mechanical ventilation. Whether hMPV infection causes RTI cannot be formally concluded from the present data. Most importantly, acute and convalescent paired sera were not available for the children from whom hMPV was isolated. However, considering that no other virus was identified in any of the 28 hMPV-infected children and that hMPV was not isolated from 400 samples taken from children under two years of age without respiratory symptoms (not shown), a causal relationship seems likely. The experimental infection of cynomolgus macaques supports this conclusion.
Based on the data here we propose the name human metapneumovirus (hMPV). hMPV proved to have a paramyxovirus-like morphology in electron microscopy, to be chloroform-sensitive and to replicate optimally in a trypsin-dependent manner in tMK cells. The clinical symptoms of the hMPV-infected children, the characteristic cytopathic effects and the lack of haemagglutinating activity further suggested that this virus might be related to hRSV; although most paramyxoviruses have hemagglutinating activity, most of the pneumoviruses do not18. Genetic analyses of the putative N, M, P and F genes revealed that hMPV showed a higher sequence homology to the recently proposed genus Metapneumovirus (average of 66%) as compared to the genus Pneumovirus (average of 30%) and a genomic organization that is most similar to APV/TRTV. In contrast to the genomic organization of the pneumoviruses (3′-NS1-NS2-N-P-M-SH-G-F-M2-L-5′), metapneumoviruses lack NS1 and NS2 genes and have a different positioning of the genes between M and L (3'-N-P-M-F-M2-SH-G-L-5′). The absence of ORFs between the M and F genes in this virus and the lack of NS1 and NS2 genes adjacent to N is in agreement with it being the first identified non-avian member of the Metapneumovirus genus. However, until sequence information becomes available for the entire genomic organization of hMPV, a definitive classification of this virus is not possible.
Of the four serotypes of APV that have been described, serotype C was most closely related to hMPV based on the N, P, M and F genes. However, for serotype D only limited sequence information for the F gene is available from Genbank and for serotype B, only M, N and F sequences are available. For both hRSV and APV, different genetic and serological subtypes have been described. We also found evidence for genetic variation between hMPV isolates. Although it is tempting to speculate that these subgroups of hMPV isolates represent different serotypes of hMPV, sequence analyses of other parts of the hMPV genome and of isolates obtained from different geographical locations as well as detailed serological studies will be needed to support this hypothesis.
Our serological survey revealed that hMPV commonly infects children. The repeated isolation of this virus from clinical samples from children with RTI indicates that hMPV infection may have considerable clinical and economical impact. New diagnostic assays based on virus detection and serology should allow a more detailed analysis of the incidence and both clinical and economic impact of this newly identified human virus. To get an idea of the potential impact of this virus for RTI we have tested nasopharyngeal aspirate samples collected during the 2000 winter season from children presenting with RTI, at Erasmus University Medical Center that were negative for RSV, influenza A and B viruses, parainfluenza virus types 1–3, human rhinovirus and Corona virus (HCV-229E and HCV-OC43). Seven of the 68 samples tested proved to be positive for hMPV by RT-PCR, indicating that for the 2000 winter season approximately 10% of the unexplained RTI may be associated with hMPV.
The minor differences between our IFA and virus neutralization results (5 samples) may be due to several factors; only IgG serum antibodies are detected in IFA, virus neutralization antibodies are generally directed against surface glycoproteins, the sensitivities of the two assays may differ and different serotypes of hMPV may exist. The latter suggestion is of particular interest, since only one isolate (00-1) was used for IFA and virus neutralization, potentially giving rise to false negative results using sera raised against genetically distinct hMPV isolates such as those shown in Fig. 5.
As hMPV seems most closely related to APV, we speculate that the human virus has originated from birds. Analysis of serum samples taken from humans in 1958 revealed that hMPV has been widespread in the human population for at least half a century, indicating that a possible zoonotic event must have taken place before 1958.
