Abstract
Over the past decade, microbiology and infectious disease research have undergone the most profound revolution since the times of Pasteur. Genomic sequencing has revealed the much-awaited blueprint of most pathogens. Screening blood for the nucleic acids of infectious agents has blunted the spread of pathogens by transfusion, the field of antiviral therapeutics has exploded and technologies for the development of novel and safer vaccines have become available. The quantum jump in our ability to detect, prevent and treat infectious diseases resulting from improved technologies and genomics was moderated during this period by the greatest emergence of new infectious agents ever recorded and a worrisome increase in resistance to existing therapies. Dozens of new infectious diseases are expected to emerge in the coming decades. Controlling these diseases will require a better understanding of the worldwide threat and economic burden of infectious diseases and a global agenda.
Main
Introduction
Human populations have been shaped by continuous interactions with the world of microbes. Textbooks document the plague of Athens described by Thucydides in the fifth century BC, the European bubonic plague of 1347 that killed one-third of the human population and represents the largest epidemic ever recorded, and measles and smallpox viruses that were introduced to the American continent by Hernán Cortes, which contributed to the disappearance of the Aztec civilization. Thousands of other examples can be found in the records, accounting for hundreds of millions of deaths, changes in the course of history and selective expansion of genetic diseases such as the thalassemias, in which the defective hemoglobins provided increased resistance to endemic malaria1. In the twentieth century, clean water, vaccination and antimicrobial therapies brought most infectious diseases under control, marking one of the greatest achievements of civilization. At the end of the century, however, infectious agents started to retaliate and we have seen the emergence of pathogens that are resistant to antimicrobials and of new pathogens that had not been previously detected in humans. This review will focus on the major changes in infectious diseases and the scientific progress in the field during the last decade.
The diseases
An overview of the principal infectious diseases of the last decade shows that they can be divided into three major groups: those against which significant progress was made during this period, those that newly emerged and those on which we had no impact (Fig. 1 and Box 1).
Figure 1. Major changes in infectious diseases during the last decade: the diseases that have been decreasing are shown on the left; the ones that emerged are in the center; those that continued without any change to their trend are shown on the right.
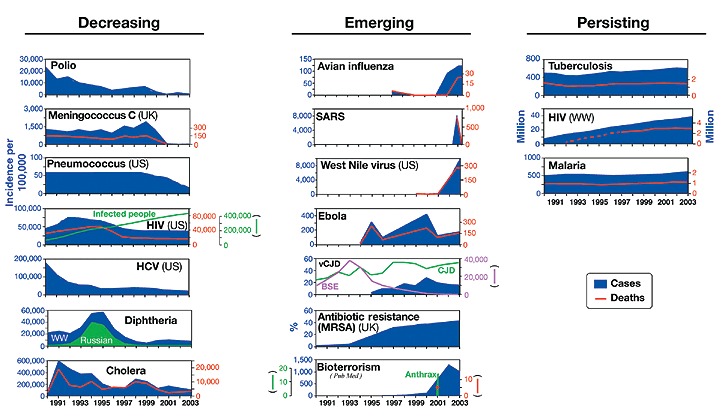
WW represents wordwide. Websites where information for each disease is available are reported in Box 1. In HIV USpanel, new cases per year are shown in blue and total infected population is shown in green. In the bioterrorism panel, publications in PubMed are shown in blue, cases in green and deaths in red.
Considerable progress during the last decade was made against human immunodeficiency virus (HIV) and hepatitis C virus (HCV) in developed countries, poliomyelitis, and meningococcal and pneumococcal disease. The campaign for the eradication of poliomyelitis, involving vaccination of more than two billion children, reduced the global incidence of polio by 99.9% from 350,000 cases per year to less than 800 during the period from 1998 to 2002 (Fig. 1)2, and today we believe that the disease will be eradicated from the world within the next few years.
Similarly, in the United States, the introduction of conjugate vaccines against seven serotypes of pneumococcus decreased the incidence of invasive pneumococcal disease from 60 to less than 20 cases per 100,000 people3, while the proof of concept of conjugate vaccines against meningococcus in early 1990s4 made possible the introduction of the conjugate vaccine against meningococcus C in the United Kingdom in 2000, virtually eliminating the disease5,6. Global elimination of bacterial meningitis may now be an achievable target when vaccines against meningococcus ACYW and B become available within the next decade. Significant progress during this period was also achieved for the control of HCV and HIV in developed countries. In 1987, the cloning of the hepatitis C genome7 enabled the development of diagnostic methods to test blood and blood derivatives, which in the early 1990s resulted in the reduction of new HCV cases from >130,000 per year to 25,000 per year in the United States alone (Fig. 1), while the introduction of a cocktail of drugs (highly active antiretroviral therapy, or HAART) as a standard of care for people with HIV8 transformed HIV in the Western world from a deadly disease into a manageable chronic disease. It is also worth mentioning that diphtheria9 and cholera10, two diseases that had re-emerged at the beginning of the 1990s have been controlled during this period (Fig. 1).
Although control was achieved for some infections, an unprecedented number of infectious diseases emerged during this period. These include avian influenza, severe acute respiratory syndrome (SARS), West Nile, Ebola and variant Creutzfeldt-Jakob disease (vCJD). The last decade also saw an increase in the prevalence of antibiotic-resistant bacteria, and the re-emergence of previously eradicated pathogens as agents of bioterror (Fig. 1). Avian influenza was reported to infect humans seven times during the last seven years—the last outbreak before this period was in 1968. The virus first reappeared in Hong Kong in 1997 as an H5N1 strain, infecting 18 humans and causing 6 deaths. Two years later, again in Hong Kong, a new avian virus, H9N2, infected two people: the same virus strain caused three infections and one death in 2003. In 2004, we witnessed an explosion of new cases, with 43 cases and 31 deaths reported in a series of independent outbreaks in Thailand and Vietnam. So far, the outbreaks have been controlled by culling more than 100 million fowl11, but the multiple, independent outbreaks of the last two years suggest the proximity of a new flu pandemic12,13. The last decade also saw the emergence of other new diseases caused by transmission of pathogens from animals to humans. The SARS coronavirus, which in the short period between February and July 2003 infected 8,098 people and caused 774 deaths14 (Fig. 1), was likely to have been transmitted by increased contact between humans and virus-carrying wild animals that are sold for food in Asian markets14. Although the SARS outbreak was quickly resolved, it generated widespread panic, paralyzed travel and threatened the global economy.
