Abstract
Precise manipulation of protein activity in living systems has broad applications in biomedical sciences. However, it is challenging to use light to manipulate protein activity in living systems without genetic modification. Here, we report a technique to optically switch off protein activity in living cells with high spatiotemporal resolution, referred to as molecular hyperthermia (MH). MH is based on the nanoscale confined heating of plasmonic gold nanoparticles by short laser pulses to unfold and photo-inactivate targeted proteins of interest. Firstly, we show that protease activated receptor 2 (PAR2), a G-protein coupled receptor and an important pathway that leads to pain sensitization, can be photo-inactivated in situ by MH without compromising cell proliferation. PAR2 activity can be switched-off in laser-targeted cells without affecting surrounding cells. Furthermore, we demonstrate the molecular specificity of MH by inactivating PAR2 while leaving other receptors intact. Secondly, we demonstrate that the photo-inactivation of a tight junction protein in brain endothelial monolayers leads to a reversible blood brain barrier opening in vitro. Lastly, the protein inactivation by MH is below the nanobubble generation threshold and thus is predominantly due to the nanoscale heating. MH is distinct from traditional hyperthermia (that induces global tissue heating) in both its time and length scales: nanoseconds versus seconds, nanometers versus millimeters. Our results demonstrate that MH enables selective and remote manipulation of protein activity and cellular behavior without genetic modification.
Keywords: plasmonic nanoparticle, nanosecond laser, protein inactivation, G-protein coupled receptor, blood-brain barrier
Graphical Abstract
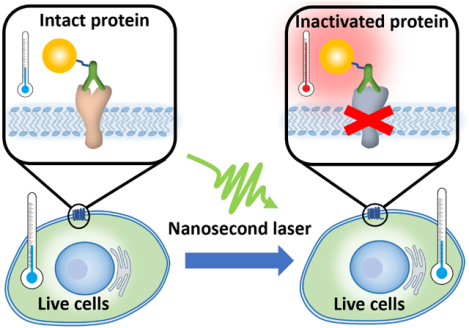
Selective and remote manipulation of protein function in living systems is important to elucidate the protein function and develop precise therapeutics for disease treatment. Advances in tool development have made significant impact in this direction. Among the various modalities, optical control of protein activity in live cells offers unparalleled spatial and temporal resolution. Current approaches include optogenetics,1, 2 chromophore-assisted light inactivation (CALI)3, 4 and synthetic photoswitches.5, 6 Optogenetics uses light to control cell activity, especially in neurons that have been genetically modified to express photo-sensitive proteins. Over the last decade, this technique has been instrumental in our understanding on how specific cell types contribute to brain activity and neurological diseases. While it has had transformative impact in the field, optogenetics requires genetic modification to enable the photosensitivity of target proteins. CALI is probably the most general method for optical control of protein activity with high spatial and temporal resolution and has been used in many areas in cell biology. CALI utilizes a photosensitizer to produce reactive oxygen species (ROS) and inactivate the adjacent proteins during light exposure. While small molecule photosensitizers (typically xanthene-based) have a high efficiency to generate ROS, it is challenging to design small molecules to target proteins in live cells. On the other hand, photosensitizers such as KillerRed are genetically encoded in target cells. Importantly, CALI efficiency is dependent on the expression level of photosensitizer when genetically encoded and on the cellular microenvironment such as oxygen concentration and endogenous reducing agent (glutathione).7 Synthetic photoswitches (e.g. based on azobenzene) have also emerged as a promising strategy to control protein activity with light, but currently there are a very limited number of photoswitches and thus narrow options of light source (i.e. wavelength). Therefore, it remains challenging to optically control protein activity in live cells without genetic modification and independent of the cellular microenvironment.
Nanoparticles provide an interface to manipulate the function of living systems from cells8, 9 to macromolecules such as proteins.10 Cellular behavior control with noninvasive stimulation such as magnetic,11, 12 acoustic13 and optical methods14 can be achieved with the assistance of nanoparticles. Among these stimulation methods, optical control of cellular behavior15, 16 using plasmonic nanoparticles provides the highest spatiotemporal resolution, including neuron firing,14, 17–20 heat shock protein expression,21 and optoporation.22–24 Continuous photothermal stimulation of plasmonic nanoparticles leads to global heating in the tissue (Figure. 1) and has been used for hyperthermia therapy of cancer. Physically, it is possible to apply short laser pulses to excite plasmonic nanoparticles and create a nanoscale hotspot, or “thermal confinement” when the pulse duration is less than the timescale of heat diffusion.25–27 The possibility of “molecular hyperthermia” (MH) was introduced recently, defined as using ultrashort laser pulses (nanosecond) to excite a plasmonic gold nanoparticle (AuNP) as a nanoheater to unfold and inactivate targeted proteins with a very well-defined impact zone (on the nanometer length scale).25, 28, 29 The heating is confined within nanometers of the nanoparticle (nanoscale hotspots) and doesn’t cause bulk heating, which allows for the possibility to control protein activity without affecting the cell viability. While it has been shown that MH can photo-inactivate isolated proteins of interest, it remains unclear whether MH works in complex cellular environments.
Figure 1.
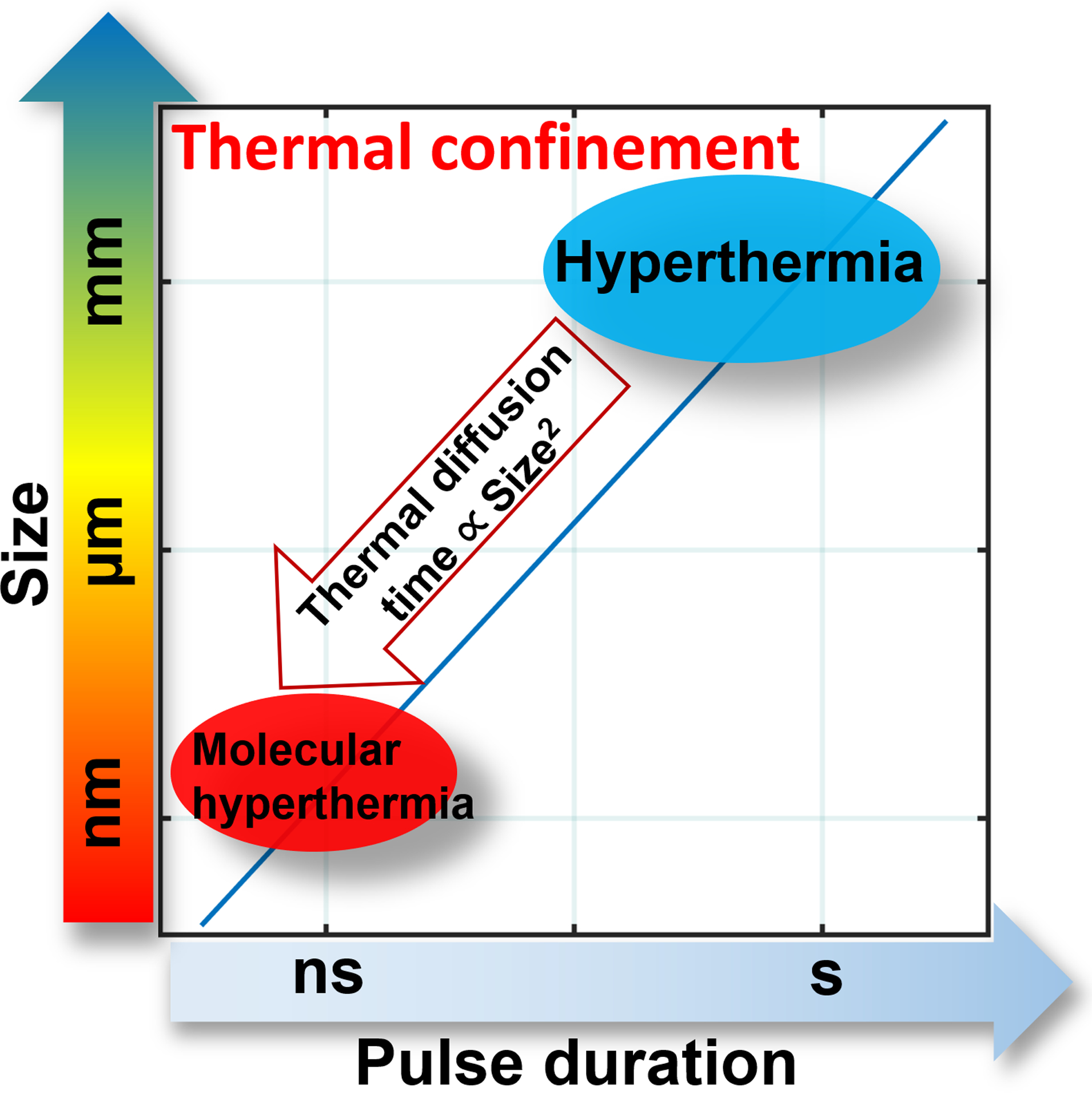
Schematic to illustrate the time and length scales of molecular hyperthermia (MH) compared with hyperthermia. Area left of the line represents thermal confinement. The thermal diffusion time is proportional to the square of size.34
In this report, we demonstrate that MH can inactivate protein in situ and control cellular activity with two examples. In the first example, we targeted a membrane receptor, protease activated receptor 2 (PAR2), which is important in pain sensitization signaling.30, 31 Ca2+ imaging analysis demonstrates that PAR2 can be inactivated in situ by MH without compromising cell proliferation. In the second example, we demonstrate the photo-inactivation of a tight junction protein in brain endothelial monolayers, which is important in maintaining the blood-brain barrier (BBB),32, 33 but also limits therapeutic delivery to the brain. MH of tight junction protein (junctional adhesion molecule A, or JAM-A) leads to a reversible BBB opening in vitro that recovers within 6 hours. We further demonstrated that the energy levels for MH is insufficient to form vapor nanobubbles around AuNP. Our results indicate that MH is a promising approach to manipulate protein activity in live cells without genetic modification and develop therapeutics with high spatiotemporal resolution.
RESULTS AND DISCUSSION
AuNP-Antibody Conjugation and Characterization.
