Abstract
Background:
Neointimal hyperplasia is a major contributor to restenosis after arterial interventions, but the genetic and environmental mechanisms underlying the variable propensity for neointimal hyperplasia between individuals, including the role of commensal microbiota, are not well understood. We sought to characterize how shifting the microbiome using cage sharing and bedding mixing between rats with differing restenosis phenotypes after carotid artery balloon angioplasty could alter arterial remodeling.
Methods and results:
We cohoused and mixed bedding between genetically distinct rats (Lewis [LE] and Sprague Dawley [SD]) which harbor different commensal microbes and which are known to have different neointimal hyperplasia responses to carotid artery balloon angioplasty. Sequencing of the 16S rRNA gene was used to monitor changes in the gut microbiome. There were significant differences in neointimal hyperplasia between non-cohoused LE and SD rats 14 days after carotid artery angioplasty (mean intima+media [I+M] area .117±.014 mm2 LE vs .275±.021 mm2 SD, P<.001), which were diminished by cohousing. Cohousing also altered local adventitial Ki67 immunoreactivity, local accumulation of leukocytes and macrophages (total and M2), and IL-17A concentration 3 days post-surgery in each strain. Non-cohoused SD and LE rats had microbiomes distinguished by both weighted (P=.012) and unweighted (P<.001) unifrac beta diversity distances, though without significant differences in alpha diversity. The difference in unweighted beta diversity between the fecal microbiota of SD and LE rats was significantly reduced by cohousing. Operational taxonomic units that significantly correlated with average I+M area include Parabacteroides distasonis, Desulfovibrio, Methanosphaera, Peptococcus, and Prevotella. Finally, serum concentrations of microbe-derived metabolites hydroxyanthranilic acid and the kynurenine/tryptophan ratio were significantly associated with I+M area in both rat strains independent of cohousing.
Conclusions:
We describe a novel mechanism for how microbiome manipulations affect arterial remodeling and the inflammatory response after arterial injury. A greater understanding of the host inflammatory-microbe axis could uncover novel therapeutic targets for the prevention and treatment of restenosis.
Table of Contents Summary
Shifting the microbiome in rats using cage sharing and bedding mixing altered the restenosis and inflammatory response after carotid artery balloon angioplasty. These results suggest a direct role for gut microbiota in the arterial remodeling response after injury that occurs through modulation of the acute inflammatory response.
Background
Neointimal hyperplasia development is a prevalent cause of restenosis after revascularization procedures such as balloon angioplasty, stenting, and bypass surgery, affecting 20-50% of patients in the first year after revascularization.1 Arterial “injury,” as manifested by creation of a surgical anastomosis, balloon angioplasty, or stent implantation, induces a wound healing response that is the consequence of endothelial disruption, platelet adherence and aggregation, and activation of an inflammatory cascade that modulates cellular migration, proliferation and extracellular matrix production.1
In experimental rodent models, differences in neointimal hyperplasia after arterial injury have been ascribed to genetic background, which determine extracellular matrix formation, vascular fibroelastic content, wall fragility, and other biological processes.2,3 Strain-related differences in phenotypes have also been observed in a wide variety of conditions, including pulmonary arterial hypertension,4 epilepsy,5 and stroke.6 We previously advanced the novel concept that restenosis is inversely associated with intestinal-derived metabolites such as the short chain fatty acid butyrate, which has anti-proliferative and anti-migratory effects on vascular smooth muscle cells.7
In this study, we hypothesize that the arterial remodeling response can be modulated by commensal gut microbes. Because of the complexity of both the microbiome and microbiome interventions, one informed avenue for interrogating this relationship would be to exchange microbes between rats with differing neointimal hyperplasia phenotypes and distinct gut microbiota. Thus, changes in the arterial remodeling phenotype could then be correlated with shifts in the microbiome.
Methods
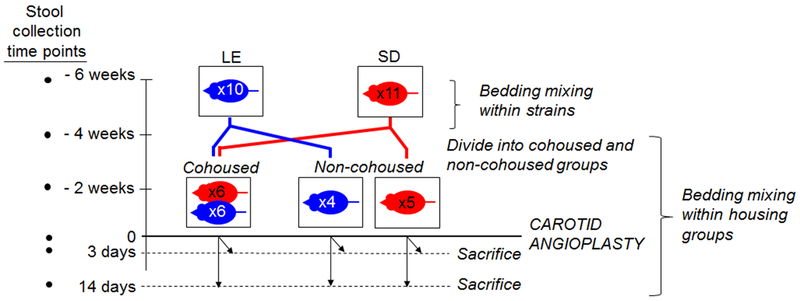
Experimental schema is shown in Fig 1. All experimental methods are provided in the Appendix.
Fig. 1.
Experimental schema. Bedding mixing among cages of similar rat strains (Lewis [LE] and Sprague-Dawley [SD]) was carried out for 2 weeks. Thereafter, rats were divided into cohoused and non-cohoused groups for 4 weeks, followed by carotid angioplasty and then sacrifice at either 3 or 14 days post-angioplasty. Bedding mixing was also carried out within housing groups during this time. Stool samples were collected at time points designated on the left.
Results
Neointimal hyperplasia development after carotid artery balloon angioplasty
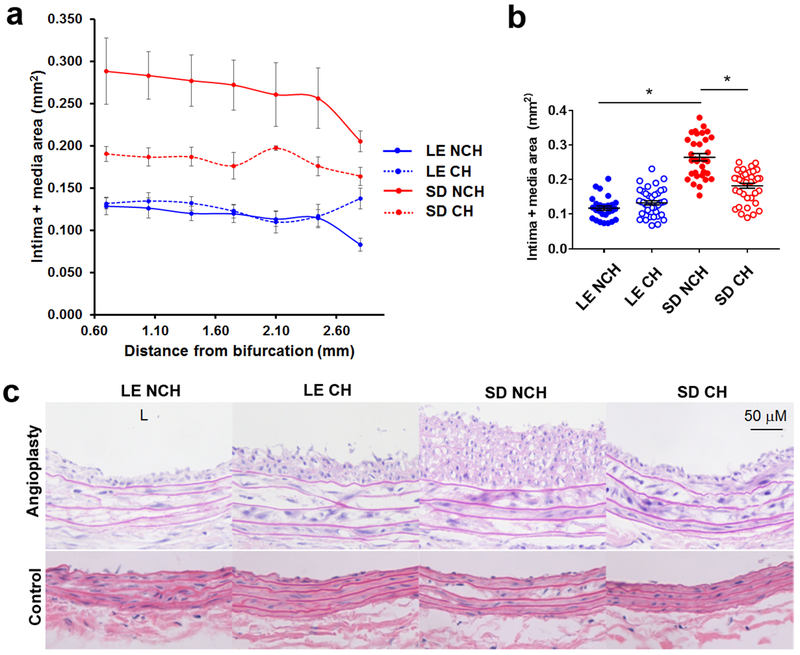
As anticipated from prior work by others demonstrating strain-dependent differences in neointimal hyperplasia development,2 arterial remodeling 14 days after carotid angioplasty differed significantly between non-cohoused (NCH) Lewis (LE) and Sprague-Dawley (SD) rats (Table I), with LE rats displaying a “resistant” phenotype to neointimal hyperplasia and SD rats displaying a “susceptible phenotype”. Cohousing modulated these differences (Fig. 2a and 2b, Table I). Cohoused (CH) LE rats had significantly more neointima area compared to NCH LE counterparts (.031±.006 mm2 NCH vs .047±.007 mm2 CH, P=.005). In contrast, CH SD rats had significantly less neointima area compared to their NCH SD counterparts (.104±.026 mm2 NCH vs .057±.001 mm2 CH, P=.02). When we compared intima+media (I+M) area as a measure of neointimal hyperplasia, there was a slight difference between LE rats by housing group that did not reach statistical significance (.117±.014 mm2 NCH vs .130±.015 mm2 CH, P=.08), but CH SD rats again had less I+M area compared to NCH SD rats (.275±.021 mm2 NCH vs .182±.018 mm2 CH; P<.001). Of note, there were no instances of complete carotid thrombosis in any group. Representative photomicrographs of hematoxylin-eosin-stained post-angioplasty carotid arteries from each rat strain and both housing groups are shown in Fig. 2c (top). Of note, there was no qualitative histological difference between the uninjured right-sided carotid arteries of LE and SD rats (Fig. 2c, bottom).