Other diseases of animal origin that emerged over the past decade were Ebola hemorrhagic fever, which, after several independent instances of virus transmission from nonhuman primates to hunters15, caused a total of 264 deaths in Zaire, the Republic of Congo and Gabon, and the variant form of Creutzfeldt-Jakob Disease (vCJD), a new type of lethal neurodegenerative disease affecting young people, which had been previously very rare and during this decade caused 143 cases (Fig. 1). The disease was transmitted to humans through consumption of the meat of cows infected by the bovine spongiform encephalitis (BSE) prion. West Nile virus, a flavivirus described for the first time in 1937 in Africa and endemic in Europe and central Asia, was reported for the first time in New York in 1999. Most likely air travel was responsible for the virus crossing continents. The virus infects birds and is transmitted from birds to mammals by the urban mosquito Culex pipiens—within four years the virus spread across most of the Unites States, infecting 9,862 people and causing 264 deaths in 2003 (Fig. 1).
Another worrisome trend over the past 10 years has been the increased resistance of bacteria to multiple antibiotics16,17. In Figure 1, methicillin-resistant Staphylococcus aureus in the United Kingdom is used as an example for this global trend, which represents a serious threat to hospitalized patients. In addition to methicillin-resistant S. aureus, multiple antibiotic resistance is a problem also for Streptococcus pneumoniae, Enterococcus faecalis, Pseudomonas aeruginosa and Mycobacterium tuberculosis18,19.
Finally, humans have also propagated the use of microbial pathogens for bioterrorism. A few days after the September 11th terrorist attack in New York, an anthrax infection ignited by the deliberate release of bacterial spores was reported in Florida. Although the powdered spores, which were delivered by mail, caused only a total of 18 infections and 5 deaths20, they changed forever the public perception of microbial pathogens. Scientific publications on bioterrorism increased to more than 1,000 per year (Fig. 1). Smallpox vaccination was started again for the first time after it had been stopped following the eradication of the virus in 1977. Finally, the fear of biological weapons capable of mass destruction was a motivation for beginning the war in Iraq in 2003.
Unfortunately, as shown in Figure 1, during the last decade, we had no impact on the three diseases which alone contribute to half of the global burden of infectious disease mortality: tuberculosis, malaria and HIV (worldwide). Indeed, we have only seen a worsening of tuberculosis and malaria in the immunocompromised population infected by HIV.
While the events summarized in Figure 1 were happening in the field, a parallel revolution was in progress in microbiology laboratories.
The genomic revolution and cellular microbiology
The sequence of the first bacterial genome in 1995 (ref. 21) and the subsequent genomic revolution represent the largest changes in microbiology since the times of Pasteur. In less than a decade, genomics has provided the blueprints of microorganisms and the ability to explore, in a culture-independent manner, the global diversity of living organisms. Today, the availability in public databases of more than 190 bacterial genomes, 1,600 viral genomes and the malaria parasite sequence is incredible progress, considering that fewer than ten years ago microbiologists spent most of their research efforts in cloning and sequencing one gene at a time. In addition to providing information about every gene of known pathogens, genome sequencing facilitated research on 'old' infectious agents difficult or impossible to grow in vitro, such as HCV or the bacillus causing Whipple disease22,23. For instance, HCV, which cannot be grown in vitro and has never been seen by electron microscopy, is impossible to study using the basic technologies of conventional microbiology. But the cloning of the genome in 1987 (ref. 7) allowed the development of diagnostic methods to test blood and blood derivatives, which in the early 1990s virtually eliminated new transmissions of HCV by transfusion (Fig. 1). In addition, the genomic sequence provided information on the proteins needed to make the virus, allowing the expression of recombinant viral proteins that have been used to develop prophylactic and therapeutic vaccines or to test antiviral compounds that are now in clinical trials24.
During the SARS outbreak, the genome sequence became available in less than a month from the identification of the virus, immediately enabling the development of nucleic acid tests for the detection of the virus, the optimization of measures to contain the spread, the understanding of the probable animal origin and the design of therapeutics, monoclonal antibodies and several effective vaccines that are moving toward clinical trials25,26. The SARS example shows how emerging pathogens can be identified, sequenced and classified in real time, confirming that modern technologies can be very effective in handling unknown emerging infections.
Another example of a pathogen that had escaped detection by conventional microbiology is human metapneumovirus. Isolated for the first time in 2001 from the nasopharyngeal aspirates of children in the Netherlands, today this negative RNA virus of the paramyxoviridae family is a recognized cause of acute respiratory infections and considered to be responsible for morbidity and mortality worldwide. Together with respiratory syncytial virus (RSV) it is believed to cause a large fraction of severe acute respiratory tract infections in infant, elderly and immunocompromised hosts27. Although metapneumovirus can be handled by conventional technologies, the virus had probably been missed because it is rather difficult to grow in conventional cell cultures—molecular technologies, such as gene hybridization and sequencing, were instrumental in facilitating its identification.
