Abstract
Objective:
This study aims to investigate the callipyge gene (CLPG) polymorphism in sheep of Edilbay, Volgograd, and Kalmyk breeds.
Materials and Methods:
The analysis was performed by the polymerase chain reaction–restriction fragment length polymorphisms method. The objects of the study were Edilbay fat-tailed sheep (n = 500) at the breeding plant Volgograd-Edilbay (Volgograd region), Volgograd fine-wool sheep (n = 500) at the breeding plant Romashkovskiy (Volgograd region), and Kalmyk fat-tailed sheep (n = 500) at the breeding plant Kirovsky (the Republic of Kalmykia, Yashkul rayon). To conduct the research, tissue samples of 1 cm² from sheep of Kalmyk and Edilbay breeds were taken from the auricle.
Results:
The allelic CLPG gene variants have been determined and genotypes of representative sampling of the three breeds of livestock grown in the steppe zone of Russia. The presented results of the CLPG gene polymorphism in these sheep breeds grown in Russia were obtained for the first time. The research study has revealed that in terms of the CLPG gene, the Edilbay, Volgograd, and Kalmyk sheep breeds have only a homozygous form.
Conclusion:
The results obtained expand the current understanding of the molecular markers that characterize the meat qualities of sheep.
Keywords: Callipyge, marker-assisted selection, PCR-RFLP, Sheep Breeding, single-nucleotide polymorphism
Introduction
In the selection of farm animals, new and traditional methods of evaluation and selection are applied [1,2]. New methods for assessing animals belong to modern techniques of the DNA technology that allow the identification of genes directly or indirectly associated with commercially important qualities [3,4]. In addition to the traditional selection of animals, the identification of the genes variants that are preferred from the point of view of selection makes it possible to breed out directly at the DNA level and conduct the so-called genomic selection [5].
Due to the lack of government contracts for wool in Russia, the demand for sheep products has recently changed substantially. The meat production has become more profitable economically, so the structure of the industry has changed dramatically. The meat sheep breeding, in turn, has achieved considerable success due to the applied genetics. In breeding programs, there were usually used stud rams selected on the basis of the quality of their offspring, relatives, or ancestors. At the same time, the use of the achievements of molecular genetics can bring such work to a new methodological level [6].
To date, the problem of establishing a reliable link between the production characters and genetic markers is still unsolved [7–9]. Breeding based on genetic production markers is aimed at work with animals with high genetic potential in terms of the live weight gain and meat quality [10,11].
One of the most promising candidate genes is the callipyge gene (CLPG). Phenotypically in sheep, the callipyge-single-nucleotide polymorphism (SNP) CLPG mutation (callipyge muscle hypertrophygen—CLPG) is manifested by muscular hypertrophy, primarily in the pelvic and hind limbs regions. The muscles of such lambs were enlarged to varying degrees, with not all muscles being hypertrophied. In callipyge lambs, some desirable commercially important traits and properties of meat quality were manifested, i.e., a higher percentage of meat yield, a larger loin and leaner meat, limbs were higher appreciated. In these lambs, the best qualities of carcass were expressed in better yield (compared to lambs of ordinary muscularity) of whole limbs, sirloin part, loin on the bone and shoulder by 11.8%, 4.7%, 2.5%, and 2.3%, respectively [12]. In this regard, the purpose of the work was to study the Callipigia gene polymorphism that affects the manifestation of muscular hypertrophy in the pelvic and posterior areas in sheep of Edilbay, Volgograd, and Kalmyk breeds. To date, very few studies have been devoted to the study of the Callipigia gene polymorphism. In Russia, the polymorphism of this kind has not been studied in the Edilbay, Volgograd, and Kalmyk sheep.
Materials and Methods
Ethical approval
Experiments were performed in accordance with the Guide for the care and use of laboratory animals [13] and the use of experimental animals completely observed the local animal welfare laws and policies. The current study is in compliance with ethical standards.
Sample collection
The objects of the study were Edilbay fat-tailed sheep (n = 500) at the breeding plant Volgograd-Edilbay in the Volgograd region, Volgograd fine-wool sheep (n = 500) at the breeding plant Romashkovskiy in the Volgograd region, and Kalmyk fat-tailed sheep (n = 500) at the breeding plant Kirovsky (the Republic of Kalmykia, Yashkul rayon). Molecular and genetic studies, as well as the processing of the results obtained, were carried out in the laboratory of molecular diagnostics and biotechnology of farm animals at Don State Agrarian University, Persianovsky (Rostov-on-Don city).