Table I.
Morphometric data of angioplastied carotid arteries at 14-day time point.
| Intima area (I) (mm2) | P value | P value | Media area (M) (mm2) | P value | P value | P value | ||
|---|---|---|---|---|---|---|---|---|
| LE NCH vs. CH | SD NCH vs. CH | LE NCH vs. CH | SD NCH vs. CH | |||||
| LE NCH | .031±.006 | <.001 | .005 | .02 | .086±.010 | <.001 | .07 | .01 |
| SD NCH | .104±.026 | .171±.009 | ||||||
| LE CH | .047±.007 | .02 | .083±.009 | <.001 | ||||
| SD CH | .057±.001 | .125±.011 | ||||||
| I+M (mm2) | P value | P value | I/I+M | P value | P value | P value | ||
| LE NCH vs. CH | SD NCH vs. CH | LE NCH vs. CH | SD NCH vs. CH | |||||
| LE NCH | .117±.014 | <.001 | .08 | <.001 | .265±.034 | .02 | .003 | .37 |
| SD NCH | .275±.021 | .378±.070 | ||||||
| LE CH | .130±.015 | <.001 | .360±.034 | .23 | ||||
| SD CH | .182±.018 | .313±.047 | ||||||
All areas expressed as mean ± SEM. N=4-6 rats/group. P values < .05 are denoted in bold.
Fig. 2.
Neointimal hyperplasia 14 days after carotid angioplasty in NCH and CH LE and SD rats. a Differences in mean intima + media area (mm2 ± SEM) along the entire length of the angioplastied carotid artery in NCH (solid lines) LE (“resistant” to neointimal hyperplasia) and SD rats (“susceptible” to neointimal hyperplasia) were attenuated upon cohousing (dotted lines). b Mean intima and media area (mm2 ± SEM) of NCH and CH LE and SD rats were compared between all groups. *, P<.001; Mann-Whitney U test. c Representative cross-sectional photomicrographs of NCH and CH LE and SD rat carotid arteries after carotid angioplasty after H+E staining (top). Un-injured right (control) carotid artery sections are grossly similar (bottom). L, orientation of the arterial lumen. Original magnification 40x. Scale bar, 50 μM. N=4-6 rats per group.
Local arterial cellular proliferation
To determine if the observed changes in neointimal hyperplasia were due to altered cellular proliferation in the arterial wall, we compared Ki67 immunoreactivity in the carotid arteries of NCH and CH LE and SD rats 3 days after angioplasty. As shown in Fig. 3a, Ki67 immunoreactivity was greatest in the adventitia and least in the intima of both LE and SD rats. There was also more Ki67 staining in the medial and adventitial layers of NCH SD rats compared to LE rats (media: 11.9±4.2% NCH LE vs 24.1±4.3% NCH SD; P=.02; adventitia: 13.0±2.7% NCH LE vs 51.5±5.6% NCH SD; P=.001). There was no significant difference in Ki67 staining in the intimal or medial layers between the NCH and CH groups of either rat strain. However, there was significantly more adventitial Ki67 staining in the CH compared to the NCH LE rats (13.0±2.7% NCH vs 28.5±4.8% CH; P=.004) and significantly more adventitial Ki67 staining in the NCH SD rats compared to the CH rats (51.5±5.6% NCH vs 35.5±5.1% CH; P=.04), corresponding to the pattern of neointimal hyperplasia at the 14-day time point.
Fig. 3.
Local arterial cell proliferation and arterial accumulation of monocytes/macrophages 3 days post-angioplasty. a Representative immunofluorescence for Ki67 in carotid artery sections of left LE NCH, LE CH, SD NCH, and SD CH rats. b Representative immunofluorescence for CD68 and CD206 in carotid artery sections of left LE NCH, LE CH, SD NCH, and SD CH rats. Negative isotype control (NEG) is also shown. Red represents positively stained cells. Blue represents DAPI nuclear counterstain. Green represents autofluorescence of elastin. Lumen of artery (L) is oriented towards the bottom for all sections. Original magnification 200x. Scale bar 50 μM.
Early local arterial monocyte/macrophage infiltration
Since early monocyte/macrophage infiltration to sites of arterial injury is associated with more severe neointimal hyperplasia,8–10 we next examined CD68 immunoreactivity in each layer of the arterial wall 3 days after carotid angioplasty. As shown in Fig. 3b, CD68 immunoreactivity was highest in the adventitial layer of both LE and SD rats regardless of housing group. There was less CD68 staining in the adventitial layer of NCH SD rats compared to NCH LE rats (34.8% NCH LE vs 18.6% NCH SD; P=.02). However, the fraction of CD68-positive cells in the adventitia did not change in the LE group after cohousing (34.8% NCH LE vs 34.2% CH LE; P=.54). In contrast, cohoused SD rats had much higher adventitial accumulation of CD68-positive cells compared to NCH SD rats (18.6% NCH SD vs 28.3% CH SD; P=.004). There was no significant difference in CD68 staining in the uninjured right CCA between LE and SD rats (not shown).
Cohousing also impacted macrophage polarization in the adventitial layers after angioplasty. M2 macrophage marker (CD206) staining was similar in the adventitia of NCH and CH LE rats (25.4% NCH vs. 30.4% CH; P=.07) (Fig. 3b). However, in the SD rats, CD206 staining was present in 15.4% of adventitial cells in the NCH group, which was significantly increased (to 34%) in the CH group (P=.003). There was minimal CD206 staining in the intimal and medial layers of both strains at the 3-day time point.
Composition of rat fecal microbiome
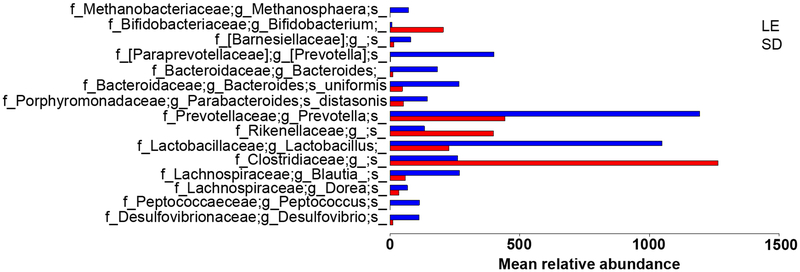
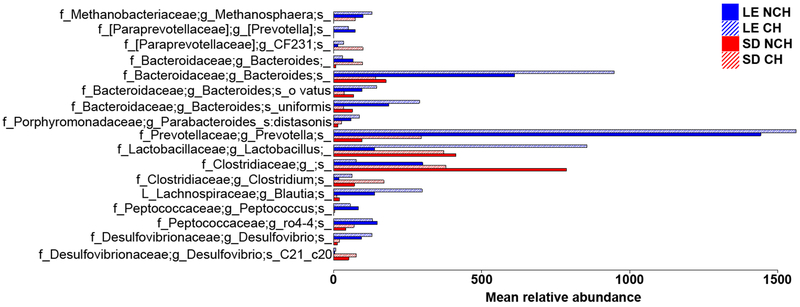
NCH SD and LE rats had microbiomes significantly differentiated by both weighted (P=.012) and unweighted (P<.001) unifrac beta diversity distances, though without significant differences in alpha diversity (P=.43). This is indicative of similarly rich microbiota that are nonetheless distinct in composition between rat strains, as would be expected due to controlled environmental conditions. The baseline between-strain microbiome composition differences are summarized in Fig. 4.
