Abstract
Oxaliplatin therapy can be complicated by chemotherapy-induced peripheral neuropathy (CIPN). Other neurotoxic chemotherapies have been linked to single nucleotide variants (SNV) in Charcot-Marie-Tooth disease (CMT) genes. Whether oxaliplatin carries increased risks of CIPN due to SNV in CMT-associated genes is unknown.
353 patients receiving oxaliplatin in NCCTG N08CB were serially evaluated for CIPN using a validated patient-reported outcome (PRO) instrument, the CIPN20 questionnaire (sensory scale). 49 canonical CMT-associated genes were analyzed for rare and common SNV by nextgen sequencing.
The 157 patients with the highest and lowest susceptibility to CIPN (cases and controls) harbored 270 non-synonymous SNV in CMT-associated genes (coding regions). 143 of these were rare, occurring only once (“singletons”). CIPN cases had 0.84 singletons per patient compared with 0.98 in controls. An imbalance in favor of cases was noted only in few genes including PRX, which was previously highlighted as a candidate CIPN gene in patients receiving paclitaxel. However, the imbalance was only modest (5 singleton SNV in cases and 2 in controls). Therefore, while singleton SNV were common, they did overall not portend an increased risk of CIPN. Furthermore, testing CMT-associated genes using recurrent non-synonymous SNV did not reveal any significant association with CIPN.
Genetic analysis of patients from N08CB provides clinical guidance that oxaliplatin chemotherapy decisions should not be altered by the majority of SNV that may be encountered in CMT-associated genes when common genetic tests are performed, such as exome or genome sequencing. Oxaliplatin’s CIPN risk appears unrelated to CMT-associated genes.
Keywords: Oxaliplatin, neuropathy, hereditary, risk factors, Charcot-Marie-Tooth disease, chemotherapy
1. INTRODUCTION
Clinical oncologists and pharmacists prescribing common cancer treatments have recently been alerted with increasing frequency to the possibility of encountering patients with a pre-existing genetic disorder termed Charcot-Marie-Tooth disease (CMT). CMT is a heterogeneous group of neuropathies with clinical features ranging from relatively mild, sensory-only neurological symptoms to severe neurological disability such as abasia. CMT ranks among the most common genetic diseases, with an incidence estimated at around ½500 individuals [1]. Most importantly, CMT may be under-diagnosed because this figure accounts only for symptomatic patients according to clinical criteria or abnormal by electrophysiological testing. The incidence of patients with occult CMT, such as those who carry a predisposition to developing CMT or a CMT-like neuropathy when exposed to neurotoxic agents, may be substantially higher.
Antineoplastic agents can exacerbate CMT causing severe debility from the resultant irreversible peripheral neuropathy. A 2017 study by Ibañez-Juliá et al. placed a spotlight on the hazard of an unrecognized CMT risk in oncology patients [2]. Integrating registry data from France with a systematic review of the literature, the study described 43 patients with severe neuropathy from chemotherapy that occurred in the setting of occult CMT. The authors concluded that rigorous screening of cancer patients for a personal CMT risk before prescribing any neurotoxic chemotherapy should be implemented widely in oncology.
The patients described by Ibañez-Juliá et al. [2] represent the extreme severity spectrum of chemotherapy induced peripheral neuropathy (CIPN). CIPN is a common side effect of vincristine (and other vinca alkaloids), paclitaxel and docetaxel (and other taxanes), bortezomib (and other novel oral agents), cisplatin, and oxaliplatin. CIPN usually worsens with each cycle of treatment, i.e., it is cumulative and can be dose-limiting. Mild CIPN such as occasional numbness and tingling in the extremities is so common that it may be expected to occur in most patients at least towards the end of a series of chemotherapy treatments. Knowing that a patient will eventually develop CIPN is therefore not a contraindication to the use of a particular neurotoxic drug. Most importantly, withholding chemotherapy on the basis of a risk of neurotoxocity may be problematic, especially if the withheld drug has a proven benefit in terms of survival or cure such as in several gastrointestinal cancers chemotherapy with oxaliplatin.
Genetic variants in CMT-associated genes may prompt concerns for CIPN risk, either if these genes are analyzed in the scope of clinical pharmacogenomic testing or if encountered incidentally by clinicians. These concerns will most commonly be raised by single nucleotide variants (SNV) that are so infrequent among populations that no information is available whether they are benign or may represent a risk of occult CMT. While such variants are individually rare, they are in aggregate common. If such SNV are encountered in CMT-associated genes , the concern may be heightened by recent studies that linked CIPN from taxanes to certain genetic variants in some of the CMT-associated genes including rare “singleton” SNV occurring in a given study population only once [3–5]. As DNA sequencing technology has recently outpaced the availability of solid clinical evidence, clinicians are increasingly confronted with the question whether to incorporate a genetic test result of uncertain significance into a treatment recommendation and how to interpret many of the incidental findings. This situation also poses a risk of denying patients a potentially life-saving treatment. The enclosed study addresses this emerging challenge, providing guidance for oncologists who prescribe oxaliplatin.