First, we designed and characterized an antibody modified AuNP to target PAR2. PAR2 is an important G-protein coupled receptor implicated in pain.30, 31 During inflammation or cancer, protease-activated receptors (PAR) are activated by proteolytic cleavage of the extracellular amino terminus and lead to diverse pathologies including pain sensitization.35 Proteolytic cleavage of the N terminus of PAR2 results in exposure of a tethered ligand that activates the receptor to induce signaling (Figure 2A). One consequence of the PAR2 activation in the periphery nervous system is sensitization of neurons responsible for transmitting noxious information to the central nervous system (CNS). Importantly, nociceptive neurons express PAR2, and PAR2 activation on these neurons leads to enhanced signaling via a variety of channels including the capsaicin and noxious heat receptor, TRPV1. For instance, PAR2 is responsible for protease sensitization of TRPV1 in vivo, leading to thermal hyperalgesia.36, 37 Thus, targeting and photo-inactivating PAR2 will allow us to control pain signaling (Figure 2A). As a routinely used cell line in the PAR2 research community,38 human embryonic kidney 293 (HEK293) cells were used in this study to investigate PAR2 activity. PAR2 activation leads to intracellular Ca2+ concentration increase through the inositol 1,4,5-trisphosphate (IP3) signaling pathway.39 Here, the FRET sensor (TN-XXL) monitors Ca2+ during PAR2 activation by a specific PAR2 agonist (Figure 2B).40, 41 We selected gold nanospheres with a diameter of 45 nm as nanoheaters due to their relatively high absorption cross section and because they facilitate protein denaturation farther than 10 nm away from themselves.28, 42
Figure 2.
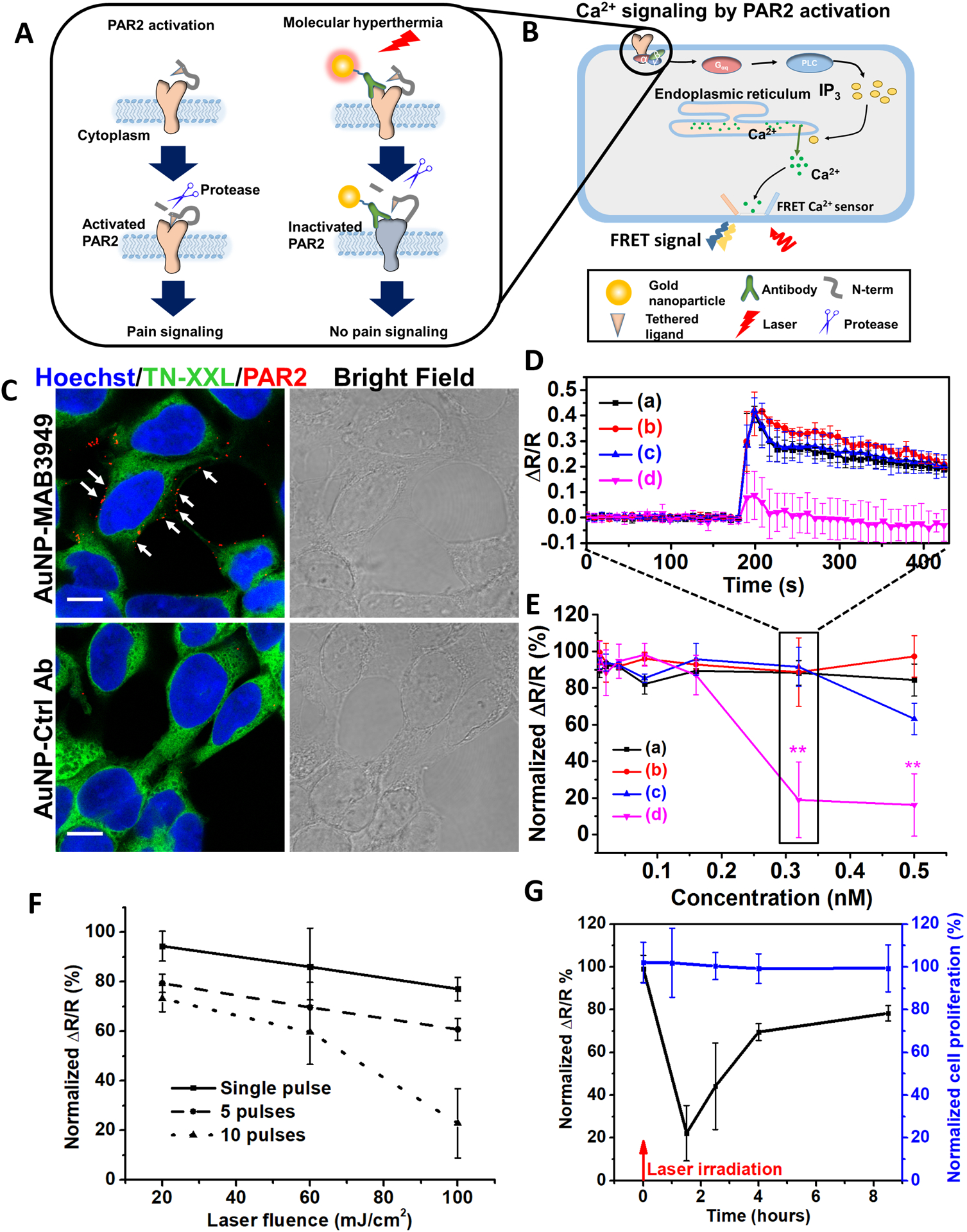
Photo-inactivation of PAR2 by molecular hyperthermia. A) Schematic of PAR2 inactivation by molecular hyperthermia. B) Schematic illustrating the Ca2+ signaling due to PAR2 activation. Gαq is heterotrimeric G protein subunit, PLC is phospholipase C, IP3 is inositol trisphosphate. C) Fluorescent immunocytochemistry imaging of HEK293 cells. Nucleus is in blue (Hoechst 33342), FRET Ca2+ sensor protein TN-XXL is in green, and PAR2-targeting antibody modified gold nanoparticle (AuNP-MAB3949) is in red (stained by Alexa 647 conjugated secondary antibody, indicated by white arrows), Control antibody modified gold nanoparticles (AuNP-Ctrl Ab) as a control show minimum signal (scale bar: 10 μm), AuNP concentration [AuNP] = 0.5 nM. D) The FRET ratio (ΔR/R) signal for different groups: (a) AuNP-Ctrl Ab, no laser; (b) AuNP-MAB3949, no laser; (c) AuNP-Ctrl Ab, with laser; (d) AuNP-MAB3949, with laser. [AuNP] = 0.32 nM. Laser conditions: 532 nm wavelength, fluence at 100 mJ/cm2 and 10 pulses. The incubation time for AuNPs is 5 minutes. E) Dose response of PAR2 activity under different particle concentrations for different groups. The groups are color-coded as in Figure 2D. F) Dose-response curve for PAR2 activity under different laser fluence and pulse numbers. [AuNP] = 0.32 nM and AuNP incubation time is 5 minutes. G) PAR2 function recovery and cell proliferation after molecular hyperthermia treatment. The experimental conditions are the same as (d) group in Figure 2D. Laser were irradiated at 0 hour time point (indicated by red arrow). The normalized ΔR/R and cell proliferation indicate normalization with the control group (without AuNP incubation or laser treatment).
The prepared AuNP was characterized by transmission electron microscope (TEM) and dynamic light scattering (DLS) to confirm its size and lack of aggregation (Figure S1B and C). Conjugation of poly(ethylene glycol) 2-mercaptoethyl ether acetic acid (Thiol-PEG-Carboxyl) onto AuNP increased the hydrodynamic diameter by 15 nm, in agreement with previous reports.43 Both PAR2 target antibody (MAB3949) and control antibody (Ctrl Ab) coating further increased the hydrodynamic size by 11–12 nm. UV-Vis measurements showed that the AuNP absorbance peak shifted slightly from 531 nm to 532 nm after conjugating with Thiol-PEG-Carboxyl, and then again to 536 nm after anti-PAR2 antibody (MAB3949) conjugation (Figure S1D). A Western blot experiment was then performed to confirm MAB3949 antibody specificity to PAR2. PAR2 was observed by MAB3949 staining in the dorsal root ganglion (DRG) sample from wild type mouse (PAR2 +/+) at around 50 kDa, with a negative staining for PAR2 knockout mouse (PAR2 −/−, Figure S1E). PAR2 expression on HEK293 cells was also tested and confirmed by immunocytochemistry (ICC) experiments with methanol-prefixed cells (Figure S1F). Furthermore, we show that MAB3949 antibody binds to PAR2 after conjugation onto the AuNP surface by ICC staining on HEK293 cells (Figure 2C). Specifically, cells incubated with AuNP-MAB3949 show a clear binding on the cell membrane, while control antibody modified AuNP (AuNP-Ctrl Ab) doesn’t bind with the cell membrane. It is worth noting that the cells incubated with AuNP-MAB3949 have relatively stronger signal than the case with MAB3949 in methanol-prefixed cells. This is because multiple antibodies (57±28 antibodies/AuNP, Figure S2) conjugated on the AuNP amplify the secondary antibody binding and the resulting fluorescent intensity.
Photoinactivation of PAR2 by Molecular Hyperthermia in Situ.
Next, we demonstrated that PAR2 can be inactivated by MH on live HEK293 cells. We first tested and confirmed that the engineered HEK293 cell with Ca2+ indicator44 is a robust model to study PAR2 activity. The addition of PAR2 agonist 2-aminothiazol-4-yl-LIGRL-NH2 (2AT, 330 nM) leads to a robust increase in the citrine channel and decrease in the ECFP channel (Figure S2B). A dose-response curve was then obtained from analyzing the FRET signal (Figure S3C–D). It is worth noting that the FRET signal (ΔR/R) was triggered within 10s after 2AT addition at all concentrations. However, the time duration to reach the maximum ΔR/R reduces with increasing 2AT concentration. Later, we consider the maximum FRET ratio change to compare PAR2 activity.