Genomic DNA isolation
To conduct the research, tissue samples of 1 cm² from sheep of Kalmyk and Edilbay breeds were taken from the auricle. The DNA was isolated using the DIAtom DNA Prep 100 reagent kit (Genlab LLC, Russia). The analysis was performed by the polymerase chain reaction–restriction fragment length polymorphism method [14].
Polymerase chain reaction (PCR) analysis and SNP genotyping
Primers were used to amplify a CLPG gene fragment: 5'-TGA AAA CGT GAA CCC AGA AGC-3', 5'-GTC CTA AAT AGG TCC TCT CG-3'. The PCR conditions were pre-denaturation at 95°C for 4 min and then 35 cycles: 94°C for 20 sec, 58°C for 30 sec, 72°C for 1 min, and final synthesis at 72°C for 10 min. The restriction of an amplified fragment was performed with endonuclease BsmF1. The size of the restriction fragments obtained was determined by 2% agarose gel electrophoresis in the presence of ethidium bromide.
Results and Discussion
The Edilbay sheep breed is characterized by outstanding meat production. The breed was selected in Kazakhstan by local inhabitants; since the 1930s, selective breeding work was carried out in the Kazakh SSR; the Birlik and Suyunduk crossbreed types were identified [2]. Sheep of both crossbreed types were brought from Kazakhstan to the Volgograd region of the Russian Federation in 1992. These animals are hardy and unpretentious and also adapt well to any climate and can tolerate both heat and severe frosts.
The Volgograd sheep was being bred in the period of 1931–78 by means of a complex reproductive crossing of fat-rumped hair sheep ewes with rams of Soissons, Caucasian, and Groznian breeds, followed by the animals of a desired type to be selected and main economic traits in the stud lines to be fixated in the Southern regions of the Soviet Union. Tribal herds of the Volgograd breed were developed by the method of accumulation cross breeding of fine-wooled crossbreeds with Volgograd rams from the “Romashkovsky” state farm. Large-scale selection to create new Volgograd breed herds was carried out by the Pallasovsky and Bykovsky breeding enterprises. Through their branches in the Volga region’s rayons, they annually inseminated more than 400 thousand ewes by rams, grown and selected in the herd of the “Romashkovsky” state farm [15].
The Kalmyk fat-tailed sheep is characterized by tall stature, strong massive bones, high legs, and brashy wool compared to fat-tailed sheep raised in the North Caucasus, Ukraine, Kazakhstan, Buryatia, and Mongolia [2].
The callipyge lambs show some desirable commercially important characteristics and properties of meat quality, i.e., a higher percentage of meat yield, a larger loin part, the meat was leaner, and their limbs were evaluated to be higher [16,17]. These lambs also had better expressed meat forms (compared to lambs of ordinary muscularity) of whole limbs, sirloin part, loin bones and shoulder by 11.8%, 4.7%, 2.5%, and 2.3%, respectively [12]. Further, the callipyge lamb was more productive in meat at less daily feed intake [17], which resulted in lower production costs. Consequently, the widespread use of the lamb described has the potential to reduce the cost of lamb for consumers and increase the profitability of sheep breeding. Unfortunately, a negative trait, even a defect of callipyge lambs is high meat rigidity [16,18,19].
The increased fleshing of the CLPG animals is mainly due to hypertrophy of muscle fibers. A histological comparison of muscle fibers [20,21] of callipyge animals with normal muscle fibers showed that the former showed, on average, a larger diameter of “fast” fibers of both type IIb and type IIx, and a smaller diameter of “slow” muscle fibers. In addition, the CLPG expressing sheep had a higher percentage of “fast” type muscle fibers using glycolysis than “slow” fibers and “fast” fibers with oxidative-glycolytic metabolism.
Thus, the changes in muscle tissue of the CLPG animals were tightly associated with “fast” fibers that used a glycolytic type of metabolism, and these were the fibers that increased in diameter and percentage of other types of muscle fibers. The described hypertrophy appeared in lamb from 8 weeks of age, but did not manifest in 2-weeks-old lamb [22], so extensive research on the postnatal development of callipyge sheep is in demand.