North Central Cancer Treatment Group (NCCTG) study N08CB, an adjuvant colorectal cancer (CRC) trial in which all patients received oxaliplatin-containing chemotherapy (FOLFOX), is the second largest oxaliplatin-induced peripheral neuropathy (OIPN) prevention trial reported to date [6]. (NCCTG is now part of the Alliance for Clinical Trials in Oncology.) While the treatment intervention tested in N08CB failed (disproving the utility of calcium and magnesium infusions), the trial was also designed as a pharmacogenomics study of OIPN. Participants consented to collection of blood for evaluation of germline genetic markers and recording of serial patient-reported neuropathy assessments. OIPN was assessed using the European Organization for Research and Treatment of Cancer (EORTC) Quality of Life Questionnaire for Chemotherapy-Induced Peripheral Neuropathy (QLQCIPN20) [7–9]. The enclosed study represents the first report of any genetic analyses derived from N08CB.
2. METHODS
2.1. Patients
The study cohort of N08CB consisted of 353 patients, who were evaluated serially for CIPN while undergoing treatment with oxaliplatin. Patient characteristics and the collection of patient-reported outcomes in trial N08CB were reported previously[6]. Patients were ineligible for N08CB if they had a pre-existing peripheral neuropathy of any grade, had received prior treatment with neurotoxic chemotherapy such as oxaliplatin, cisplatin, a taxane, or a vinca alkaloid, were receiving any treatment for neuropathy at study entry, or had a family history of neuropathy. Written IRB-approved informed consent was obtained from all patients for the serial CIPN phenotype assessment and for collection of blood for genetic analysis (Mayo IRB# 10–001801). Data collection was performed by the Alliance Statistics and Data Center.
2.2. OIPN quantification by patient-reported outcomes
OIPN was quantified using the nine sensory items that are included in the EORTC QLQ-CIPN20, a dedicated patient-reported neuropathy instrument that has been well validated[7,9]. Patients were asked to complete the CIPN20 instrument at the beginning of each oxaliplatin chemotherapy cycle, thereby providing a measure of symptoms as a function of cumulative oxaliplatin dose. The rate of symptom progression with increasing cumulative dose was used as an OIPN susceptibility score. It was computed using a study-wide, Rasch-type statistical model[10], which provided an estimate and standard error (SE) of the rate for each patient. The SE tended to be smaller when there were more CIPN20 measurements and when the cycle-to-cycle change was steady; conversely, the SE was larger if only few measurements were available or if the cycle-to-cycle change was unsteady (i.e., alternating up and down).
2.3. Selection of OIPN cases and controls
Using the estimate and SE for the rate of symptom progression, a confidence interval for the rate was formed for each patient. With patients ranked by the upper bound of the confidence interval, the lowest-ranking 25% of patients were selected as controls. Then, ranking patients by the lower bound of the confidence interval, the highest-ranking 25% of patients were selected as cases. Thus, patients whose symptoms worsened least with increasing cumulative dose were selected as controls, and patients who showed the most rapid worsening of symptoms were selected as cases. The purpose of using the upper and lower bounds, respectively, of the confidence interval was to account for uncertainty in the rate estimates. Focusing on 157 patients with the highest susceptibility to CIPN (cases) and the lowest susceptibility (controls), we sequenced and statistically analyzed all genetic variants in 49 canonical CMT-associated genes. Our patient selection method is described in detail in SI Methods 1.
2.4. CMT-associated genes
49 CMT-associated genes were selected for this study, which are listed in SI Table 1. While the number of proven or candidate CMT may be even larger (approximately up to twice as many), we decided to focus on these genes for three reasons. First, they account for those accepted by most as canonical CMT-associated genes ; second, we studied the same set in a previous report on paclitaxel-induced CIPN [4]; and, most importantly, we had defined this gene set at the outset of the present study as the primary endpoint for statistical analysis.
2.5. Genotyping
Genomic DNA was extracted from peripheral blood leukocytes, and sequencing libraries were prepared using SureSelect (Agilent) exome target enrichment. Sequencing was performed on a HiSeq 2000 system (Illumina) at the Genomics Core Facility sequencing laboratory at Mayo Clinic. BBDuk (https://sourceforge.net/projects/bbmap/), Picard (http://broadinstitute.github.io/picard/), BWA[11], and Genome Analysis Toolkit (GATK)[12] were used to process sequencing reads and obtain SNV genotypes. Predicted SNV effects were obtained using Ensembl VEP[13]. Details of the data processing and variant filtering steps are provided in SI Methods 2.