Then, we tested the effect of MH on PAR2 activity. Three experimental groups were used: MH including PAR2 targeting AuNP (AuNP-MAB3949), control antibody modified AuNP (AuNP-Ctrl Ab), and cells alone with no AuNP treatment (Ctrl). The AuNP was incubated with cells at 4°C to prevent cell uptake. Our results suggest that laser irradiation alone (Ctrl group, 532 nm, 100 mJ/cm2, and 10 pulses) does not cause a significant difference in either PAR2 activity or cell proliferation (Figure S4A), and the antibody alone (MAB3949) does not change PAR2 activity (Figure S4B). Comparing AuNP-MAB3949 and AuNP-Ctrl Ab under the same laser exposure (532 nm, 100 mJ/cm2, 10 pulses), a significant drop (>80 %) in PAR2 activity was observed for the AuNP-MAB3949 group at concentrations above 0.32 nM (Figure 2D&E) while there is no significant change for the AuNP-Ctrl Ab group (except at higher AuNP concentration 0.5 nM). Without laser irradiation, neither the AuNP-MAB3949 group nor the AuNP-Ctrl Ab group showed an obvious drop in PAR2 activity (Figure 2D&E). Further investigation shows a gradual drop in PAR2 activity as the pulse number and intensity were increased (Figure 2F). Also, increasing the incubation time of AuNP to 30 mins does not dramatically change the PAR2 inactivation (Figure S5A). At conditions that lead to a significant PAR2 activity drop in the AuNP-MAB3949 group, we found no significant differences in the cell proliferation between AuNP-Ctrl Ab and AuNP-MAB3949 groups regardless of laser fluence (Figure S5B). This suggests a specific molecular level photo-inactivation by MH. Also, we measured the bulk medium temperature after laser irradiation and confirmed that there is no obvious temperature rise, indicating that the temperature change is limited to the nanoparticle and its immediate surroundings (Figure S5D). Next, propidium iodide (PI) staining was performed to test for possible cellular membrane damage and necrosis (Figure S6). The MH group does not show significant PI signal increase compared with the control groups. This result demonstrates that the MH does not cause membrane damage or necrosis. Finally, we investigated the PAR2 function recovery and cell proliferation at different time points. The PAR2 activity drops to 20% at 1.5 hours after MH treatment, and gradually increase to 80% at 8.5 hours (Figure 2G). The reversible PAR2 activity does not directly imply that photo-inactivation process is reversible. The cell is recycling the protein and possibly synthesizing new proteins during this time. As a result, even if the protein inactivation is irreversible, we can still observe protein function recovery after several hours.
PAR2 Inactivation in Laser-Targeted Cells.
Furthermore, we demonstrated that MH can switch off protein activity in laser-targeted cells. By incubating cells with AuNP-MAB3949 and exposing only half of the cells to laser pulses, we compared the cellular PAR2 response with and without laser irradiation in the same petri-dish (Figure 3A). By perfusing 2AT and imaging under 2-photon microscopy (Figure 3B–C), the cell response was imaged at real time. The results suggest that the region without laser irradiation had an obvious Ca2+ release after 2AT loading while the region with laser irradiation had a much attenuated Ca2+ release (Figure 3D). The 10s delay of Ca2+ release after 2AT loading is probably due to the drug diffusion from the loading pipette to cells. This result suggests MH allows laser targeting of selected cells to photo-inactivate PAR2 activity.
Figure 3.
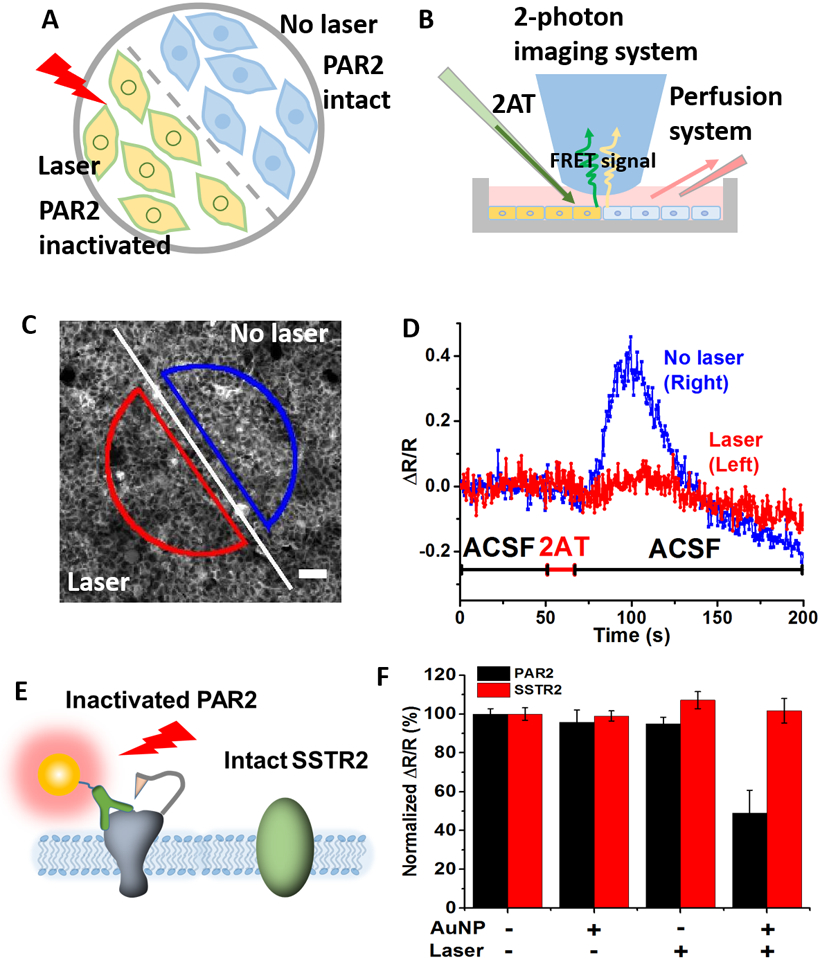
High spatial resolution and molecular specificity for PAR2 photo-inactivation by molecular hyperthermia. A) Experimental design: after targeting PAR2 with AuNP-MAB3949, half of the cells were blocked from laser irradiation. B) Experimental setup including 2-photon imaging of cytoplasmic Ca2+ release. C) Fluorescence images of cells after laser irradiation (scale bar: 50 μm). The line indicates the border of laser irradiation, with the bottom left side receiving laser irradiation (ROI in red) and the top right side without laser irradiation (ROI in blue). D) Ca2+ signals for different ROIs. The red line is the FRET ratio from the cells in the red semicircle (with laser) and the blue line indicates the FRET ratio from the blue semicircle (no laser). ACSF refers to artificial cerebral spinal fluid. E) Schematic to illustrate the photo-inactivation of PAR2 without compromising somatostatin receptor 2 (SSTR2) activity. F) PAR2 and SSTR2 activity after molecular hyperthermia treatment. [AuNP] = 0.32 nM and AuNP incubation time is 5 minutes. Laser conditions: 532 nm wavelength, fluence at 100 mJ/cm2 and 10 pulses.
Molecular Selectivity of PAR2 Inactivation by MH.
Finally, we tested the molecular specificity of PAR2 inactivation. We used HEK293 cells transferred with somatostatin receptor 2 (SSTR2), another GPCR protein that can be activated by somatostatin (SST) (Figure 3E). We then inactivated PAR2 by MH and measured SSTR2 and PAR2 activities. The results show that the SSTR2 activity remains the same as the control groups while the PAR2 activity reduces by 50% (Figure 3F and Figure S7). This demonstrates that MH can selectively switch off the activity of targeted cell receptors while not affecting other cell membrane receptors. We note that the molecular specificity is not due to laser focusing on specific receptors since this is well below the diffraction limit and not physically possible. Instead, the nanoparticles target specific receptors and the laser excites the targeted nanoparticles to inactivate the receptor, while leaving other non-targeted receptors intact.
BBB opening by disruption of tight junction protein induced by MH.
Targeting of Junctional Adhesion Molecule-A (JAM-A) by Surface Modified AuNPs.
JAM-A is one of the tight junction proteins that are located between brain endothelial cells,32 and is an important component of the blood-brain barrier (BBB). Here we test whether targeting and photo-inactivation of JAM-A leads to BBB opening (Figure 4A). To target JAM-A, we first modified AuNPs with BV16 (antibody to human JAM-A)32 and observed similar changes in hydrodynamic size and absorbance peak shift (Figure S8) as with PAR2 antibody conjugation. JAM-A expression was confirmed between cells by immunocytochemistry staining (Figure S9). The immunocytochemistry staining of the AuNP-BV16 group showed a clear signal between cell boundaries while the AuNP-PEG group did not show any obvious signal, confirming antibody binding activity after conjugation to AuNP (Figure 4B).
Figure 4.