Fig 4.
Relative abundances of differentially abundant operational taxonomic units (OTUs) at baseline, prior to cohousing. Relative abundances were defined by the Benjamini-Hochberg multiple comparison corrected Kruskal-Wallis test with FDR P < .05 using samples collected prior to any microbiome manipulations.
Reshaping the fecal microbiome by cohousing and bedding mixing
Bedding mixing and cohousing of rats in the same treatment group reduces fecal microbial variation between individuals via coprophagy and direct animal-to-animal contact, resulting in increased similarity in microbial composition.11 To probe the role of commensal microbes in regulating neointimal hyperplasia, we cohoused LE and SD rats for 4 weeks before performing carotid angioplasty.
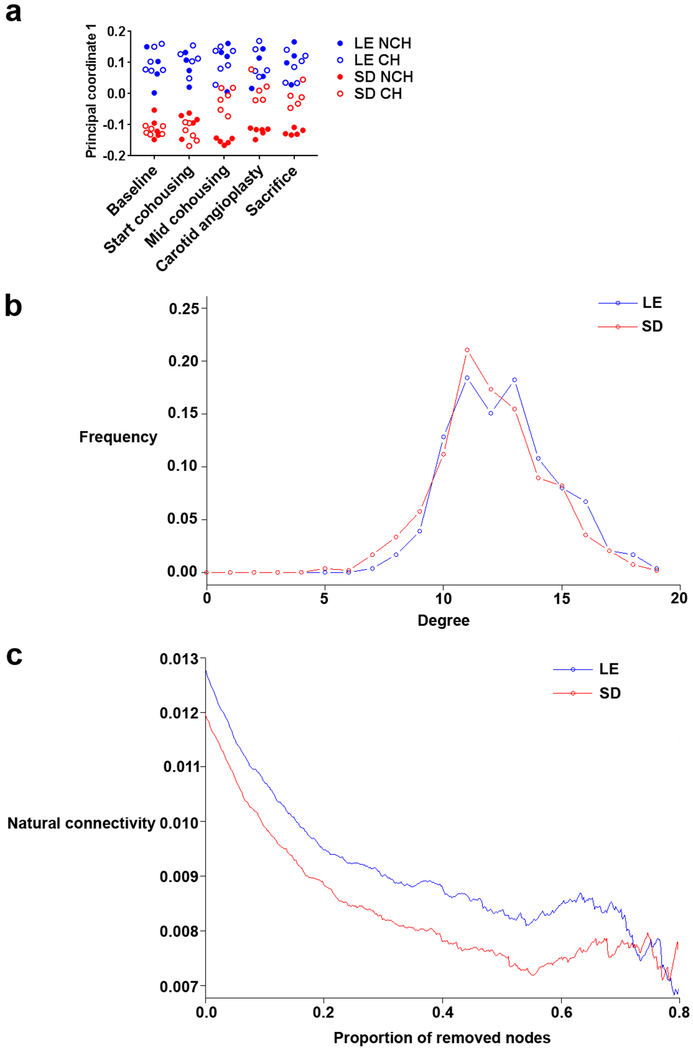
Over the course of the experiment, the microbial community shifted by cohousing. Although alpha diversity and weighted unifrac beta diversity was not significantly influenced by cohousing, unweighted beta diversity differed significantly between all groups (PERMANOVA P<.05). This is indicative of changes in relatively rare microbes after cohousing, rather than changes in the relative abundance of dominant taxa. Significant differences between strains at baseline and consistently across the experiment in NCH groups were observed in both weighted and unweighted beta diversity, indicating a distinct microbial makeup of the 2 strains (P<.05). The difference in unweighted beta diversity between the fecal microbiota of SD and LE rats was significantly reduced by cohousing (see principal component analysis in Fig. 5a), showing a convergence in the microbiota along the first principle axis (13% of variance explained). Strikingly, while an SD rat cohoused with an LE rat became significantly more similar to LE rats, an LE rat cohoused with an SD rat stayed more similar to LE rats than SD (Fig. 5a). This asymmetry might be attributed to a more stable LE microbiome, where new taxa cannot as easily take residence, and/or behavioral differences (primarily coprophagia). To test this hypothesis, operational taxonomic unit (OTU) co-occurrence networks were constructed via SPIEC-EASI12 for the 2 rat strains at baseline. While the 2 networks were similarly dense, measured by degree distribution, (Fig. 5b), progressively removing nodes revealed a lower natural connectivity in SD rats (Fig. 5c). This is indicative of an interaction network of microbes that is relatively similar but less robust to change; a result that corroborates the observation that the SD rat microbiome shifts more dramatically due to cohousing.
Fig 5.
Microbial diversity shifts between experimental timepoints and possible basis in baseline microbiome stability. a Principal coordinate analysis of unweighted unifrac beta diversity of microbiome samples in both strains and housing groups across sampling times. The first component (principal coordinate 1) is shown and explained 13% of the total variance. b Degree distribution of the OTU co-occurrence graph created by SPIEC-EASI on baseline time point samples. Both SD and LE rats show similar distributions, indicating similar network density. c Natural connectivity as a function of proportion of removed nodes of OTU co-occurrence network of SD and LE rats at baseline time point. The connectivity, as defined by the eigenvalue of the adjacency matrix of the shrinking graph, accesses the stability of the network, which becomes lower in the SD rats as nodes are removed.
A total of 17 OTUs were significantly different between the four animal groups (CH or NCH LE and SD; Fig. 6), and as expected from beta diversity analysis, the majority were low abundance taxa. Of these, Lactobacillus increased the most in CH LE rats and Methanosphaera increased the most in CH SD rats.
Fig 6.
Relative abundances of OTUs differentially abundant across experimental groups at the time of sacrifice. Relative abundances were defined by raw 16S read counts and differences between groups found by the Benjamini-Hochberg multiple comparison corrected Kruskal-Wallis test with FDR P<.05.
Correlation of relative abundance with neointimal hyperplasia
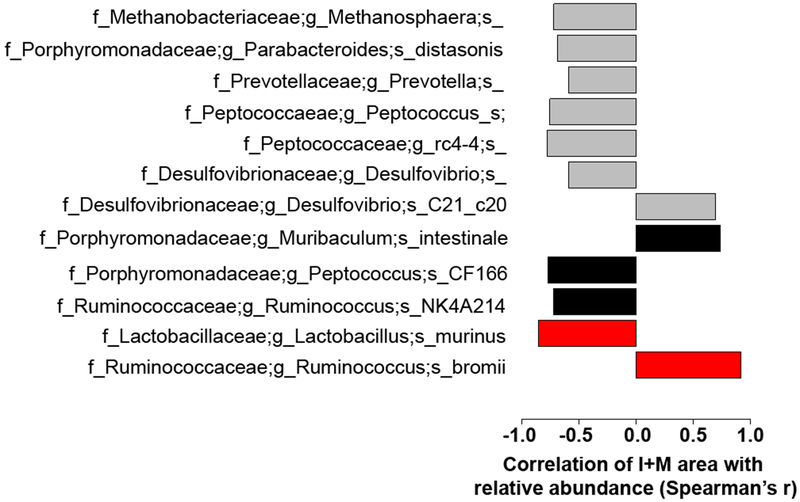
Mantel tests of average I+M areas with beta diversity were performed, and concomitant with above, only unweighted beta diversity was significantly differentiated by cohousing (R=.38; P<.001). Correlation between average I+M area and the relative abundances of OTUs on the day of surgery and day of sacrifice were performed; importantly, no significant difference in microbial composition was found between the day of surgery and the day of sacrifice, indicating recovery of the microbiome from possible perturbation due to surgery. At sacrifice, the relative abundances of 7 OTUs were significantly different between groups and highly significantly correlated with average I+M area per rat (R>.55, P<.05; Fig. 7), including Parabacteroides distasonis, Desulfovibrio, Methanosphaera, Peptococcus, and Prevotella. Stratifying the analysis by comparing only within rat strains across housing conditions resulted in no significant correlations at the OTU level, likely due to a loss of statistical power and the low relative abundance of the important OTUs. Following OTU denoising (DADA2), 5 subOTUs significantly correlated with I+M area and were identifiable at >96% 16S sequence similarity via Basic Local Alignment Search Tool (BLAST) (R>.7; P<.01; Fig. 7). These subOTUs represent individual 16S sequences instead of multiple sequences clustered into OTUs based on similarity, which increases the specificity of results and in some cases lead to identification of an exact microbe to culture, at the expense of often reduced statistical power. Three of these, Muribaculum intestinale, Peptococcus CF166, and Ruminococcus NK4A214 correlated across all groups. Two subOTUs correlated significantly with I+M area in SD rats alone: Ruminococcus bromii (R=.92;P=.001) and Lactobacillus murinus (R=−.85; P=.03).