2.6. Orthogonal validation of genotyping
Sequencing results for a subset of SNV were validated for 471 genotype calls using SNV genotyping by mass spectrometry (Sequenom MassARRAY), an old, well-established genotyping method that we used as described previously [14,15].
2.7. Statistical analysis
The primary analysis was per-gene statistical testing for association between singleton SNV and CIPN susceptibility using a burden test in which the number of singletons found in a patient served as the explanatory variable in a univariate logistic regression of case vs. control status. The burden test p-value corresponds to the test of β=0, where β is the coefficient on the number of singletons. The co-primary analysis was testing for association between recurrent SNV and CIPN susceptibility using the Sequence Kernel Association Test (SKAT) [16] with a linear kernel. As a secondary analysis, SKAT was repeated with a linear-weighted kernel. For the linear-weighted kernel, the default weighting scheme was used; this scheme gives higher weight to rarer variants, based on the sample minor allele frequency. As an additional secondary analysis, genes were tested for association with CIPN using SKAT-O [17], which combines test statistics from the SKAT and a burden test to maximize the power to detect an association. Recurrent SNV were also tested individually for association with CIPN using Fisher’s exact test. All statistical analyses were performed using R version 3.4.2[18] with the SKAT package[19] for association testing.
3. RESULTS
3.1. Selection of OIPN cases and controls
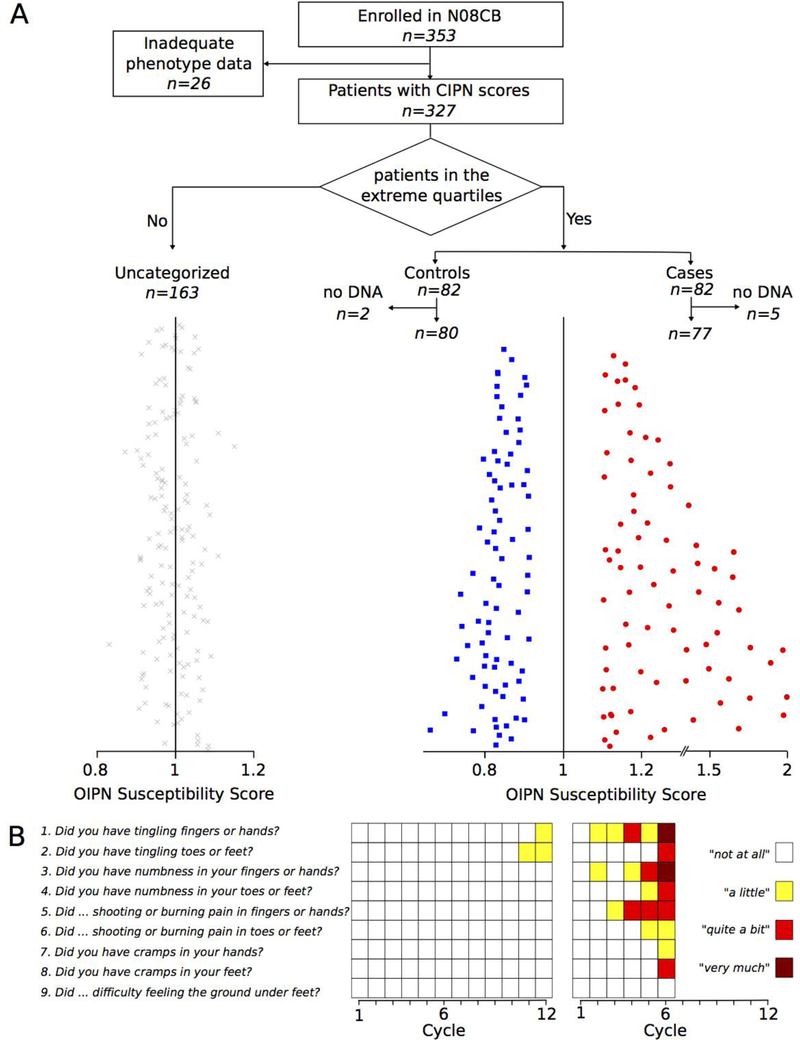
A CONSORT diagram showing patient selection is depicted in Figure 1A. The groups of cases and controls selected based on CIPN20 data consisted of 82 patients each. DNA was available for analysis of 77 cases and 80 controls, which represented the final groups for statistical analysis. The OIPN symptom rates (susceptibility scores) of all cases and controls are shown on the right side in Figure 1A. The left side depicts patients excluded from analysis because they had moderate symptom progression. Questionnaire responses for two patients are also shown, illustrating that a typical control patient did not experience worsening of symptoms while receiving a full course of chemotherapy; in contrast, a typical case patient reported worsening of symptoms with each chemotherapy re-administration and had early termination of therapy, after 6 cycles (Figure 1B).