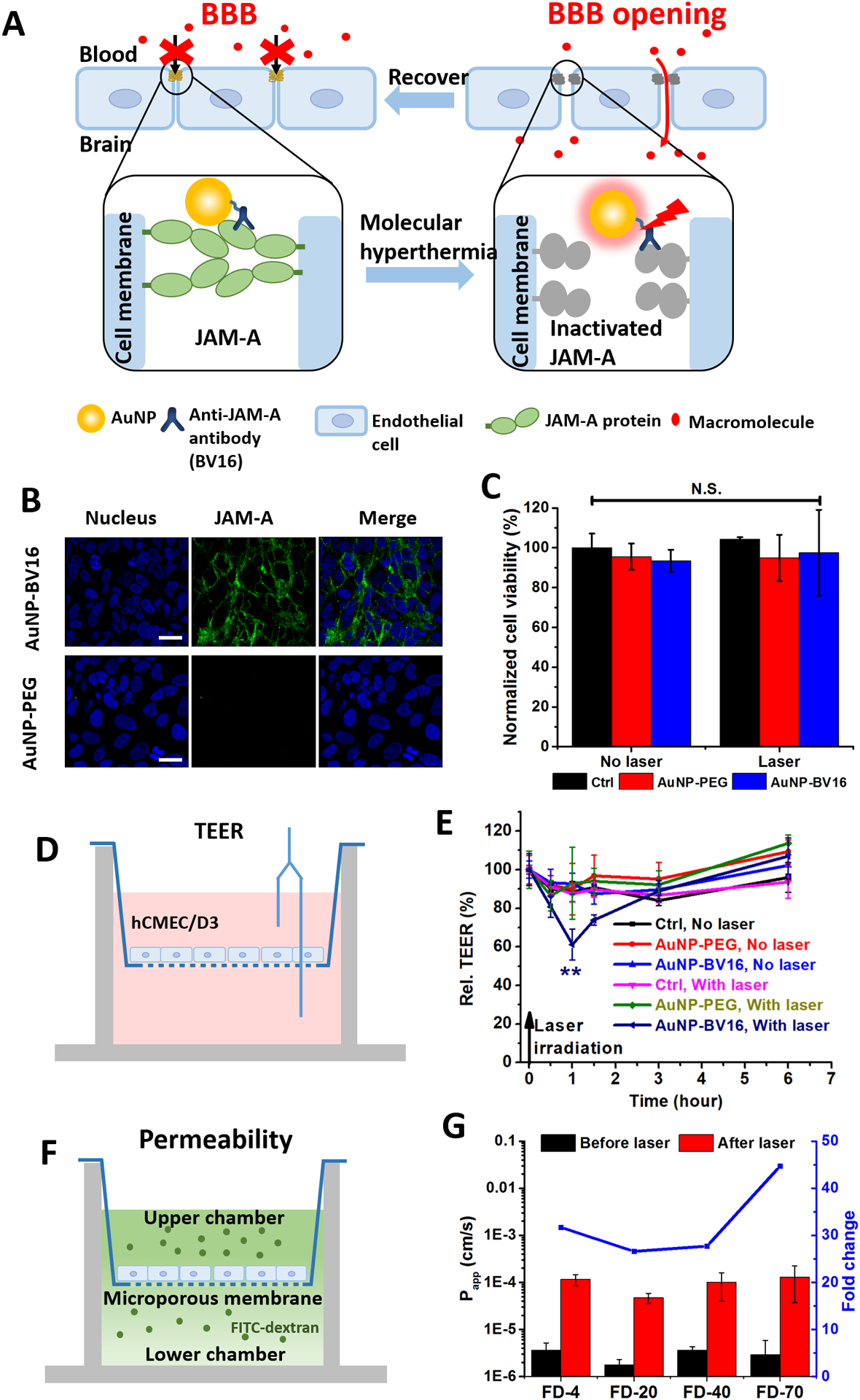
Photo-inactivation of a tight junction protein (JAM-A) by molecular hyperthermia to manipulate the blood-brain barrier (BBB). A) Schematic illustrating the JAM-A photo-inactivation by molecular hyperthermia. B) Fluorescent immunocytochemistry staining for brain endothelial cells (hCMEC/D3) targeted by AuNP-BV16. Nucleus is in blue and JAM-A is in green (Alexa 488) (scale bar: 20 μm). C) Cell proliferation measurements with and without laser irradiation and AuNP incubation. Ctrl indicates control group (no AuNP). D) Schematic illustrating the transendothelial electrical resistance (TEER) measurement. E) TEER measurements before and after molecular hyperthermia. F) Schematic of permeability measurement. G) Permeability measurements of different sized macromolecules diffusing cross the cellular barrier. FD-4 indicates 4 kDa FITC-labeled dextran.
Molecular Hyperthermia Inactivation of JAM-A.
Next, we investigated MH to photo-inactivate JAM-A and potentially manipulate BBB. First, we showed that the AuNP accumulation and laser irradiation does not affect cell proliferation (Figure 4C). To characterize the changes in the BBB, the transendothelial electrical resistance (TEER) was measured (Figure 4D–E). With MH, the TEER value dropped to 60% in 30 minutes (AuNP-BV16 group) while no significant drop was observed for AuNP-PEG groups and control groups (without AuNP incubation). Also, with absence of laser irradiation, we did not observe any significant drop in TEER values for all three groups. After 6 hours, the TEER value for the MH group recovers to the original level comparable with the control groups. This is important since it indicates that MH treatment leads to a reversible BBB opening. The endothelial barrier function was also assessed by measuring the permeability of macromolecules (Figure 4F). The results show that the permeability of FITC-labeled dextran increased more than 25 times (molecule weight from 4 kDa to 70 kDa) (Figure 4G).
Comparison of Laser Fluence for MH and Vapor Nanobubble Generation.
The heating of plasmonic nanoparticles by a pulsed laser can lead to vapor nanobubble formation around the nanoparticle. To determine whether there is vapor nanobubble formation under MH, we measured the AuNP under a pump-probe system that detects the presence of vapor nanobubble scattering. The laser fluence (100 mJ/cm2) that induces PAR2 inactivation is much lower than the nanobubble generation threshold for 45 nm AuNP (295 mJ/cm2 for 5% probability and 415 mJ/cm2 for 50% probability, Figure 5A). The nanobubble threshold (835 mJ/cm2) for 30 nm AuNP is higher than that for 45 nm AuNP. The measured nanobubble threshold agrees with previous study.45 For comparison, we also included our previous results of photo-inactivation of an enzyme (α-chymotrypsin, α-Cht) by MH induced by 30 nm AuNP (figure 5A).28 The α-Cht was linked directly to 30 nm AuNP by a PEG linker, and the activity of α-Cht can be monitored directly through an enzyme colorimetric activity assay. Similar to PAR2 inactivation, α-Cht inactivation is dependent on laser fluence. The laser fluences required to induce protein inactivation are lower than the nanobubble generation threshold for 30 nm AuNP (Figure 5A). Further comparison shows a large difference in the laser fluence for 50% protein inactivation and nanobubble generation probability (Figure 5B). Therefore, we have confirmed that the MH does not lead to nanobubble generation.
Figure 5.
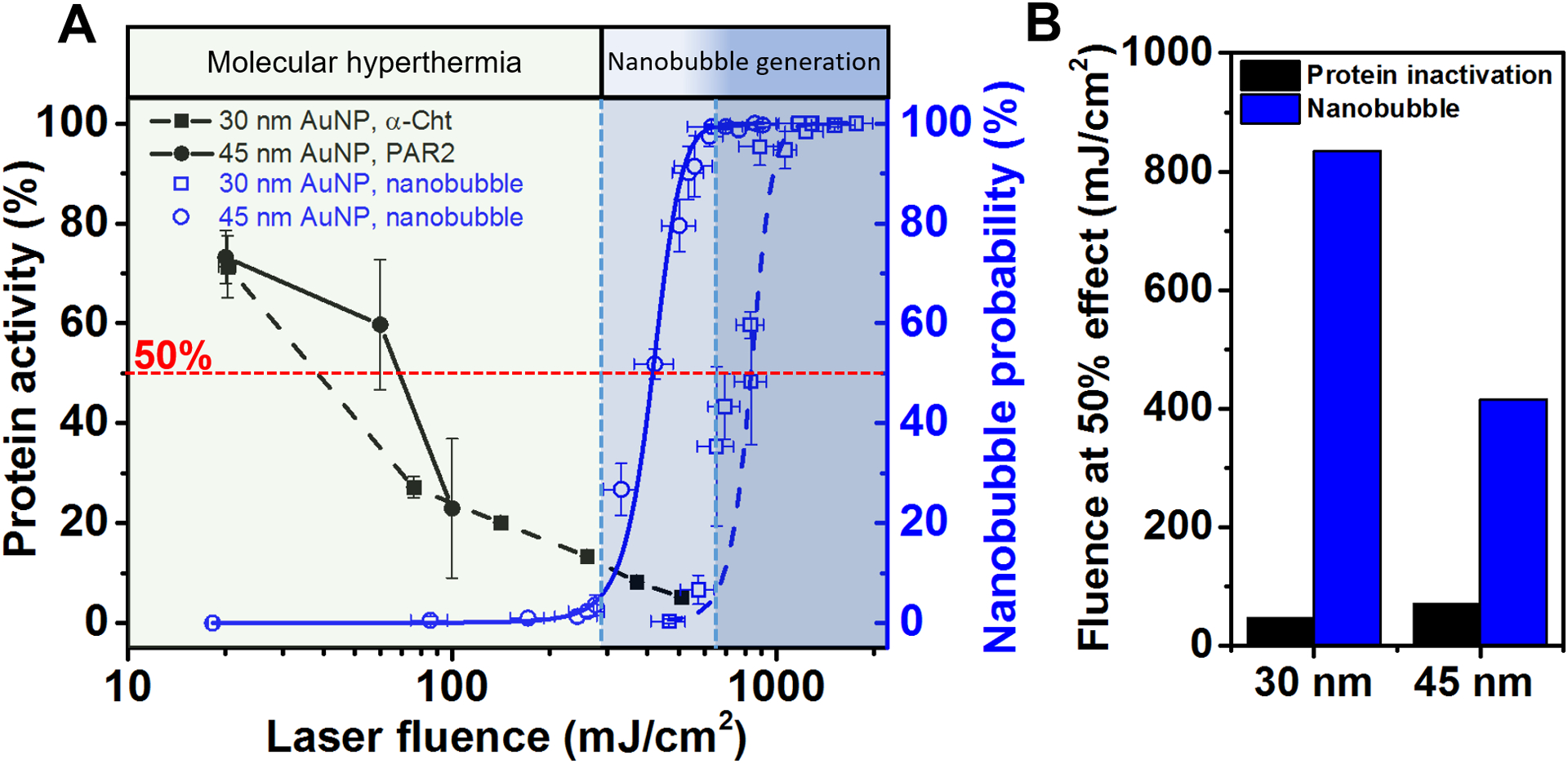
Protein inactivation by molecular hyperthermia (MH) does not lead to nanobubble generation. (A) Protein inactivation and nanobubble generation probability as a function of laser fluence. Photo-inactivation of α-chymotrypsin (α-Cht) by 30 nm AuNP (in solution) is adapted with permission from ref 28. Copyright 2017 John Wiley and Sons. The blue dash and solid lines are Boltzmann fitting for nanobubble probability measurements for 30 and 45 nm AuNP respectively. The blue shaded zone indicates >5% bubble generation probability. (B) Laser fluence at 50% protein inactivation and nanobubble probability.
Estimation of AuNP and Surrounding Medium Heating.