The question then arises: Why has this virus remained unidentified for so many years? First of all, many virology laboratories use continuous cell lines for virus isolation, in which hMPV does not seem to replicate efficiently. Second, hMPV displays very slow replication kinetics in vitro, and may therefore have remained undetected. Third, replication of hMPV in-vitro appeared to be trypsin dependent and many of the earlier studies to find respiratory pathogens did not use trypsin. Finally, the apparent lack of cross-reactivity in the serologic assays we performed and the low nucleotide sequence homology to known human viral pathogens make unlikely the detection of hMPV with standard serological or molecular techniques. Pringle and Eglin have described serological evidence for an unidentified 'PVM-like' pneumovirus that frequently infects humans19. We feel it is unlikely that hMPV is the PVM-like-virus described by Pringle and Eglin for a number of reasons. First, high-titer sera specific for several paramyxoviruses, including PVM and RSV, failed to react in immunofluorescence studies with hMPV-infected tMK cells as targets (data not shown). Second, most of our hMPV–positive human serum samples did not have PVM-specific antibodies as measured by ELISA. Furthermore, PVM (and supposedly a PVM-like virus) has hemagglutinating activity, whereas we have no evidence that hMPV does. Finally, the genomic organization of PVM is similar to that of RSV and thus different from that of hMPV.
The identification of this tentative new member of the Metapneumovirus genus may aid in the development of diagnostic assays, vaccines and antiviral agents for viral RTIs.
Methods
Virus isolation.
Over the past decades our virus diagnostic hospital-based laboratory has collected nasopharyngeal aspirates from children suffering from RTI. These samples were routinely tested for the presence of viruses by direct immune fluorescence assays (DIF) using antibodies against viruses known to cause RTI (hPIV types 1–3, hRSV, influenza virus types A and B) and by virus isolation using rapid shell-vial techniques on tMK cells and CEF, as well as various cell lines such as Vero, A549 and MDCK cells20,21. Samples showing cytopathic effects after 2 or 3 passages that were negative in DIF were tested by indirect IFA using virus-specific antibodies against influenza virus types A, B and C, hRSV, hPIV types 1–4, measles virus, mumps virus, Sendai virus, SV-5, and NDV. Although for many cases the etiological agent could be identified, some specimens yielded cytopathic effects in cell cultures but were negative for all viruses tested.
Animal immunization.
To raise virus-specific antisera, 1.0 ml volumes of virus-infected tMK cell-culture supernatants of 2 different virus isolates containing approximately 1 × 104 TCID50/ml were used to inoculate guinea pigs and ferrets intranasally without prior purification. Samples were taken at day 0 and at day 14 or 28 after infection, the animals were bled by cardiac puncture and their pre- and post-infection sera were used as negative and positive reference sera respectively. The sera were tested for reactivity against all previously described viruses with DIF and IFA as described above.
Virus characterization.
We cultured the virus isolates in tMK cells in Eagle's MEM with Hanks' salt, supplemented with 0.02% trypsin and without serum. After inoculation, the plates were incubated at 37 °C for a maximum of 14 d during which time the medium was changed once a week and cultures were checked daily for cytopathic effects. Because the degree of cytopathic effects differed slightly for each isolate, all cultures were tested at days 10–14 with indirect IFA using ferret antisera raised against one of the isolates. For electron microscopy analyses, virus was concentrated from infected cell-culture supernatants in a micro-centrifuge at 4 °C at 17,000g, after which the pellet was resuspended in PBS and after potassium tungstat acid (PTA) staining, inspected by negative contrast electron microscopy22. Standard hemagglutination assays, chloroform sensitivity tests, DIF and IFA were performed as described11,20,21,23. Virus neutralization assays were performed on tMK cells with serial 2-fold dilutions of human or animal sera (starting at 1:8) and with 100 TCID50 of hMPV, following procedures as described23. Since cytopathic effects were not always easy to detect, IFA was used to determine the virus neutralization titer. The virus neutralization titer was defined as the reciprocal of the highest dilution of the serum sample resulting in negative IFA and inhibition of cytopathic effects in cell cultures.
RAP-PCR and RT-PCR.