Figure 1. Selection of OIPN cases and controls.
OIPN case- and control groups of equal sizes were selected by extreme phenotype sampling.
A) A CONSORT diagram shows the selection of patients without OIPN (controls) and with OIPN (cases) identified by the rate of progression of OIPN symptoms with increasing cumulative oxaliplatin dose. Equal number of cases and controls were selected from the tail ends of the distribution of OIPN rates. Specifically, the OIPN rates were ordered by the upper bound of their rate confidence interval and the bottom 25% were taken as controls. Similarly, the cases were obtained by ordering the OIPN slopes by the lower bound of their CI and taking the top 25%. The red, blue, and grey colored points in the panel correspond to cases, controls and unselected patients.
B) Two examples showing extreme CIPN20 phenotypes. The left graph shows the data from the sensory subscale of the CIPN20 questionnaire for a patient who did not develop OIPN throughout his/her 12 cycles of chemotherapy. The right graph shows the example of a patient who quickly developed the symptoms of OIPN and had to be taken off the drug after 6 cycles.
Patients in the control group were slightly older than those in the case group (mean age 59 vs. 56 years, p=0.08). There was no noteworthy difference between cases and controls in sex (53% vs. 58% female, p=0.71) or race (82% vs. 85% Caucasian, p=0.36). Patients in the case group received less oxaliplatin than patients in the control group (mean cumulative dose 670 mg/m2 vs. 829 mg/m2, p<0.0001), an expected result reflecting the clinical practice of dose reduction or ending adjuvant chemotherapy early in patients who experience toxicity. Table 1 summarizes clinical characteristics of the case and control groups.
Table 1.
Patient characteristics
| Characteristic | Cases (n=77) | Controls (n=80) | p-value |
|---|---|---|---|
| Mean age (SD) | 56 (10) years | 59 (12) years | 0.08 |
| Mean cumulative dose of oxaliplatin (SD) | 670 (214) mg/m2 | 829 (192) mg/m2 | <0.0001 |
| Race | 0.37 | ||
| White | 63 (82%) | 68 (85%) | |
| Black or African American | 10 (13%) | 11 (14%) | |
| Other | 4 (5%) | 1 (1%) | |
| Gender | 0.71 | ||
| Female | 41 (53%) | 46 (58%) | |
| Male | 36 (47%) | 34 (42%) | |
| EORTC CIPN21 “Sensitivity touching cold items” (Higher scores are better) Month 1 Month 3 Month 6 Month 12 Month 18 |
78.4 (29.7) 81.8 (30.6) 87.3 (24.1) 91.1 (17.5) 78.7 (27.6) |
91.9 (18.4) 93.0 (19.8) 96.2 (11.8) 95.6 (9.2) 93.3 (11.4) |
3.2. Study-wide detection and categorization of coding SNV
We identified 548 unique SNV study-wide in the coding region of the 49 CMT-associated genes (SI Figure 1). Of these, 270 were non-synonymous SNV: 268 missense SNV and two splice donor variants. 278 synonymous SNV were found, i.e., variants predicted to be silent in regards to the encoded protein. Of the non-synonymous SNV, 143 were found only in a single patient in the study cohort, termed “singleton” SNV. Singleton SNV were thus—by definition—rare in the context of the present study. These are the “uncommon variants” to which the title of this report refers. They are the object of the primary analysis of the study which supports the main conclusion. The remaining 127 SNV were identified in several patients in the study cohort, termed “recurrent” SNV. Of the synonymous SNV, 96 were singleton SNV and 182 were recurrent SNV in the study population.
3.3. Genotype validation
For a subset of 471 SNV calls (rs9038, rs17722209, rs6875902), the sequencing-based genotype was compared with the genotype obtained previously with a non-sequencing method on the same patients. Of these, validation data were missing in 4 instances for the SNV rs9038. 467 calls were informative by both methods. 467 of 467 of these instances agreed between the genotyping methods, providing orthogonal validation for the sequencing methodology employed.
3.4. Per-patient frequency of SNV
On average, each patient harbored 0.91 singleton non-synonymous SNV and 17.6 recurrent non-synonymous SNV across the 49 genes (SI Figure 1). The 49 CMT-associated genes could be categorized according to whether they harbored singleton and/or recurrent SNV (SI Figure 2). 38 genes harbored ≥1 non-synonymous singleton SNV and 34 genes harbored ≥1 non-synonymous recurrent SNV across the population.