To understand the local temperature change during MH, we simulated the laser plasmonic heating process of AuNP and its surrounding medium. The optical properties of 45 nm AuNP was obtained by Mie theory and the simulation accuracy was confirmed by comparing with experimental measurement (Figure S10A).46 A nanosecond laser pulse (Full width half maximum, FWHM=6 ns) is applied to the AuNP (Figure S10B). Finite element modeling (FEM) of the 3D temperature profile (at 8 ns, Figure S11A) shows that the particle temperature increases dramatically and the immediate surrounding medium is heated. The heating process by the nanosecond laser pulse is fast and lasts around 40 ns (Figure S11B). Although the temperature increases of AuNP and surrounding medium is dramatic, the heating area is confined to tens of nanometers from the AuNP surface (Figure S11C). Next, we compared the FEM with molecular dynamics (MD) simulation for a smaller 3 nm AuNP. A small AuNP is modeled here due to the computational limitation of MD (Figure S11D). The gold temperature given by MD is much higher than the FEM result while the water temperature at the interface is lower than the FEM result (Figure S8E&F). This indicates that there may be some size-dependent properties that are not considered in the FEM simulation. Further studies are required to elucidate the temperature changes during MH combining computational and experimental approaches.
Protein Manipulation by Plasmonic Nanoparticles and Short Laser Pulses:
In this study, we show that molecule hyperthermia (MH) allows optical switch-off of protein activity in live cells with two distinct examples (PAR2 and JAM-A). First, PAR2 is a G-protein coupled receptor (GPCR) and is implicated in disease conditions such as allergic asthma, cancer, arthritis, and chronic pain.31, 47 PAR2 can be activated in response to various exogenous and endogenous proteases, usually elevated in inflammation or cancer. Proteolytic cleavage of the N terminus of PAR2 results in exposure of a tethered ligand that activates the receptor to induce signaling. In the periphery nervous system, PAR2 has become an important target to understand and treat diseases such as chronic pain. Second, JAM-A is part of the tight junction protein complex, which is a key component of the blood-brain barrier (BBB). BBB is an important mechanism to protect brain functions from toxins and pathogens in blood circulation. However, it also presents a major challenge and leads to insufficient drug delivery to treat brain diseases. Optical switch-off of JAM-A by MH causes temporary disruption of BBB and offers a promising way to enhance drug delivery to the brain for a variety of brain diseases including gliomas.
In contrast to photothermal heating that leads to global tissue heating including cancer hyperthermia, optical control neuron firing and optical control of cell behavior with thermally sensitive ion channels, molecular hyperthermia utilizes the thermal confinement enabled by ultrashort nanosecond lasers. Specifically, the nanoscale confined heating of MH operates in a fundamentally different time and spatial scale (10−9s, 10−9 m, Figure 1) when compared with global heating (1–100 s, 10−3 – 10−2 m). Excitation of plasmonic nanoparticles with even shorter laser pulses (picosecond or femtosecond) leads to fundamentally different mechanisms from photothermal effects, and has been explored for optoporation of cells and the release DNA/RNA from AuNP surface.48, 49
Nanosecond laser excitation can lead to nanobubble generation at higher laser fluence than MH (Figure 5). Several recent studies have reported using nanosecond laser excitation of membrane-targeted AuNP for optoporation. For example, Xiong et al. utilized 2040 mJ/cm2 (70 nm AuNP, 7 ns laser) to generate nanobubble and efficiently trigger optoporation.50 Yao et al. reported the cell-membrane permeabilization at 400~600 mJ/cm2 laser fluence (30 nm AuNP, 103 NPs/cell, 4 ns laser) for adhesive cells.51 Comparison of the laser fluence from Yao et al. with Figure 5 shows that the laser fluence is close to nanobubble generation threshold and much higher than MH. Therefore, we attribute the protein inactivation by MH to the nanoscale heating while the membrane optoporation by AuNP and nanosecond laser excitation is possibly a result of vapor nanobubble generation.
There are several unanswered questions in this area. First, what is the temperature of the nanoparticle and surrounding water during plasmonic heating of gold nanoparticles? It is not trivial to obtain temperature values of both gold nanoparticles and water directly from experiment. The numerical model in this study used thermal properties of bulk materials and thermal interface conductance measured by femtosecond excitation. The thermal interface conductance on the nanoscale is still an emerging area with many unknowns. The nanoscale thermal properties, including the thermal conductivity of water, are reported to have a recognizable difference from their bulk values.52 It remains a challenge to develop new methods to reliably predict and validate temperature changes of nanoparticles during short laser excitations (e.g. nanosecond pulses). Second, what is the fate of AuNP? There have been some reports on nanosecond pulse induced particle reshaping and fragmentation.45 It is known that nanosecond laser pulses can lead to nanoparticle reshaping towards a more spherical geometry (for example 20–30 mJ/cm2 for 40 nm AuNP). The fluence we use in this paper is higher than this value and we anticipate some particle reshaping and even fragmentation. Further investigation will be necessary if it is desirable to avoid particle fragmentation. For example, it may be possible to reduce the chance of particle fragmentation by further stretching the pulse duration to reduce the local temperature and extend the nanoscale heating duration.53 Third, how does the protein structure react to such intense heating? Previous studies suggest not only changes in the secondary structures of proteins but also in the primary structure.54–56 However, the mechanism of this protein inactivation phenomenon remains unclear although there have been some molecular dynamics simulations in this area.57 The fate of the protein under MH needs further investigation. Lastly, how does the protein expression level affect the MH? The exact number of PAR2 and JAM-A expressed on the cell surface remains to be quantified. An important next step would be to determine the receptor density on the cell membrane and number of AuNP targeted on the membrane. Towards this, MH may be a useful tool for precisely probing and manipulating protein activity at a single-cell level.58,59 Another question is how the cell reacts to MH in real time. It may be an interesting topic to investigate cellular responses such as intracellular pathways60, 61 in real time after MH.
Broad Applicability of MH:
There has been significant interest in the use of plasmonic nanoparticles to control cell behavior.14, 17, 18 However, most of the techniques require thermally sensitive ion channels such as expressing TRPV-1 in cells, thus limiting their applications. Here MH utilizes the intrinsic properties of nanoscale heating to inactive neighboring proteins, thus allowing broad applications as demonstrated here. Furthermore, MH doesn’t require genetic modification and thus removes a barrier as a more general method to optically switch off protein function and control cell behaviors. Furthermore, MH utilizes plasmonic nanoparticles that can be engineered to strongly absorb in the near-infrared window for in vivo applications such as using gold nanorods62–64 or nanoshells.65 This is a significant advantage compared with other approaches such as CALI and synthetic photoswitches where few photosensitizers and photoswitches work in the near-infrared window.
Intracellular Proteins as Targets:
The two proteins in this work are both located on cell surfaces for ease of targeting. GPCRs are a class of the most important transmembrane proteins that transfer extracellular messages to intracellular signaling. More than 40% of all modern drugs and almost 25% of the top 200 best-selling drugs target GPCRs.66 While the importance of membrane proteins can’t be overstated, there are potential methods to deliver nanoparticles to target intracellular proteins. For example, one study suggests that hydrophobic surface modification of silicon nanoparticle conjugates enhances intracellular protein delivery to target intracellular protein machinery.67 In addition, it is possible to use other targeting methods besides antibodies as was used in this report. In particular, antibodies may have limited applicability for intracellular targets due to their large size, complex structures and disulfide bonds.68 Some promising methods utilize relatively smaller molecules such as aptamers69 and nanobodies70 to provide more flexibility for intracellular protein targeting. Also, the majority of therapeutics utilize small molecules due to their small size and amphiphilic properties. Thus, an alternative is to use a small molecule as a target ligand instead of using antibodies to achieve intracellular MH.71
CONCLUSIONS
In this study, we demonstrate that molecular hyperthermia (MH) allows us to optically switch off protein activity in live cells without genetic modification. MH is based on the nanoscale plasmonic heating of AuNPs to inactivate targeted proteins of interest. To demonstrate the broad applicability of MH, we show that MH can inactivate protease activated receptor-2 (PAR2), a G-protein coupled receptor implicated in pain, and junctional adhesion molecule A (JAM-A), one of the tight junction proteins in the blood-brain barrier. In both cases, the photo-inactivation of the target protein did not compromise cell proliferation. Furthermore, the laser fluence used for MH is not sufficient to generate vapor nanobubble around AuNPs. MH is a promising method with broad applicability to switch off protein activity without genetic modification and will find many applications in biomedical sciences.
METHODS
Reagents and Cells.
Tetrachloroauric (III) trihydrate (HAuCl4·3H2O: 99.9%), sodium citrate tribasic dehydrate (≥99%) and hydroquinone (≥99%) were purchased from Sigma-Aldrich. Heterobifunctional polyethylene glycol OPSS-PEG-SVA (3.4 kDa, OPSS-PEG-SVA-3400) were purchased from Laysan Bio Inc. Poly(ethylene glycol) 2-mercaptoethyl ether acetic acid (Thiol-PEG-Carboxyl, 600 Da, PG2-CATH-600) were purchased from Nanocs Inc. Fluorescein isothiocyanate–dextran (FITC-dextran) and WST-1 cell proliferation reagent (05015944001) was purchased from Sigma-Aldrich. Propidium iodide (PI) is purchased from Cayman chemical (14289). Mouse IgG isotype control (Ctrl Ab, 31901) was purchased from Invitrogen. Anti-PAR2 antibody (MAB3949) was purchased from R&D systems. Anti-JAM-A antibody BV16 was kindly provided by Dr. Elisabetta Dejana’s lab.32 Human cerebral microvessel endothelial cells (hCMEC/D3) were purchased from EMD Millipore. HEK293 cell with Ca2+ indicator TN-XXL was kindly provided by Dr. Paul Slesinger’s lab.44 PAR2 agonist 2-aminothiazol-4-yl-LIGRL-NH2 (2AT) was provided by Dr. Theodore Price’s lab.40 Somatostatin (SST) was purchased from Sigma-Aldrich (S9129). Pierce modified lowery protein assay kit was obtained from ThermoFisher Scientific (23240).
Gold Nanoparticle Synthesis.
