Abstract
Antibiotic-associated diarrhea (AAD) is a common and unintended adverse effect of antibiotic treatment. It is characterized by the disruption of the gut microbiota, decreased intestinal short chain fatty acid (SCFA) concentrations, accumulation of luminal carbohydrates and colonic bile acids, altered water absorption, and ultimately diarrhea. Probiotics were shown to prevent AAD in numerous clinical trials. This review examines what is currently known about how probiotics reduce the risk for AAD via modulating the gut microbiota, altering nutrient and bile acid metabolism, inducing epithelial solute transporter activity, supporting intestinal barrier function, and influencing the immune system. Although probiotics are frequently prescribed with antibiotic use, mechanistic evidence verifying how they confer protection against AAD is extremely limited. This information is urgently needed for improving recommendations for sustaining probiotic development and for implementing probiotics in clinical settings.
Keywords: antibiotics, gut microbiome, probiotic, Lactobacillus, Bifidobacterium, diarrhea, AAD, SCFA
Graphical Abstract
Introduction
Antibiotic-associated diarrhea (AAD) is an important morbidity resulting from antibiotic use. AAD is more than a bothersome adverse event of antibiotic treatment; it is associated with prescription noncompliance and overuse of second-line antibiotics. Although all age groups are affected by AAD, children are particularly at risk because they are often placed on antibiotics, and the rate of diarrhea associated with antibiotic usage among children is between 20 to 35% [1]. AAD is defined as clinically unexplained diarrhea that occurs in connection with antibiotic administration. Any antibiotic could potentially cause AAD, but broad-spectrum antibiotics that predominantly target anaerobes and are poorly absorbed (such as clindamycin, cephalosporins (cefixime and ceftriaxone), and amoxicillin-clavulanate) have a higher AAD incidence [2].
One of the most commonly prescribed uses of probiotics is for the prevention of antibiotic-associated diarrhea (AAD). Strains from numerous bacterial species have been tested in clinical studies for mitigating AAD including members of the Bacillus, Bifidobacterium, Clostridium, Lactobacillus, Lactococcus, Leuconostoc, and Streptococcus genera. Among the fungi, Saccharomyces boulardii has also been examined. Lactobacillus rhamnosus strain GG and S. boulardii strain CNCM I-745 have been most frequently studied [3,4]. A recent (2019) Cochrane review of probiotics for the prevention of pediatric AAD found 33 randomized clinical trials with 6352 participants meeting the inclusion criteria [5••]. This review reported that probiotics conferred a moderate beneficial effect for AAD prevention (number needed to treat for an additional beneficial outcome (NNTB) 9, 95% CI 7 to 13). Consistent with a prior Cochrane review (2015), the risk ratio of developing AAD was significantly reduced when ≥ 5 billion colony forming units (CFUs)/day were consumed [6]. It was suggested that 5 to 40 billion CFU/day of L. rhamnosus or S. boulardii, the two most commonly applied species, were the most appropriate for preventing AAD in children receiving antibiotics [6]. Nonetheless, the certainty of evidence in the Cochrane review was ranked as moderate because of minor issues with the risk of bias and inconsistency between probiotic strains used [6]. New large, well-designed, multi-centered, randomized trials were recommended [6]. Such studies will be difficult to design because many questions remain on which strains are most effective and on the appropriate timing and duration of use. Understanding the underlying molecular mechanisms of probiotic effects in the gastrointestinal (GI) tract would help to address those questions. Therefore, this review examines what is presently known about the mechanistic basis for probiotic prevention of AAD.
Modulation of gut microbiota composition
Antibiotics cause significant disruptions to the normal composition and functional attributes of the gut microbiome [7]. Such deficits can persist well after the cessation of antibiotic administration [7] and are associated with the development of obesity, asthma, and inflammatory bowel disease (IBD) [8]. Among the numerous impacts of antibiotics on the gut microbiome are reductions in microbial taxonomic richness, diversity, and evenness in the GI tract [7]. Those drastic changes result in a depletion of the normal gut bacterial residents, and opportunities for colonization by pathogens such as Clostridium difficile. Presently, C. difficile is predicted to account for about 20% of all AAD cases [9]. However, other opportunistic pathogens, such as Clostridium perfringens, Klebsiella oxytoca, Klebsiella pneumonia, Staphylococcus aureus, and Candida species have also been associated with AAD [10].
Probiotics are assumed to benefit human health by their direct actions on the composition and function of the human gut microbiota [11,12]. However, very few studies on antibiotic use have addressed this possibility (Table 1). In a four-week trial with patients treated for C. difficile infection, administration of a four-strain capsule of Lactobacillus and Bifidobacterium together with antibiotics was associated with significant reductions in the duration of C. difficile diarrhea [13]. Examination of fecal contents showed that subjects consuming the probiotic capsules contained lower proportions of Verrucomicrobiaceae in their stools compared to those given placebo (empty) capsules [14•] (Table 1). Although few other probiotic-induced differences in bacterial fecal composition were found [14•], the reduced levels of Verrucomicrobiaceae were consistent with the positive association that this family has with susceptibility to C. difficile infection [15]. In another study initiated after completion of antibiotic treatment for Helicobacter pylori infection, there were fewer antibiotic-induced changes to fecal bacterial and fungal composition among subjects taking a multi-strain mixture of Bacillus subtilis and Enterococcus faecium compared to subjects on a placebo product [16] (Table 1). A similar finding was reported in another study on H. pylori treatment, but in that case, the putative probiotic strains were taken during antibiotic use [17].
Table 1.