3.5. Non-synonymous singleton SNV in cases/controls
143 non-synonymous singleton SNV were found in the 157 patients analyzed. This corresponds to an average of 0.91 singleton SNV per patient highlighting that such genetic findings can be expected in the majority of patients seen in clinical practice. 65 of these SNV were detected in the 77 cases and 78 in the 80 controls. The average was thereby lower in cases, 0.84 per patient, compared with controls, 0.98 per patient. Taken in aggregate, a rare mutation in a CMT gene as captured in this analysis as singleton non-synonymous SNV is therefore not only common but, importantly, appears not to portend an increased risk of CIPN
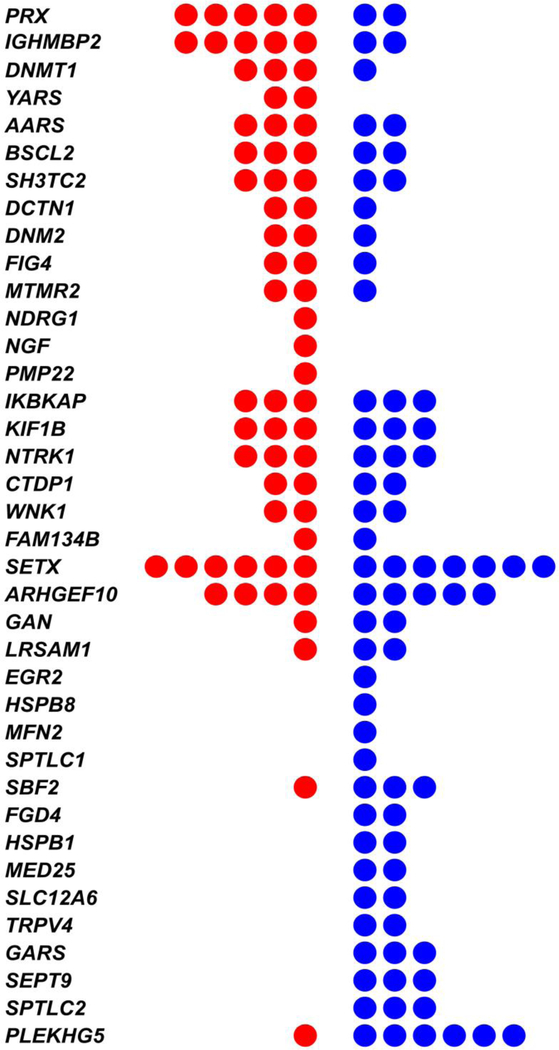
Next we considered the frequency of singleton SNV in each CMT gene (Figure 2). PRX and IGHMBP2 were the genes with the greatest excess of singleton SNV in cases and PLEKHG5 was the gene with the greatest excess seen in controls, but none of the differences were significant study-wide. SI Table 1 provides uncorrected per-gene p-values (Burden P-value, column J). Singleton SNV were most commonly found in SETX, 0.083 on average per patient, where they were evenly balanced between cases and controls.
Figure 2. Uncommon (rare) “singleton” SNV found in cases (red) or controls (blue) across 49 canonical CMT-associated genes.
Genes are ordered according to the excess of mutations found in cases (top) to excess in controls (bottom).
3.6. Per-gene SKAT testing of non-synonymous recurrent SNV for an association with cases/controls
Testing of each gene by SKAT using either a linear kernel (the co-primary analysis of the study) or a linear weighted kernel (a secondary analysis) did not find any significant association with case/control status (SI Table 1, columns F and G).
3.7. Individual testing of non-synonymous recurrent SNV
Recurrent SNV were also tested individually as another secondary analysis. A complete list of all SNV with p-values are provided as SI Table 2 (column Q). None of the SNV was statistically associated with CIPN when appropriately corrected for multiple testing.
3.8. Synonymous SNV
Of the synonymous SNV found in the study, 96 were singleton SNV and 182 were recurrent SNV. The most marked imbalance of these singletons was found in SBF2, 0 in the cases and 5 in the controls, but the significance level for this candidate association (p=0.04) was within the range of stochastic variation expected in the exploration of multiple secondary endpoints across 49 genes and therefore was deemed inconsequential.