AuNP seeds were synthesized following the modified Frens’ method.72 Briefly, 1 mL HAuCl4 (25 mM) was added to 98 mL pure water and allowed to boil on a hot plate with vigorous stirring. 1 mL sodium citrate (112.2 mM) was then added swiftly and reacted for 10 minutes. After cooling the solution to room temperature, water was added to bring the volume to 100 mL. The AuNP seeds had a concentration of 2.23 nM and average size of 15 nm in diameter. AuNP seeds were stored in room temperature and used within one week.
To synthesize 45 nm AuNPs, we first mixed 94.4 mL pure water with 973 μL HAuCl4 (25 mM), 973 μL sodium citrate (15 mM) and 3.7 mL AuNP seeds (2.23 nM). With vigorous stirring, 973 μL hydroquinone solution (25 mM) was injected rapidly. The solution quickly switched color to purple and then to red in a few minutes. The reaction was done overnight at room temperature. Then the 45 nm AuNPs were concentrated by centrifugation (1300 g, 30 minutes) and stored at 4 °C for use within one month.
Surface Modification of AuNPs and Characterization.
Antibody conjugation to AuNP was performed following a previously reported protocol.73 First, the antibody for PAR2 or JAM-A was dissolved in phosphate buffer saline (PBS, 1X, pH=7.4) with a concentration of 0.5 mg/mL. To modify the PEG onto the antibody, the antibody solution was then added in OPSS-PEG-SVA solution (0.03mg/mL in 2 mM borate buffer, pH=8.5) with molar ratio as 1:132.73 The tubes were slowly rotated for 3 hours at room temperature. To remove extra PEG molecules, the solution was dialyzed (20 kDa dialysis bag, Spectra/Pro Biotech, 131342T) overnight at 4 °C. To modify the AuNP surface with antibody, 45 nm AuNP colloidal solution was added to PEG-antibody conjugate solution with molar ratio of 1:150 and reacted at room temperature for 3 hours. Then 100 μL Thiol-PEG-Carboxyl (4 mg/mL, in 2 mM borate buffer, pH = 8.0) was added into 1 mL AuNP solution to backfill the AuNP surface. For the AuNP-PEG sample, Thiol-PEG-Carboxyl solution (2 mg/mL, in 2 mM borate buffer, pH=8.0) was added into bare AuNP solution with the molar ratio 1:150. Particles were then washed with borate buffer 3 times (1300 g, 30 minutes, with 0.5% tween-20) and then re-suspended in cell medium for further experiments. To confirm the antibody conjugation efficiency, we measured protein concentration in the supernatant after centrifugation by the Pierce-modified Lowry assay after antibody conjugation to AuNPs. The standard curve was obtained by measuring standard BSA dilutions. After considering that the signal ratio between mouse IgG and BSA is 1.2 from the user manual in the Pierce-modified Lowry assay, a standard curve of IgG was obtained. The numbers of antibodies/AuNP was calculated by subtracting the free antibody number from the initial total antibody quantity and divided by the AuNP concentration.
Dynamic light scattering (DLS) (Malvern instruments, Nano ZS) and UV-vis spectrometer (BioTek Synergy 2) were used to characterize the nanoparticle conjugation. The AuNP concentration was determined with the Beer-Lambert law:
| (1) |
where C is concentration of AuNPs, A is absorbance, b is path length and ε is molar attenuation coefficient which was determined with the following equation:
| (2) |
Here, size of the particle (D) was determined by transmission electron microscope (TEM) (JEOL 1400+, 100 keV). The AuNP solution was carefully dropped on a copper mesh (PELCO 160, PELLA Inc.) and air-dried for 2 hours before TEM imaging.
Western Blotting.
Western blotting experiments were performed following the procedure reported before.74 Male mice (C57BL/6) were sacrificed by decapitation under anesthesia as approved by IACUC at University of Texas at Dallas. The DRG neuron tissues were collected and flash frozen on dry ice. The lysis solution (50 mM Tris-HCl buffer, 150 mM NaCl, 1 mM Ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100, pH=8.0, with protease and phosphatase inhibitors) was used to homogenize frozen DRG tissues. The sample was then centrifuged at 14000 rpm for 15 minutes at 4 °C. Protein was then separated into 10% SDS-PAGE gel, and transferred onto 0.45 PVDF membrane (Millipore, IPVH00010) at 30V overnight at 4 °C. The concentration of protein was checked with BCA assay to confirm the protein quantity was the same for different samples. To reduce the non-specific binding, the membrane was blocked with 5% non-fat milk powder (NFDM, Bio-rad, 1706404) dissolved in 1X Tris-HCl buffer containing 1% Tween-20 for 3 hours. Subsequently, membranes were washed with 1X Tris buffered saline with 0.1% Tween-20 (TTBS, pH=7.5, VWR) 3 times for 5 minutes each. Then primary antibody (MAB3949, 5 μg/mL) was used to incubate overnight at 4 °C. The membrane was then washed with 1X TTBS 3 times for 5 minutes each and incubated with secondary antibody at room temperature for 1 hour. Antibody for Glyceraldehyde 3-phosphate dehydrogenase (GADPH) was used to stain GADPH to check that the same number of cells were added for both samples. The membrane was then washed with 1X TTBS 6 times for 5 minutes each. The band signal was then generated with immobilon western chemiluminescent HRP substrate (Millipore, WBKLS0500) and detected by Bio-Rad ChemiDoc Touch (Hercules, CA).
Ca2+ Measurement with Plate Reader.
HEK393 cells with Ca2+ indicator were cultured in 75 mL cell flask with cell medium (1% Pen Strep, 10% FBS in DMEM) in 37°C and 5% CO2. The day before the experiments, cells were seeded on a collagen coated 96 well plate (Costar, 3603). Cell culture medium was replaced by 100 μL artificial cerebral spinal fluid (ACSF; 125 mM NaCl, 5 mM KCl, 10 mM D-glucose, 10 mM HEPES, 3.1 mM CaCl2, 1.3 mM MgCl2, pH=7.4) for each well at 30 minutes before Ca2+ measurements. The signal was then monitored in a plate reader (Biotek Synergy H4), with light excitation of 436 nm, and the cyan (ECFP) and yellow (Citrine) signals were collected at 485 ± 10 nm and 527 ± 12 nm with 9 s interval. After reading baseline for 3 minutes, 50 μL of agonist in ACSF was mixed into each well, and the signal was then collected for another 4 minutes. The fluorescence resonance energy transfer (FRET) ratio change ΔR/R was calculated with the following equation:
| (3) |
The maximum FRET ratio change (Max. ΔR/R) indicates the PAR2 activity. SSTR2 activity was measured following the same procedure as PAR2.
Transendothelial Electrical Resistance Measurement.
The hCMEC/D3 cells were incubated in EndoGRO-MV complete media with FGF-2 supplement (Millipore, SCME004) on a collagen-coated porous membrane in a transwell filter insert (Falcon, 353097, 6.4 mm membrane diameter). Cells were cultured for 6 to 7 days at 37 °C and 5% CO2 to form a tight monolayer.
TEER measurements were performed to monitor the cellular monolayer formation as well as the BBB opening by MH. Briefly, Millicell ERS-2 Voltohmmeter (Millipore, MERS00002) was used to measure the electrical resistance across the cellular monolayer. The resistance of collagen-coated culture membrane (no cells) was used as baseline. The TEER value was obtained by subtracting baseline from the resistance of the endothelial monolayer.
Permeability Measurement
Permeability measurement was performed to quantify the molecular transport across the cell monolayer barrier. The culture insert was placed in the serum free cell medium. Cells were incubated in 37 °C and 5% CO2 for 30 minutes. FITC-dextran solution with different molecular weights (300 μL/well, 1 mg/mL, in cell medium) was added into the well. Culture medium from the bottom well was sampled at different times to obtain the concentration of FITC-dextran that had crossed the cellular barrier. The permeability (Papp, cm/s) was calculated with the following equation:
| (4) |
Here Q is the quantity (mg) of molecules that have diffused through the cell monolayer, t is time duration (s), A is the membrane area (cm2), and C is the initial concentration of FITC-dextran (mg/mL).
Immunocytochemistry Staining.
Cells were first seeded on a glass bottom Petri dish (MatTek, P35G-0–10-C) one day before the experiment. To check the AuNP targeting site on live cells, the AuNP samples (0.5 nM) were then incubated with cells for 15 mins at 4 °C. The cells were rinsed by ACSF 3 times. Then 100% methanol was used to fix the cell. The secondary antibody (Alexa 488 or Alexa 647 conjugated donkey anti-mouse IgG, 0.2 μg/mL) was then incubated with cells for 1 hours under room temperature. Free secondary antibody was removed by washing with PBS. Then cells were washed carefully with PBS buffer. Cell nuclei were stained with Hoechst 33342 (5 μg/mL) for 7 minutes. Cells were finally washed with PBS 3 times and imaged under a confocal microscope (Olympus FV3000RS). We also conducted ICC experiments on prefixed cells. Briefly, 100% methanol was used to fix the cell first. After 5 minutes and washing with PBS (1X, pH=7.4), cells were incubated with blocking buffer (2% BSA, 5% donkey serum, 0.05% Tween-20) for 2 hours at room temperature. The primary antibodies or AuNP samples were then incubated with cells (MAB3949 (10 μg/ml), BV16 (5 μg/mL), AuNP samples (0.5 nM)) overnight at 4 °C. The secondary antibody staining steps and Hoechst staining steps are same as the procedure described before. The green channel (FITC filter) shows TN-XXL in HEK293 cell transferred with TN-XXL. The red channel (Alexa 647 filter) shows that secondary antibodies (conjugated with Alexa 647) bind to MAB3949.
Molecular Hyperthermia Inactivation in Situ.
Cells were first incubated in 40 μL antibody-modified AuNP in cell culture medium for 30 minutes. To remove free particles, cells were then carefully washed with bath solutions (ACSF for HEK293 cells, PBS for hCMEC/D3 cells). Then laser pulses (Quantel Q-smart Nd:YAG laser, FWHM = 6 ns, 532 nm, diameter of laser beam is 10 mm (Intensity = 1/e2)) were used to irradiate directly to each well. The laser energy was monitored with a laser energy meter (FieldMaxII, Coherent USA). After laser irradiation, cells were incubated in the working solution (ACSF for HEK 293 cells, cell medium for hCMEC/D3 cells) for further experiments. To check whether MAB3949 affects PAR2 activity, we incubate HEK293 cells with MAB3949 (0.02–67 nM in ACSF, 100 μL/well) 30 minutes before the Ca2+ assay.