Fig 7.
Benjamini-Hochberg multiple comparison corrected Spearman’s r correlations of specific OTUs and subOTUs with average I+M area with FDR P<.05. Gray bars indicate correlations at the OTU level, black at the subOTU level as defined by DADA2 denoised, Basic Local Alignment Search Tool (BLAST) assigned individual sequence variants, and red at the subOTU level only comparing within groups of SD rats. All other correlations are across all rat groups.
Microbe-dependent metabolites and neointimal hyperplasia
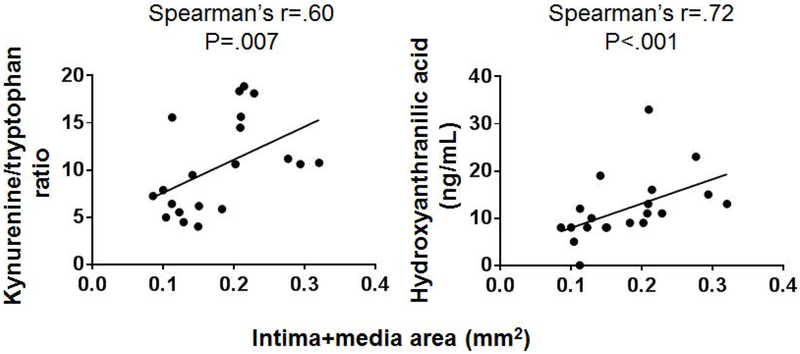
Microbe-derived components and metabolic products function as signaling molecules in a variety of inflammatory and immune pathways. An untargeted metabolomics study comparing plasma present in conventional and germ-free mice revealed distinct plasma metabolite profiles between the 2 sample sets,13 implying a role for bacterial-dependent metabolism on host-microbial interactions. Some of these metabolites were indole and phenyl derivatives, which we previously have found to be associated with presence of advanced atherosclerosis14 and which, like neointimal hyperplasia, is driven by chronic systemic inflammation. Thus, we assessed potential associations between indole and phenyl-derived metabolites and neointimal hyperplasia. As shown in Table II, there were significant differences in mean serum concentration of multiple indole-derived metabolites between the NCH LE and SD rats, including serotonin (2.8±.1 μM LE vs 4.8±.2 SD μM; P=.02), kynurenine (.65±.03 μM LE vs 1.47±.18 μM SD; P=.02), the kynurenine/tryptophan ratio (6.7±.5 LE vs 13.0±1.5 SD; P=.02), and hydroxyanthranilic acid (58.6±.0 nM LE vs 97.8±13.7 nM SD; P=.02). Cohousing and bedding mixing did not significantly alter the serum concentration of any of the assessed metabolites (Table II) despite the significant changes in gut microbial communities described in the Results. However, when we performed a correlation analysis between metabolite concentrations and I+M area in the entire rat cohort, we found significant positive correlations between the kynurenine/tryptophan ratio (Spearman r=.60, P=.007) and 3-hydroxyanthranilic acid (Spearman r=.72; P<.001) (Fig 8). Since the SD rats demonstrated decreased mean I+M area upon cohousing, we next examined whether these 3 metabolites had any correlation with I+M area in the subgroups of NCH and CH SD rats. There was no correlation between 3-hydroxyanthranilic acid concentration and I+M area in either the NCH or the CH SD rats. However, in both NCH and CH SD rats, there was a trend towards a correlation between kynurenine/tryptophan ratio and I+M area (Spearman’s r=.81, P=.1 NCH; r=.78,P=.1 CH).
Table II.
Serum metabolite concentrations in NCH and CH LE and SD rats.
| Metabolite | LE | P value | SD | P value | P value LE NCH vs. SD NCH | ||
|---|---|---|---|---|---|---|---|
| NCH | CH | NCH | CH | ||||
| Indole (μM) | .35±.07 | .34±.03 | .99 | .44±.08 | .52±.03 | .45 | .78 |
| Tryptophan (mM) | 98.2±4.3 | 112.3±5.8 | .06 | 116.2±17.7 | 143.9±23.5 | .43 | .91 |
| Serotonin (μM) | 2.8±.1 | 2.4±.1 | .06 | 4.8±.2 | 4.4±.5 | .50 | .02 |
| Kynurenine (μM) | .65±.03 | .58±.06 | .78 | 1.47±.18 | 2.00±.21 | .08 | .02 |
| Kynurenine/tryptophan ratio | 6.7±.5 | 5.1±.5 | .11 | 13.0±1.5 | 14.8±1.6 | .66 | .02 |
| Indole-3-propionic acid (μM) | 5.1±.6 | 4.8±.7 | .90 | 6.2±.7 | 7.9±.8 | .18 | .56 |
| 3-Hydroxyanthranilic acid (nM) | 58.6±0 | 41.7±13.0 | .99 | 97.8±13.7 | 108.9±23.4 | .37 | .02 |
| Hippuric acid (μM) | 3.8±.2 | 4.6±.7 | .19 | 5.6±1.9 | 4.4±.7 | .66 | .29 |
| Indoxyl sulfate (μM) | 4.6±.2 | 5.4±.5 | .19 | 5.1±.5 | 7.0±.9 | .25 | .56 |
| P-Cresyl sulfate (μM) | 1.18±.36 | 1.20±.27 | .90 | .92±.32 | .92±.46 | .79 | .56 |
All areas expressed as mean ± SEM. P values < .05 are denoted in bold.
Fig. 8.
Spearman’s r correlations of microbe-derived metabolites with neointimal hyperplasia (mean intima+media area [mm2]) in all rats irrespective of strain or housing group. N=16-20 samples.
Discussion
This is a descriptive study of how cohousing genetically different rat strains resulted in altered adult host phenotypic differences in neointimal hyperplasia development and in local arterial inflammatory cell infiltration after carotid angioplasty as well as significant shifts in fecal microbiota communities. These results are important because they suggest a direct role for gut microbiota in the arterial remodeling response after injury that occurs through modulation of the acute inflammatory response.
A striking modulation of I+M area after angioplasty was observed in response to cohousing and bedding mixing, especially in SD rats. While the relative contributions of genetics and microbiome compositions to neointimal hyperplasia susceptibility are unknown, this work demonstrates that the microbiome likely plays a strong role. The finding that CH LE rats exhibit a relatively small change in neointimal hyperplasia as well as a relatively minor shift in gut microbial composition supports this idea.