4. DISCUSSION
CIPN is a serious adverse effect of several important chemotherapeutic drugs including oxaliplatin. The etiology and molecular mechanisms of CIPN are only incompletely understood and no preventive measures or effective treatment are currently available except not administering the offending drug. While CIPN is dose-dependent (in a cumulative fashion), its severity varies markedly amongst patients undergoing similar treatments indicating that individuals differ in terms of their susceptibility. CIPN has therefore been conceptualized and investigated as a candidate pharmacogenomic trait. The earliest pharmacogenomic studies of CIPN examined and found associations of SNV in drug metabolism genes [20,21]. Over the current decade numerous other genes have been linked to CIPN through candidate SNV genotyping, genome-wide association studies (GWAS), and, most recently, the use of the high throughput sequencing technology employed in the current study. Commonalities across this literature could be viewed as follows. First, genetic associations appear to be different between drug classes, e.g., taxanes or vinca alkaloids or platinum drugs, and possibly also between individual drugs within a class such as cisplatin or oxaliplatin. Second, cross-validation between different studies has been only moderate (at best) [22]; lack of cross-validation may be due to patient phenotyping, study populations shortcomings in statistical analyses such as retrospective identification of tests and endpoints (and therefore inflation of significance), or a combination of poor statistical power with expected stochastic variation. Third, any genetic effects are modest at best (typical hazard ratios and/or odds ratios are often close to 2); no single SNV is strongly predictive in a diagnostic sense. While genetic scores have been proposed (and used as retrospective endpoint) as a way out, no clinically convincing genetic test for CIPN exists today.
Alliance N08CB was a clinical trial conducted with the primary objective to investigate a prophylactic intervention to prevent OIPN. Unlike treatment studies concentrating on cancer response to oxaliplatin, this protocol was focused solely on the prospective assessment of OIPN, thereby creating one of the largest OIPN cohorts available to date with state-of-the-art clinical phenotypes. OIPN symptoms were assessed serially in all patients—up to 12 times during chemotherapy—allowing the present study to determine the OIPN susceptibility of individual patients from the rate of symptom change (usually worsening), which we describe as an OIPN susceptibility score in the present report. This method for quantifying OIPN, which we have used in previous genetic association studies[3,4], has the advantages of being intuitive and making full use of patients’ reported symptom trajectories. In the interest of transparency and reproducibility, we have provided a detailed description of our method as part of the supplementary material.
CMT-associated genes have been linked to CIPN from taxanes by multiple groups including ours [3,5]. The rationale seems compelling because the symptomatology and neuropathology of CMT and CIPN certainly overlap markedly. Moderately strong genetic evidence supporting the link has therefore not been surprising. At the same time, the genetic studies may have contributed to a renewed emphasis to screen for the possibility of occult CMT in cancer patients who are candidates for neurotoxic drugs [2]. This confluence of factors poses two (related) new questions for oncologists: First, should genetic testing be performed for CMT-associated genes in the knowledge that most patients will have genetic variants of uncertain significance in those genes (and only a small minority will have unknown CMT)? Second, how should CMT gene variants of unknown significance be interpreted if found coincidentally or when they are provided as a matter of routine in the medical record? The latter situation may become increasingly common as genome sequencing will become more and more common as a matter of routine in health and illness.
DNA sequencing technology has advanced markedly over the past decade. Sequencing all (or most) of the (roughly) 6 billion base pairs of a patient’s DNA, i.e., whole genome sequencing, can now be performed for one to two thousand US dollars, less than the analysis of a single gene a decade earlier. For that reason alone, many patients will have CMT gene sequences available in the future regardless of whether a neurotoxic chemotherapy is considered. However, correlative clinical studies in clinical cohorts have not kept pace with the increasing ease of obtaining genetic data for individual patients. In another area of oncology, tumor genetics, clinicians are already confronted with a similar situation: reports of numerous mutations (from tumor “panels” or whole exome- or whole genome sequencing) for which no clinical guidance is available. In the area applicable to CIPN pharmacogenomics and CMT, germline genetics, the challenge may be compounded by the fact that there are many more genetic variations among humans than “somatic” mutations that make up the difference between a tumor and its host (by a factor of about 1000-fold more). The present study addresses this challenge for the case of oxaliplatin-induced CIPN and CMT-associated genes by providing a CMT gene sequencing dataset from an oxaliplatin treated patients who were enrolled in a study that included dedicated serial phenotyping for CIPN.
The present study supports the impression that the connection is sufficiently weak to suggest that modifying clinical treatment decisions based on the studied genetic variants would be premature at this time. As expected, across the 49 canonical CMT-associated genes studied we found at least one uncommon (singleton) non-synonymous SNV and many more common (recurrent) SNV in a majority of patients. Upon testing of our prospectively defined endpoints, there was no statistically significant association of any of these genes with oxaliplatin CIPN. While an imbalance of singleton SNV was observed in some genes, including one that was previously highlighted as a candidate CIPN gene in patients receiving paclitaxel [4], the total number of singleton SNV was small (n=7) and the imbalance, 5 cases and 2 controls, would indicate only moderate risk and may be a chance event. In aggregate, the likelihood of encountering (at least) one singleton SNV in a case was the same as in a control patient, 57% versus 59%.