Nanobubble Signal Measurement.
AuNP solution was flowed through a glass capillary tube (VitroTube, 82400–050). The pump laser (532 nm, FWHM=6 ns, frequency=50 Hz) and the probe laser (MKS R-30989, 633 nm, continuous wave) were focused and co-localized at the center of the capillary tube. The probe laser signal was recorded by a photodetector (Menlo systems, FPD510). The probe laser energy was adjusted by an optical filter (Thorlab, NDC-25C-2M). The data and nanobubble probability was processed by MATLAB 2016a. The Boltzmann fitting of the probability was calculated in Origin 9.1 software.
Propidium Iodide Staining.
HEK 293 cells were seeded on a circular glass bottom dish for 24 hours before the experiment. The cells were first incubated with AuNP-MAB3949 (0.32 nM) or cell culture medium for 30 minutes. Extra AuNPs were washed away by ACSF. The positive control group was obtained by killing cells with 100% methanol (10 min incubation). Cells of the MH group and laser only group are irradiated with the laser (100 mJ/cm2, 10 pulses). After laser irradiation, cells are incubated in ACSF for 30 minutes followed by staining with PI for 10 minutes (10 μg/mL in ACSF). Cells are then washed by ACSF. The cells are then fixed with 100% methanol for 10 minutes (including positive control). Hoechst 33324 was used to stain the cell nucleus for counting purposes. Cells were finally washed with PBS 3 times and imaged under a confocal microscope (Olympus FV3000RS).
Ca2+ Imaging with 2-photon Imaging System.
HEK293 cells were seeded on a circular glass slide in a 24 well plate 2 days before the experiment. On the day of the experiment, glass slides were transferred to petri dishes. AuNP-MAB3949 (0.2 nM, 70μL) was added carefully on top of the slide to cover the cells for 30 minutes under 4 °C. Then extra particles were removed from cells and ACSF was used to wash away free particles. We used a black marker to mark the outside of the petri dish and the laser irradiation partially compromises the marker, a convenient method to indicate the laser treated cells. Half of the petri dish was covered with cardboard to block the laser beam (6 mm in diameter) while leaving the other half exposing to the laser. The laser fluence is 100 mJ/cm2 and 10 pulses were used. A multichannel perfusion system (DWV, 64–1940) was used to load ACSF and drugs (1 μM 2AT in ACSF). To image the fluorescent signal on the 2-photon system (Olympus MPE-RS twin), the laser excitation wavelength was set at 820 nm. The cyan (ECFP) and yellow (Citrine) emission was set at 485 nm and 527 nm respectively. ACSF was perfused to cells for the first 52s to obtain the baseline followed by perfusing 2AT for 16 seconds. ImageJ was used to analyze the images.75 The regions of interest (ROI) were drawn near the edge of laser irradiation. ΔR/R was calculated using equation (3).
WST-1 Cell Proliferation Measurement.
Cells were cultured in a 96 well plate with 50,000 cells per well. Cells of the experimental groups were incubated with 35 μL AuNP-MAB3949 (in cell medium) for 5 to 30 minutes as indicated in the figures, while the control group was incubated with the same amount of cell medium. Extra AuNPs were washed away with ACSF. Then cells were irradiated with the laser (marked as 0 hour time point). ACSF was replaced with 0.1 mL cell culture medium and the cells were then incubated at 37 °C and 5% CO2. At various time points (30 mintues to 8.5 hours) after laser irradiation, 10 μL WST-1 stocking solution was added to each well. The cells were further incubated at 37 °C and 5% CO2 for 2.5 hrs. The absorbance was read by a plate reader at 450 nm. The signals were normalized with signals from control samples, i.e. cells without laser treatment or AuNP incubation.
Simulation of AuNP Plasmonic Heating and Protein Inactivation.
Absorption cross section area (Cabs) was calculated by Mie theory and the Discrete dipole approximation (DDA) method. Then the temperature profile was obtained by simulating the heating process of AuNPs by a one nanosecond laser pulse (45 nm AuNP: fluence F = 100 mJ/cm2, full width at half maximum or FWHM τ = 6 ns; 3 nm AuNP: F = 280 mJ/cm2, τ = 1 ns,) using COMSOL 5.3. We assume a homogeneous heating source inside the particle and the volumetric heat generation of the laser pulse with Gaussian shape is defined by equation (5)
| (5) |
where VNP is AuNP volume, t is time and t0 = 7.64 ns which is the center of the pulse. Gv has unit of W/m3. The thermal interface conductance of the AuNP surface was set as 105 MW/(m2·K).76
Molecular Dynamics Simulation of AuNP Heating:
Molecular dynamics (MD) simulation was performed using the LAMMPS software package. The interactions between gold atoms are described by an embedded atom method (EAM) potential,77 and the CHARMM TIP3P water model was utilized in the simulation.78 The interaction between water and gold atoms was considered by prescribing a Lennard-Jones (LJ) 12–6 interaction between oxygen and gold atoms. A cutoff radius of 10 Å was used in the LJ potential to reduce the computational cost. Table S3 reports the parameters for the non-bonded interactions used in the MD simulation. The water domain was set as a cubic box with side length 15 nm. The AuNP was placed in the center of the water domain with a diameter of 3 nm. The water molecules between the box domain and the outside of a spherical domain of 6.5 nm radius were maintained at a constant temperature of 300 K to serve as a heat sink. The laser pulse was treated as a heating source of the gold atoms. The absorption cross-section of the 3 nm AuNP was calculated by Mie theory.79 During the laser heating, an isoenthalpic-isobaric ensemble (NPH) was prescribed to maintain the overall pressure of the remaining water region at 1 atm.
Statistical Information.
Except where otherwise noted, values are reported as mean ± standard deviation (s.d.). Statistical significance analysis was calculated using the two-tailed student t test in MATLAB R2016a. A statistically significant value was denoted with two asterisks (**) for p < 0.005.
Supplementary Material
ACKNOWLEDGMENT
We acknowledge the support of Cancer Prevention and Research Institute of Texas (RP160770, RP180846, RP190278), National Science Foundation (1631910), National Institutes of Health (GM133653), and startup fund from The University of Texas at Dallas to Z. Qin, and National Institutes of Health (NS098826) to T. J. Price. We thank Dr. Paul A. Slesinger (Icahn School of Medicine) for providing the HEK293 cells, and Dr. Jamie Moy for helpful discussions on Ca2+ imaging. We dedicate this paper to Dr. John Pearce’s 70th birthday.
Footnotes
Supporting Information. Supporting Information is available The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.9b01993.
Additional figures and tables (https://pubs.acs.org/doi/suppl/10.1021/acsnano.9b01993/suppl_file/nn9b01993_si_001.pdf)
The authors declare no competing financial interest.
REFERENCES
- (1).Boyden ES; Zhang F; Bamberg E; Nagel G; Deisseroth K, Millisecond-Timescale, Genetically Targeted Optical Control of Neural Activity. Nat. Neurosci 2005, 8, 1263–1268. [DOI] [PubMed] [Google Scholar]
- (2).Muir J; Lopez J; Bagot RC, Wiring the Depressed Brain: Optogenetic and Chemogenetic Circuit Interrogation in Animal Models of Depression. Neuropsychopharmacology 2018, 44, 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Bulina ME; Chudakov DM; Britanova OV; Yanushevich YG; Staroverov DB; Chepurnykh TV; Merzlyak EM; Shkrob MA; Lukyanov S; Lukyanov KA, A Genetically Encoded Photosensitizer. Nat. Biotechnol 2006, 24, 95–99. [DOI] [PubMed] [Google Scholar]
- (4).Jay DG, Selective Distruction of Protein Function by Chromophore-Assisted Laser Inactivation. Proc. Natl. Acad. Sci. U. S. A 1988, 85, 5454–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Gomez-Santacana X; de Munnik SM; Vijayachandran P; Da Costa Pereira D; Bebelman JPM; de Esch IJP; Vischer HF; Wijtmans M; Leurs R, Photoswitching the Efficacy of a Small-Molecule Ligand for a Peptidergic GPCR: from Antagonism to Agonism. Angew. Chem., Int. Ed. Engl 2018, 57, 11608–11612. [DOI] [PubMed] [Google Scholar]
- (6).Fehrentz T; Huber FME; Hartrampf N; Bruegmann T; Frank JA; Fine NHF; Malan D; Danzl JG; Tikhonov DB; Sumser M; Sasse P; Hodson DJ; Zhorov BS; Klocker N; Trauner D, Optical Control of L-Type Ca2+ Channels Using a Diltiazem Photoswitch. Nat. Chem. Biol 2018, 14, 764–767. [DOI] [PubMed] [Google Scholar]
- (7).Jacobson K; Rajfur Z; Vitriol E; Hahn K, Chromophore-Assisted Laser Inactivation in Cell Biology. Trends Cell Biol. 2008, 18, 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ali MRK; Wu Y; Tang Y; Xiao H; Chen K; Han T; Fang N; Wu R; El-Sayed MA, Targeting Cancer Cell Integrins Using Gold Nanorods in Photothermal Therapy Inhibits Migration Through Affecting Cytoskeletal Proteins. Proc. Natl. Acad. Sci. U. S. A 2017, 114, E5655–E5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Jiang W; Kim BY; Rutka JT; Chan WC, Nanoparticle-Mediated Cellular Response Is Size-Dependent. Nat. Nanotechnol 2008, 3, 145–150. [DOI] [PubMed] [Google Scholar]
