CTCF sufficiency depends on various combinations of architectural proteins.
Abstract
In mammals, a C2H2 zinc finger (C2H2) protein, CTCF, acts as the master regulator of chromosomal architecture and of the expression of Hox gene clusters. Like mammalian CTCF, the Drosophila homolog, dCTCF, localizes to boundaries in the bithorax complex (BX-C). Here, we have determined the minimal requirements for the assembly of a functional boundary by dCTCF and two other C2H2 zinc finger proteins, Pita and Su(Hw). Although binding sites for these proteins are essential for the insulator activity of BX-C boundaries, these binding sites alone are insufficient to create a functional boundary. dCTCF cannot effectively bind to a single recognition sequence in chromatin or generate a functional insulator without the help of additional proteins. In addition, for boundary elements in BX-C at least four binding sites for dCTCF or the presence of additional DNA binding factors is required to generate a functional insulator.
INTRODUCTION
Chromosomes in multicellular eukaryotes are organized into a series of discrete, topologically independent domains (TADs) (1). Within these domains, dynamic interactions can be observed between regulatory elements (enhancers and silencers) and the promoters for their gene targets (2). In contrast, regulatory interactions between enhancers/silencers located in one TAD and potential gene targets in neighboring TADs are greatly suppressed. Special elements, called chromatin boundaries or insulators, are thought to be responsible for restricting regulatory interactions to enhancers/silencers and genes that reside within the same TAD (3).
In mammals and other vertebrates, a single protein, CTCF, has been implicated in boundary function (3). CTCF is a highly conserved, polydactyl, C2H2 zinc finger DNA binding protein. It contains 11 C2H2 zinc fingers and binds to a recognition sequence of 15 base pairs (bp). The CTCF protein localizes to the insulators that define many of the TAD boundaries in vertebrates, and genome-wide studies have shown that TAD endpoints are frequently correlated with the presence of two convergently oriented CTCF recognition sequences (4–6). In murine embryonic stem cells, for example, more than 60% of the CTCF-delimited TADs have convergently oriented CTCF sites at their borders (6). TAD formation is thought to involve a loop extrusion mechanism (7). In this mechanism, a cohesin complex encircles the end of a small loop protrusion and then proceeds along the chromatin fiber, extruding a chromatin loop until the cohesin complex encounters CTCF proteins bound to their convergent recognition sequences. This assembly generates a looped domain that insulates the regulatory elements and genes located within the CTCF-cohesin-CTCF–delimited loop from the actions of regulatory elements and genes on either side of the loop. This model makes several predictions regarding the properties of the CTCF-dependent insulators that define the boundaries of TADs. The first prediction is that CTCF-binding sites are necessary for insulator activity. The second prediction is that the CTCF-binding sites are sufficient for insulator activity. To satisfy the criteria of sufficiency, the CTCF protein must share properties with the so-called “pioneer” class of DNA binding proteins, including the ability to access its cognate binding sites within chromatin, without the assistance of accessory DNA binding proteins. Moreover, it must remain stably bound not only during TAD formation but also as long as the CTCF-cohesin-CTCF loop persists. In addition to this pioneer activity, the presence of a single CTCF protein should be sufficient to generate a fully functional insulator that is capable of blocking regulatory element/gene interactions on either side of the bound protein.
In the studies reported here, we used a boundary replacement strategy in the Drosophila bithorax complex (BX-C) to test whether these predictions are true for the fly dCTCF protein and for two other members of the polydactyl C2H2 zinc finger DNA binding protein family, Pita and Su(Hw). Similar to the vertebrate Hox clusters, the fly dCTCF protein localizes to boundaries in the BX-C (8). These insulators play a central role in determining the chromosomal architecture and regulatory activities of the parasegment-specific regulatory domains that control the expression of the three BX-C homeotic genes, Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B) (9). In addition, as in vertebrates, dCTCF is one of dozens of polydactyl C2H2 zinc finger proteins encoded in the fly genome (10, 11). However, unlike vertebrates, several of these polydactyl C2H2 zinc finger protein family members [Su(Hw), Pita, Zipic, Zw5, CLAMP, and Opbp] have been shown to play roles in chromosomal architecture in flies, and it is likely that other members of this large protein family will have similar functions (12–17). Moreover, like dCTCF, two of these zinc finger proteins, Su(Hw) and Pita, are components of boundaries in BX-C and are important in the proper regulation of the BX-C homeotic genes (18–20).
The 300-kb BX-C is subdivided into a series of functionally autonomous regulatory domains (9). As shown in Fig. 1, two of these domains, abx/bx and bxd/pbx, are responsible for regulating the expression of Ubx in parasegments PS5 (segment T3) and PS6 (segment A1). The infra-abdominal (iab) domains regulate the transcription of abd-A and Abd-B. The abd-A gene is controlled by iab-2, iab-3, and iab-4 in parasegments PS7 (A2), PS8 (A3), and PS9 (A4), respectively. Four domains, iab-5, iab-6, iab-7, and iab-8, regulate Abd-B expression in PS10 (A5), PS11 (A6), PS12 (A7), and PS13 (A8), respectively. These parasegment-specific regulatory domains are activated sequentially in successive parasegments along the anterior-posterior axis. For example, iab-6 is turned on in PS11, where it specifies PS11 identity by controlling Abd-B expression in an appropriate parasegment-specific pattern. The adjacent regulatory domain, iab-7, is silenced in PS11 by a Polycomb-dependent mechanism, as is iab-8. In PS12, iab-7, but not iab-8, is activated, and it controls Abd-B expression in this parasegment. A similar sequential pattern of activation can be found elsewhere in the complex. In PS6, for example, the bxd/pbx regulatory domain is activated and controls Ubx expression, while the adjacent regulatory domain, iab-2, is silenced. In PS7, iab-2 is activated and regulates abd-A expression, instead of Ubx expression.
Fig. 1. Fragments of Fub (Fab-2), Mcp, and Fab-8 that were used for replacements.
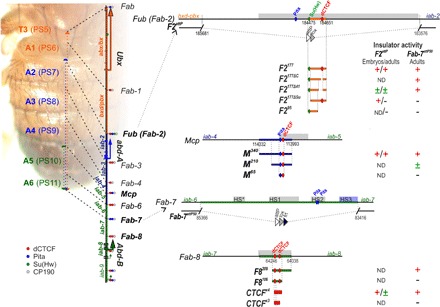
The organization of the genes and regulatory domains in BX-C is shown on the left. Orange, blue, and green arrows represent transcripts of Ubx, abd-A, and Abd-B genes, respectively. Abx/bx, bxd/pbx, and iab-2–iab-8 are responsible for the regulation of these three genes and for the development of the 5 to 13 parasegments (PS)/segments (T3-A8). The lines with colored circles mark characterized (Fub, Mcp, Fab-6, Fab-7, and Fab-8) and predicted (Fab, Fab-1, Fab-3, and Fab-4) boundaries. dCTCF-, Pita-, and Su(Hw)-binding sites at the boundaries are shown as red, blue, and green circles/ovals, respectively. On the right side of the figure, molecular maps of the Fub (Fab-2), Mcp, Fab-7, and Fab-8 boundaries are shown, including their deletions and the fragments used in the replacement experiments. Deoxyribonuclease I hypersensitive sites are shown as light gray boxes above the coordinate bar. The proximal and distal deficiency endpoints of the Fab-2 and Fab-7 deletions used in the replacement experiments are indicated by breaks in the black line. The attP, lox, and frt sites used in genome manipulations are shown as white, gray, and black triangles, respectively. On the right side, summarizing data for insulator activity with the various fragments in the F2attP and Fab-7attP50 insertion sites are shown in embryos and adults. Signs +, ±, and − indicate complete, moderate, and lack of the insulator activity, respectively. ND, not determined.