This difference in resistance of the microbiome to change after cohousing between SD and LE rats may be caused by higher stability of the LE microbiome and/or a stronger selective pressure by the host towards certain microbial consortia. The stability to loss of OTUs was tested through attack on the occurrence networks of the baseline SD and LE microbiomes, revealing that SD microbial networks fell apart more quickly as nodes were removed. This is indicative of a less stable microbiome; however, further experimental tests (e.g., longitudinal data as the microbiomes are being shifted) are needed to expand on this result. Other investigators have demonstrated that co-housing mice of different strains led bacterial communities to be more similar to each other, but strain-specificity was retained,15 as we also observed. Though there have been some experiments on comparisons and manipulations of LE and SD microbiomes,16 the compositional and functional differences of these strains’ microbiota continues to be underexplored given their potentially large effects on study outcomes. This is further confounded by handling and cage effects, which also remain understudied.17 Nonetheless, by focusing on changes in microbiota from a known starting point and following perturbations such as cohousing longitudinally, these confounders can be partly mitigated.
We observed several notable associations between specific microbial strains, neointimal hyperplasia, and inflammation that can serve as hypothesis-generators for future studies. There was increased M2 macrophage infiltration in the adventitia of CH SD rats compared to NCH SD rats at the 3-day time point (P=.003), which corresponded with increased relative abundance of Parabacteroides distasonis in the CH group and with attenuated neointimal hyperplasia. Notably, P. distasonis relative abundance in all groups strongly negatively correlated with I+M area (r=−.69). Prior work by others has shown that oral administration of P. distasonis has an anti-inflammatory effect in a mouse model of acute colitis, and that its antigens decrease pro-inflammatory cytokine production macrophages in vitro.18 Thus, this commensal organism may modulate innate immunity mechanisms to attenuate neointimal hyperplasia, a process initiated and potentiated by inflammation. Alternatively, P. distasonis may exert its effect via adaptive immunity mechanisms. Treatment of mice with P. distasonis increased the number of FoxP3+ regulatory T cells (Tregs),18 which may represent a change in the balance of Treg-TH17 cells required for commensalism.19 TH17 cells produce IL-17A, which mediates pro-inflammatory responses and was observed in our study to be significantly higher in NCH SD than in NCH LE SD rats at 3 days (P=.04) (data not shown), corresponding with greater neointimal hyperplasia in the NCH SD rats. Furthermore, IL-17A was significantly lower in CH SD rats compared to NCH SD rats (P=.04) (data not shown), corresponding to attenuated neointimal hyperplasia. IL-17A has also been shown to stimulate vascular smooth muscle cell migration and MMP9 expression in vitro,20 cellular processes which are relevant to neointimal hyperplasia.
We observed a negative correlation between Prevotella relative abundance and I+M area across all groups (R=−.59). Prevotella is one of the predominant genera in the Bacteroidetes phyla. Prevotella relative reduction has been associated with atherosclerotic disease in humans, another chronic inflammatory disease,21 and with hypertension.22 However, Prevotella abundance has also been associated with increased TH17 immune responses, mucosal inflammation, and systemic dissemination of inflammatory mediators.23 These conflicting data suggest a likely complex host-Prevotella relationship, especially since species-specific roles of Prevotella exist.
We also found a significant negative association between Methanosphaera (R=−.72) and conflicting associations between Desulfovibrio (R=.70 and =−.59) and I+M area across all groups. Desulfovibrio reduce sulfate to produce hydrogen sulfide,24 while Methanosphaera utilize hydrogen to reduce methanol to methane.25 Hydrogen sulfide has been shown to have a protective effect on restenosis in atherosclerotic rabbits26 and in balloon-injured rats,27 but given the complex relationship of hydrogen balance to host homeostasis,28 it is not surprising that different Desulfovibrio OTUs correlate with I+M area in different directions and magnitude.
Ruminococcus bromii, which had a strong positive correlation with I+M area in the SD rats, is a specialized “keystone” organism that can degrade certain forms of resistant starch to short chain fatty acids.29 We previously demonstrated an inverse relationship between serum butyrate concentration in antibiotic-treated LE rats and neointimal hyperplasia after carotid angioplasty and a potential direct anti-migratory and anti-proliferative effect on vascular smooth muscle cells.7 Others have observed attenuated LPS-induced neuroinflammation in rats given supplementary dietary acetate,30 another major short chain fatty acid. In contrast, acetate is a pro-inflammatory mediator in hepatitis31 and cultured gastric adenocarcinoma cells.32 These data suggest a tissue-specific effect for acetate whose role in inflammatory processes needs further clarification.
Finally, Muribaculum intestinale, which also had a strong positive correlation with I+M area (R=.8), is a relatively newly isolated member of mouse gut bacteria33 whose functional effect on the host is not yet known. Similarly, the functional significance of the genus Peptococcus, gram-positive anaerobes that negatively correlated across all groups with I+M area, is unknown. However, glycosulfatase-like enzymes have been found in Peptococcus niger, pointing towards another possible sulfur-driven role in neointimal hyperplasia reduction. L. murinus, which had a negative correlation with I+M area, has been observed to modulate Th17 cells in salt-sensitive hypertension.34
We observed strong correlations between neointimal hyperplasia and microbe-derived metabolites kynurenine/tryptophan ratio and 3-hydroxyanthranilic acid. However, given the diverse effects of gut microbes on host physiology,35 it is not surprising that there may be multiple mechanistic pathways which are both overlapping and distinct. The kynurenine pathway accounts for the catabolism on 99% of ingested tryptophan not used for protein synthesis. 36 Upon entering the kynurenine pathway, tryptophan is converted to N-formyl-l-kynurenine by indole-2,3-deoxygenase (IDO1) (ubiquitously expressed) and then to kynurenine. IDO1 is known to be upregulated by interferon-gamma, IL-2, and IL-10 activity. The kynurenine/tryptophan ratio, which is an index of IDO1 activity, has been found to be elevated in states of immune stimulation, such as infection, and endotoxin administration.37,38 Downstream kynurenine catabolites including 3-hydroxyanthranilic acid reduce Th1 and Th17 responses and affect T cell apoptosis.39,40 Thus, as neointimal hyperplasia an inflammatory process, it is conceivable that there is a strong positive correlation with kynurenine/tryptophan ratio and 3-hydroxyanthranilic acid, as we observed.
This study’s strengths lie in its experimental design, which demonstrates that exchange of microbes in adult rats is sufficient to incur changes in acute arterial inflammation and downstream neointimal hyperplasia. Our findings also add to a growing body of literature linking gut microbiota to the peripheral vasculature, specifically atherosclerosis, arterial stiffness, blood pressure, and endothelial function.41–44 While we did not obtain direct mechanistic data linking specific microbial strains with macrophage polarization or neointimal hyperplasia development, we identified several bacterial strains that correlated highly with neointimal hyperplasia severity that can be the subjects of future focused interventions. Furthermore, certain biomarkers and microbe-derived metabolites such as hydrogen sulfide compounds might also be targeted based on our data. For example, metagenomic studies that could differentiate and categorize the Desulfovibrio strains and correlate marker genes with metabolites are in the immediate future plan.