In addition to better detection, the new genome-based technologies (complemented by great advances in confocal microscopy, fluorescent proteins, total body imaging, microarrays and a number of animal models supported by genetic techniques such as signature-tagged mutagenesis and in vivo expression technology28) have enabled the study of pathogens while they interact with their hosts. This is such a novel approach to microbial pathogenesis that a new discipline, 'cellular microbiology'29, has been created to differentiate modern microbiology, which studies pathogens interacting with their hosts in their native environments, from classical microbiology, which studies pathogens grown in rich media under artificial conditions in the laboratory. These new approaches have contributed to the discovery of virulence factors not necessary to pathogens grown in the laboratory but essential or preferentially expressed only in vivo during infection. Perhaps the most intriguing among these are the bacterial Type III and Type IV secretion systems, molecular syringes that inject proteins into host cells to facilitate infection and survival within the host30,31.
Similarly, the human genome and the clustering of the human populations using genetic tools, such as the single nucleotide polymorphisms32, are expected to provide the genetic basis for the susceptibility to infectious diseases and the pathologies deriving from infections. We expect to be able to predict who will be most likely to succumb to or resist infections, or respond to a therapy or vaccination. Preliminary examples of what may become routine in the future are the use of gene expression profiles to study the genes which are upregulated during clinical leprosy and define the clinical forms of the diseases33 and the identification of a gene expression signature, which defines a successful vaccination and protection from infection from the Helicobacter pylori pathogen34.
More and better vaccines
For over a century, vaccines were developed according to Pasteur's principles of isolating, inactivating and injecting the causative microorganisms. These principles provided the killed, live attenuated and subunit vaccines in use today (Fig. 2) and are responsible for a large part of the control of infectious diseases achieved to date. But Pasteur's principles did not allow the development of vaccines against those microorganisms that cannot be cultivated in vitro such as HCV, papillomavirus types 16 and 18 and M. leprae. Also, the principles of Pasteur did not provide vaccines for antigenically hypervariable microorganisms such as serogroup B meningococcus, gonococcus, malaria and HIV, and did not teach how to induce cytotoxic T cells, which kill infected host cells and therefore contribute to the control of viral replication. During the last decade, we have overcome most of these technical limitations, even if we are still unable to use them effectively enough to solve challenges such as HIV. The availability of genomes allowed the identification of novel vaccine candidates without the need to cultivate microorganisms, a process named 'reverse vaccinology'35. In this approach, the genomic sequences of viral, bacterial or parasitic pathogens are used to select by computer analysis, microarrays, proteomics and other genome-based systematic approaches those antigens that are likely to confer protective immunity and eliminate potentially dangerous antigens, such as those showing homology to human proteins, which are therefore potentially able to induce autoimmunity. The predicted antigens are then expressed by recombinant DNA and tested in an animal model. This genome-based vaccine approach allowed the development of vaccines against HCV and human papillomavirus 16 and 18, which, being unable to grow in vitro, were not approachable following the principles of Pasteur. But once the genome sequence became available, it was used to predict the antigens coding for the surface proteins (E1 and E2 for HCV and VP L1 for papillomavirus), express them and develop vaccines that are presently in phase 1 and 3 clinical trials, respectively24,36.
Figure 2.
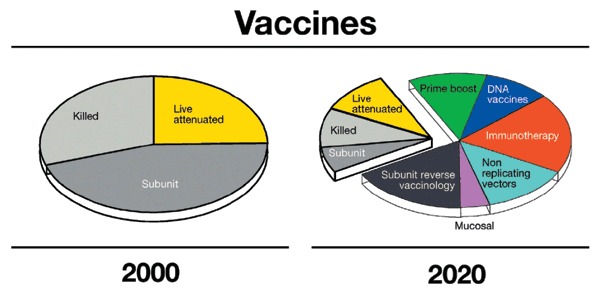
Type of vaccines licensed in the year 2000 and type of vaccines predicted to be available in the year 2020.
The most quoted example for reverse vaccinology is meningococcus B. At the end of the 1990s it was apparent that a universal meningococcal vaccine was beyond the reach of conventional vaccinology for two reasons: first, the polysaccharide antigen used successfully to make conjugate vaccines against other meningococci was not immunogenic (and, indeed, was a potential trigger of autoimmunity, being identical to a self antigen), and second, the best protein-based vaccines induced immunity against antigenically variable proteins and had been shown to induce protective immunity mostly against the strain used to make the vaccine, without substantial crossprotection. In this case, the sequence of the bacterial genome allowed the computer prediction of approximately 600 novel vaccine candidates, 350 of which were expressed in Escherichia coli and tested for their ability to elicit protective immunity. Remarkably, the approach identified 29 novel protective antigens, some of which are conserved in all meningococcal strains and are now being tested in clinical trials. The meningococcus B example has been followed for many other bacterial pathogens, including pneumococcus, group B streptococcus and chlamydia37.
Although most vaccines available today work by inducing antibodies, it is believed that conquering the most difficult diseases, such as HIV, malaria, other chronic diseases and cancer, may require involvement of the T-cell arm of the immune system. During this period, several methods have been developed to induce cytotoxic T-cell responses following vaccination and, in the case of HIV, extensive proof of concept has been obtained in nonhuman primates that effective stimulation of cytotoxic T cells is able to contain the viremia. The most effective methods to induce cytotoxic T cells in nonhuman primates and in humans are engineered nonreplicating viral vectors, such as modified vaccinia Ankara/(MVA), replication-incompetent adenoviruses and DNA vaccines38,39. Mixed regimes, where DNA priming is followed by a viral vector boost, have been shown to further increase the cytotoxic response. Successful induction of malaria-specific CD8+ T cells has been obtained in humans using a DNA-prime, MVA boost strategy40. An example of effective protection obtained in nonhuman primates by adenoviral vectors is the one-dose vaccine recently described against Ebola virus41.