Human and animal antibiotic studies measuring the effects of probiotics on intestinal responses
| Responses to probiotics compared to antibiotic treatment alone |
||||||
|---|---|---|---|---|---|---|
| Antibiotic | Strain(s) and study design | Gut microbiota and metabolome | Intestinal protein and gene expression | Physiologica l responses | ||
| Humans | Vancomycin or metronidazole; C. Difficile treatment | L. acidophilus ncfm, L. paracasei Lpc-37, B. lactis Bi-07, and B. lactis B1–04; concurrent with antibiotics | No change in bacterial alpha- or beta-diversity ↓ verrucomicrobiaceae ↑ bacteroides |
ND | ↓ Duration and total diarrhea No change in C. difficile recurrence |
[13,14•] |
| Clarithromycin, amoxicillin and esomeprazole; H. pylori treatment | B. subtilis and E. faecium (strain designations not provided); post-antibiotic treatment | ↓ Changes in bacterial and fungal composition over time | ND | No change in H. pylori eradication; ↓ Reduced incidence of side-effects (e.g. diarrhea) |
[16] | |
| Clarithromycin, amoxicillin, lansoprazole for H. pylori treatment | B. subtilis and Streptococcus faecium (strain designations not provided); concurrent with antibiotics | ↑ Bacterial alpha-diversity ↓ Changes in bacterial composition compared to baseline ↑ Genis for amino acid and sugar metabolism ↓ Genes for seleno-compound metabolism |
ND | ND, small sample size | [17] | |
| Amoxicillin-clavulanate; Healthy volunteers | S. boulardii CNCM I-745’ conc. Tern with antibiotics | ↓ Escherichia
↓ Parabacteroides and Ralstonia ↑ Fecal secondary bile acids |
ND | ↓AAD scores | [18,45•] | |
| Ciprofloxacin and metronidazole; Healthy volunteers | 11-strain mixture ith species of Lactobacillus, Bifidobacterium, Lactococcus, and Streptococcus a; post-antibiotic treatment | ↓ Recovery of the indigenous microbiome (fecal and mucosal), including: ↓ Bacterial alpha-diversity; ↓ Bacterial load; ↓ Clostridiales | ↑ REG3G, IL1B transcripts | ND | [19] | |
| Metronidazole; C. difficile treatment | L. plantarum 299V; concurrent with antibiotics | Non-significant reductions in SCFA | ND | Slight reduction in recurrence of clinical symptoms (small sample size) | [26] | |
| Piglets | Aureomycin | Bacillus amyloliquefaciens SC06; concurrent with antibiotics | ND | ↑ IL-6, MyD88, NOD-1, TLR4, and ↓ TNFα transcripts in jejunum ↑ TNFα protein in serum ↑ TNFα, IFN-γ, IL-6, and IL-10 in liver |
↑ Intestinal villus height ↓ crypt depth ↓ Gt permeability |
[51] |
| Rats | Clindamycin, ampicillin. streptomycin | B. fragilis ZY-312 in different doses (107, 108, 109 CFU/day) and/or a mixture of Bifidobacterium longum, L. acidophilus, and E. faecalis post-antibiotic treatment | No change in bacterial alpha-diversity ↑ Akkermansia ↓ Escherichia |
↑ Colon Aqpi, Aqp3 and Aqp8 transcripts ↑ Mucin 2 transcripts, colon ZO-1, occludin and ↑ Ki-67 positive cells with the highest dose of B. fragilis ZY-312 |
↓ Fecal water and ↑ fecal consistency score with the highest dose of B. fragilis ZY-312 | [25•] |
| Mice | Clindamycin (subcutaneous) and E. faecalis V583 | L. paracasei CNCM I-3689; concurrent with antibiotics | ↓ Vancomycin-resistant enterococci ↑ Bacteroidetes |
↑ Ki67 and PCNA-positive cells ↑ camp (cathelicidin LL-37) transcripts |
ND | [20] |
| Metronidazole, neomycin sulfate and vancomycin | L. rhamnosus GG; concurrent with antibiotics | ND | ↑ GPR109a, SLC5A8, SLC26A3, AQP4, and NHE3 transcripts | No effect on mouse weight | [34] | |
| Cefixime | Two strain mixtures with L. plantarum, L. casei, L. rhamnosus, and Lactobacillus helveticus b; post-antibiotic treatment | ↑ Recovery of microbiota composition: ↑ Firmicutes ↓ Bacteroidetes ↑ Proteobacteria ↑ SCFA |
↓ CRP, C3, and IgG | ↓ Cecum size ↓ Inflammatory infiltrate |
[22] | |
| Ampicillin, streptomycin, clindamycin | L. rhamnosus A191, L. acidophilus, Bifidobacterium breve and B. longum (no designations); post-antibiotic treatment | No change in bacterial alpha-diversity ↓ Enterobacteriaceae, ↑ Firmicutes |
ND | ND | [21] | |
| Ampicillin | Separate groups given L. casei CGMCC 12435 (LacC), L. plantarum CGMCC 12436 (LacP), or L. rhamnosus GG (LacG); post-antibiotic treatment | ↑ Recovery of microbiota composition: ↑ alpha-diversity ↑ Bacteroidetes ↓ Proteobacteria (LacC and LP) ↑ SCFA (LacC) |
↑ Ileal ZO-1, occludin (LacC), Claudin- 1(LacP) transcripts ↓ NF-κB (LacC, LacP) ↓ IL-1β, IFN-γ ↓ (LacC) ↓ Reg-3γ ↓ (LacG), ↓ sIgA (LacC, LacG) |
↓ Gut permeability (LacC) ↓ Serum endotoxin and diamine oxidase |
[24•] | |
| Ampicillin | Mixture of L. casei CCFM2710, L. plantarum CCFM2602, L. rhamnosus CCFM492, and L. helveticus CCFM671; post-antibiotic treatmen. |
↑ Recovery of microbiota composition: ↓ Firmicutes and ↑ Bacteroidetes ↓ Proteobacteria in stools |
↑ Ileum and colon ZO-1, occludin and claudin-1 transcripts ↓ Ileum TNF-α, IL-6, MCP-1and IFN-γ |
↓ Gut permeability ↓Serum endotoxin and D-lactate levels |
[23] | |
ND: Not Determined
Abbreviations: C3: Complement 3, sIgA: secretory immunoglobulin A, aqp: aquaporin, GPR109A: G-protein-coupled receptor 109A, SLC5A8: Solute Carrier Family 5 Member 8, SLC26A3: Solute Carrier Family 26 Member 3, and NHE3: Na+/H+ exchanger 3
For reference [19], a multistrain mixture was used as follows: L. acidophilus ATCC4356, L. rhamnosus (strain designation not provided), L. casei ATCC393, L. paracasei ATCC BAA-52, L. plantarum ATCC8014, Bifidobacterium longum subsp infantis ATCC15697, Bifidobacterium bifidum ATCC29521, Bifidobacterium breve ATCC15700, Bifidobacterium longum subsp. longum ATCC15707, Lactococcus lactis (strain designation not provided), Streptococcus thermophilus ATCC BAA-491
For reference [22], a multistrain mixture was used as follows: L. plantarum including CCFM4, CCFM10, CCFM595, CCFM602, and CCFM605; L. casei CCFM5, CCFM30, CCFM236, CCFM2710 and CCFM2711; L. rhamnosus LGG, CCFM237, CCFM311, CCFM319 and CCFM492; and L. helveticus CCFM6, CCFM672 and CCFM673.