Limitations of this study include its descriptive nature, which precludes any inference of causality by gut microbiota or microbe-derived metabolites, since correlative microbiota and metabolite data from organismal systems will not give a true picture of which microbiota contribute to specific metabolic pathways. While we examined differences in local cell proliferation and inflammatory cell accumulation between the NCH and CH groups, we did not characterize differences in cell-specific proliferation, vascular smooth muscle cell phenotypes, extracellular matrix content, thrombosis, endothelial function, and oxidative stress, although all these measures would deepen our understanding of poorly understood microbial-driven responses in the peripheral vasculature. In addition, while our use of CD68 as a pan-macrophage cell marker and CD206 as a marker of M2 macrophages is supported by recent literature,45–47 we also recognize that macrophage polarization is likely not binary and is controlled by multiple genes.48 Thorough characterization of macrophage polarization in relation to our observed microbiome changes would require detailed analysis of source of macrophages, definition of activators, and description of multiple cellular markers,49 which would be beyond the scope of this exploratory study but well within our plans for future mechanistic studies. This detailed assessment of macrophage activation would also include assessment of alternatively activated adventitial macrophages.50 Finally, we did not explore the possibility that other microbiomes, such as the oral, skin, or urogenital microbiomes, and not just the fecal microbiome, have a role in the vascular and inflammatory phenotypes that we observed.
Conclusions
In summary, these data presented in this exploratory study argue for in-depth mechanistic studies into a novel mechanism for how microbiome manipulations affect arterial remodeling after injury and for disentanglement of the genetic versus microbial mechanisms for neointimal hyperplasia and, more generally, for immune function. Furthermore, we demonstrate that cohousing and bedding mixing, which are relatively easily undertaken experimental methods, are a valuable starting point for focused interventions.
Article Highlights.
Type of Research:
basic science
Key Findings:
Exchange of fecal microbiota resulted in alteration of neointimal hyperplasia phenotype and acute arterial inflammatory response after carotid angioplasty in rats
Take home Message:
Microbiome manipulation can affect arterial remodeling and the inflammatory response after arterial injury. A greater understanding of the host inflammatory8microbe axis could uncover novel therapeutic targets for the prevention and treatment of restenosis.
Clinical Relevance.
Neointimal hyperplasia as a cause of arterial restenosis after vascular interventions such as balloon angioplasty, stenting, and bypass surgery is a highly prevalent problem. Our goal is to ascertain the role of gut microbiota in mediating inflammatory and cell proliferative pathways that drive neointimal hyperplasia. The purpose of this study is to determine if the exchange of microbes between rats with differing neointimal hyperplasia phenotypes and distinct intestinal microbiota could alter the arterial remodeling phenotype after angioplasty. Our observations provide a starting point for focused microbiota-related interventions to prevent or treat neointimal hyperplasia.
Appendix
Methods
Experimental rats
Eight-week-old male Lewis Inbred (LE) and Sprague-Dawley (SD) male rats obtained from Envigo (Indianapolis, IN) were housed in a barrier facility at Northwestern University under a 12-hour light cycle. Standard irradiated rat chow and autoclaved drinking water were provided ad libitum.
Study design
In order to equilibrate initial animal housing conditions, bedding (including stool pellets) from all cages within one strain were intermixed 3 times per week for 2 weeks. A cohort of LE and SD rats were then cohoused (CH) (separated and then re-caged with the opposing strain), while a separate cohort of LE and SD rats (non-cohoused [NCH]) remained in cages segregated by rat strain (see experimental schema in Fig. 1). Bedding intermixing continued thereafter among cages in the same housing groups. After four weeks, all animals underwent left carotid artery balloon angioplasty; the right carotid artery served as the uninjured control artery. Animals were sacrificed for tissue and blood collection either 3 or 14 days after carotid angioplasty. Serum was immediately prepared from blood using serum separator tubes (BD, Franklin Lakes, NJ) and frozen at −80°C in single use aliquots until use. Animals were weighed on arrival and weekly thereafter. Stool samples were collected upon arrival to the animal facility, prior to the cohousing period, 2 weeks after the start of the cohousing period, on the day of carotid angioplasty, and on the day of sacrifice. Freshly collected stool pellets were immediately frozen and stored at −80°C until use.
Rat carotid artery balloon angioplasty, tissue processing, and morphometric analysis
After induction of general anesthesia with isoflurane, all animals underwent left carotid artery balloon angioplasty using a 2-French Fogarty catheter (Edwards Lifesciences, Irvine, CA) as previously described.1 The balloon catheter is positioned in the distal common carotid artery just proximal to the carotid bifurcation. At the appropriate time point, rats were euthanized and in situ perfusion fixation with phosphate-buffered saline (PBS) and 2% paraformaldehyde was performed prior to harvest of both carotid arteries. Arteries were fixed in 2% paraformaldehyde at 4°C for 1 hour followed by overnight cryoprotection in 30% sucrose at 4°C and then embedded in OCT compound (Tissue-Tek, Sakura Finetek, Torrance, CA). Whole blood was collected by cardiac puncture prior to perfusion fixation at the time of sacrifice. Serum was isolated and stored at −80°C until use.
Serial 5-μm frozen sections were collected on Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA) from the carotid bifurcation to the distal common carotid artery over a total distance of approximately 4 mm (corresponding to the entire angioplastied segment), air dried, and stored at −20°C. Carotid arteries were examined for neointimal hyperplasia after hematoxylin-eosin staining of artery sections at evenly-spaced 350-μM intervals. Digital images of stained sections were obtained using a Leica DM2000 light microscope with a 20x objective and camera (W. Nuhsbaum, Inc., McHenry, IL). Intima and media area were measured using ImageJ software (NIH, Bethesda, MD) after calibration.
Antibodies
Rabbit polyclonal antibody to Ki67 (ab15580) and CD206 (ab64693) were purchased from Abcam (Boston, MA). Mouse monoclonal antibody to CD68 (MCA341R) was purchased from Bio-Rad (Hercules, CA). Secondary antibodies were Alexa Fluor 555 goat anti-rabbit IgG and AlexaFluor 555 goat anti-mouse IgG from Invitrogen (Waltham, MA).
Immunofluorescence and quantification of staining
Frozen sections were outlined using ImmEdge hydrophobic barrier pen (Vector Labs, Burlingame, CA), rehydrated, fixed in 2% paraformaldehyde for 20 minutes, washed with PBS, incubated with the primary antibodies diluted in IHC-Tek diluent pH 7.4 (IHC-World, Woodstock, MD) for 1 hour at room temperature, washed in PBS, incubated with the appropriate secondary antibody diluted in IHC-Tek diluent pH 7.4 for 1 hour at room temperature, washed in PBS, and mounted with ProLong Gold Antifade mountant with DAPI (ThermoFisher Scientific, Waltham, MA). Primary antibodies were used at the following concentrations: Ki67, 2 μg/mL; CD68, 1 μg/mL; CD206, 1 μg/mL. AlexaFluor 555 goat anti-rabbit IgG secondary antibody was used at 4 μg/mL for Ki67 and CD206 and AlexaFluor 555 goat anti-mouse IgG secondary antibody was used at 2 μg/mL for CD68.
Digital images were acquired using a Nikon Eclipse 50i microscope (Nikon Instruments, Inc., Melville, NY) with a 20x objective and equipped with DAPI, Texas red, and FITC emission filters, SPOT Advanced Imaging Software (Diagnostic Instruments, Sterling Heights, MI), and RTTM KE Color 3-Shot digital microscope camera (Diagnostic Instruments). Positively stained cells and DAPI-stained nuclei in the intimal, medial, and adventitial layers of each high-powered field were manually counted in a blinded fashion and a ratio of stained cells per total cells was calculated. At least six high-powered fields per animal sampled from evenly-spaced sections and five or six animals per treatment group were examined.
Genomic DNA extraction from fecal samples
Stool pellets were collected and stored at −80°C until processing. 100 mg of stool pellets were used for microbial DNA extraction with the DNeasy Powersoil HTP 96 kit (Qiagen, Germantown, MD). The following modifications were made to the manufacturer’s protocol: after samples, the bead solution, and the C1 solution were added to the bead plate, the plate was partially submerged in a water bath for 20 minutes at 60°C, followed by a 20-minute shaking step on a MM 400 plate shaker (Retsch, Haan, Germany). The manufacturer’s protocol was then followed to obtain clean DNA.