The CLPG animals are a new mode of inheritance called polar superdominance [23]. The CLPG phenotype was manifested in progeny heterozygous in terms of the CLPG mutations, with this progeny’s gene being obtained from the father (i.e., the one having the +M/CLPGP genotype, where M and P denote inheritance of the maternal and paternal lines, respectively). The other three possible genotypes (+M/+P, CLPGM/+P, or CLPGM/CLPGP) are phenotypically normal. Although hybrid dysgenesis in Drosophila [24] and polar lethality in mice [25] manifested, when there was a mutation in the heterozygous state, depending on whether the allele was inherited from the father or the mother, the callipyge in sheep is the only example of strict polar superdominance known today. The polar superdominance pattern for the callipyge sheep phenotype was confirmed in studies of various unrelated herds [26], thereby demonstrating that polar superdominance of the CLPG mutations in sheep is a new mode of inheritance. The results of positional cloning that aimed to identify the CLPG mutation and lasted for a decade have been recently obtained. The SNPCLPG A->-G point was found, which determined the occurrence of the CLPG allele [26,27]. This polymorphic region, denoted as SNP CLPG, was located within the conservative 12-bp sequence. The ram that first had a mutation characteristic of a callipyge trait was mosaic for this mutation [27], but passed it on to its descendants.
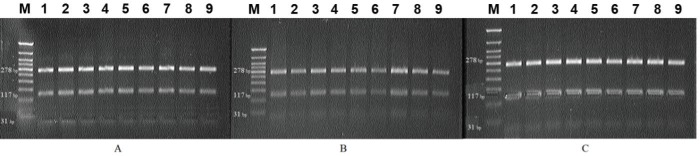
The CLPG gene polymorphism still has not been studied in the Edilbay, Volgograd, and Kalmyk sheep. The results for these breeds were obtained for the first time. Molecular genetic studies in sheep of Edilbay, Volgograd, and Kalmyk breeds resulted in the identification of allelic variants of the CLPG gene and the determination of genotypes. Our study detected only homozygous AA genotype in the sheep populations of these breeds (Fig. 1 A–C). The AG and GG genotypes were not identified. The CLPG locus appeared to be monomorphic in these populations.
Figure 1. Electropherograms of the PCR-RFLP result of the CLPG/BsmF1 gene in 2% agarose gel. (A) Edilbay fat-tailed sheep breed; (B) Volgograd fine-wool sheep breed; (C) Kalmyk fat-tailed sheep breed; M is DNA marker (50+ bp DNA Ladder, Evrogen, Russia).
Nanekarani et al. [14] in their studies also observed only the AA genotype in Iranian sheep of the Lori breed, which suggested that sheep were also monomorphic in the CLPG locus. Alakilli et al. [28] also reported that CLPG locus is a monomorphic for population sheep kept in Saudi. This was also mirrored by Dimitrova and Bozhilova [29], who identified the genetic polymorphism in CLPG in sheep from Karakachan breed. It was detected only homozygous genotype AA, respectively, CLPG locus was found to be monomorphic in this population too.
The studies of the CLPG by Jackson et al. [17] however found out the presence of a mutant G allele in Dorset, Ramboulee, and Hampshire sheep breeds. Similar results were reported by Shah et al. [30], where it was founded that Thalli breed has mutant homozygous (GG, 0.02), heterozygous (AG, 0.10), and normal homozygous (AA, 0.88) forms of CLPG. However Kajli and Lohi breeds were found normal for this genetic variation of CLPG. There was no polymorphism or variation in amplified fragments of the CLPG gene in Egyptian small ruminant breeds [31]. Similar results were observed by Meena et al. [32] in Indian sheep. Pomitun et al. [33] reported that the mutation of this gene may be built into the genome of domestic breeds of sheep only via cross-breeding with foreign breeds, in which this trait is manifested. In that study (in Prydniprovska meat sheep), locus CLPG was also monomorphic, only allele A was determined (278, 117 and 31 b.p.). Allele G with the mutation, manifested in muscle hypertrophy phenotype, was not detected, all the animals under investigation had genotype AA.
The results obtained by Jackson et al. [17] showed the prospects of the gene as a marker of sheep meat production. One of the candidate genes that affect the meat production is the CLPG gene. According to many scientists, the CLPG gene contributes to the intensive development of muscle tissue and is recommended as a DNA marker affecting higher meat yields [12]. Due to mutation, the CLPG sheep are noted for hypertrophy of certain muscle groups on the thighs and minimal amount of fat [34]. Compared to the lambs of ordinary muscularity, these lambs had the best carcass qualities with respect to the best yield of whole limbs, sirloin, loin bones and shoulders by 11.8%, 4.7%, 2.5%, and 2.3%, respectively [12]. Jawasreh et al. [35] reported that the callipyge mutation with 25% Rambouillet genes can provide efficient improvements in growth and meat characteristics, with the exception of tenderness in Awassi sheep. Freking et al. [36] showed a synergistic interaction between Myostatin gene and CLPG.