To generate the appropriate parasegment-specific patterns of Hox gene expression, the nine regulatory domains must be able to function autonomously. Autonomy is conferred by the boundary elements (Fig. 1) that bracket each parasegment-specific regulatory domain (9, 18, 21–26). Like boundary elements elsewhere in the fly genome, all known BX-C boundaries contain sites for one or several architectural proteins, such as dCTCF, Su(Hw), and Pita (8, 12, 20, 22). The most thoroughly characterized BX-C boundaries are Fab-7 and Fab-8, which bracket the iab-7 regulatory domain (9, 21, 25–27). Deletion of Fab-7 fuses the iab-6 and iab-7 regulatory domains, enabling parasegment-specific initiation elements in iab-6 to ectopically activate iab-7 (25, 27). As a consequence, iab-7 drives Abd-B expression in PS11 (and PS12), transforming PS11 (A6) into a copy of PS12 (A7). An equivalent gain-of-function (GOF) transformation of PS12 (A7) into PS13 (A8) is observed when Fab-8 is deleted. Functional studies indicate that Fab-7 and Fab-8 have two distinct activities. The first activity is blocking cross-talk, and it is this activity that is needed to ensure that adjacent regulatory domains can function autonomously. The second activity is a bypass function. The bypass function is needed so that regulatory domains distal to one or more of the Abd-B boundaries (e.g., iab-6 and Fab-7/Fab-8) can “jump over” the intervening boundaries and contact the Abd-B promoter (28). In the case of Fab-8, blocking activity depends on two dCTCF-binding sites, whereas bypass activity requires recognition sequences for a large multiprotein complex, called the large boundary complex (LBC) (19). Similar to the deletions of Fab-7 and Fab-8, deletions of Fab-6 (which separates iab-5 and iab-6), Mcp (which separates iab-4 and iab-5), and Fub (which separates bxd/pbx and iab-2) result in GOF transformations in the parasegment (segment) specified by the centromere proximal regulatory domain (22, 24, 25). Mcp and Fub differ from Fab-6, Fab-7, and Fab-8 because they correspond to the border between the sets of regulatory domains that control different BX-C genes. Mcp separates the regulatory domains for abd-A and Abd-B, whereas Fub separates the regulatory domains controlling Ubx and abd-A (Fig. 1). Because of their roles in demarking the limits of the regulatory domains that control abd-A versus Abd-B and Ubx versus abd-A, Mcp and Fub require blocking but not bypass activity.
To address the questions of necessity and sufficiency, we used two BX-C boundary replacement platforms, Fab-7 (27) and Fub. In both platforms, the endogenous BX-C boundary was deleted, and an attP site was introduced in its place. This attP site can then be used to insert any sequences of interest to test for insulator functions. For both of these BX-C chromatin neighborhoods, we find that single binding sites for polydactyl zinc finger DNA binding proteins are insufficient for the cognate protein to access its binding site and/or to reconstitute a functional insulator. Instead, other sequences, recognized by known or unknown factors, are required to generate boundary activity.
RESULTS
The two dCTCF sites in the Fab-8 boundary are insufficient for blocking activity
The Fab-8 boundary is included in an approximately 400-bp nuclease hypersensitive region and contains two divergently oriented dCTCF sites that are separated by 29 bp (8, 21). In previous boundary replacement experiments, we found that a 337-bp fragment, which spans most of the Fab-8 nuclease hypersensitive site (F8337), was sufficient to fully rescue a Fab-7 boundary deletion, Fab-7attP50 (18). While males carrying the starting Fab-7attP50 deletion lack both A6 and A7 (Fig. 2A), males carrying the F8337 replacement have an A6 segment that has a morphology identical to wild-type (wt) males. This result indicates that F8337 has both blocking and bypass functions and can fully substitute for the endogenous Fab-7 boundary. We also found that the two dCTCF-binding sites in Fab-8 are essential for boundary function in the replacement experiments, and male flies carrying a Fab-8 boundary with mutated dCTCF sites lack the A6 segment, just like starting Fab-7attP50 deletion platform (18).
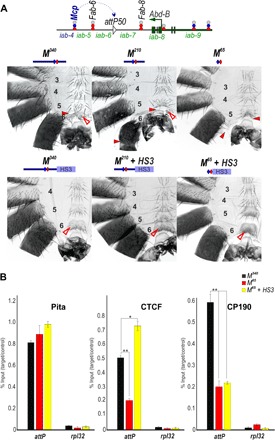
Fig. 2. The number of dCTCF sites is critical for boundary function.
(A) Top: Schematic presentation of Fab-7 substitution. CTCF×4 and CTCF×3 represent multimerized proximal CTCF sites from Fab-8. All designations are the same as described in Fig. 1. Bottom: Morphology of the male abdominal segments (numbered) in F8209, F8106, F8106 + HS3, CTCF×4, CTCF×3, and CTCF×3 + HS3. The filled red arrowheads show morphological features indicative of GOF transformations. The empty red arrowheads show the signs of the LOF transformation, which is directly correlated with the boundary functions of tested DNA fragments. wt, wild type. (B) Binding of dCTCF and CP190 with F8209, F8106, and F8106 + HS3. (C) Binding of dCTCF and CP190 with CTCF×4, CTCF×3, and CTCF×3 + HS3. The results of ChIPs are presented as a percentage of the input DNA and normalized against a positive genomic site, the 59F5 region, for dCTCF and CP190 binding. The negative control is the rpl32 promoter region. Error bars indicate SDs of quadruplicate polymerase chain reaction (PCR) measurements from two independent biological samples of chromatin. Asterisks indicate significance levels: *P < 0.05 and **P < 0.01.
Although these findings demonstrate the necessity of the dCTCF sites, they do not address the question of sufficiency. To test for sufficiency, we started with a previously characterized Fab-8 derivative, F8209, in which 128 bp from the proximal side of F8337 was deleted (Fig. 1). This deletion removes the recognition sequence for the LBC but retains the two dCTCF-binding sites. As previously reported (19), F8209 blocks cross-talk between iab-6 and iab-7, similar to F8337; however, it does not support bypass (Fig. 2A). Unlike the starting Fab-7attP50 platform, an A6-like segment is present in F8209 males, indicating that this truncated boundary is able to prevent cross-talk between iab-5 and iab-6. However, the identity of this “A6-like” segment is A5, not A6. Thus, instead of the banana-shaped A6 sternite observed in wild-type flies, the A6 sternite in F8209 flies has a quadrilateral shape and is covered in bristles just like the A5 sternite. A similar loss-of-function (LOF) A6→A5 transformation is evident in the A6 tergite, which is covered in trichomes like A5 tergite (fig. S1). To further define the sequences needed for blocking, we deleted a 103-bp sequence from the distal side of F8209, leaving the two dCTCF sites (plus their immediate flanking sequences: 26 bp on the proximal side and 10 bp on the distal side). In contrast to F8209 males, the A6 segment is almost completely absent in F8106 males (Figs. 1 and 2A). This nearly complete A6→A7 transformation indicates that the two Fab-8 dCTCF sites are not sufficient to effectively block cross-talk between iab-6 and iab-7.
dCTCF association in vivo is compromised by F8106
In vivo, the Fab-8 boundary is marked by an approximately 400-bp nucleosome-free region (21). Thus, one plausible reason why the F8106 element does not have blocking activity is that additional sequences/factors are necessary to facilitate dCTCF binding to chromatin. To test this idea, we used chromatin immunoprecipitation (ChIP) with material from 3-day adult males to examine the in vivo association of dCTCF association with F8106 and F8209. Figure 2B shows that dCTCF binding to F8106 in vivo is reduced nearly sixfold compared with dCTCF binding with F8209. In accordance with the role played by dCTCF in the recruitment of the architectural protein CP190 to chromatin, we also observe a reduction in CP190 association with F8106 compared to F8209. These results suggest that additional proteins associated with the distal portion of the F8209 element contribute to dCTCF association and/or boundary activity.