When antibiotics were administered to healthy volunteers, ingestion of S. boulardii CNCM I-745 together with a seven-day regime of amoxicillin-clavulanate was associated with attenuation of microbiota shifts, including less Escherichia coli overgrowth [18]. A very different outcome of putative probiotic-mediated effects on the gut microbiome was found in another study wherein healthy volunteers were given a course of ciprofloxacin and metronidazole for 7 days and then administered a 28-day course of an 11-strain mixture or placebo [19]. Consumption of the microbial preparation resulted in potentially negative and persistent consequences on the mucosal and fecal gut microbiome composition (i.e. delayed post-antibiotic reconstitution of the indigenous mucosal microbiome composition and the host GI transcriptome) and a similar effect was observed in mice [19]. However, it should be noted that no clinical endpoints were measured to establish whether that particular study design or strain preparation had measurable consequences on human health.
Animal models have also been used to investigate the extent to which the administration of (putatively) probiotic microorganisms alter antibiotic-mediated changes to the gut microbiome (Table 1). The fecal contents of rats given Lactobacillus paracasei CNCM I-3689 prior to, during, and after subcutaneous challenges with clindamycin and oral vancomycin-resistant Enterococcus faecalis contained significantly lower levels of vancomycin-resistant enterococci (VRE) and showed a better recovery of members of the phylum Bacteroidetes after antibiotic treatment was ended [20]. Other studies focused specifically on administration of the (putative) probiotic microorganisms after antibiotic use [21–23,24•,25•]. Most consistent among the findings was a reduction in the proportions of Proteobacteria [21–23, 24•, 25•,] and an increase in Bacteroides when strains of Lactobacillus were consumed [23,24•]. Although several reports noted that Lactobacillus consumption did not result in significant changes to the diversity of bacteria in the distal GI tract [21,25•], other studies described an improved recovery of the gut microbiota towards pre-treatment composition [23,24•,26].
The numerous ways that probiotics could modulate gut microbiome composition have been reviewed elsewhere [11]. Specific molecular mechanisms include inhibition of intestinal pathogens by the production of anti-bacterial compounds, competitive exclusion either by the consumption of limited nutrient resources or adherence to the epithelium, or stimulation of indigenous microbial activity (Figure 1). Recently, probiotic E. coli Nissle 1917 was found to produce a class of bacteriocins that limit the expansion of competing Enterobacteriaceae during intestinal inflammation [27]. The importance of bacteriocins for ecological fitness of bacteria in the GI tract was also shown for Gram-positive bacteria such as E. faecalis [27]. Pathogen outgrowth could also be inhibited by competitive exclusion by probiotics for intestinal binding sites. This capacity was indicated by the inhibition of Campylobacter jejuni infection by a Lactobacillus gasseri SBT2055 (LG2055) cell surface-associated, aggregation-promoting factor which binds to extracellular matrix proteins on intestinal cells [28]. Alternatively, end-products of probiotic metabolism could be consumed by members of the gut microbiota in cross-feeding interactions. For example, a propionogenic bacterial consortium was recently shown to restore fecal propionate levels and alter bacterial composition after antibiotic treatment in a model of the human intestinal microbial ecosystem (M-SHIME) [29•].
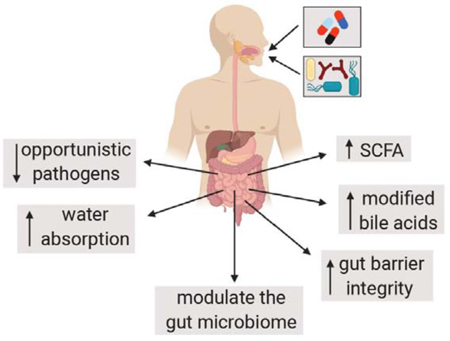
Figure 1. Schematic model of the potential molecular mechanisms responsible for probiotic prevention of AAD.