Conclusion
The genetic analysis of the sheep population with respect to the CLPG gene found the AA genotype in the Edilbay, Volgograd, and Kalmyk sheep. No homozygotes for the type G allele were detected; the AG genotype was not found. All sheep breeds under consideration were characterized by the A allele predominant. To optimize and monitor the breeding processes in sheep breeding, make a decision on the conservation and improvement of Edilbay, Volgograd, and Kalmyk sheep breeds, as well as determine the genetic status and breeding resource of the breed, it is advisable to use the data on the CLPG genotypes frequency in the breed. All this again confirms the need to search and study the DNA markers associated with the productive qualities of sheep for greater efficiency of breeding and increasing the profitability of the sheep industry [37].
Acknowledgment
The authors are grateful to the Russian Science Foundation for financial support in the implementation of this research according to the scientific project # 15-16-10000, NIIMMP.
Conflict of interests
The authors declare that they have no conflict of interest.
Authors’ contribution
Conceptualization, IFG, YuAK, NVSh; Data curation, AYuK, LVG; Formal analysis, MIS; Funding acquisition, NIM, EYuA, VVP; Investigation, NVSh, AYuK, LVG, VVP; Methodology, IFG, YuAK; Resources, MIS, YuAK; Supervision, IFG, YuAK; Writing-original draft, EYuA.
References
- [1].Sulimova GE, Voronkova VN, Perchun AV, Gorlov IF, Randelin AV, Slozhenkina MI, et al. Characterization of the Russian beef cattle breed gene pools using inter simple sequence repeat DNA analysis (ISSR analysis) Russ J Genet. 2016;52:963–8. https://doi.org/10.1134/S1022795416090143. [PubMed] [Google Scholar]
- [2].Deniskova TE, Dotsev AV, Selionova MI, Kunz E, Medugorac I, Reyer H, et al. Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genet Sel Evol. 2018;50:29. doi: 10.1186/s12711-018-0399-5. https://doi.org/10.1186/s12711-018-0399-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gorlov IF, Fedunin AA, Randelin DA, Sulimova GE. Polymorphisms of bGH, RORC, and DGAT1 genes in Russian beef cattle breeds. Russ J Genet. 2014;50:1302–7. https://doi.org/10.1134/S1022795414120035. [PubMed] [Google Scholar]
- [4].Zinovieva NA, Selionova MI, Gladyr EA, Petrovic MP, Caro Petrovic V, Ruzic Muslic D, et al. Investigation of gene pool and genealogical links between sheep breeds of Southern Russia by blood groups and DNA microsatellites. Genetika-Belgrade. 2015;47:395–404. https://doi.org/10.2298/GENSR1502395Z. [Google Scholar]
- [5].Deniskova T, Dotsev AV, Selionova M, Wimmers K, Reyer H, Kharzinova VR, et al. Whole-genome single nucleotide polymorphism study of Romanov sheep. J Anim Sci. 2017;95:339–40. https://doi.org/10.2527/asasann.2017.696. [Google Scholar]
- [6].Deniskova TE, Sermyagin AA, Bagirov VA, Okhlopkov IM, Gladyr EA, Ivanov RV, et al. Comparative analysis of the effectiveness of STR and SNP markers for intraspecific and interspecific differentiation of the genus Ovis. Russ J Genet. 2016;52:79–84. https://doi.org/10.1134/S1022795416010026. [PubMed] [Google Scholar]
- [7].Gorlov IF, Shirokova NV, Randelin AV, Voronkova VN, Mosolova NI, Zlobina EY, et al. CAST/MspI gene polymorphism and its impact on growth traits of Soviet Merino and Salsk sheep breeds in the South European part of Russia. Turk J Vet Anim Sci. 2016;40:399–405. https://doi.org/10.3906/vet-1507-101. [Google Scholar]
- [8].Gorlov IF, Kolosov YA, Shirokova NV, Getmantseva LV, Slozhenkina MI, Mosolova NI, et al. Association of the growth hormone gene polymorphism with growth traits in Salsk sheep breed. Small Rum Res. 2017;150:11–4. https://doi.org/10.1016/j.smallrumres.2017.02.019. [Google Scholar]