Three dCTCF sites are not sufficient to generate blocking activity
Although the two dCTCF sites in Fab-8 alone are not sufficient to block cross-talk between iab-6 and iab-7, we found that a multimer containing four dCTCF (CTCF×4) sites displayed near-complete blocking activity (19). To determine the minimal number of dCTCF sites necessary to block cross-talk between iab-6 and iab-7 activity, we generated a CTCF×3 replacement (Fig. 2A). CTCF×3 males have a GOF phenotype just like the Fab-7attP50 platform, indicating that three dCTCF sites are not sufficient to provide insulating activity. The loss of insulating activity is probably due, at least in part, to a failure to access the dCTCF-binding sites as ChIP experiments show that dCTCF association with CTCF×3 is reduced approximately twofold compared with CTCF×4 (Fig. 2B). In contrast, the binding of CP190 did not change significantly between CTCF×3 and CTCF×4.
Fab-7 HS3 can rescue defective CTCF insulators
The Fab-7 boundary spans four chromatin-specific nuclease hypersensitive regions: HS*, HS1, HS2, and HS3 (26). There are two LBC recognition sequences in Fab-7 (16). One corresponds to an ~200-bp sequence, dHS1, on the centromere distal side of the 400-bp hypersensitive region HS1, while the other spans the 200-bp HS3 sequence. Alone, each of these LBC recognition sequences has limited blocking activity; however, when they are combined (dHS1 + HS3), they are sufficient to reconstitute a fully functional Fab-7 boundary (16, 29). Transgene assays (26) showed that, in addition to its boundary function, HS3 is also a Polycomb response element (PRE). The Polycomb-silencing activity of HS3 depends on several GAGAG motifs and binding sites for the Polycomb protein Pleiohomeotic (Pho) (30). In contrast, the boundary function of HS3, in combination with dHS1, requires the GAGAG motifs but not the Pho sites (16). Consistent with these observations, LBC binding in nuclear extracts is largely eliminated by mutations in the GAGAG motifs, whereas mutations in the Pho sites have no effects on LBC binding.
Because the dHS1 + HS3 combination has full boundary functionality, we wondered whether HS3 would also be able to complement the boundary defects of F8106 and CTCF×3. Much like F8106, HS3 alone has only limited boundary activity: The A6 tergite is greatly reduced in size, while the sternite is missing altogether (Fig. 2A). In contrast, the F8106+ HS3 combination has significant boundary function. In F8106+ HS3 males, the A6 tergite is similar in size to that of wild-type males, indicating that the GOF transformations observed for A6 (PS11) in either F8106 or HS3 males are almost completely suppressed in the dorsal cuticle. However, the tergite is covered in trichomes (fig. S1). This phenotype indicates that the A6 tergite is transformed into A5 (PS10), which would be expected if the F8106+ HS3 combination does not support bypass. Blocking activity appears to be somewhat weaker in the ventral sternite as this cuticular structure is significantly reduced in size, as expected for a GOF transformation. On the other hand, the residual tissue is covered in bristles, consistent with an LOF transformation caused by a lack of bypass activity. HS3 also rescued the blocking defects of CTCF×3, with the morphologies of both the tergite and the sternite, indicating that A6 (PS11) is transformed into A5 (PS10).
Although HS3 rescued the insulating activity of both F8106 and CTCF×3, it did not enhance dCTCF binding (Fig. 2B). ChIP experiments show that the dCTCF association with F8106 and CTCF×3 when they are combined with HS3 is essentially indistinguishable from that observed with each element alone. The same is true for CP190. Note that little or no dCTCF and no CP190 binding are observed for HS3 either alone or in combination with F8106 or CTCF×3.
The Mcp Pita and dCTCF sites are necessary, but not sufficient, for boundary function
We next turned our attention to the Mcp boundary, which separates the regulatory domains controlling abd-A and Abd-B expression and has closely spaced (9 bp) binding sites for dCTCF and Pita (25). In previous replacement experiments, we showed that a 340-bp sequence, M340, in combination with HS3, blocks cross-talk between iab-6 and iab-7 when inserted into the Fab-7attP50 replacement platform (20). Blocking activity in this assay was disrupted by mutations in either the dCTCF- or Pita-binding sites. To test whether these two binding sites are sufficient for boundary function, we generated two truncated replacements, M210 and M65, which include the Mcp dCTCF and Pita sites. We also tested M340 without HS3.
Unlike the M340 + HS3 combination, M340 alone has a weak GOF phenotype, indicating that it is unable to completely block regulatory interactions between iab-6 and iab-7 (Fig. 3A). In transgene assays, the smaller M210 sequence functioned much like the larger M340 sequence; it had enhancer blocking activity and was able to support long-distance boundary:boundary interactions (31). However, as a Fab-7 replacement, the boundary activity of the M210 sequence is less than that of M340 and is also tissue specific. Figure 3A shows that the sternite is completely absent in M210 males, indicating that M210 lacks boundary functions in the PS11 cells that give rise to the ventral A6 cuticle. In contrast, M210 is able to block cross-talk between iab-6 and iab-7 in the PS11 cells that form the dorsal A6 tergite; however, the tergite is smaller than normal, indicating that blocking activity is insufficient to prevent iab-6 initiation elements from activating iab-7 in a subset of PS11 cells. The smaller M65 replacement has no apparent blocking activity, and males carrying this replacement exhibit a complete GOF transformation of A6 into A7. We also tested whether the boundary defects of M210 and M65 could be rescued when combined with HS3. In both cases, blocking activity was largely, if not completely, reconstituted. Figure 3A and fig. S2 show that A6 is transformed into a copy of A5, as would be expected for boundary elements that are able to block cross-talk but fail to mediate bypass.
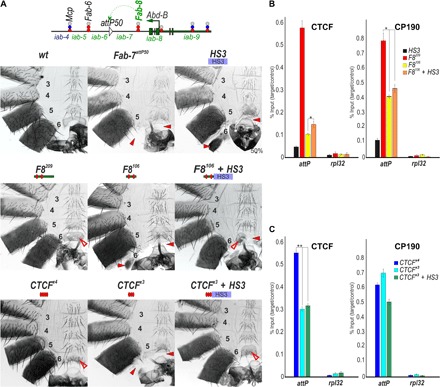
Fig. 3. Testing boundary activities of Mcp sequences.
(A) Top: Schematic presentation of Fab-7 substitution. Bottom: Morphology of the male abdominal segments (numbered) in M340, M210, M210 + PRE M65, and M65 + PRE. Other designations are the same as those in Figs. 1 and 2. (B) Binding of dCTCF, Pita, and CP190 with M210, M210 + PRE M65, and M65 + PRE. The results of ChIPs are presented as percentages of the input DNA normalized against a positive genomic site: 100C, for Pita binding; and the 59F5 region, for dCTCF and CP190 binding. The negative control is the rpl32 promoter region. Error bars indicate SDs of quadruplicate PCR measurements from two independent biological samples of chromatin. Asterisks indicate significance levels: *P < 0.05 and **P < 0.01. Other designations are the same as in Fig. 2.