RNA was isolated from the supernatant of infected cell cultures or sucrose gradient fractions using a High Pure RNA Isolating kit according to instructions from the manufacturer (Roche Diagnostics, Almere, the Netherlands). RT-PCR assays were performed according to established techniques. Primers specific for the panel of paramyxoviruses tested and conditions are available upon request. For RAP-PCR, virus was concentrated from infected tMK-cell supernatants by ultracentrifugation on a 20–60% sucrose gradient. Gradient fractions were inspected for the presence of virus-like particles by electron microscopy and poly-acrylamide gel electrophoresis followed by silver staining. The approximately 50% sucrose fractions that appeared to contain nucleocapsids were used for RNA isolation and RAP-PCR. RAP-PCR was performed essentially as described12,13 and primer sequences used are available upon request. Differentially displayed fragments specific for hMPV were purified from the gel with a Qiaquick Gel Extraction kit (Qiagen, Leusden, the Netherlands) and cloned in vector pCR2.1 (Invitrogen, Groningen, the Netherlands) according to instructions from the manufacturer.
Sequence analysis.
RAP-PCR products cloned in vector pCR2.1 were sequenced with M13-specific oligonucleotides. DNA fragments obtained by RT-PCR were purified from agarose gels using a Qiaquick Gel Extraction kit (Qiagen, Leusden, the Netherlands) and sequenced directly with the same oligonucleotides used for RT-PCR assays. Sequence analyses were performed using a Dyenamic ET terminator sequencing kit (Amersham Pharmacia Biotech, Roosendaal, the Netherlands) and an ABI 373 automatic DNA sequencer (PE Applied Biosystem, Nieuwerkerk aan den TJssel, the Netherlands), according to the instructions of the manufacturer.
Phylogenetic analyses.
For all phylogenetic trees, DNA sequences were aligned using the ClustalW software package and maximum likelihood trees were generated using the DNA-ML software package of the Phylip 3.5 program using 100 bootstraps and 3 jumbles. Bootstrap values were computed for consensus trees created with the consense package24. hMPV sequences are available from Genbank under accession numbers AF371330 through AF371367. All other sequences used here are available from Genbank under accession numbers NC001781 (hRSV all ORFs); NC001989 (bRSV all ORFs); D10331 (PVM N);U39295 (APV-A N); U39296 (APV-B N); AF176590 (APV-C N); U09649 (PVM P); U22110 (APV-A P); AF176591 (APV-C P); U66893 (PVM M); X58639 (APV-A M); U37586 (APV-B M; AF262571 (APV-C M); D11128 (PVM F); D00850 (APV-A F); Y14292 (APV-B F); AF187152 (APV-C F).
Acknowledgements
We thank M.H. Malim and R. Oakey for providing oligonucleotides for and stimulating discussions on RAP-PCR and J. Habova, T. Bestebroer, B. Niemeyer, G. Aaron and C. Burghoorn for excellent technical assistance. R.F. is a fellow of the Royal Dutch Academy of Arts and Sciences.
References
- 1.Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses (eds. van Regenmortel, M.H., Fauquet, C.M. & Bishop, D.H.) 551, 557, 559–560 (Academic, San Diego, 2000).
- 2.Domachowske JB, Rosenberg HF. Respiratory syncytial virus infection: immune response, immunopathogenesis, and treatment. Clin. Microbiol. Rev. 1999;12:298–309. doi: 10.1128/CMR.12.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selwyn BJ. The epidemiology of acute respiratory tract infection in young children: comparison of findings from several developing countries. Rev. Infect. Dis. 1990;12:S870–888. doi: 10.1093/clinids/12.Supplement_S870. [DOI] [PubMed] [Google Scholar]
- 4.Giraud P, Bennejean G, Guittet M, Toquin D. Turkey rhinotracheitis in France: preliminary investigations on a ciliostatic virus. Vet. Rec. 1986;119:606–607. [PubMed] [Google Scholar]
- 5.Cook JK. Avian rhinotracheitis. Rev. Sci. Tech. 2000;19:602–613. doi: 10.20506/rst.19.2.1233. [DOI] [PubMed] [Google Scholar]