Antibiotic treatment disrupts the composition of the GI tract microbiota, leading to increased growth of opportunistic pathogens, the accumulation of undigested carbohydrates, and reduced levels of SCFAs and modified bile acids. Probiotics might counter antibiotic-induced effects in the GI tract by directly impairing pathogen growth or by inducing other alterations to gut microbiota composition via SCFA synthesis, production of other secreted metabolites such as bacteriocins, or by reducing lumenal pH and O2 levels. Probiotics might also cause changes to bile acid composition as well as directly interact with the intestinal epithelium and immune system to result in increased gut barrier function and modulation of water and solute transport.
Alter nutrient metabolism in the intestine
Antibiotic-mediated, gut microbiome remodeling also results in significant alterations to intestinal metabolomes [30]. Perhaps most important among lumenal metabolite changes is the reduction in short-chain fatty acid (SCFA) levels [31,32]. SCFAs butyrate, propionate, and acetate are the primary end-products of bacterial carbohydrate metabolism in the GI tract and constitute approximately 10% of the human daily caloric requirement [33]. Reductions in SCFA biosynthesis might lead to AAD because these compounds promote NaCl and water absorption [31]. SCFA are rapidly absorbed by the colon and stimulate Na-dependent fluid absorption via a cyclic AMP-independent process with Na-H, SCFA-HCO3, and Cl-SCFA exchanges [31].
Evidence of probiotic-mediated effects on intestinal SCFA was provided in a human trial whereby Lactobacillus plantarum 299V prevented a decrease in SCFA during metronidazole use [26] (Table 1). In mice, L. rhamnosus GG was as effective as the butyrate derivative tributyrin at preventing antibiotic-induced intestinal injury and reductions in SCFA receptor (GPR109a) and transporter (SLC5A8) levels [34]. Because lactobacilli lack the pathways necessary for butyrate production, the effect of L. rhamnosus GG was likely the result of probiotic-induced, cross-feeding with the gut microbiota to result in increased lumenal butyrate levels or via a butyrate-independent mechanism. In another study, Lactobacillus acidophilus ATCC4537-secreted compounds were able to prevent enteropathogenic E. coli inhibition of butyrate uptake by Caco-2 cells due to a mechanism that involved prevention of monocarboxylate transporter isoform 1 (MCT1) endocytosis [35]. Notably, this activity was not found for heat-killed L. acidophilus or for three other Lactobacillus strains tested [35].
Probiotics could contribute directly to intestinal SCFA by the production of organic acids such as lactate and acetate or by providing a more hospitable environment for SCFA-producing bacteria (Figure 1). Acetate production by probiotic Bifidobacterium in the GI tract was shown to reduce the risk for enteropathogenic E. coli infection [36]. Probiotic metabolism and production of organic acids (e.g. lactate and acetate) in situ could lower lumenal pH and oxygen levels as well as provide substrates used for butyrate and propionate synthesis by other bacterial GI inhabitants [37]. Additionally, growth of probiotic bacteria in the intestine could lead to lower concentrations of undigested carbohydrates, thereby reducing the risk of diarrhea caused by disruptions in osmogradients [31].
Direct modulation of solute secretion and absorption
The lack of solute absorption and/or active solute secretion by the intestinal epithelium results in watery diarrhea. Solute levels are controlled by a variety of basolateral and apical channels and transporters which are responsible for Cl− secretion and the active transport of Na+ across the epithelium with parallel Cl− or HCO3− absorption [38,39]. In mice, B. subtilis CU1 (CNCM I2745), but not L. plantarum CNCM I-4547, reduced the risk of AAD by inducing the expression of higher quantities of the epithelial Na+/H+ exchanger 3 protein NHE3, a protein that promotes fluid absorption, and lower levels of cystic fibrosis transmembrane conductance regulator (CFTR), a protein with a major role in Cl− secretion [40]. In another study, L. acidophilus ATCC4357 prevented Citrobacter rodentium-induced diarrhea in mice by counteracting the inhibition of NHE3 [41]. The Cl−/HCO3− exchanger protein DRA also remained active with L. acidophilus administration [41]. Additionally, Bacteroides fragilis ZY-312 [25•] and L. rhamnosus GG [34] resulted in the increased expression of genes coding for aquaporin water-channel membrane proteins (Table 1).
Because of the importance of lumenal solute concentrations in diarrhea development, probiotic-mediated alterations to intestinal electrolyte transporters could be a potent mechanism for AAD prevention (Figure 1). Besides SCFAs (discussed above), other compounds might confer similar effects as was recently shown for gassericin A, a bacteriocin made by L. gasseri and Lactobacillus frumenti [42••]. Wild-type L. gasseri, but not an isogenic mutant deficient in gassericin A synthesis, was able to prevent diarrhea in piglets. Testing of the purified bacteriocin in vitro showed that it increased intestinal fluid absorption as a result of inducing higher cellular cyclic nucleotide levels in epithelial cells via mechanism involving binding to the membrane protein Keratin 19 (KRT19) and activating mTOR (mechanistic Target of Rapamycin) phosphodiesterase activity [42].
Increase secondary bile acid concentrations
In healthy individuals, approximately 95% of lumenal bile acids are reabsorbed in the distal ileum [43]. The remaining amounts are modified by intestinal bacteria and then are either excreted or passively absorbed [43]. Antibiotics disrupt this process and result in increases in colonic primary bile acids, compounds that inhibit epithelial ion transport proteins [43]. Reductions in microbially-modified, secondary bile acids also increase susceptibility to C. difficile infection [44].