16S rRNA gene amplification and sequencing
The V4 region of the bacterial 16S rRNA gene was amplified using the Earth Microbiome Project primer set (515f-806r).2 Each 25 μl PCR reaction contained 12.5 μl of AccuStart II PCR ToughMix (Quantabio, Beverly, MA), 1 μl of 5 μM forward primer, 1 μl of 5 μM reverse primer, 9.5 μl of water, and 1 μl of DNA extraction. The PCR program was 94°C for 3 minutes to denature the DNA, with 35 cycles at 94°C for 45 seconds, 50°C for 60 seconds, 72°C for 90 seconds, and then a final 72°C step. Amplification was quantified using Picogreen (Invitrogen, Carlsbad, CA), and each sample was pooled at 70 ng per sample. Pools were cleaned using Agencourt AMPure XP beads (Beckman Coulter, Indianapolis, IN), and the clean pools combined. This final pool was quantified and sent to the Environmental Sample Preparation and Sequencing Facility (ESPSF) at Argonne National Laboratory for sequencing on an Illumina Miseq using V3 chemistry, following the Earth Microbiome Project protocol.3
Sequence data handling and analysis
Raw 16S reads were demultiplexed, chimera and phiX filtered through the Qiime2 pipeline2 and denoised with DADA2.4 The resulting representative sequences were aligned and masked with MAFFT.5 A phylogenetic tree was constructed via FastTree2.6 Samples were subsequently clustered and taxonomically classified into operational taxonomic units (OTUs) with Qiime2 utilizing the greengenes 99% database.7 Samples were filtered by globally removing OTUs with minimum count fractions of .001. Differences in relative abundances were calculated via bootstrapped Kruskal-Wallis tests (999 permutations) for multiple group comparisons. Correlations of metadata with OTU abundances were calculated as Spearman’s rank correlations with raw P values determined via bootstrapping (1000 permutations). All reported P values are Benjamini-Hochberg false discovery rate (FDR) corrected for multiple comparisons. Diversity metrics were calculated after rarefying reads at a depth of 20,000 sequences. Beta diversity was calculated as unweighted8 and weighted9 unifrac distances. Faith’s Phylogenetic Diversity was used to measure alpha diversity.10 Beta diversity correlations with metadata were calculated by Mantel test (999 permutations). OTU co-occurrence networks were created with SPEIC-EASI.11 SubOTUs were taxonomically assigned via the NCBI Basic Local Alignment Search Tool (BLAST) web interface.12
Detection and quantification of serum metabolites by high performance liquid chromatography (HPLC)-tandem mass spectrometry
P-cresyl sulfate (PCS), hippuric acid, indole-3-propionic acid (I3P), tryptophan (trp), kynurenine (kyn), indoxyl sulfate (IS), and serotonin. Fifty μL of serum and internal standards (n-methyl serotonin and p-toluene sulfonic acid) were prepared as previously described and analyzed using tandem mass spectrometry (MS/MS) using an Agilent HPLC-MS/MS system with a 6400-series triple quadrupole (QQQ) mass spectrometer described in detail previously.13 Indole, indole-3-aldehyde (I3A), and 3-hydroxyanthranilic acid. Samples were prepared from 50 μL of serum by solid-phase extraction and analyzed by an API 3000 HPLC–MS/MS system (Applied Biosystems, Foster City, CA) equipped with an Agilent 1100 series HPLC system (Agilent Technologies) operating in positive ion mode as described in detail previously.13
Statistical analysis
Morphometric and metabolite data analysis was carried out using GraphPad Prism 6 (GraphPad Software, La Jolla, CA). Student’s t-test and Mann-Whitney U-tests were used as appropriate. Spearman’s rank-order correlations were used to measure the strength and direction of the association between metabolite concentrations and neointimal hyperplasia. P<.05 was considered statistically significant.
References for Appendix
- 1.Ho KJ, Xiong L, Hubert NJ, Nadimpalli A, Wun K, Chang EB, et al. Vancomycin treatment and butyrate supplementation modulate gut microbe composition and severity of neointimal hyperplasia after arterial injury. Physiological reports. 2015;3(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6(8):1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature methods. 2016;13(7):581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution. 2013;30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PloS one. 2010;5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72(7):5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71(12):8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Applied and environmental microbiology. 2007;73(5):1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation. 1992;61(1): 1–10. [Google Scholar]
- 11.Kurtz ZD, Muller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. Sparse and compositionally robust inference of microbial ecological networks. PLoS computational biology. 2015;11(5):e1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, et al. BLAST: a more efficient report with usability improvements. Nucleic acids research. 2013;41(Web Server issue):W29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cason CA, Dolan KT, Sharma G, Tao M, Kulkarni R, Helenowski IB, et al. Plasma microbiome-modulated indole- and phenyl-derived metabolites associate with advanced atherosclerosis and postoperative outcomes. Journal of vascular surgery. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jukema JW, Verschuren JJ, Ahmed TA, Quax PH. Restenosis after PCI. Part 1: pathophysiology and risk factors. Nat Rev Cardiol. 2011;9(1):53–62. [DOI] [PubMed] [Google Scholar]
- 2.Assadnia S, Rapp JP, Nestor AL, Pringle T, Cerilli GJ, Gunning WT 3rd, et al. Strain differences in neointimal hyperplasia in the rat. Circulation research. 1999;84(11):1252–1257. [DOI] [PubMed] [Google Scholar]
- 3.Harmon KJ, Couper LL, Lindner V. Strain-dependent vascular remodeling phenotypes in inbred mice. The American journal of pathology. 2000;156(5):1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang B, Deng Y, Suen C, Taha M, Chaudhary KR, Courtman DW, et al. Marked Strain-Specific Differences in the SU5416 Rat Model of Severe Pulmonary Arterial Hypertension. American journal of respiratory cell and molecular biology. 2016;54(4):461–468. [DOI] [PubMed] [Google Scholar]
- 5.Langer M, Brandt C, Loscher W. Marked strain and substrain differences in induction of status epilepticus and subsequent development of neurodegeneration, epilepsy, and behavioral alterations in rats. [corrected]. Epilepsy research. 2011;96(3):207–224. [DOI] [PubMed] [Google Scholar]
- 6.Bardutzky J, Shen Q, Henninger N, Bouley J, Duong TQ, Fisher M. Differences in ischemic lesion evolution in different rat strains using diffusion and perfusion imaging. Stroke; a journal of cerebral circulation. 2005;36(9):2000–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho KJ, Xiong L, Hubert NJ, Nadimpalli A, Wun K, Chang EB, et al. Vancomycin treatment and butyrate supplementation modulate gut microbe composition and severity of neointimal hyperplasia after arterial injury. Physiological reports. 2015;3(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furukawa Y, Matsumori A, Ohashi N, Shioi T, Ono K, Harada A, et al. Anti-monocyte chemoattractant protein-1/monocyte chemotactic and activating factor antibody inhibits neointimal hyperplasia in injured rat carotid arteries. Circulation research. 1999;84(3):306–314. [DOI] [PubMed] [Google Scholar]
- 9.Moreno PR, Bernardi VH, Lopez-Cuellar J, Newell JB, McMellon C, Gold HK, et al. Macrophage infiltration predicts restenosis after coronary intervention in patients with unstable angina. Circulation. 1996;94(12):3098–3102. [DOI] [PubMed] [Google Scholar]
- 10.Danenberg HD, Fishbein I, Epstein H, Waltenberger J, Moerman E, Monkkonen J, et al. Systemic depletion of macrophages by liposomal bisphosphonates reduces neointimal formation following balloon-injury in the rat carotid artery. Journal of cardiovascular pharmacology. 2003;42(5):671–679. [DOI] [PubMed] [Google Scholar]