We used ChIP with material from 3-day adult males to examine Pita, dCTCF, and CP190 interactions with M340 and M65. Reducing the length of the Mcp sequence had no apparent effect on the Pita association in vivo. In contrast, dCTCF and CP190 binding to M65 were more than twofold less than their binding with M340. The addition of HS3 restored dCTCF binding, whereas it had no effects on CP190 association (Fig. 3B). Together, these results suggest that unknown proteins associated with the larger M340 and M210 sequences assist in recruiting dCTCF (and CP190) and probably also contribute to boundary activity.
Insulator function of the Fub boundary requires the Su(Hw) site, but not the dCTCF site
The Fub (Front–ultra-abdominal) boundary is located between the bithoraxoid (bxd) domain, which activates the Ubx gene in PS6 (A1), and the iab-2 domain, which controls abd-A expression in PS7 (A2). Bender and Lucas (22) isolated a 4328-bp Fub deletion that resulted in the ectopic activation of abd-A in PS6 (A1). Within this large deletion (which included a jockey transposable element), there is an approximately 1.3-kb nuclease hypersensitive region, HSFub. HSFub contains binding sites for Pita, Su(Hw), and dCTCF, and in ChIP experiments, prominent peaks for these zinc finger proteins are observed (Fig. 1) (8, 20, 32).
In the first experiment, we used the Fab-7attP50 replacement platform to test the blocking activity of a 177-bp sequence, F2177, spanning the Fub, Su(Hw), and dCTCF sites. Figure 4 and fig. S3 show that the A5 segment is duplicated in F2177 male flies, indicating that F2177 is able to block cross-talk between iab-6 and iab-7 (but does not support bypass). To assess the contributions of dCTCF and Su(Hw) to boundary function, we generated F2177 mutant variants by deleting either the Su(Hw) (F2177ΔSu)– or the dCTCF (F2177ΔC)–binding sites (Fig. 4A and fig. S3). Using electrophoretic mobility shift assays, we found that the Su(Hw) protein shifts a wild-type F2177 DNA probe but not the mutant F2177ΔSu probe (fig. S4). Similarly, F2177 is shifted by the dCTCF protein, whereas the F2177ΔC mutant is not. These two mutations had very different effects on the insulating activity of the F2177 element. The deletion of the Su(Hw)-binding site completely disrupts its insulating activity. Figure 4 shows that A6 is absent, indicating that cells in PS11 have assumed a PS12 identity. By contrast, the dCTCF deletion has no apparent effects on boundary function, and like the wild-type F2177 boundary, A6 is transformed into a copy of A5 in the presence of F2177ΔC. Thus, Su(Hw), but not dCTCF, is critical for the insulating activity of Fub177.
Fig. 4. Testing the role of dCTCF and Su(Hw) in boundary activity of F2177.
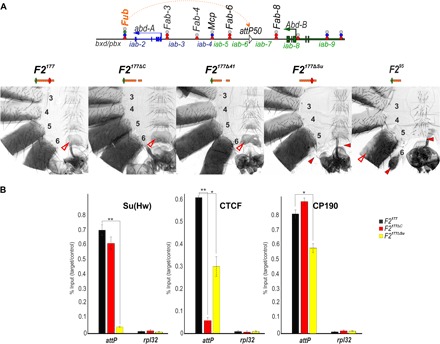
(A) Top: Schematic presentation of Fab-7 substitution. Bottom: Morphology of the abdominal segments (numbered) in F2177, F2177ΔC, F2177Δ41, F2177ΔSu, and F295 flies. Other designations are the same as those in Figs. 1 and 2. (B) Binding of dCTCF, Pita, and CP190 with F2177, F2177ΔC, and F2177ΔSu. The results of ChIPs are presented as the percentage of input DNA, normalized against a positive genomic site: 62D, for Su(Hw) binding; and the 59F5 region, for dCTCF and CP190 binding. The negative control is the rpl32 promoter region. Error bars indicate SDs of quadruplicate PCR measurements from two independent biological samples of chromatin. Asterisks indicate significance levels: *P < 0.05 and **P < 0.01. Other designations are the same as in Fig. 2.
We used ChIP experiments to compare Su(Hw) and dCTCF binding with the wild-type and mutant versions of F2177 (Fig. 4B). In F2177ΔC males, dCTCF binding was not detected, whereas no changes were observed for Su(Hw) binding. In contrast, the deletion of the Su(Hw) site in F2177ΔSu males not only results in the complete loss of Su(Hw) binding but also strongly reduces dCTCF enrichment. These findings suggest that Su(Hw) binding is required for the efficient recruitment of dCTCF to F2177. The binding of CP190 was only moderately reduced in F2177ΔSu males, suggesting that additional, unknown proteins that bind to F2177 recruit CP190.
We generated two additional F2177 mutants, one in which we deleted an internal 41-bp sequence that includes the dCTCF-binding site and one in which we deleted an 82-bp sequence (including the dCTCF site) from the centromere distal side of the F2177 element. The 41-bp deletion mutant, F2177Δ41, displayed full boundary activity, and the A6 segment resembled A5 (Fig. 4A). In contrast, the insulating activity of the 82-bp deletion, F295, was substantially compromised. The A6 sternite was absent, and only a residual A6 tergite was observed. Thus, like the other BX-C boundaries that we have tested using the Fab-7attP50 replacement platform, the presence of a single binding site for a polydactyl zinc finger DNA binding protein, Su(Hw), is not sufficient for boundary function.
F2177 blocks cross-talk in its endogenous location
When introduced into a heterologous context between iab-6 and iab-7, F2177 blocks cross-talk between these two Abd-B regulatory domains. This finding raises the question of whether F2177 would fulfill the same function in its native context, between the bxd and iab-2 regulatory domains (Fig. 1). To address this question, we used the CRISPR-Cas9 system to delete a 2106-bp DNA segment (183,576 to 185,681 in SEQ89E numbering) that spans the Fub nuclease hypersensitive site (32), and in its place, we introduced a 3×P3-DsRed reporter, flanked by lox sites and containing an attP site (F2attP; fig. S5).
Flies heterozygous or homozygous for the F2attP deletion showed evidence of a GOF transformation from a PS6 to a PS7 identity. In wild-type flies, the A1 (PS6) tergite has a distinct shape (pinched along the anterior margin) and is covered in thin, short hairs. On the ventral side, the A1 sternite is absent (Fig. 5). In hetero- or homozygous F2attP flies, the A1 (PS6) segment is transformed toward A2 (PS7). The anterior margin of the tergite is wider than that in wild-type flies, while the posterior margin is pigmented as is the case in A2. Also similar to A2, the A1 tergite is covered in large bristles instead of fine hairs. The ventral cuticle also differs from wild type in that a sternite is present. It is covered in bristles and resembles the sternite in A2. In addition, all F2attP homozygous flies have crumpled or unfolded wings (Fig. 5B). F2attP homozygotes also display additional phenotypes, including low viability and sterility. Approximately 30% of homozygotes have extra tissues along the upper margin of A1 (Fig. 5) and lack one or both halters and/or third legs. The F2attP chromosome is also lethal in combination with the TM6 balancer (which carries a Ubx mutation). The transformations observed in the adult cuticle are reflected in the pattern of abd-A expression in the embryo. In wild-type embryos, abd-A is off in PS5 and PS6, whereas it is active in PS7 (A2) and in the more posterior parasegments PS8–PS12 (Fig. 6). In F2attP embryos, we detect Abd-A protein expression in PS6.
Fig. 5. Testing the activity of dCTCF and Su(Hw) in F2177, when inserted in place of the Fub boundary.