One of the surprises of this decade has also been the understanding that living organisms have a conserved 'innate' immune defense against pathogens, which is mediated by Toll-like receptors and Nod proteins42, which sense molecules that have a signature of microbes, such as DNA containing unmethylated CpG sequences, double-stranded RNA, lipopolysaccharide, etc. The understanding of the molecular basis of the innate immune response provides further potential for manipulating the immune system. This represents one of the most intriguing developments that we expect in the next decade. Among the early results in this new area is the development of adjuvants based on unmethylated CpG motifs, which, being prevalent in bacterial but not in vertebrate genomic DNA, are seen as the signature of bacterial infection by the innate immune system. Synthetic CpG-containing oligonucleotides have been shown to stimulate Toll-like receptor 9 and to induce TH1-like proinflammatory cytokines and to be effective adjuvants for many vaccines in animal models and in some preliminary clinical trials43. Figure 2 provides an example of what we predict will happen in the field of vaccines during the next decades. Reverse vaccinology, DNA vaccination, nonreplicating vectors, the understanding of the innate immune system and the development of new, rationally designed adjuvants for mucosal and systemic delivery of vaccines are likely to be useful in driving the development of many novel vaccines to prevent and treat infectious diseases.
Ultimately, it should be mentioned that the safety of vaccines has improved enormously during the last decade. Very efficacious vaccines, such as the oral poliovirus and the whole-cell pertussis vaccines, have been removed from the vaccination schedules of Western countries because of very rare or perceived untoward effects and replaced with less reactogenic vaccines. All new vaccines are based on technologies that deliver vaccines with a very high standard of safety.
The era of antivirals
The large-scale screening for natural compounds able to kill bacteria in vitro, which was the basis for the boom of antibiotics in the 1950s, was not successful for antivirals. In fact, whereas bacteria are distinct free-living organisms that can be targeted by drugs, viruses are parasitic entities that invade healthy host cells and hijack their reproductive machinery, making it very difficult to kill viruses without also killing the cells they have infiltrated. 5-iodo-2'-deoxyuridine, the first antiviral, was described in 1959; by 1990, only four were licensed (amantadine, ribavirin, acyclovir and derivatives and AZT, against influenza, RSV, herpes and HIV, respectively)44. Since then, more than 35 new antivirals have been approved, most of which combat HIV and others that combat hepatitis B virus, HCV, influenza and herpes (Fig. 3).
Figure 3. Antivirals approved by FDA in 1990 and 2004.
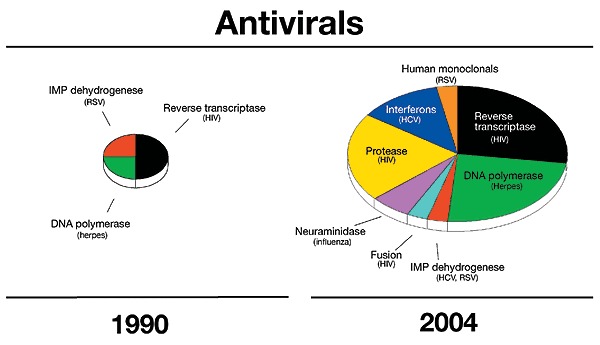
The area is proportional to the number of drugs approved: a total of four in 1990 and 40 in 2004.
The driving force for the boom of antivirals in this period has been the pressure to contain the HIV pandemic, combined with the increased understanding of the molecular mechanisms of viral life cycles, which has allowed the identification of new targets for therapeutic intervention. At the beginning of the 1990s, HIV was growing in the US and worldwide. In the US, 49,000 new HIV cases and 31,000 deaths were recorded in 1990, growing to 72,000 and 50,000, respectively, in 1994 (Fig. 1). Worldwide, there were 8 million cases in 1990 and 18 million in 1994 (Fig. 1). The first effective therapy for HIV came in 1992 with the approval of the first combination of reverse transcriptase inhibitors, 3′-azido-2',3′-dideoxythymidine (AZT) and 2',3′-dideoxycytidine (ddC), which prevent viral DNA synthesis by acting as chain terminators. The approval of (−)-B-L-3′-thia-2'-3′-dideoxycytidine (3TC), a safer inhibitor of reverse transcriptase in 1993 and of the first protease inhibitor in 1995, together with the ability to use the polymerase chain reaction to measure the amount of virus present in a given volume of blood (viral load), allowed the introduction of a cocktail of drugs as a standard of care for people with HIV8. HAART increased life expectancy and transformed HIV in the Western world from a deadly disease into a more manageable chronic disease, which postpones, but unfortunately does not solve, the HIV problem. New infections and deaths from HIV started to plateau and decrease for the first time in the mid-1990s. Today in the United States there are approximately 30,000 new cases and 15,000 deaths per year. But the number of people living with HIV has increased dramatically, from 30,000 in 1992 to more than 400,000 (Fig. 1 and Box 1), and currently we have no long-term solution for them. The management of the population now chronically infected by HIV provides new challenges, such as the continuous development of new antivirals active against the resistant HIV isolates selected by the present therapy and the control of side effects resulting from chronic treatment.
Today the target enzymes for development of antiviral compounds are the viral enzymes necessary for the replication of the viral genome, such as the retroviral reverse transcriptases, DNA- and RNA-dependent polymerases and helicases, the proteases necessary to cleave viral polyproteins, the influenza neuraminidase necessary for the release of the virus from the cell, and even cellular enzymes involved in the generation of the nucleotide pools, such as the inosine 5′-monophosphate deydrogenase45, that for unknown reasons are important for viral replication. The availability of validated target enzymes used for high-throughput screening of natural and combinatorial libraries is today complemented by the ability to cocrystalize target proteins and their inhibitors, allowing the use of structure-based drug design for the generation and optimization of more and better leads. Recently, the understanding of the molecular mechanisms of viral entry into host cells has allowed the development of a new class of antiviral compounds, known as fusion inhibitors, one of which (enfuvirtide) is already in use for the therapy of AIDS46. Enfurvitide is a synthetic peptide which binds to a region of the HIV-1 envelope and prevents a conformational change necessary to drive the fusion of the viral membrane with that of the host cell. Very effective in vitro, although not yet clinically relevant, are the small interfering RNAs (siRNAs), which are double-stranded RNA oligonucleotides that use a cell-defense enzyme pathway to drive the selective degradation of viral RNA products47. siRNAs have been shown to be effective in vitro against virtually all medically important viruses, including HIV and HCV; however, their development as drugs needs to meet the challenge of delivering effectively large and unstable molecules such as oligonucleotides to infected tissues. In conclusion, whereas antibiotics were the hot drugs of the past century, antivirals will be the hot drugs of the twenty-first century during which time we expect to have effective drugs against most viral infections.