- 6.Ling R, Easton AJ, Pringle CR. Sequence analysis of the 22K, SH and G genes of turkey rhinotracheitis virus and their intergenic regions reveals a gene order different from that of other pneumoviruses. J. Gen. Virol. 1992;73:1709–1715. doi: 10.1099/0022-1317-73-7-1709. [DOI] [PubMed] [Google Scholar]
- 7.Randhawa JS, Marriott AC, Pringle CR, Easton AJ. Rescue of synthetic minireplicons establishes the absence of the NS1 and NS2 genes from avian pneumovirus. J. Virol. 1997;71:9849–9854. doi: 10.1128/jvi.71.12.9849-9854.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Q, Davis PJ, Li J, Cavanagh D. Cloning and sequencing of the matrix protein (M) gene of turkey rhinotracheitis virus reveal a gene order different from that of respiratory syncytial virus. Virology. 1992;186:426–434. doi: 10.1016/0042-6822(92)90007-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans AS. Viral Infections of Humans. Epidemiology and Control. 1989. Epidemiologic concepts and methods; pp. 22–28. [Google Scholar]
- 10.Lamb RA, Kolakofsky D. Virology. 1996. Paramyxoviridae: the viruses and their replication; pp. 1177–1204. [Google Scholar]
- 11.Osterhaus AD, et al. The isolation and partial characterization of a highly pathogenic herpesvirus from the harbor seal (Phoca vitulina) Arch. Virol. 1985;86:239–251. doi: 10.1007/BF01309828. [DOI] [PubMed] [Google Scholar]
- 12.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ralph D, McClelland M, Welsh J. RNA fingerprinting using arbitrarily primed PCR identifies differentially regulated RNAs in mink lung (Mv1Lu) cells growth arrested by transforming growth factor beta 1. Proc. Natl. Acad. Sci. USA. 1993;90:10710–10714. doi: 10.1073/pnas.90.22.10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayon-Auboyer MH, Arnauld C, Toquin D, Eterradossi N. Nucleotide sequences of the F, L and G protein genes of two non-A/non-B avian pneumoviruses (APV) reveal a novel APV subgroup. J. Gen. Virol. 2000;81:2723–2733. doi: 10.1099/0022-1317-81-11-2723. [DOI] [PubMed] [Google Scholar]
- 15.Cook JK, Huggins MB, Orbell SJ, Senne DA. Preliminary antigenic characterization of an avian pneumovirus isolated from commercial turkeys in Colorado, USA. Avian Pathology. 1999;28:607–617. doi: 10.1080/03079459994407. [DOI] [PubMed] [Google Scholar]
- 16.Mulder J, Masurel N. Pre-epidemic antibody against 1957 strain of asiatic influenza in serum of older people living in the Netherlands. Lancet. 1958;1:810–814. doi: 10.1016/S0140-6736(58)91738-0. [DOI] [PubMed] [Google Scholar]
- 17.Masurel N. Relation between Hong Kong virus and former A2 isolates and the A/EQU12 virus in human sera collected before 1957. Lancet. 1969;1:907–910. doi: 10.1016/S0140-6736(69)92544-6. [DOI] [PubMed] [Google Scholar]
- 18.Pringle CR. The Paramyxoviruses. 1991. The genetics of paramyxoviruses; pp. 1–39. [Google Scholar]
- 19.Pringle CR, Eglin RP. Murine pneumonia virus: seroepidemiological evidence of widespread human infection. J. Gen. Virol. 1986;67:975–982. doi: 10.1099/0022-1317-67-6-975. [DOI] [PubMed] [Google Scholar]
- 20.Brandenburg AH, et al. Respiratory syncytial virus-specific serum antibodies in infants under six months of age: limited serological response upon infection. J. Med. Virol. 1997;52:97–104. doi: 10.1002/(SICI)1096-9071(199705)52:1<97::AID-JMV16>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 21.Rothbarth PH, Groen J, Bohnen AM, de Groot R, Osterhaus AD. Influenza virus serology—a comparative study. J. Virol. Methods. 1999;78:163–169. doi: 10.1016/S0166-0934(98)00174-8. [DOI] [PubMed] [Google Scholar]
- 22.Madeley CR, Field AM. Virus Morphology. 1988. [Google Scholar]
- 23.Lennette DA. Diagnostic Procedures for Viral, Ricketssial and Chlamydial Infections. 1995. [Google Scholar]
- 24.Felsenstein J. PHYLIP - Phylogeny Inference Package. Cladistics. 1989;5:164–166. [Google Scholar]