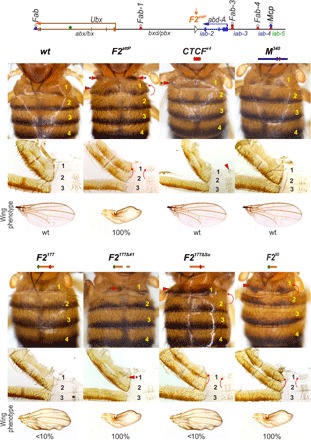
(Top) Schematic presentation of the Ubx and abd-A regulatory regions with F2attP platform. (Bottom) Morphology of the abdominal segments of different Fub replacements. The red arrows show the signs of the GOF phenotype: the appearance of A1 sternite and the appearance of bristles on the A1 tergite. Asterisks at the arrow show that this sign is not full penetrance. The additional tissues in F2attP and the split of A1 tergite in F2Δ41 are present in approximately 30% of homozygous flies. The several additional bristles on A1 tergite of F2Δ41 are present in approximately 80% of homozygous flies. Wing phenotypes in Fub mutants are shown under cuticle images. “<10%” means that about 10% of the flies do not have fully spread wings, while the remaining 90% are of the wild type. “100%” designates that all homozygous flies have wing phenotypes, but the wings can be both slightly spread and completely crumpled. Other designations are the same as in Fig. 2.
Fig. 6. Expression of the abd-A gene in embryos with different F2attP replacements.
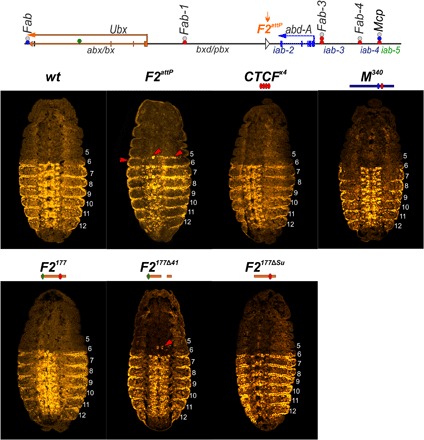
Each panel shows a confocal image of the embryo at stage 14, stained with Abd-A (yellow). Parasegments are numbered from 5 to 12, on the right side of each embryo image. Red arrowheads indicate the ectopic expression of Abd-A.
We used the attP site in the F2attP deletion to test the boundary function of F2177 and several of its mutant derivatives. We found that F2177 rescues the lethality and sterility associated with the much larger F2attP deletion, and as shown in Fig. 6, the morphology of A1 resembles that of wild-type flies. Similarly, unlike F2attP, Abd-A expression is absent in PS6 (Fig. 6). Curiously, a small fraction (<10%) of homozygous F2177 flies display a weak, crumpled wing phenotype. With the exception of this anomaly, these observations suggest that the small F2177 fragment is able to effectively block cross-talk between the bxd and iab-2 regulatory domains.
The Su(Hw)-binding site in F2177 appears to be critical for blocking activity, as F2177ΔSu flies display a strong transformation of A1 into A2 (Fig. 5). Like F2attP, the A1 tergite in F2177ΔSu flies is covered in bristles, and there is a ventral sternite whose morphology resembles the A2 sternite. However, unlike homozygous F2attP flies, homozygous F2177ΔSu flies display near-normal viability and usually have a wild-type wing phenotype. Unexpectedly, Abd-A expression in F2177ΔSu embryos resembles that of wild-type embryos (Fig. 6), raising the possibility that the blocking activity of the Su(Hw)-binding site mutant is tissue and/or stage specific. The phenotype of the larger dCTCF deletion (F2177Δ41) suggests that it retains significant insulating activity. Similar to wild-type flies, no A1 sternite is observed, and in approximately half of the F2attP flies, the morphology of the A1 tergite is similar to wild type. In the remaining flies, the tergite is slightly deformed (see Fig. 6). In embryos, a few speckles of Abd-A expression can be detected in PS6, suggesting that F2177Δ41 does not completely block bxd/iab-2 cross-talk at this stage of development (Fig. 6). Similar to F2attP, all F2177Δ41 homozygotes displayed a crumpled wing phenotype. Because the dCTCF-binding site is not so critical for the blocking activity of F2177, we tested the smaller F295 sequence in F2attP. As was the case when this truncated element was used as a Fab-7 replacement, the presence of the Su(Hw)-binding site in F295 is not sufficient for boundary function (Fig. 6). In F295 flies, A1 is completely transformed into A2, and we also observe a strong wing phenotype. However, unlike homozygous F2attP flies, homozygous F295 flies have normal viability. These results indicate that besides Su(Hw), additional unknown architectural proteins are required for the boundary activity of the F2177 fragment.
As CTCF×4 and M340 were able to block cross-talk between iab-6 and iab-7 when inserted in place of Fab-7, we tested whether they could insulate bxd from iab-2 when inserted in F2attP. For CTCF×4, we found that it has insulating activity in this context, although this activity is not complete. On the dorsal side, patches of tissue in the A1 tergite have an A2-like morphology, while on the ventral side, a partially developed sternite is typically observed. On the other hand, flies carrying the M340 replacement are indistinguishable from wild-type flies. At the same time, both CTCF×4 and M340 completely restore boundary activity in F2attP embryos (Fig. 6 and figs. S6 and S7).
DISCUSSION
In mammals, insulator activity has been ascribed to the polydactyl C2H2 zinc finger DNA binding protein, CTCF (3). A substantial portion of the TADs in mammals is bracketed by single convergently oriented binding sites for CTCF, and this observation has suggested that single CTCF-binding sites are sufficient to generate a fully functional chromatin boundary (1). Although this model is consistent with available genome-wide data in mammals, there have been few, if any, attempts to directly demonstrate sufficiency.
To address the questions of necessity and sufficiency, we used two boundary replacement platforms, Fab-7 and Fub, in the Drosophila BX-C. In both platforms, the endogenous boundary was deleted and replaced with an attP site, which can be used to introduce sequences of interest. Like boundary deletions elsewhere within the BX-C, the Fab-7 and Fub deletions fuse neighboring regulatory domains (22, 26). In the Fab-7 deletion, the fused iab-6 and iab-7 domains misregulated Abd-B expression in PS11/A6, whereas in the Fub deletion, the fused bxd and iab-2 domains misregulated abd-A expression in PS6/A1. In both cases, the misexpression of homeotic genes induces readily identifiable alterations in segmental morphology, providing a very sensitive assay for boundary function.
Although the regulatory consequences of deleting the Fab-7 and Fub boundaries were similar, with neighboring regulatory domains fusing to induce predominantly GOF transformations in parasegment/segment identities, these two boundaries function at different levels of chromosome organization. In tissue culture–based Hi-C experiments, Fub marks the boundary between two large TADs, one that encompasses the Ubx gene and its two regulatory domains and one that encompasses the abd-A gene and its three regulatory domains (33). The Fab-7 boundary, in contrast, is located within a large TAD that includes the four Abd-B regulatory domains, iab-5, iab-6, iab-7, and iab-8, plus the Abd-B gene and its various upstream promoters. Thus, the Fab-7 boundary defines a “sub-TAD” level of chromosomal organization, and similar to the other boundaries within the larger Abd-B TAD, it has both insulating and bypass activities.