Human monoclonal antibodies
Treatment of infectious diseases made a quantum leap in 1890 when Behring and Kitasato discovered that immunization with low doses of sterile culture supernatants of tetanus or diphtheria induced sera that, after transfer to other animals, could cure them of symptoms of disease48. During the following century, sera were one of the most important life-saving tools to treat bacterial infections such as diphtheria, tetanus, anthrax, botulism, pneumococcal disease and viral infections such as rabies. But animal sera have largely disappeared from the anti-infection toolbox during the last 20 years, mostly because of the toxic reactions (serum sickness) caused by injecting humans with horse sera, while the decreased popularity of blood derivatives has reduced the use of human gammaglobulins to only a few applications such as cytomegalovirus and parvovirus B19. The development of the technology to commercialize human monoclonal antibodies by humanizing mouse antibodies allowed the licensure of the first human monoclonal antibody that neutralizes RSV in vitro and prevents RSV infection in newborns49. Today this represents an important tool for newborns but it is still the only licensed humanized antibody for infectious diseases. The high doses required (10–15 mg/kg, that is >600 mg/adult person) are expensive and make administration impractical in adults. Recently, a new technology to efficiently clone human memory B cells from infected or immunized people may transform the field26. This technology allows screening for very high-affinity human monoclonal antibodies against viruses and the isolation of clones that produce antibodies may be able to be therapeutically effective at 0.1–1.0 mg/kg, making industrial development possible. Therefore, we expect that human monoclonal antibodies will become an increasingly important tool for the passive prevention of and therapy for infectious diseases50.
Diagnosis and screening
The availability of the sequence of viral, bacterial and parasite genomes provided the information necessary for the expression of recombinant microbial antigens that could be used to detect antibodies in patient sera, and the development of specific assays, based on nucleic acid amplification technologies, able to directly detect a few genome copies of infectious agents. These new technologies allowed a timely response to the emergence of new pathogens, as in the case of SARS, or the development of assays to monitor and control the spread of infectious agents. The virtual elimination of serious infections transmitted by blood transfusion51, which is one of the milestones in the history of medicine that has been achieved during the last decade, was made possible by these technologies (Fig. 4). In the early 1980s the risk of being infected by HIV or HCV by blood transfusion was in the range of 1 × 10−2. The introduction of screening for antibodies against HIV in 1984 and for HCV in 1990 tremendously increased the safety of blood and blood products reducing the risk to less than 1 × 10−5. Finally, the introduction of nucleic acid testing in 1999 allowed the detection of those recently infected donors that had not had the time yet to develop antibodies, reducing the risk for both to one in two million52. Encouraged by the dramatic success of testing blood for HIV and HCV, new assays have been recently introduced to test blood for hepatitis B and West Nile viruses, virtually eliminating most common diseases that can be transmitted by blood transfusion. Similar technologies can be applied to the detection of other infectious agents such as parvovirus, malaria, leishmania, vCJD, pathogenic bacteria or agents spread by bioterrorists.
Figure 4. Risk of infection of human immunodeficiency virus (HIV), hepatitis C virus (HCV) and hepatitis B virus (HBV) per unit of blood transfusion during the last twenty years.
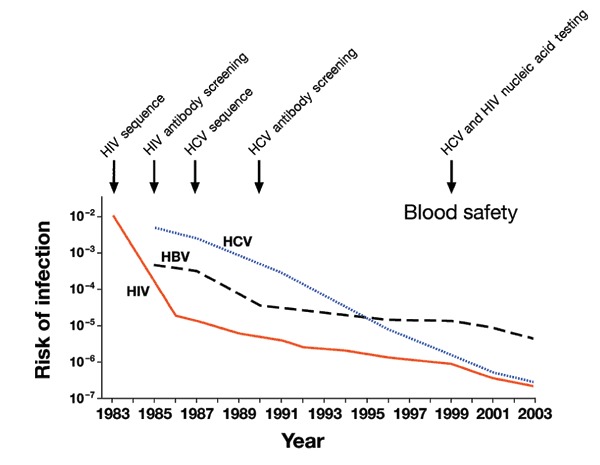
The figure is revised from reference 51.
Conclusions
In the late 1960s the optimism deriving from the successes of vaccines and antibiotics had generated the belief that infectious diseases had been conquered and were no longer a problem for humanity. We now know that the optimism was premature and that infectious diseases are here to stay. Indeed, human history has never seen so many new infectious diseases as in recent times. The shotgun sequencing of microorganisms captured by filtering hundreds of liters of water of the Sargasso sea, which identified 1.2 million new genes, 1,800 species and 148 novel bacterial phylotypes53, showed that microbial diversity is orders of magnitude larger than had been previously estimated. This implies that in the environment and the animal world there is a huge reservoir of microbes which, under the appropriate circumstances, may recombine, jump species and generate new human pathogens. Modern society provides the most fertile environment for this to happen; the accelerated changes of human behavior and ecosystems continuously disturb the equilibrium between microbes and their hosts, and exposes them to new environments where their behavior is unpredictable sometimes, resulting in diseases.