The potential for probiotics to alter bile acid composition concurrent with antibiotic consumption was shown in healthy volunteers given S. boulardii CNCM I-745 [45•] (Table 1). Fecal samples from individuals on amoxicillin-clavulanate contained higher quantities of cholic acid, a primary bile acid, and lower levels of secondary bile acids. Those changes were reversed in subjects taking S. boulardii CNCM I-745 [45•]. Although more studies are needed to assess the relative importance of bile acid metabolism on probiotic prevention of AAD, such outcomes might occur either by direct modification of bile acids by probiotic microbes or by broader effects which result in the maintenance/enrichment of certain members of the gut microbiome (Figure 1). Direct modification of bile acids with bile salt hydrolases (BSH) by Lactobacillus, Bifidobacterium, and Clostridium species is already well known. BSHs deconjugate bile acids and the resulting compounds can then be further modified to secondary and tertiary bile acids by other intestinal bacteria [43]. A functional role for intestinal BSH was demonstrated in mice whereby it was shown that higher BSH activity resulted in systemic responses mitigating cardiometabolic impacts of a high fat diet [46]. Recently, it was determined that bsh genes are enriched among vertebrate-associated Lactobacillus species [47]. An intestine-associated BSH phylotype with the highest enzymatic activity was only found in Lactobacillus and not other members of the human gut microbiome [48•].
Improve intestinal barrier function
Intestinal epithelial barrier integrity is increasingly understood to be important for the pathology of a number of intestinal and systemic diseases [49]. Antibiotics induce deficits in barrier function, or a “leaky gut”, according to studies with rodent models (Table 1) but the severity of the barrier losses appears to vary depending on the antibiotic used [50]. The capacity of certain, administered bacteria to prevent antibiotic-induced disruptions to the intestinal epithelium was demonstrated in animal studies (Table 1). A strain of Bacillus amyloliquefaciens was associated with improved structural and functional aspects of small intestine tissues in piglets given aureomycin [51]. Similar findings were reported for Lactobacillus casei CGMCC 12435 and a mixture of Lactobacillus and Bifidobacterium strains given to mice after ampicillin treatment [23,24]. Those results were supported by the observed increases in transcripts for tight junction proteins [23,24,51]. A high dose of B. fragilis ZY-312 (daily administration of 109 CFU) was associated with colonic increases in ZO-1 and occludin tight junction proteins, mucin synthesis, and cell markers for epithelial cell proliferation [25•]. Currently, specific compounds responsible for probiotic-induced changes to epithelial barrier function are largely unknown [52]. Recently, we have shown that the bacteriocin Plantaricin EF produced by L. plantarum can prevent pro-inflammatory cytokine mediated deficits to barrier integrity in in vitro and that wildtype L. plantarum, but not a plantaricin-deficient mutant strain, increases intestinal ZO-1 synthesis in obese mice [53•]. Other extracellular bacterial proteins such as the outer membrane pilus-associated protein synthesized by Akkermansia mucinophilia can also confer improvements to the intestinal barrier [54].
Alter intestinal immune responses
Antibiotics also affect immune homeostasis. In human subjects, antibiotic use resulted in impaired vaccine responses among individuals with low pre-existing antibody titers [55]. Antibiotics were also found to induce long-term, macrophage-dependent increases in inflammatory T helper 1 (Th1) responses in mice and heightened susceptibility to some infections [56].
Putative and established probiotic bacteria and yeast were found to counter antibiotic activation of inflammatory pathways in humans [19], piglets [51], and mice [23,24] (Table 1). Reductions in C-reactive protein, Complement C3, and IgG with administered Lactobacillus strains [24] indicate that those microbes might be able to limit the systemic effects of antibiotics (Table 1). These findings are consistent with strain-specific immunomodulation capacities of probiotics in healthy human subjects and individuals with chronic immune-associated diseases (e.g. allergy, asthma) [57,58]. Although the specific probiotic cell products able to directly alter immune cell function during antibiotic use are not yet known, recent reports show that certain extracellular compounds, such as Bifidobacterium exopolysaccharides [59] and Lactobacillus Slayer proteins [60] are immunomodulatory. Therefore, there are likely multiple secreted compounds made by probiotics which could influence immune system during antibiotic administration (Figure 1).
Conclusions
Results from clinical trials support the use of probiotics for preventing AAD. Therefore, it is notable that very few studies have investigated the molecular basis for probiotic AAD prevention (Table 1). Most reports have focused on strains that are not commercially available and are poorly characterized. Moreover, very few mechanistic studies with humans and animal models have directly examined L. rhamnosus GG or S. boulardii CNCM I-745, the strains most commonly tested in human trials [6] (Table 1). For these reasons, it is not yet possible to report which gut-modulating activities of probiotic microorganisms are the most important for protecting against AAD. Just as AAD is result of multiple factors connected with antibiotic administration (e.g. disruption of the gut microbiota, decreased intestinal SCFA concentrations, accumulation of luminal carbohydrates and colonic bile acids, altered water absorption), it is most likely that probiotic effects are multi-factorial and are both strain- and host-background dependent. It is also expected that there will be mechanistic overlap between strains (e.g. effects due to the production of organic acids) as well as strain-specific, host-microbe interactions (e.g. effects due to secretion of strain-specific enzymes and proteins) [61]. Such effects could be assessed in well-controlled, multi-center clinical trials for which intestinal and gut microbiota responses are measured and combined with complementary animal model studies using the same protocols. Elucidating the molecular mechanisms of probiotic action in the gut is extremely important for developing recommendations for existing strains such as the recommended dose, frequency, and duration of a probiotic intervention, the value of using multi-strain formulations, and the optimal protocols and ingredients for probiotic manufacture and carrier delivery. This knowledge is also needed for designing the appropriate assays to select the next-generation probiotics.
Highlights:
Clinical studies support the use of probiotics for preventing AAD.
AAD prevention by probiotics is multifactorial and strain dependent.
Probiotics are associated with gut microbiome modulation upon antibiotic use.
Intestinal bile acids and SCFAs are implicated in probiotic prevention of AAD.
Probiotic effector compounds may regulate intestinal fluid secretion and absorption.