We tested sequences derived from three endogenous BX-C boundaries, Fab-8, Mcp, and Fub, for their insulating activity using these two platforms (Fig. 1). Like boundaries in the mammalian Hox complexes (34–36), all three boundaries contain binding sites for the dCTCF protein. Fab-8 has two binding sites, while there is one in Mcp and Fub. Mcp and Fub have binding sites for at least one other polydactyl zinc finger DNA binding protein: Pita in Mcp, and Su(Hw) in Fub. Previous studies have shown that the two dCTCF-binding sites in Fab-8 are essential for insulating activity, as are the sites for Pita and dCTCF in Mcp (12, 18, 20). However, as we show here, these binding sites alone are not sufficient for boundary function. Although a 209-bp Fab-8 sequence that contains both dCTCF sites is able to block cross-talk between iab-6 and iab-7, a smaller 106-bp truncation has no insulating activity, even though both dCTCF-binding sites are present. A similar result was obtained for the Pita and dCTCF combination in Mcp. Although the full-length 340-bp Mcp sequence has full boundary function, a 65-bp sequence spanning the Pita and dCTCF sites has no detectable activity.
One reason why the shorter Fab-8 and Mcp sequences lack insulator activity is that dCTCF binding is reduced. These findings indicate that dCTCF requires the assistance of accessory DNA binding factors to bind to its recognition sequence in chromatin. For Fab-8, these accessory factors would presumably interact with sequences distal to the two dCTCF-binding sites. Although the identities of the factors in Fab-8 that promote dCTCF binding remain unknown, previous studies have suggested that Pita may play this role in Mcp (20). When we mutated the Pita site in M340, we found that dCTCF binding was reduced by more than fivefold compared with wild-type boundary. However, Pita is likely not the only factor that facilitates dCTCF binding to Mcp. Whereas Pita association with the truncated M65 replacement is similar to that observed for M340, dCTCF binding is still reduced more than threefold. These findings indicate that the fly dCTCF is unable to bind to a single site in chromatin without the assistance of other factors. In this respect, the fly protein must differ from the mammalian CTCF, which is believed to function like a pioneer protein and be able to bind to single cognate recognition sequence without the assistance of other accessory DNA binding proteins.
This is not the only difference between the fly and mammalian CTCF proteins. According to the loop extrusion model, single (convergent) CTCF-binding sites are sufficient to generate a boundary element that is capable of subdividing the chromosome into functionally autonomous domains, insulating the genes and regulatory elements on one side of the boundary from the genes and regulatory elements on the other side of the boundary (4, 35, 37). By contrast, single dCTCF sites are not sufficient for boundary function in flies at least in the context of BX-C. In Fab-7 replacements, four copies of the CTCF-binding site are required for boundary function, whereas three copies have no insulating activity. While dCTCF binding to CTCF×3 is less than that observed for the CTCF×4 element, this is probably not the only factor contributing to the difference in boundary function. In particular, the insulating activity of CTCF×3 can be substantially enhanced by combining it with one of the Fab-7 LBC elements, HS3; however, despite this increased functionality, there is little, if any, change in the level of dCTCF association. A similar result was observed for the defective F8106 element: dCTCF association remained low in the F8106 + HS3 combination, even though insulating activity was significantly enhanced. These observations suggest that, although single sites for dCTCF might be necessary for insulating activity, they are not in themselves sufficient. Instead, multiple dCTCF-binding sites or other factors such as Pita or the LBC must be deployed to generate an element that can function as an insulator.
In addition, we found that the dCTCF association was not always necessary for insulating activity. For the Fub boundary sequence F2177, the deletion of either just the dCTCF site or a 41-bp sequence spanning the dCTCF site had no apparent effects on its ability to block cross-talk between iab-6 and iab-7 when inserted in the place of Fab-7. The dCTCF site also appears to be largely dispensable for the insulating activity of F2177 in its native location, between the bxd and iab-2 regulatory domains. Although the blocking activity of F2177Δ41 was not fully equivalent to that of F2177, flies carrying the 41-bp deletion were nearly indistinguishable from wild-type flies. In contrast, mutation of the single Su(Hw)-binding sequence disrupted the insulating activity of F2177 in both the Fab-7 and Fub replacements. In these two BX-C replacement contexts, the phenotype of the F2177ΔSu mutant was essentially indistinguishable from the initial attP deletions. Moreover, as was observed for the Pita mutations in Mcp, mutating the Su(Hw)-binding site in F2177ΔSu reduced dCTCF binding.
Although the association of Pita and Su(Hw) with their binding sites in the Mcp and Fub boundaries was not greatly affected by mutations in the dCTCF sites, whether either of these proteins would be able to access their respective single cognate binding sites without the help of accessory DNA binding proteins remains unclear. However, single Pita- or Su(Hw)-binding sites alone are not sufficient to generate insulator activity. In previous studies on the Fab-7 boundary, we found that the two Pita sites in HS2 were unable to confer boundary activity, even in the presence of HS3 (27). Moreover, they are also unnecessary, as a fully functional Fab-7 boundary can be reconstituted from other Fab-7 sequences, without including the Pita sites in HS2 (16). In the case of Su(Hw), enhancer-blocking transgene experiments showed that similar to dCTCF, three copies of the Su(Hw)-binding site resulted in little, if any, insulator activity, whereas four copies were sufficient to block enhancer-promoter interactions (38).
The experiments described above demonstrate that single recognition sequences for fly polydactyl C2H2 zinc finger DNA binding proteins, such as dCTCF, are insufficient to confer insulator functionality. Although these results indicate that popular models for the assembly and functioning of insulators in vertebrates are unlikely to be applicable in flies, it is not clear to what extent our findings are relevant in vertebrates as the DNA binding and insulator activities of vertebrate CTCF sites and the cognate CTCF protein remain largely unexplored. Consistent with the idea that the insulators at TAD boundaries in vertebrates are generated by a pair of single convergent CTCF-binding sites, mutations in CTCF sites at TAD borders or the deletions of these sites plus their surrounding sequences have been found to disrupt boundary function and/or change the patterns of gene regulation (6, 35–37, 39). However, the correlation between convergently oriented CTCF sites and insulator formation may only tell part of the story. A substantial fraction of the TADs in murine stem cells are not delimited by convergent CTCF sites (6), and in several cases, mutations in single or even multiple CTCF sites have no apparent impact on boundary functions or regulatory interactions (6, 40). In addition, there are thousands of CTCF sites in vertebrate chromosomes that do not correspond to TAD boundaries, and the factors that distinguish these sites from sites that are thought to delimit TADs remain unknown (4). Last, as we have found here, sufficiency and necessity are not always equivalent.
MATERIALS AND METHODS
Chromatin immunoprecipitation
Chromatin for the immunoprecipitations was prepared from 3-day-old adult flies as described in (12). Aliquots of chromatin were incubated with rabbit antibodies against Pita (1:500) (12), Su(Hw) (1:1000), dCTCF (1:500), and CP190 (1:500) (12) or with nonspecific rabbit immunoglobulin G (control). At least two independent biological replicates were made for each chromatin sample. The results of the ChIP experiments are presented as a percentage of the input genomic DNA after triplicate polymerase chain reaction (PCR) measurements. The RpL32 coding region (devoid of binding sites for the test proteins) was used as negative control; 59F5, 100C, and 62D regions were used as positive controls.
Electrophoretic mobility shift assay
Recombinant proteins for the binding assays were expressed and purified as described in (12). Fluorescently labeled DNA fragments were generated by PCR amplification with the corresponding fluorescein amidite (FAM)– or Cy5-labeled primers. Aliquots of purified recombinant proteins (10 to 15 μg) were incubated with the fluorescently labeled DNA fragments in the presence of nonspecific binding competitor poly(dI dC). Incubations were performed in phosphate-buffered saline (pH 8.0) containing 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM dithiothreitol, 0.1% NP-40, and 10% glycerol at room temperature for 30 min. The mixtures were resolved by nondenaturing 5% polyacrylamide gel electrophoresis in 0.5× TBE (tris-borate EDTA) buffer at 5 V/cm. Signals were detected using the Kodak Image System for the FAM-labeled fragments at excitation (Ex) 500 nm/emission (Em) 535 nm and for the Cy5-labeled fragments at the Ex 630 nm/Em 700 nm.