The factors involved are the increased global population, overcrowded cities, increased travel, intensive animal farming and food production, sexual practices, poverty, global warming, and the breakdown of public health measures. Some believe that humans are primarily responsible for this54. Most infectious diseases are zoonoses (animal infections transferred to man) promoted by human behavior (Fig. 5 and Table 1), such as bush-meat hunting (Ebola and probably HIV), the sale of live wild animals in open markets (SARS55) and intensive farming and sale of different species next to each other in open markets (avian influenza finds a favorable environment in the open markets in Asia where migratory aquatic birds, in which the influenza virus grows without causing disease, are sold next to chickens and pigs, the cultivation of which has expanded enormously to feed the growing Asian population12).
Figure 5.
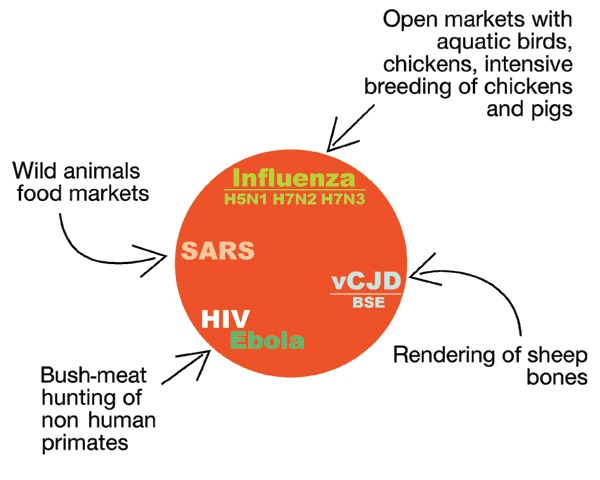
Most emerging diseases are of animal origin.
Table 1.
Human behavior and ecosystems linked to the emergence of new infectious diseases
| Disease | Human behavior/ecosystem | Year of first appearance | Affected people/deaths | Ref. |
|---|---|---|---|---|
| vCJD | Feeding of cows with 'rendered' carcasses of cows and scrapie-infected sheep | 1996 | 143/141 | 59,60 |
| HCV | Unscreened blood products before 1992, medical practices involving unsterilized tools, including syringe reuse | 1987 HCV discovered | 170 million | 61 |
| SARS | Live animals food markets, air travel | 2003 | 8,098/774 in 2002–2003 | 14 |
| Legionella pneumophila | Air-conditioning cooling towers | 1976 | 221/34 in 1976 outbreak | 62 |
| Ebola | Bush-meat hunting | 1976, 1994 | 313/264 during last 2 years | 15 |
| HIV | Bush-meat hunting, sex, travel | 1981 | 40 million/3 million per year | 63 |
| West Nile virus | Air travel, climatic change | 1999 USA | 9,858/262 | 64 |
| Avian influenza | Open markets with aquatic birds, chickens, intensive cultivation of chickens and pigs | 1997 | 43/31 in 2004 | 11 |
| Antibiotic resistance | Widespread use of available antibiotics and spread in the environment | Few years after the introduction of each class of antibiotics | Not available | 65 |
It is therefore easy to predict that the next few decades will see dozens of new infectious agents emerging in humans. The question is whether they will be contained or cause new global pandemics such as HIV, influenza or the Black Death in the Middle Ages, which have each caused several dozens of million deaths, and destroyed economies and societies.
Today, the most compelling new threat seems to be avian influenza. The multiple independent outbreaks of the last two years, involving several different virus strains and outbreaks in Asia, Canada and the Netherlands, are dire warnings of a new flu pandemic; however, it is impossible to predict which virus is going to cause it and when it is going to happen12,13.
Fortunately, today's technologies can provide adequate tools to detect, manage, control and prevent emerging infections. But, technologies are not sufficient on their own to contain the impact of infectious diseases and we are likely to fail unless all governments increase the priority of infectious diseases in the political agenda and promote a coordinated, collaborative global action against them, both in developed and developing countries. A list of measures necessary for the global control of infectious diseases include increased global priority, global legislation and collaboration, coordination of regulatory agencies, development of vaccines to prevent all diseases and a sustainable economic system to support these initiatives (Box 2). An example showing how effective a global approach can be is represented by the campaign for eradication of poliomyelitis, which is the largest public health initiative ever tried. Launched in 1985 for South America, it was taken up by the World Assembly, the governing body of the World Health Organization, which in 1988 committed to the global eradication of poliovirus by the year 2000. During the period from 1998 to 2002, more than two billion children were immunized using the national immunization days and a budget of approximately four billion56. Progress towards eradication of the virus was fast. In 1988 125 countries in 5 continents reported endemic poliovirus. By 1994, 2000 and 2002 the American continent, the Western Pacific Region and the European regions, respectively, were certified polio free (three years without a case of polio). In 2003 only six polio-endemic countries were reported (Niger, Nigeria, and Egypt in Africa and India, Afghanistan and Pakistan in Asia). During the same period, cases were reduced by 99.9% from 350,000/year to less than 800 (Fig. 1). Unfortunately, in 2004 the virus was imported from these countries to eight African countries that were previously polio free2. Although the polio eradication missed the deadline of the year 2000 and has seen a setback in 2004, the initiative is one of the best examples of how a global effort can, within a short period of time, reduce the incidence of an infectious disease by more than 99.9%. The final effort in the remaining endemic regions will eliminate the disease from the world within the next few years and leave us with the problem of whether and how long we should continue to vaccinate against a disease that no longer exists. Hopefully, the success against polio will be a catalyst to trigger similar global priority and collaboration against all infectious diseases.
Box 1: Websites for infectious diseases.

Box 2: Measures necessary for the control of infectious diseases.
Increased global priority. The global threat and the economic burden of infectious diseases need to be visible to the public and to policymakers both in developed and developing countries. Developing countries need to be aware that although they do not spend enough money on prevention, the cost of not doing so is very high57. Scientists and the media need to make an effort to improve effective communication so that infectious diseases will receive high priority in the political and economic agendas of every country in the world.