Oxaliplatin has not been linked to CMT before. The present study supports the impression that any connection, if it exists, may be weak. With this perspective, it may be notable that the recent in-depth registry and literature-based clinical report on CMT and CIPN by Ibañez-Juliá et al. [2] reported that vincristine was the cause of 83% of the presented cases (n=34), taxanes 10% (n=4), cisplatin 5% (n=2), and vinorelbin 2% (n=1). Oxaliplatin was not identified as cause of even a single case of exceptionally severe CIPN due to occult CMT. These insights deepen the current understanding of the pharmacogenomics of oxaliplatin that are a very active area of investigation [23–25] due to its imminent importance for clinical oncology. There are a few limitations to the present study. Genetic markers such as ancestry informative markers are more accurate than self-reported race to account for population differences. In the present study we relied on self-reported race only. Secondly, although our extreme phenotype method has created two clearly separate groups, and N08CB is the second largest OPIN study to date, the sample size of N=157 is indeed small based on SKAT power simulation studies [16].
The present study does not question the principle that neurotoxic drugs, including oxaliplatin, should be avoided in any patient with a clinically proven diagnosis of CMT, or with one of the uniquely diagnostic CMT mutations documented in the neurogenomics literature, or with clinically marked suspicion of occult CMT, unless the diagnosis can be ruled out by a specialty evaluation including electrophysiological testing by a peripheral nerve neurologist. For the large majority of patients, however, for whom these finding do not apply, the presented study provides reassurance especially for the large number of patients in whom SNV of uncertain significance may be encountered in CMT gene as a result of widely available genome sequencing.
Supplementary Material
HIGHLIGHTS.
Life-saving oxaliplatin therapy should not be withheld if genetic variants in Charcot-Marie-Tooth disease (CMT) genes are encountered.
Oxaliplatin, unlike other neurotoxic chemotherapy drugs, appears not to be linked to CMT-associated genes as shown in this first exome sequencing study from the NCCTG/Alliance oxaliplatin induced peripheral neuropathy (OIPN) prevention trial N08CB.
The results provide practical and evidence-based guidance to clinicians prescribing oxaliplatin for a large number of different solid tumor types with palliative of curative intent.
ACKNOWLEDGEMENT
The authors wish to acknowledge the accrual of patients to this study by Dr. Shaker R. Dakhil, Wichita NCI Community Oncology Research Program, Wichita, KS; and Dr. Patrick L. Gomez, Heartland Cancer Research NCI Community Oncology Research Program, Decatur, IL. Research reported in this publication was supported by the National Institute of Nursing Research (NINR) of the National Institutes of Health (NIH) under Award Number R01NR015259 (to A.S.B.) and the National Cancer Institute of the NIH under Award Numbers UG1CA189823 (to the Alliance for Clinical Trials in Oncology NCORP Grant) and U10CA180790. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
SUPPORT
Research reported in this publication was supported by the National Institute of Nursing Research (NINR) of the National Institutes of Health (NIH) under Award Number R01NR015259 (to A.S.B.) and the National Cancer Institute of the NIH under Award Numbers UG1CA189823 (to the Alliance for Clinical Trials in Oncology NCORP Grant), U24CA196171 (Alliance NCTN Biorepository and Biospecimen Resource Grant) and U10CA180790. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
ClinicalTrials.gov Identifier: NCT01099449
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Barreto LCLS, Oliveira FS, Nunes PS, França Costa IMP, Garcez CA, Goes GM, et al. Epidemiologic Study of Charcot-Marie-Tooth Disease: A Systematic Review. Neuroepidemiology [Internet]. 2016. [cited 2018 Jan 13];46:157–65. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26849231 [DOI] [PubMed] [Google Scholar]
- 2.Ibañez-Juliá MJ, Berzero G, Reyes-Botero G, Maisonobe T, Lenglet T, Slim M, et al. Antineoplastic agents exacerbating Charcot Marie tooth disease: red flags to avoid permanent disability. Acta Oncol (Madr) [Internet]. 2017. [cited 2018 Jan 13];1–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29243538 [DOI] [PubMed] [Google Scholar]