Generation of F2attP by CRISPR-Cas9–induced homologous recombination
For generating double-stranded DNA donors for homology-directed repair, we used pHD-DsRed vector that was a gift from K. O'Connor-Giles (Addgene plasmid no. 51434). The final plasmid contains genetic elements in the following order: [bxd proximal arm]-[attP]-[lox]-[3×P3-dsRed-SV40polyA]-[lox]-[iab-2 distal arm]. Homology arms were PCR amplified from yw genomic DNA using the following primers: ATAGCGGCCGCCGTTGAATGAATCCCC and ATACATATGCTTGGCTTGATCTTGGCAG for the proximal arm (995-bp fragment), and ATAAGATCTGGGGCAAAGTTTTGATTG and ATACTCGAGCGTTGCGGTTTCGGATTAC for the distal arm (914-bp fragment). Targets for Cas9 were selected using “CRISPR optimal target finder”—the program from O'Connor-Giles Laboratory. The recombination plasmid was injected into embryos of y[1] M[Act5C-Cas9.P.RFP-]ZH-2A w[1118] DNAlig4[169] (58492 from the Bloomington Drosophila Stock Center) together with two single-guide RNAs containing the following guides: GATTTGTAATGAAACTGTTC and GATTTCGGACTAATGTTGCT. Injectees were grown to adulthood and crossed with y w; TM6/MKRS line. Flies with dsRed-signal in eyes and the abdomens were selected into a new separate line. The successful integration of the recombination plasmid was verified by PCR and corresponds to the removal of 2106 bp within the Fub region (genome release R6.22: 3R:16,797,757..16,799,862; or complete sequence of BX-C in SEQ89E numbering: 183,576 to 185,681).
Generation of the replacement lines
The strategy of the Fab-7 replacement lines is described in detail in (27). For the F2attP replacement, the recombination plasmid was designed de novo and contains several genetic elements in the following order: [attB]-[pl]-[lox]-[3P3-mCherry]-[mini-y] (fig. S5). All elements were assembled within the pBluescript SK vector. loxP site is located after polylinker [pl] and in combination with the second site, which is located in the platform, use for excision of marker genes and plasmid body. DNA fragments used for the replacement experiments were generated by PCR amplification and verified by sequencing (presented in the Supplementary Methods).
Cuticle preparations
Adult abdominal cuticles of homozygous enclosed 3-day-old flies were prepared essentially as described in (28). Photographs in the bright or dark field were taken on the Nikon SMZ18 stereomicroscope using a Nikon DS-Ri2 digital camera, processed with ImageJ 1.50c4 and Fiji bundle 2.0.0-rc-46.
Embryo immunostaining
Primary antibodies were mouse monoclonal anti-Ubx at 1:30 dilution (FP3.38, generated by R. White, deposited to the Developmental Studies Hybridoma Bank), anti–abd-A at 1:50 dilution (sc-390990, purchased from Santa Cruz Biotechnology), and polyclonal rabbit anti-engrailed at 1:2000 dilution (a gift from J. Kassis). Secondary antibodies were goat anti-mouse Alexa Fluor 546 and anti-rabbit Alexa Fluor 488 (Thermo Fisher Scientific) at 1:500 dilution. Stained embryos were mounted in the following solution: 23% glycerol, 10% Mowiol 4-88, 0.1 M tris-HCl (pH 8.3). Images were acquired on a Nikon A1 HD25 confocal microscope and processed using ImageJ 1.50c4.
Supplementary Material
Acknowledgments
We thank F. Hasanov and A. Parshikov for the fly injection. We thank A. Golovnin for the rabbit Su(Hw) antibodies and J. Kassis for the rabbit anti-engrailed antibodies. This study was performed using the equipment of the IGB RAS facilities supported by the Ministry of Science and Education of the Russian Federation. Funding: This work was supported by the Russian Science Foundation project no. 19-74-30026 (to P.G.). CRISPR-Cas9–directed editing and embryo immunostaining were supported by grant 075-15-2019-1661 from the Ministry of Science and Higher Education of the Russian Federation. Confocal imaging was supported by GM R35GM126975 (to P.S.). Author contributions: O.K., O.M., A.I., V.S., N.P., and M.L., performed different experiments. O.K., O.M., P.S., and P.G. designed the study, interpreted the data, and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/13/eaaz3152/DC1
Supplementary Methods
Fig. S1. Morphology of the abdominal segments (numbered) in males carrying different variants of the Fab-8 or CTCF site replacements in Fab-7attP50 in the dark field.
Fig. S2. Morphology of the abdominal segments (numbered) in males carrying different variants of Mcp in Fab-7attP50 in the dark field.
Fig. S3. Morphology of the abdominal segments (numbered) in males carrying different variants of F2177 in Fab-7attP50 in the dark field.
Fig. S4. In vitro binding of dCTCF and Su(Hw) to F2177and its derivatives.
Fig. S5. Strategy for creating Fub replacement lines.
Fig. S6. A lateral view of abd-A expression patterns in stage 14 embryos carrying different substitutions in F2attP.
Fig. S7. Ubx expression in F2attP, F2177, and F2177DC, exhibiting wing phenotypes.