Global legislation and collaboration. SARS, HIV and avian influenza have clearly shown that whatever happens in one part of the world may very quickly become a global problem. An international law to regulate infectious diseases should be implemented58. International teams with power to oversee the global interest above local concerns, and promote effective international collaboration should be in place.
Coordination of regulatory agencies. Agencies that regulate human health, animal health, trade, hunting, food and travel have so far been acting independently. They need to talk to each other and coordinate their actions because we have seen more than one example (BSE, SARS) in which decisions by one agency resulted in strong implications in the spread of infectious diseases in other areas.
Development of vaccines to prevent all diseases. Vaccines should be studied, developed and licensed against all infectious agents, including those that are economically uninteresting. Smallpox and avian influenza have shown that orphaned infectious agents can become of global interest at any time, and that once they are a problem it is too late to develop measures to control them.
Sustainable economic system. We must develop a sustainable system for protecting the health of the world population. Public-private partnerships such as the Global Fund, the Gates Foundation in the short-term seem to be the best way to make rapid progress. They triggered enormous progress never seen before. But in the long-term, global health cannot rely on the good will of charities.
Acknowledgements
The author is grateful to M. Lattanzi and R. Santonini for supplying the statistical data, G. Corsi for the artwork, and C. Mallia for manuscript editing.
Competing interests
The author is an employee of Chiron Corporation.
References
- 1.McNeill WH. Plagues and peoples. 1998. p. 365. [Google Scholar]
- 2.Roberts L. Health workers scramble to contain African epidemic. Science. 2004;305:24–25. doi: 10.1126/science.305.5680.24. [DOI] [PubMed] [Google Scholar]
- 3.Flannery B, et al. Impact of childhood vaccination on racial disparities in invasive Streptococcus pneumoniae infections. JAMA. 2004;291:2197–2203. doi: 10.1001/jama.291.18.2197. [DOI] [PubMed] [Google Scholar]
- 4.Costantino P, et al. Development and phase 1 clinical testing of a conjugate vaccine against meningococcus A and C. Vaccine. 1992;10:691–698. doi: 10.1016/0264-410X(92)90091-W. [DOI] [PubMed] [Google Scholar]
- 5.Ramsay ME, Andrews N, Kaczmarski EB, Miller E. Efficacy of meningococcal serogroup C conjugate vaccine in teenagers and toddlers in England. Lancet. 2001;357:195–196. doi: 10.1016/S0140-6736(00)03594-7. [DOI] [PubMed] [Google Scholar]
- 6.Trotter CL, Andrews NJ, Kaczmarski EB, Miller E, Ramsay ME. Effectiveness of meningococcal serogroup C conjugate vaccine 4 years after introduction. Lancet. 2004;364:365–367. doi: 10.1016/S0140-6736(04)16725-1. [DOI] [PubMed] [Google Scholar]
- 7.Choo QL, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 8.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 9.Markina SS, Maksimova NM, Vitek CR, Bogatyreva EY, Monisov AA. Diphtheria in the Russian Federation in the 1990s. J. Infect. Dis. 2000;181:S27–S34. doi: 10.1086/315535. [DOI] [PubMed] [Google Scholar]
- 10.Sack DA, Sack RB, Nair GB, Siddique AK. Cholera. Lancet. 2004;363:223–233. doi: 10.1016/S0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 11.Normile D, Enserink M. Infectious diseases. Avian influenza makes a comeback, reviving pandemic worries. Science. 2004;305:321. doi: 10.1126/science.305.5682.321a. [DOI] [PubMed] [Google Scholar]
- 12.Guan Y, et al. H5N1 influenza: a protean pandemic threat. Proc. Natl. Acad. Sci. USA. 2004;101:8156–3161. doi: 10.1073/pnas.0402443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li KS, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 14.Stadler K, et al. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leroy EM, et al. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science. 2004;303:387–390. doi: 10.1126/science.1092528. [DOI] [PubMed] [Google Scholar]
- 16.Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Invest. 2003;111:1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt FR. The challenge of multidrug resistance: actual strategies in the development of novel antibacterials. Appl. Microbiol. Biotechnol. 2004;63:335–343. doi: 10.1007/s00253-003-1344-1. [DOI] [PubMed] [Google Scholar]
- 18.Reacher MH, et al. Bacteraemia and antibiotic resistance of its pathogens reported in England and Wales between 1990 and 1998: trend analysis. BMJ. 2000;320:213–216. doi: 10.1136/bmj.320.7229.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper BS, et al. Methicillin-resistant Staphylococcus aureus in hospitals and the community: stealth dynamics and control catastrophes. Proc. Natl. Acad. Sci. USA. 2004;101:10223–10228. doi: 10.1073/pnas.0401324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 22.Relman DA, Schmidt TM, MacDermott RP, Falkow S. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 23.Relman DA. The search for unrecognized pathogens. Science. 1999;284:1308–1310. doi: 10.1126/science.284.5418.1308. [DOI] [PubMed] [Google Scholar]
- 24.Houghton M, Abrignani S. New Generation Vaccines. 2004. Vaccination against the hepatitis C viruses; pp. 593–606. [Google Scholar]
- 25.Bisht H, et al. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. USA. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Traggiai E, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamelin ME, Abed Y, Boivin G. Human metapneumovirus: a new player among respiratory viruses. Clin. Infect. Dis. 2004;38:983–990. doi: 10.1086/382536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDaniel TK, Valdivia RH. Cellular Microbiology. 2000. Promising new tools for virulence gene discovery; pp. 333–345. [Google Scholar]