- [9].Gorlov IF, Kolosov YA, Shirokova NV, Getmantseva LV, Slozhenkina MI, Mosolova NI, et al. GDF9 gene polymorphism and its association with litter size in two Russian sheep breeds. Rend Lincei Sci Fis Nat. 2018a;29:61–6. https://doi.org/10.1007/s12210-017-0659-2. [Google Scholar]
- [10].Trukhachev V, Skripkin V, Kvochko A, Kulichenko A, Kovalev D, Selionova M, et al. Sequencing of the NFE2l1 gene in sheep and evaluation influence of gene polymorphisms on meat production. J Anim Plant Sci. 2016;26:1262–7. [Google Scholar]
- [11].Trukhachev V, Dzhailidy G, Skripkin V, Kulichenko A, Kovalev D, Selionova M, et al. The polymorphisms of MyoD1 gene in Manych Merino sheep and its influence on body conformation traits. J Hell Vet Med Soc. 2017;68:319–26. https://doi.org/10.12681/jhvms.15476. [Google Scholar]
- [12].Busboom JR, Wahl TI, Snowder GD. Economics of callipyge lamb production. J Anim Sci. 1999;77:243–8. doi: 10.2527/1999.77suppl_2243x. https://doi.org/10.2527/1999.77suppl_2243x. [DOI] [PubMed] [Google Scholar]
- [13].Committee for the Update of the Guide for the Care and Use of Laboratory Animals; Institute for Laboratory Animal Research (ILAR); Division on Earth and Life Studies (DELS); National Research Council of the national academies. Guide for the care and use of laboratory animals. 8th. The National Academies Press; Washington, D.C: 2011. p. 246. [Google Scholar]
- [14].Nanekarani S, Goodarzi M, Mahdavi M. Analysis of polymorphism of Callipyge gene in Lori Sheep by PCR-RFLP Method. APCBEE Procedia. 2014;8:65–9. https://doi.org/10.1016/j.apcbee.2014.03.002. [Google Scholar]
- [15].Gorlov IF, Filatov AS, Natyrov AK, Mosolova NI, Nikolaev DV, Nelepov YN, et al. Meat productivity of volgograd breed ram hogs of different genotypes. Res J Pharm Biol Chem Sci. 2018b;9:2152–61. [Google Scholar]
- [16].Koohmaraie M. The role of Ca(2+)-dependent proteases (calpains) in post mortem proteolysis and meat tenderness. Biochimie. 1992;74:239–45. doi: 10.1016/0300-9084(92)90122-u. https://doi.org/10.1016/0300-9084(92)90122-U. [DOI] [PubMed] [Google Scholar]
- [17].Jackson SP, Miller M, Green R. Phenotypic characterization of Rambouillet sheep expressing the callipyge gene: I. Inheritance of the condition and production characteristics. J Anim Sci. 1997;75:14–8. doi: 10.2527/1997.75114x. https://doi.org/10.2527/1997.75114x. [DOI] [PubMed] [Google Scholar]
- [18].Shackelford SD, Wheeler TL, Koohmaraie M. Effect of the callipyge phenotype and cooking method on tenderness of several major lamb muscles. J Anim Sci. 1997;75:2100–5. doi: 10.2527/1997.7582100x. https://doi.org/10.2527/1997.7582100x. [DOI] [PubMed] [Google Scholar]
- [19].Moore RK, Shimasaki S. Molecular biology and physiological role of the oocyte factor, BMP-15. Mol Cell Endocrinol. 2005;234:67–73. doi: 10.1016/j.mce.2004.10.012. https://doi.org/10.1016/j.mce.2004.10.012. [DOI] [PubMed] [Google Scholar]
- [20].Cockett N, Jackson S, Shay T, Nielsen D, Moore S, Steele M, et al. Chromosomal localization of the callipyge gene in sheep (Ovisaries) using bovine DNA markers. Proc Natl Acad Sci USA. 1994;91:3019–23. doi: 10.1073/pnas.91.8.3019. https://doi.org/10.1073/pnas.91.8.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cockett NE, Jackson SP, Shay TL, Farnir F, Berghmans S, Snowder GD, et al. Polar overdominance at the Ovine callipyge locus. Science. 1996;273:236–8. doi: 10.1126/science.273.5272.236. https://doi.org/10.1126/science.273.5272.236. [DOI] [PubMed] [Google Scholar]
- [22].Carpenter CE, Cockett NE. Histology of longissimus muscle from 2-week-old and 8-week-old normal and callipyge lambs. Can J Anim Sci. 2000;80:511–4. https://doi.org/10.4141/A00-009. [Google Scholar]