REFERENCES AND NOTES
- 1.Szabo Q., Bantignies F., Cavalli G., Principles of genome folding into topologically associating domains. Sci. Adv. 5, eaaw1668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furlong E. E. M., Levine M., Developmental enhancers and chromosome topology. Science 361, 1341–1345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali T., Renkawitz R., Bartkuhn M., Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 37, 17–26 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Rao S. S., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., Robinson J. T., Sanborn A. L., Machol I., Omer A. D., Lander E. S., Aiden E. L., A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Y., Xu Q., Canzio D., Shou J., Li J., Gorkin D. U., Jung I., Wu H., Zhai Y., Tang Y., Lu Y., Wu Y., Jia Z., Li W., Zhang M. Q., Ren B., Krainer A. R., Maniatis T., Wu Q., CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162, 900–910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Wit E., Vos E. S. M., Holwerda S. J. B., Valdes-Quezada C., Verstegen M. J. A. M., Teunissen H., Splinter E., Wijchers P. J., Krijger P. H. L., de Laat W., CTCF binding polarity determines chromatin looping. Mol. Cell 60, 676–684 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Fudenberg G., Abdennur N., Imakaev M., Goloborodko A., Mirny L. A., Emerging evidence of chromosome folding by loop extrusion. Cold Spring Harb. Symp. Quant. Biol. 82, 45–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holohan E. E., Kwong C., Adryan B., Bartkuhn M., Herold M., Renkawitz R., Russell S., White R., CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLOS Genet. 3, e112 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeda R. K., Karch F., The open for business model of the bithorax complex in Drosophila. Chromosoma 124, 293–307 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedotova A. A., Bonchuk A. N., Mogila V. A., Georgiev P. G., C2H2 zinc finger proteins: The largest but poorly explored family of higher eukaryotic transcription factors. Acta Naturae 9, 47–58 (2017). [PMC free article] [PubMed] [Google Scholar]
- 11.Najafabadi H. S., Mnaimneh S., Schmitges F. W., Garton M., Lam K. N., Yang A., Albu M., Weirauch M. T., Radovani E., Kim P. M., Greenblatt J., Frey B. J., Hughes T. R., C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 33, 555–562 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Maksimenko O., Bartkuhn M., Stakhov V., Herold M., Zolotarev N., Jox T., Buxa M. K., Kirsch R., Bonchuk A., Fedotova A., Kyrchanova O., Renkawitz R., Georgiev P., Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 25, 89–99 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zolotarev N., Fedotova A., Kyrchanova O., Bonchuk A., Penin A. A., Lando A. S., Eliseeva I. A., Kulakovskiy I. V., Maksimenko O., Georgiev P., Architectural proteins Pita, Zw5,and ZIPIC contain homodimerization domain and support specific long-range interactions in Drosophila. Nucleic Acids Res. 44, 7228–7441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zolotarev N., Maksimenko O., Kyrchanova O., Sokolinskaya E., Osadchiy I., Girardot C., Bonchuk A., Ciglar L., Furlong E. E. M., Georgiev P., Opbp is a new architectural/insulator protein required for ribosomal gene expression. Nucleic Acids Res. 45, 12285–12300 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baxley R. M., Bullard J. D., Klein M. W., Fell A. G., Morales-Rosado J. A., Duan T., Geyer P. K., Deciphering the DNA code for the function of the Drosophila polydactyl zinc finger protein suppressor of hairy-wing. Nucleic Acids Res. 45, 4463–4478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyrchanova O., Kurbidaeva A., Sabirov M., Postika N., Wolle D., Aoki T., Maksimenko O., Mogila V., Schedl P., Georgiev P., The bithorax complex iab-7 Polycomb response element has a novel role in the functioning of the Fab-7 chromatin boundary. PLOS Genet. 14, e1007442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaye E. G., Kurbidaeva A., Wolle D., Aoki T., Schedl P., Larschan E., Drosophila dosage compensation loci associate with a boundary-forming insulator complex. Mol. Cell. Biol. 37, e00253-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyrchanova O., Mogila V., Wolle D., Deshpande G., Parshikov A., Cléard F., Karch F., Schedl P., Georgiev P., Functional dissection of the blocking and bypass activities of the Fab-8 boundary in the drosophila bithorax complex. PLOS Genet. 12, e1006188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyrchanova O., Sabirov M., Mogila V., Kurbidaeva A., Postika N., Maksimenko O., Schedl P., Georgiev P., Complete reconstitution of bypass and blocking functions in a minimal artificial Fab-7 insulator from Drosophila bithorax complex. Proc. Natl. Acad. Sci. U.S.A. 116, 13462–13467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyrchanova O., Zolotarev N., Mogila V., Maksimenko O., Schedl P., Georgiev P., Architectural protein Pita cooperates with dCTCF in organization of functional boundaries in Bithorax complex. Development 144, 2663–2672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barges S., Mihaly J., Galloni M., Hagstrom K., Müller M., Shanower G., Schedl P., Gyurkovics H., Karch F., The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127, 779–790 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Bender W., Lucas M., The border between the ultrabithorax and abdominal-A regulatory domains in the Drosophila bithorax complex. Genetics 193, 1135–1147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowman S. K., Deaton A. M., Domingues H., Wang P. I., Sadreyev R. I., Kingston R. E., Bender W., H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife 3, e02833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iampietro C., Gummalla M., Mutero A., Karch F., Maeda R. K., Initiator elements function to determine the activity state of BX-C enhancers. PLOS Genet. 6, e1001260 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karch F., Galloni M., Sipos L., Gausz J., Gyurkovics H., Sched P., McpandFab-7: Molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 22, 3138–3146 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mihaly J., Hogga I., Gausz J., Gyurkovics H., Karch F., In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124, 1809–1820 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Wolle D., Cleard F., Aoki T., Deshpande G., Schedl P., Karch F., Functional requirements for Fab-7 boundary activity in the bithorax complex. Mol. Cell. Biol. 35, 3739–3752 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Postika N., Metzler M., Affolter M., Müller M., Schedl P., Georgiev P., Kyrchanova O., Boundaries mediate long-distance interactions between enhancers and promoters in the Drosophila bithorax complex. PLOS Genet. 14, e1007702 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cleard F., Wolle D., Taverner A. M., Aoki T., Deshpande G., Andolfatto P., Karch F., Schedl P., Different evolutionary strategies to conserve chromatin boundary function in the bithorax complex. Genetics 205, 589–603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishra R. K., Mihaly J., Barges S., Spierer A., Karch F., Hagstrom K., Schweinsberg S. E., Schedl P., The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol. 21, 1311–1318 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H. B., Muller M., Bahechar I. A., Kyrchanova O., Ohno K., Georgiev P., Pirrotta V., Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol. Cell. Biol. 31, 616–625 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas S., Li X.-Y., Sabo P. J., Sandstrom R., Thurman R. E., Canfield T. K., Giste E., Fisher W., Hammonds A., Celniker S. E., Biggin M. D., Stamatoyannopoulos J. A., Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 12, R43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuettengruber B., Oded Elkayam N., Sexton T., Entrevan M., Stern S., Thomas A., Yaffe E., Parrinello H., Tanay A., Cavalli G., Cooperativity, specificity, and evolutionary stability of Polycomb targeting in Drosophila. Cell Rep. 9, 219–233 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Narendra V., Bulajić M., Dekker J., Mazzoni E. O., Reinberg D., CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 30, 2657–2662 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narendra V., Rocha P. P., An D., Raviram R., Skok J. A., Mazzoni E. O., Reinberg D., CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo H., Wang F., Zha J., Li H., Yan B., du Q., Yang F., Sobh A., Vulpe C., Drusbosky L., Cogle C., Chepelev I., Xu B., Nimer S. D., Licht J., Qiu Y., Chen B., Xu M., Huang S., CTCF boundary remodels chromatin domain and drives aberrant HOX gene transcription in acute myeloid leukemia. Blood 132, 837–848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren G., Jin W., Cui K., Rodrigez J., Hu G., Zhang Z., Larson D. R., Zhao K., CTCF-mediated enhancer-promoter interaction is a critical regulator of cell-to-cell variation of gene expression. Mol. Cell 67, 1049–1058.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott K. C., Taubman A. D., Geyer P. K., Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics 153, 787–798 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanssen L. L. P., Kassouf M. T., Oudelaar A. M., Biggs D., Preece C., Downes D. J., Gosden M., Sharpe J. A., Sloane-Stanley J. A., Hughes J. R., Davies B., Higgs D. R., Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat. Cell Biol. 19, 952–961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barutcu A. R., Maass P. G., Lewandowski J. P., Weiner C. L., Rinn J. L., A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nat. Commun. 9, 1444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/13/eaaz3152/DC1
Supplementary Methods
Fig. S1. Morphology of the abdominal segments (numbered) in males carrying different variants of the Fab-8 or CTCF site replacements in Fab-7attP50 in the dark field.
Fig. S2. Morphology of the abdominal segments (numbered) in males carrying different variants of Mcp in Fab-7attP50 in the dark field.
Fig. S3. Morphology of the abdominal segments (numbered) in males carrying different variants of F2177 in Fab-7attP50 in the dark field.
Fig. S4. In vitro binding of dCTCF and Su(Hw) to F2177and its derivatives.
Fig. S5. Strategy for creating Fub replacement lines.
Fig. S6. A lateral view of abd-A expression patterns in stage 14 embryos carrying different substitutions in F2attP.
Fig. S7. Ubx expression in F2attP, F2177, and F2177DC, exhibiting wing phenotypes.