- 29.Cossart P, Boquet P, Normark S, Rappuoli R. Cellular microbiology emerging. Science. 1996;271:315–316. doi: 10.1126/science.271.5247.315. [DOI] [PubMed] [Google Scholar]
- 30.Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 31.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 2003;1:137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clifford RJ, et al. Bioinformatics tools for single nucleotide polymorphism discovery and analysis. Ann. NY Acad. Sci. 2004;1020:101–109. doi: 10.1196/annals.1310.011. [DOI] [PubMed] [Google Scholar]
- 33.Bleharski JR, et al. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science. 2003;301:1527–1530. doi: 10.1126/science.1087785. [DOI] [PubMed] [Google Scholar]
- 34.Mueller A, et al. Protective immunity against Helicobacter is characterized by a unique transcriptional signature. Proc. Natl. Acad. Sci. USA. 2003;100:12289–12294. doi: 10.1073/pnas.1635231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rappuoli R. Reverse vaccinology. Curr. Opin. Microbiol. 2000;3:445–450. doi: 10.1016/S1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 36.Koutsky LA, et al. A controlled trial of a human papillomavirus type 16 vaccine. N. Engl. J. Med. 2002;347:1645–1651. doi: 10.1056/NEJMoa020586. [DOI] [PubMed] [Google Scholar]
- 37.Mora M, Veggi D, Santini L, Pizza M, Rappuoli R. Reverse vaccinology. Drug Discov. Today. 2003;8:459–464. doi: 10.1016/S1359-6446(03)02689-8. [DOI] [PubMed] [Google Scholar]
- 38.Shiver JW, et al. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature. 2002;415:331–335. doi: 10.1038/415331a. [DOI] [PubMed] [Google Scholar]
- 39.Shiver JW, Emini EA. Recent advances in the development of HIV-1 vaccines using replication-incompetent adenovirus vectors. Annu. Rev. Med. 2004;55:355–372. doi: 10.1146/annurev.med.55.091902.104344. [DOI] [PubMed] [Google Scholar]
- 40.McConkey SJ, et al. Enhanced T-cell immunogenicity of plasmid DNA vaccines boosted by recombinant modified vaccinia virus Ankara in humans. Nat. Med. 2003;9:729–735. doi: 10.1038/nm881. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan NJ, et al. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature. 2003;424:681–684. doi: 10.1038/nature01876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Athman R, Philpott D. Innate immunity via Toll-like receptors and Nod proteins. Curr. Opin. Microbiol. 2004;7:25–32. doi: 10.1016/j.mib.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 43.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 44.Field HJ, De Clercq E. Antiviral drugs–a short history of their discovery and development. Microbiology Today. 2004;31:58–61. [Google Scholar]
- 45.De Clercq E. Strategies in the design of antiviral drugs. Nat. Rev. Drug Discov. 2002;1:13–25. doi: 10.1038/nrd703. [DOI] [PubMed] [Google Scholar]
- 46.Matthews T, et al. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 2004;3:215–225. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 47.Howard K. Unlocking the money-making potential of RNAi. Nat. Biotechnol. 2003;21:1441–1446. doi: 10.1038/nbt1203-1441. [DOI] [PubMed] [Google Scholar]
- 48.Behring EA, Kitasato S. Ueber das zustandekommen der diphtherie-immunitat und der tetanus-immunitat bei thieren. Deutch Med Woch. 1890;49:1113–1114. [Google Scholar]
- 49.Johnson S, et al. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 1997;176:1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 50.Casadevall A, Dadachova E, Pirofski L. Passive antibody therapy for infectious diseases. Nat. Rev. Microbiol. 2004;2:695–703. doi: 10.1038/nrmicro974. [DOI] [PubMed] [Google Scholar]
- 51.Busch MP, Kleinman SH, Nemo GJ. Current and emerging infectious risks of blood transfusions. JAMA. 2003;289:959–962. doi: 10.1001/jama.289.8.959. [DOI] [PubMed] [Google Scholar]
- 52.Stramer SL, et al. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid-amplification testing. N. Engl. J. Med. 2004;351:760–768. doi: 10.1056/NEJMoa040085. [DOI] [PubMed] [Google Scholar]
- 53.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 54.Walters MJ. Six Modern Plagues. 2003. pp. 19–46. [Google Scholar]
- 55.Guan Y, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 56.Minor PD. Polio eradication, cessation of vaccination and re-emergence of disease. Nat. Rev. Microbiol. 2004;2:473–482. doi: 10.1038/nrmicro906. [DOI] [PubMed] [Google Scholar]
- 57.Bloom BR, Murray CJ. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 58.Gostin LO. International infectious disease law: revision of the World Health Organization's International Health Regulations. JAMA. 2004;291:2623–2627. doi: 10.1001/jama.291.21.2623. [DOI] [PubMed] [Google Scholar]
- 59.Harris DA. Mad cow disease and related spongiform encephalopathies. 2004. p. 219. [Google Scholar]
- 60.Giles J. Tissue survey raises spectre of 'second wave' of vCJD. Nature. 2004;429:331. doi: 10.1038/429331a. [DOI] [PubMed] [Google Scholar]
- 61.McHutchison JG. Understanding hepatitis C. Am. J. Manag. Care. 2004;10:S21–S9. [PubMed] [Google Scholar]
- 62.Roig J, Sabria M, Pedro-Botet ML. Legionella spp.: community acquired and nosocomial infections. Curr. Opin. Infect. Dis. 2003;16:145–151. doi: 10.1097/00001432-200304000-00011. [DOI] [PubMed] [Google Scholar]
- 63.Steinbrook R. The AIDS epidemic in 2004. N. Engl. J. Med. 2004;351:115–117. doi: 10.1056/NEJMp048156. [DOI] [PubMed] [Google Scholar]
- 64.Granwehr BP, et al. West Nile virus: where are we now? Lancet Infect. Dis. 2004;4:547–556. doi: 10.1016/S1473-3099(04)01128-4. [DOI] [PubMed] [Google Scholar]
- 65.Levy SB. Antibiotic resistance: consequences of inaction. Clin. Infect. Dis. 2001;33:S124–S129. doi: 10.1086/321837. [DOI] [PubMed] [Google Scholar]