- 3.Boora GK, Kulkarni AA, Kanwar R, Beyerlein P, Qin R, Banck MS, et al. Association of the Charcot-Marie-Tooth disease gene ARHGEF10 with paclitaxel induced peripheral neuropathy in NCCTG N08CA (Alliance). J Neurol Sci. 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutler AS, Kulkarni A a, Kanwar R, Klein CJ, Therneau TM, Qin R, et al. Sequencing of Charcot-Marie-Tooth disease genes in a toxic polyneuropathy. Ann Neurol [Internet]. 2014. cited 2014 Oct 30];76:727–37. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25164601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldwin RM, Owzar K, Zembutsu H, Chhibber A, Kubo M, Jiang C, et al. A genome-wide association study identifies novel loci for paclitaxel-induced sensory peripheral neuropathy in CALGB 40101. Clin Cancer Res. 2012;18:5099–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loprinzi CL, Qin R, Dakhil SR, Fehrenbacher L, Flynn KA, Atherton P, et al. Phase III randomized, placebo-controlled, double-blind study of intravenous calcium and magnesium to prevent oxaliplatin-induced sensory neurotoxicity (N08CB/Alliance). J Clin Oncol. 2014;32:997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Postma TJ, Aaronson NK, Heimans JJ, Muller MJ, Hildebrand JG, Delattre JY, et al. The development of an EORTC quality of life questionnaire to assess chemotherapy-induced peripheral neuropathy: the QLQ-CIPN20. Eur J Cancer. 2005;41:1135–9. [DOI] [PubMed] [Google Scholar]
- 8.Cavaletti G, Cornblath DR, Merkies ISJ, Postma TJ, Rossi E, Frigeni B, et al. The chemotherapy-induced peripheral neuropathy outcome measures standardization study: from consensus to the first validity and reliability findings. Ann Oncol. 2013;24:454–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavoie Smith EM, Barton DL, Qin R, Steen PD, Aaronson NK, Loprinzi CL. Assessing patient-reported peripheral neuropathy: the reliability and validity of the European Organization for Research and Treatment of Cancer QLQ-CIPN20 Questionnaire. Qual Life Res. 2013;22:2787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasch G Probabilistic models for some intelligence and attainment tests. Univ. of Chicago Press; 1980. [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinforma. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boora GK, Kanwar R, Kulkarni AA, Abyzov A, Sloan J, Ruddy KJ, et al. Testing of candidate single nucleotide variants associated with paclitaxel neuropathy in the trial NCCTG N08C1 (Alliance). Cancer Med [Internet]. 2016. [cited 2016 Nov 1];5:631–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26763541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulkarni AA, Boora G, Kanwar R, Ruddy KJ, Banck MS, Le-Lindqwister N, et al. RWDD3 and TECTA variants not linked to paclitaxel induced peripheral neuropathy in North American trial Alliance N08C1. Acta Oncol [Internet]. 2015. [cited 2016 Nov 1];54:1227–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25549536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Michael C., Lee S, Cai T, Li Y, Boehnke M, Lin X Rare Variant Association Testing for Sequencing Data Using the Sequence Kernel Association Test (SKAT). Am J Hum Genet. 2011;89:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austri: a; 2017. [Google Scholar]
- 19.Lee S, Miropolsky L, Wu M. SKAT: SNP-Set (Sequence) Kernel Association Test. 2017. [Google Scholar]
- 20.Gamelin L, Capitain O, Morel A, Dumont A, Traore S, Anne LB, et al. Predictive factors of oxaliplatin neurotoxicity: the involvement of the oxalate outcome pathway. Clin Cancer Res. 2007;13:6359–68. [DOI] [PubMed] [Google Scholar]
- 21.Lecomte T, Landi B, Beaune P, Laurent-Puig P, Loriot MA. Glutathione S-transferase P1 polymorphism (Ile105Val) predicts cumulative neuropathy in patients receiving oxaliplatin-based chemotherapy. Clin Cancer Res. 2006;12:3050–6. [DOI] [PubMed] [Google Scholar]
- 22.Cavaletti G, Alberti P, Marmiroli P. Chemotherapy-induced peripheral neurotoxicity in the era of pharmacogenomics. Lancet Oncol. 2011. page 1151–61. [DOI] [PubMed] [Google Scholar]
- 23.Kanai M, Kawaguchi T, Kotaka M, Shinozaki K, Touyama T, Manaka D, et al. Large-scale prospective pharmacogenomics study of oxaliplatin-induced neuropathy in colon cancer patients enrolled in the JFMC41–1001-C2 (JOIN Trial). Ann Oncol [Internet]. 2016. [cited 2018 Jun 24];27:1143–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27069012 [DOI] [PubMed] [Google Scholar]
- 24.Custodio A, Moreno-Rubio J, Aparicio J, Gallego-Plazas J, Yaya R, Maurel J, et al. Pharmacogenetic predictors of severe peripheral neuropathy in colon cancer patients treated with oxaliplatin-based adjuvant chemotherapy: a GEMCAD group study. Ann Oncol. 2014;25:398–403. [DOI] [PubMed] [Google Scholar]
- 25.Abad A, Martínez-Balibrea E, Viéitez JM, Alonso-Orduña V, García Alfonso P, Manzano JL, et al. Genotype-based selection of treatment of patients with advanced colorectal cancer (SETICC): a pharmacogenetic-based randomized phase II trial. Ann Oncol [Internet]. 2018. [cited 2018 Jun 24];29:439–44. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29145602 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.