- 11.Moore RJ, Stanley D. Experimental design considerations in microbiota/inflammation studies. Clin Transl Immunology. 2016;5(7):e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurtz ZD, Muller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. Sparse and compositionally robust inference of microbial ecological networks. PLoS computational biology. 2015;11(5):e1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(10):3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cason CA, Dolan KT, Sharma G, Tao M, Kulkarni R, Helenowski IB, et al. Plasma microbiome-modulated indole- and phenyl-derived metabolites associate with advanced atherosclerosis and postoperative outcomes. Journal of vascular surgery. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell JH, Foster CM, Vishnivetskaya T, Campbell AG, Yang ZK, Wymore A, et al. Host genetic and environmental effects on mouse intestinal microbiota. The ISME journal. 2012;6(11):2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, et al. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome research. 2010;20(10):1411–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Servick K Of mice and microbes. Science. 2016;353(6301):741–743. [DOI] [PubMed] [Google Scholar]
- 18.Kverka M, Zakostelska Z, Klimesova K, Sokol D, Hudcovic T, Hrncir T, et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clinical and experimental immunology. 2011;163(2):250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806. [DOI] [PubMed] [Google Scholar]
- 20.Cheng G, Wei L, Xiurong W, Xiangzhen L, Shiguang Z, Songbin F. IL-17 stimulates migration of carotid artery vascular smooth muscle cells in an MMP-9 dependent manner via p38 MAPK and ERK1/2-dependent NF-kappaB and AP-1 activation. Cellular and molecular neurobiology. 2009;29(8): 1161–1168. [DOI] [PubMed] [Google Scholar]
- 21.Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nature communications. 2017;8(1):845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151(4):363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Postgate JR, Campbell LL. Classification of Desulfovibrio species, the nonsporulating sulfate-reducing bacteria. Bacteriological reviews. 1966;30(4):732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller TL, Wolin MJ. Methanosphaera stadtmaniae gen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen. Archives of microbiology. 1985;141(2):116–122. [DOI] [PubMed] [Google Scholar]
- 26.Ma B, Liang G, Zhang F, Chen Y, Zhang H. Effect of hydrogen sulfide on restenosis of peripheral arteries after angioplasty. Molecular medicine reports. 2012;5(6):1497–1502. [DOI] [PubMed] [Google Scholar]
- 27.Meng QH, Yang G, Yang W, Jiang B, Wu L, Wang R. Protective effect of hydrogen sulfide on balloon injury-induced neointima hyperplasia in rat carotid arteries. The American journal of pathology. 2007;170(4):1406–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carbonero F, Benefiel AC, Gaskins HR. Contributions of the microbial hydrogen economy to colonic homeostasis. Nature reviews. Gastroenterology & hepatology. 2012;9(9):504–518. [DOI] [PubMed] [Google Scholar]
- 29.Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. The ISME journal. 2012;6(8):1535–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reisenauer CJ, Bhatt DP, Mitteness DJ, Slanczka ER, Gienger HM, Watt JA, et al. Acetate supplementation attenuates lipopolysaccharide-induced neuroinflammation. Journal of neurochemistry. 2011;117(2):264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kendrick SF, O’Boyle G, Mann J, Zeybel M, Palmer J, Jones DE, et al. Acetate, the key modulator of inflammatory responses in acute alcoholic hepatitis. Hepatology. 2010;51(6):1988–1997. [DOI] [PubMed] [Google Scholar]
- 32.Sun J, Bi L, Chi Y, Aoki K, Misumi J. Effect of sodium acetate on cell proliferation and induction of proinflammatory cytokines: a preliminary evaluation. Food and chemical toxicology : an international journal publishedfor the British Industrial Biological Research Association. 2005;43(12):1773–1780. [DOI] [PubMed] [Google Scholar]
- 33.Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N, et al. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nature microbiology. 2016;1(10):16131. [DOI] [PubMed] [Google Scholar]
- 34.Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017;551(7682):585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slingerland AE, Schwabkey Z, Wiesnoski DH, Jenq RR. Clinical Evidence for the Microbiome in Inflammatory Diseases. Frontiers in immunology. 2017;8:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peters JC. Tryptophan nutrition and metabolism: an overview. Advances in experimental medicine and biology. 1991;294:345–358. [DOI] [PubMed] [Google Scholar]
- 37.Taylor MW, Feng GS. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 1991;5(11):2516–2522. [PubMed] [Google Scholar]
- 38.Zelante T, Fallarino F, Bistoni F, Puccetti P, Romani L. Indoleamine 2,3-dioxygenase in infection: the paradox of an evasive strategy that benefits the host. Microbes and infection. 2009;11(1):133–141. [DOI] [PubMed] [Google Scholar]
- 39.Platten M, Ho PP, Youssef S, Fontoura P, Garren H, Hur EM, et al. Treatment of autoimmune neuroinflammation with a synthetic tryptophan metabolite. Science. 2005;310(5749):850–855. [DOI] [PubMed] [Google Scholar]
- 40.Terness P, Bauer TM, Rose L, Dufter C, Watzlik A, Simon H, et al. Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: mediation of suppression by tryptophan metabolites. The Journal of experimental medicine. 2002;196(4):447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim S, Goel R, Kumar A, Qi Y, Lobaton G, Hosaka K, et al. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clinical science. 2018;132(6):701–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marques FZ, Nelson E, Chu PY, Horlock D, Fiedler A, Ziemann M, et al. High-Fiber Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in Hypertensive Mice. Circulation. 2017;135(10):964–977. [DOI] [PubMed] [Google Scholar]
- 43.Natarajan N, Hori D, Flavahan S, Steppan J, Flavahan NA, Berkowitz DE, et al. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiological genomics. 2016;48(11):826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang WH, Kitai T, Hazen SL. Gut Microbiota in Cardiovascular Health and Disease. Circulation research. 2017;120(7):1183–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raggi F, Pelassa S, Pierobon D, Penco F, Gattorno M, Novelli F, et al. Regulation of Human Macrophage M1-M2 Polarization Balance by Hypoxia and the Triggering Receptor Expressed on Myeloid Cells-1. Frontiers in immunology. 2017;8:1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scodeller P, Simon-Gracia L, Kopanchuk S, Tobi A, Kilk K, Saalik P, et al. Precision Targeting of Tumor Macrophages with a CD206 Binding Peptide. Scientific reports. 2017;7(1):14655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertani FR, Mozetic P, Fioramonti M, Iuliani M, Ribelli G, Pantano F, et al. Classification of M1/M2-polarized human macrophages by label-free hyperspectral reflectance confocal microscopy and multivariate analysis. Scientific reports. 2017;7(1):8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abumaree MH, Al Jumah MA, Kalionis B, Jawdat D, Al Khaldi A, Abomaray FM, et al. Human placental mesenchymal stem cells (pMSCs) play a role as immune suppressive cells by shifting macrophage differentiation from inflammatory M1 to anti-inflammatory M2 macrophages. Stem cell reviews. 2013;9(5):620–641. [DOI] [PubMed] [Google Scholar]
- 49.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu C, Zhao Y, Xiao X, Fan Y, Kloc M, Liu W, et al. Graft-Infiltrating Macrophages Adopt an M2 Phenotype and Are Inhibited by Purinergic Receptor P2X7 Antagonist in Chronic Rejection. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2016;16(9):2563–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi G, Field DJ, Long X, Mickelsen D, Ko KA, Ture S, et al. Platelet factor 4 mediates vascular smooth muscle cell injury responses. Blood. 2013;121(21):4417–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]