Acknowledgements
This work was supported by awards from the National Institutes of Health, National Institute of Child and Human Development (1R01HD088428-01A1) and Office of Dietary Supplements (3R01HD088428-02S1).
Footnotes
Declaration of interest
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.McFarland LV, Ozen M, Dinleyici EC, Goh S: Comparison of pediatric and adult antibiotic-associated diarrhea and Clostridium difficile infections. World J Gastroenterol 2016, 22: 3078–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agamennone V, Krul CAM, Rijkers G, Kort R: A practical guide for probiotics applied to the case of antibiotic-associated diarrhea in The Netherlands. BMC Gastroenterol 2018, 18:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capurso L: Thirty years of Lactobacillus rhamnosus GG: a review. J Clin Gastroenterol 2019, 53:S1–S41. [DOI] [PubMed] [Google Scholar]
- 4.Czerucka D, Rampal P: Diversity of Saccharomyces boulardii CNCM I-745 mechanisms of action against intestinal infections. World J Gastroenterol 2019, 25:2188–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Q, Goldenberg JZ, Humphrey C, El Dib R JB: Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev 2019, 4: CD004827.•• This is the latest update of clinical evidence on probiotic prevention of antibiotic associated diarrhea since 2015. The authors concluded that probiotics were at least moderately protective against AAD even though a wide-variety of microbial strains and species were tested.
- 6.Goldenberg JZ, Lytvyn L, Steurich J, Parkin P, Mahant S: Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane database Syst Rev 2015, 12:CD004827. [DOI] [PubMed] [Google Scholar]
- 7.Willing BP, Russell SL, Finlay BB: Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol 2011, 9:233–243. [DOI] [PubMed] [Google Scholar]
- 8.Becattini S, Taur Y, Pamer EG: Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 2016, 22:458–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasiri MJ, Goudarzi M, Hajikhani B, Ghazi M, Goudarzi H, Pouriran R: Clostridium difficile infection in hospitalized patients with antibiotic-associated diarrhea: a systematic review and meta-analysis. Anaerobe 2018, 50:32–37. [DOI] [PubMed] [Google Scholar]
- 10.Larcombe S, Hutton ML, Lyras D: Involvement of bacteria other than Clostridium difficile in antibiotic-associated diarrhoea. Trends Microbiol 2016, 24:463–476. [DOI] [PubMed] [Google Scholar]
- 11.Derrien M, van Hylckama Vlieg JET: Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol 2015, 23:354–366. [DOI] [PubMed] [Google Scholar]
- 12.Lebeer S, Bron PA, Marco ML, Van Pijkeren JP, O’Connell Motherway M, Hill C, Pot B, Roos S, Klaenhammer T: Identification of probiotic effector molecules: present state and future perspectives. Curr Opin Biotechnol 2018, 49:217–223. [DOI] [PubMed] [Google Scholar]
- 13.Barker AK, Duster M, Valentine S, Hess T, Archbald-Pannone L, Guerrant R, Safdar N: A randomized controlled trial of probiotics for Clostridium difficile infection in adults (PICO). J Antimicrob Chemother 2017, 72:3177–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Wolfe TJ, Eggers S, Barker AK, Kates AE, Dill-McFarland KA, Suen G, Safdar N: Oral probiotic combination of lactobacillus and bifidobacterium alters the gastrointestinal microbiota during antibiotic treatment for Clostridium difficile infection. PLoS One 2018, 13:e0204253.• A multi-strain preparation of Lactobacillus and Bifidobacterium was given for four weeks to subjects being treated with antibiotics for an episode of C. difficile. Time-dependent changes in fecal microbiota composition were found among subjects given the probiotic supplement. The finding that those subjects also had improved diarrhea outcomes indicated that probiotic modulation of the gut microbiota is an important factor for attentuating AAD.
- 15.Bassis CM, Theriot CM, Young VB: Alteration of the murine gastrointestinal microbiota by tigecycline leads to increased susceptibility to Clostridium difficile infection. Antimicrob Agents Chemother 2014, 58:2767–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L, Wang Z, Sun G, Peng L, Lu Z, Yan B, Huang K, Yang Y: Effects of anti-H. pylori triple therapy and a probiotic complex on intestinal microbiota in duodenal ulcer. Sci Rep 2019, 9:12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh B, Kim JW, Kim BS. Changes in the functional potential of the gut microbiome following probiotic supplementation during Helicobacter pylori treatment. Helicobacter 2016. 21:493–503. [DOI] [PubMed] [Google Scholar]
- 18.Kabbani TA, Pallav K, Dowd SE, Villafuerte-Galvez J, Vanga RR, Castillo NE, Hansen J, Dennis M, Leffler DA, Kelly CP: Prospective randomized controlled study on the effects of Saccharomyces boulardii CNCM I-745 and amoxicillin-clavulanate or the combination on the gut microbiota of healthy volunteers. Gut Microbes 2017, 8:17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, Zur M, Regev-Lehavi D, Ben-Zeev Brik R, Federici S, et al. : Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 2018, 174:1406–1423. [DOI] [PubMed] [Google Scholar]
- 20.Crouzet L, Derrien M, Cherbuy C, Plancade S, Foulon M, Chalin B, Van Hylckama Vlieg JET, Grompone G, Rigottier-Gois L, Serror P: Lactobacillus paracasei CNCM I-3689 reduces vancomycin-resistant Enterococcus persistence and promotes Bacteroidetes resilience in the gut following antibiotic challenge. Sci Rep 2018, 8:5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grazul H, Kanda LL, Gondek D: Impact of probiotic supplements on microbiome diversity following antibiotic treatment of mice. Gut Microbes 2016, 7:101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y, Zhai Q, Li D, Mao B, Liu X, Zhao J, Zhang H, Chen W: Restoration of cefixime-induced gut microbiota changes by Lactobacillus cocktails and fructooligosaccharides in a mouse model. Microbiol Res 2017, 200:14–24. [DOI] [PubMed] [Google Scholar]
- 23.Shi Y, Zhao X, Zhao J, Zhang H, Zhai Q, Narbad A, Chen W: A mixture of Lactobacillus species isolated from traditional fermented foods promote recovery from antibiotic-induced intestinal disruption in mice. JAppl Microbiol 2018, 124:842–854. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, Kellingray L, Le Gall G, Zhao J, Zhang H, Narbad A, Zhai Q, Chen W: The divergent restoration effects of Lactobacillus strains in antibiotic-induced dysbiosis. J Funct Foods 2018, 51:142–152.• Several Lactobacillus strains were compared for their capacity to alter GI tract function after antibiotic administration to mice. The findings indicated potential strain-dependent and strain-independent effects of probiotics on recovery from antibiotic administration. A strain of L. casei was more effective in reducing antibiotic-induced GI tract disruptions, possibly through a mechanism involving increased intestinal SCFA levels.