- [23].Cockett NE, Smit MA, Bidwell CA, Segers K, Hadfield TL, Snowder GD, et al. The callipyge mutation and other genes that affect muscle hypertrophy in sheep. Genet Sel Evol. 2005;37:S65–81. doi: 10.1186/1297-9686-37-S1-S65. https://doi.org/10.1051/gse:2004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kidwell MG, Kidwell JF, Sved JA. Hybrid dysgenesis in DROSOPHILA MELANOGASTER: a syndrome of aberrant traits including mutation, sterility and male recombination. Genetics. 1977;86:813–33. doi: 10.1093/genetics/86.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tomita T. Oneside cross sterility between inbred strains of mice. Idengaku Zasshi. 1960;35:291. [Google Scholar]
- [26].Freking BA, Murphy SK, Wylie AA, Rhodes SJ, Keele JW, Leymaster KA, et al. Identification of the single base change causing the callipyge muscle hypertrophy phenotype, the only known example of polar overdominance in mammals. Genome Res. 2002;12:1496–506. doi: 10.1101/gr.571002. https://doi.org/10.1101/gr.571002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Smit M, Segers K, Carrascosa LG, Shay T, Baraldi F, Gyapay G, et al. Mosaicism of solid gold supports the causality of a noncoding A-to-G transition in the determinism of the callipyge phenotype. Genetics. 2003;163:453–6. doi: 10.1093/genetics/163.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Alakilli SYM. Analysis of polymorphism of caplstatin and callipyge genes in Saudi sheep breeds using PCR-RFLP technique. Int J Pharm Sci Rev Res. 2015;30:340–4. [Google Scholar]
- [29].Dimitrova I, Bozhilova-Sakova M. PCR-RFLP analysis of callipyge gene (CLPG) in karakachan sheep breed. Bulg J Agric Sci. 2016;22:482–4. [Google Scholar]
- [30].Shah A, Aziz A, Ghafoor A, Zahur AB, Yousaf M, Ishaq R, et al. Molecular Analysis of Callipyge Gene Mutation (C.267A > G) in Kajli, Lohi and Thalli Sheep Breeds of Pakistan. Russ J Genet. 2018;54:848–52. https://doi.org/10.1134/S1022795418070141. [Google Scholar]
- [31].Othman OE, Balabel EA, Mahfouz ER. Genetic characterization of myostatin and callipyge genes in Egyptian small ruminant breeds. Biotechnology. 2016;15:44–51. https://doi.org/10.3923/biotech.2016.44.51. [Google Scholar]
- [32].Meena AS, Kumari R, Jyotsana B, Kumar R, Prince LLL, Kumar V, et al. Absence of overdominace phenotype of Callipyge gene in Indian sheep. Indian J Anim Sci. 2018;88:112–3. [Google Scholar]
- [33].Pomitun IA, Rossokha VI, Boyko YA, Guzevatyi OE, Shpilka MV, Kulibaba RO. Analysis of calpastatin and callipyge genes polymorphism in Prydniprovska meat sheep. Agricultural Science and Practice. 2019;6:58–65. https://doi.org/10.15407/agrisp6.02.058. [Google Scholar]
- [34].Selionova MI, Skorykh LN, Fominova IO, Safonova NS. Proceedings of the All-Russian Research Institute of Sheep and Goat Breeding. Vol. 1. All-Russian Research Institute of sheep and goat breeding; Russia, Stavropol: 2017. Genome selection in sheep breeding; pp. 275–80. [Google Scholar]
- [35].Jawasreh KI, Alamareen AH, Obeidat MD, Aad PY. Growth performance and meat characteristics of the first awassi-rambouillet callipyge backcross. Animals. 2019;9:517. doi: 10.3390/ani9080517. https://doi.org/10.3390/ani9080517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Freking BA, King DA, Shackelford SD, Wheeler TL, Smith TPL. Effects and interactions of myostatin and callipyge mutations: I. growth and carcass traits. J Anim Sci. 2018;96:454–61. doi: 10.1093/jas/skx055. https://doi.org/10.1093/jas/skx055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hu ZL, Park CA, Reecy JM. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 2019;47:D701–10. doi: 10.1093/nar/gky1084. https://doi.org/10.1093/nar/gky1084. [DOI] [PMC free article] [PubMed] [Google Scholar]