- 25.Zhang W, Zhu B, Xu J, Liu Y, Qiu E, Li Z, Li Z, He Y, Zhou H, Bai Y, et al. : Bacteroides fragilis protects against antibiotic-associated diarrhea in rats by modulating intestinal defenses. Front Immunol 2018, 9:1040.• This comprehensive study examined the use of a gut commensal for preventing diarrhea in a rat model of AAD. A dose dependent response was found and high doses (109 CFU/day) ofB. fragilis were more effective than a commercial preparation of Lactobacillus acidophilus, Bifidobacterium longum, and Enterococcus faecalis in reducing fecal water levels.
- 26.Wullt M, Johansson Hagslätt ML, Odenholt I, Berggren A: Lactobacillus plantarum 299v enhances the concentrations of fecal short-chain fatty acids in patients with recurrent Clostridium difficile-associated diarrhea. Dig Dis Sci 2007, 52:2082–2086. [DOI] [PubMed] [Google Scholar]
- 27.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M: Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 2016, 540:280–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishiyama K, Nakazato A, Ueno S, Seto Y, Kakuda T, Takai S, Yamamoto Y, Mukai T: Cell surface-associated aggregation-promoting factor from Lactobacillus gasseri SBT2055 facilitates host colonization and competitive exclusion of Campylobacter jejuni. Mol Microbiol 2015, 98:712–726. [DOI] [PubMed] [Google Scholar]
- 29.El Hage R, Hernandez-Sanabria E, Arroyo MC, Props R, Van De Wiele T: Propionate-producing consortium restores antibiotic-induced dysbiosis in a dynamic in vitro model of the human intestinal microbial ecosystem. Front Microbiol 2019, 10:1206.• A multi-strain consortium selected for their distinct propionate biosynthetic capacities was tested in a mucosal simulator of the human intestinal microbial ecosystem (M-SHIME). Administration of multiple doses of the consortium corresponded with increases in propionate levels but not butyrate or acetate after antibiotic treatment. A partial recovery of the epithelial mitochondrial membrane potential was also noted as well as modifications to the microbiota composition.
- 30.Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, Eismann K, Otto W, Rojo D, Bargiela R, von Bergen M, et al. : Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 2013, 62:1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Binder HJ: Role of Colonic Short-chain fatty acid transport in diarrhea. Annu Rev Physiol 2010, 72:297–313. [DOI] [PubMed] [Google Scholar]
- 32.Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li JZ, Young VB: Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 2014, 5:3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Beek CM, Dejong CHC, Troost FJ, Masclee AAM, Lenaerts K: Role of short-chain fatty acids in colonic inflammation, carcinogenesis, and mucosal protection and healing. Nutr Rev 2017, 75:286–305. [DOI] [PubMed] [Google Scholar]
- 34.Cresci G, Nagy LE, Ganapathy V: Lactobacillus GG and tributyrin supplementation reduce antibiotic-induced intestinal injury. JPEN J Parenter Enteral Nutr 2013, 37:763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar A, Alrefai WA, Borthakur A, Dudeja PK: Lactobacillus acidophilus counteracts enteropathogenic E. coli-induced inhibition of butyrate uptake in intestinal epithelial cells. Am J Physiol - Gastrointest Liver Physiol 2015, 309:G602–G607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. : Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469:543–547. [DOI] [PubMed] [Google Scholar]
- 37.Louis P, Flint HJ: Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 2017, 19:29–41. [DOI] [PubMed] [Google Scholar]
- 38.Camilleri M, Sellin JH, Barrett KE: Pathophysiology, evaluation, and management of chronic watery diarrhea. Gastroenterology 2017, 152:515–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiagarajah JR, Donowitz M, Verkman AS: Secretory diarrhoea: mechanisms and emerging therapies. Nat Rev Gastroenterol Hepatol 2015, 12:446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urdaci MC, Lefevre M, Lafforgue G, Cartier C, Rodriguez B, Fioramonti J: Antidiarrheal action of Bacillus subtilis CU1 CNCM I-2745 and Lactobacillus plantarum CNCM I-4547 in mice. Front Microbiol 2018, 9:1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar A, Anbazhagan AN, Coffing H, Chatterjee I, Priyamvada S, Gujral T, Saksena S, Gill RK, Alrefai WA, Borthakur A, et al. : Lactobacillus acidophilus counteracts inhibition of NHE3 and DRA expression and alleviates diarrheal phenotype in mice infected with Citrobacter rodentium. Am J Physiol - Gastrointest Liver Physiol 2016, 311:G817–G826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, Ma L, Nie Y, Chen J, Zheng W, Wang X, Xie C, Zheng Z, Wang Z, Yang T, et al. : A microbiota-derived bacteriocin targets the host to confer diarrhea resistance in early-weaned piglets. Cell Host Microbe 2018, 24:817–832.•• Comparisons of diarrhea resistant and sensitive pigs lead to the identification of a Lactobacillus bacteriocin (gassericin A) as a probiotic effector that binds to a receptor on the membranes of intestinal epithelial cells to increase fluid absorption and reduce secretion. This paper indicates that probiotics might directly influence the intestinal epithelium to prevent AAD.
- 43.Winston JA, Theriot CM: Diversification of host bile acids by members of the gut microbiota. Gut Microbes 2019, doi: 10.1080/19490976.2019.1674124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. : Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517:205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kelly CP, Nguyen CC, Palmieri LJ, Pallav K, Dowd SE, Humbert L, Seksik P, Bado A, Coffin B, Rainteau D, et al. : Saccharomyces boulardii CNCM I-745 modulates the fecal bile acids metabolism during antimicrobial therapy in healthy volunteers. Front Microbiol 2019, 10:336.• Healthy volunteers receiving amoxicillin-clavulanate and S. boulardii CNCM I-745 had higher levels of fecal secondary bile acids and lower levels of primary bile acids as compared to individuals who received amoxicillin-clavulanate alone. This finding highlights the potential of probiotics to reduce the risk of AAD due to bile acid-associated changes in the GI tract.
- 46.Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CGM: Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A 2014, 111:7421–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Flaherty S, Briner Crawley A, Theriot CM, Barrangou R: The Lactobacillus bile salt hydrolase repertoire reveals niche-specific adaptation. mSphere 2018, 3. pii: e00140–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song Z, Cai Y, Lao X, Wang X, Lin X, Cui Y, Kalavagunta PK, Liao J, Jin L, Shang J, et al. : Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome 2019, 7:9.• This comprehensive informatics survey of bile salt hydrolase enzymes in the human gut microbiome associated concluded that the bile salt hydrolase phylotype with high enzymatic activity is made exclusively by Lactobacillus and not in other members of the human gut microbiome.
- 49.König J, Wells J, Cani PD, Garcia-Rodenas CL, MacDonald T, Mercenier A, Whyte J, Troost F, Brummer RJ: Human intestinal barrier function in health and disease. Clin Transl Gastroenterol 2016, 7:e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tulstrup MVL, Christensen EG, Carvalho V, Linninge C, Ahrne S, Højberg O, Licht TR, Bahl MI: Antibiotic treatment affects intestinal permeability and gut microbial composition in wistar rats dependent on antibiotic class. PLoS One 2015, 10:e0144854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du W, Xu H, Mei X, Cao X, Gong L, Wu Y, Li Y, Yu D, Liu S, Wang Y, et al. : Probiotic Bacillus enhance the intestinal epithelial cell barrier and immune function of piglets. Benef Microbes 2018, 9:743–754. [DOI] [PubMed] [Google Scholar]
- 52.Bron PA, Kleerebezem M, Brummer RJ, Cani PD, Mercenier A, MacDonald TT, Garcia-Rodenas CL, Wells JM. Can probiotics modulate human disease by impacting intestinal barrier function? Br J Nutr 2017, 117:93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heeney DD, Zhai Z, Bendiks Z, Barouei J, Martinic A, Slupsky C, Marco ML: Lactobacillus plantarum bacteriocin is associated with intestinal and systemic improvements in diet-induced obese mice and maintains epithelial barrier integrity in vitro. Gut Microbes 2019, 10:382–397.• The capacity for probiotics to improve intestinal barrier function was indicated by the finding that the L. plantarum bacteriocin Plantaricin EF was able to prevent pro-inflammatory cytokine mediated deficits to barrier integrity in cell culture and that Plantaricin EF is associated with L. plantarum induced increases in ZO-1 levels in the mouse ileum.
- 54.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, et al. : A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 2017, 23:107–113. [DOI] [PubMed] [Google Scholar]
- 55.Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS, Das J, Wang H, Guthmiller J, Zheng N- Y, et al. : Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 2019, 178:1313–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott NA, Andrusaite A, Andersen P, Lawson M, Alcon-Giner C, Leclaire C, Caim S, Le Gall G, Shaw T, Connolly JPR, et al. : Antibiotics induce sustained dysregulation of intestinal T cell immunity by perturbing macrophage homeostasis. Sci Transl Med 2018, 10:eaao4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maldonado Galdeano C, Cazorla SI, Lemme Dumit JM, Velez E, Perdigon G. Beneficial effects of probiotic consumption on the immune system. Ann Nutr Metab 2019,74:115–124. [DOI] [PubMed] [Google Scholar]
- 58.Peters VBM, van de Steeg E, van Bilsen J, Meijerink M. Mechanisms and immunomodulatory properties of pre- and probiotics. Benef Microbes 201910:225–236. [DOI] [PubMed] [Google Scholar]
- 59.Schiavi E, Gleinser M, Molloy E, Groeger D, Frei R, Ferstl R, Rodriguez-Perez N, Ziegler M, Grant R, Moriarty TF, et al. : The surface associated exopolysaccharide of Bifidobacterium longum 35624 plays an essential role in dampening host proinflammatory responses and repressing local TH17 responses. Appl Environ Microbiol 2016, 82:7185–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lightfoot YL, Selle K, Yang T, Goh YJ, Sahay B, Zadeh M, Owen JL, Colliou N, Li E, Johannssen T, et al. : SIGNR 3- dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J 2015, 34:881–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. : Expert consensus document: the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 2014, 14:491–502. [DOI] [PubMed] [Google Scholar]