- (10).Dennison JM; Zupancic JM; Lin W; Dwyer JH; Murphy CJ, Protein Adsorption to Charged Gold Nanospheres as a Function of Protein Deformability. Langmuir 2017, 33, 7751–7761. [DOI] [PubMed] [Google Scholar]
- (11).Huang H; Delikanli S; Zeng H; Ferkey DM; Pralle A, Remote Control of Ion Channels and Neurons Through Magnetic-Field Heating of Nanoparticles. Nat. Nanotechnol 2010, 5, 602–606. [DOI] [PubMed] [Google Scholar]
- (12).Chen R; Romero G; Christiansen MG; Mohr A; Anikeeva P, Wireless Magnetothermal Deep Brain Stimulation. Science 2015, 347, 1477–1480. [DOI] [PubMed] [Google Scholar]
- (13).Rojas C; Tedesco M; Massobrio P; Marino A; Ciofani G; Martinoia S; Raiteri R, Acoustic Stimulation Can Induce a Selective Neural Network Response Mediated by Piezoelectric Nanoparticles. J. Neural. Eng 2018, 15, 036016. [DOI] [PubMed] [Google Scholar]
- (14).Carvalho-de-Souza JL; Treger JS; Dang B; Kent SB; Pepperberg DR; Bezanilla F, Photosensitivity of Neurons Enabled by Cell-Targeted Gold Nanoparticles. Neuron 2015, 86, 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Miller IC; Gamboa Castro M; Maenza J; Weis JP; Kwong GA, Remote Control of Mammalian Cells with Heat-Triggered Gene Switches and Photothermal Pulse Trains. ACS Synth. Biol 2018, 7, 1167–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Thompson SA; Paterson S; Azab MM; Wark AW; de la Rica R, Light-Triggered Inactivation of Enzymes with Photothermal Nanoheaters. Small 2017, 13, 1603195–1603200. [DOI] [PubMed] [Google Scholar]
- (17).Nakatsuji H; Numata T; Morone N; Kaneko S; Mori Y; Imahori H; Murakami T, Thermosensitive Ion Channel Activation in Single Neuronal Cells by Using Surface-Engineered Plasmonic Nanoparticles. Angew. Chem., Int. Ed. Engl 2015, 54, 11725–111729. [DOI] [PubMed] [Google Scholar]
- (18).Yoo S; Hong S; Choi Y; Park J; Nam Y, Photothermal Inhibition of Neural Activity with Near-Infrared Sensitive Nanotransducers. ACS Nano 2014, 8, 8040–8049. [DOI] [PubMed] [Google Scholar]
- (19).Paviolo C; Haycock JW; Cadusch PJ; McArthur SL; Stoddart PR, Laser Exposure of Gold Nanorods Can Induce Intracellular Calcium Transients. J. Biophotonics 2014, 7, 761–765. [DOI] [PubMed] [Google Scholar]
- (20).de Boer WDAM; Hirtz JJ; Capretti A; Gregorkiewicz T; Izquierdo-Serra M; Han S; Dupre C; Shymkiv Y; Yuste R, Neuronal Photoactivation Through Second-Harmonic Near-Infrared Absorption by Gold Nanoparticles. Light: Sci. Appl 2018, 7, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Robert HML; Savatier J; Vial S; Verghese J; Wattellier B; Rigneault H; Monneret S; Polleux J; Baffou G, Photothermal Control of Heat-Shock Protein Expression at the Single Cell Level. Small 2018, 14, e1801910. [DOI] [PubMed] [Google Scholar]
- (22).Maier CM; Huergo MA; Milosevic S; Pernpeintner C; Li M; Singh DP; Walker D; Fischer P; Feldmann J; Lohmueller T, Optical and Thermophoretic Control of Janus Nanopen Injection into Living Cells. Nano Lett. 2018, 18, 7935–7941. [DOI] [PubMed] [Google Scholar]
- (23).Wilson AM; Mazzaferri J; Bergeron E; Patskovsky S; Marcoux-Valiquette P; Costantino S; Sapieha P; Meunier M, In Vivo Laser-Mediated Retinal Ganglion Cell Optoporation Using KV1.1 Conjugated Gold Nanoparticles. Nano Lett. 2018, 18, 6981–6988. [DOI] [PubMed] [Google Scholar]
- (24).Li X; Kang P; Chen Z; Lal S; Zhang L; Gassensmith JJ; Qin Z, Rock the Nucleus: Significantly Enhanced Nuclear Membrane Permeability and Gene Transfection by Plasmonic Nanobubble Induced Nanomechanical Transduction. Chem. Commun. (Cambridge, U. K.) 2018, 54, 2479–2482. [DOI] [PubMed] [Google Scholar]
- (25).Huttmann G; Radt B; Serbin J; Birngruber R, Inactivation of Proteins by Irradiation of Gold Nanoparticles with Nano- and Picosecond Laser Pulses. Proc. SPIE 2003, 5142, 88–95. [Google Scholar]
- (26).Qin Z; Bischof JC, Thermophysical and Biological Responses of Gold Nanoparticle Laser Heating. Chem. Soc. Rev 2012, 41, 1191–1217. [DOI] [PubMed] [Google Scholar]
- (27).Anderson RR; Parrish JA, Selective Photothermolysis: Precise Microsurgery by Selective Absorption of Pulsed Radiation. Science 1983, 220, 524–527. [DOI] [PubMed] [Google Scholar]
- (28).Kang P; Chen Z; Nielsen SO; Hoyt K; D’Arcy S; Gassensmith JJ; Qin Z, Molecular Hyperthermia: Spatiotemporal Protein Unfolding and Inactivation by Nanosecond Plasmonic Heating. Small 2017, 13, 1700841–1700847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Huttmann G; Birngruber R, On the Possibility of High-Precision Photothermal Microeffects and the Measurement of Fast Thermal Denaturation of Proteins. IEEE J. Sel. Top. Quant 1999, 5, 954–962. [Google Scholar]
- (30).Tillu DV; Hassler SN; Burgos-Vega CC; Quinn TL; Sorge RE; Dussor G; Boitano S; Vagner J; Price TJ, Protease-Activated Receptor 2 Activation Is Sufficient to Induce the Transition to a Chronic Pain State. Pain 2015, 156, 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Boitano S; Hoffman J; Flynn AN; Asiedu MN; Tillu DV; Zhang Z; Sherwood CL; Rivas CM; DeFea KA; Vagner J; Price TJ, The Novel PAR2 Ligand C391 Blocks Multiple PAR2 Signalling Pathways In Vitro and In Vivo. Br. J. Pharmacol 2015, 172, 4535–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Giannotta M; Benedetti S; Tedesco FS; Corada M; Trani M; D’Antuono R; Millet Q; Orsenigo F; Galvez BG; Cossu G; Dejana E, Targeting Endothelial Junctional Adhesion Molecule-A/ EPAC/ Rap-1 Axis as a Novel Strategy to Increase Stem Cell Engraftment in Dystrophic Muscles. EMBO Mol. Med 2014, 6, 239–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Dejana E, Endothelial Cell-Cell Junctions: Happy Together. Nat. Rev. Mol. Cell Biol 2004, 5, 261–270. [DOI] [PubMed] [Google Scholar]
- (34).Baffou G; Quidant R, Thermo-Plasmonics: Using Metallic Nanostructures as Nano-Sources of Heat. Laser Photonics Rev. 2012, 7, 171–187. [Google Scholar]
- (35).Dai Y; Wang S; Tominaga M; Yamamoto S; Fukuoka T; Higashi T; Kobayashi K; Obata K; Yamanaka H; Noguchi K, Sensitization of TRPA1 by PAR2 Contributes to the Sensation of Inflammatory Pain. J. Clin. Invest 2007, 117, 1979–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Amadesi S; Cottrell GS; Divino L; Chapman K; Grady EF; Bautista F; Karanjia R; Barajas-Lopez C; Vanner S; Vergnolle N; Bunnett NW, Protease-Activated Receptor 2 Sensitizes TRPV1 by Protein Kinase Cepsilon- and A-Dependent Mechanisms in Rats and Mice. J. Physiol 2006, 575, 555–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hazan A; Kumar R; Matzner H; Priel A, The Pain Receptor TRPV1 Displays Agonist-Dependent Activation Stoichiometry. Sci. Rep 2015, 5, 12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Suen JY; Gardiner B; Grimmond S; Fairlie DP, Profiling Gene Expression Induced by Protease-Activated Receptor 2 (PAR2) Activation in Human Kidney Cells. PLoS One 2010, 5, e13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Vetter I; Lewis RJ, Characterization of Endogenous Calcium Responses in Neuronal Cell Lines. Biochem. Pharmacol 2010, 79, 908–920. [DOI] [PubMed] [Google Scholar]
- (40).Boitano S; Flynn AN; Schulz SM; Hoffman J; Price TJ; Vagner J, Potent Agonists of the Protease Activated Receptor 2 (PAR2). J. Med. Chem 2011, 54, 1308–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lacin E; Muller A; Fernando M; Kleinfeld D; Slesinger PA, Construction of Cell-based Neurotransmitter Fluorescent Engineered Reporters (CNiFERs) for Optical Detection of Neurotransmitters In Vivo. J. Vis. Exp 2016, e53290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Jain PK; Lee KS; El-Sayed IH; El-Sayed MA, Calculated Absorption and Scattering Properties of Gold Nanoparticles of Different Size, Shape, and Composition: Applications in Biological Imaging and Biomedicine. J. Phys. Chem. B 2006, 110, 7238–7248. [DOI] [PubMed] [Google Scholar]
- (43).Perry JL; Reuter KG; Kai MP; Herlihy KP; Jones SW; Luft JC; Napier M; Bear JE; DeSimone JM, PEGylated Print Nanoparticles: The Impact of PEG Density on Protein Binding, Macrophage Association, Biodistribution, and Pharmacokinetics. Nano Lett. 2012, 12, 5304–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Muller A; Joseph V; Slesinger PA; Kleinfeld D, Cell-Based Reporters Reveal In Vivo Dynamics of Dopamine and Norepinephrine Release in Murine Cortex. Nat. Methods 2014, 11, 1245–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Fales AM; Vogt WC; Pfefer J; Ilev IK, Size-Dependent Thresholds for Melting and Nanobubble Generation Using Pulsed-Laser Irradiated Gold Nanoparticles. Proc. SPIE 2019, 10509, 105090C. [Google Scholar]
- (46).Waxengger J; Trugler A; Hohenester U, Plasmonics Simulations with the MNPBEM Toolbox: Consideration of Substrates and Layer Structures. Comput. Phys. Comm 2015, 193, 138–150. [Google Scholar]
- (47).Vergnolle N; Bunnett NW; Sharkey KA; Brussee V; Compton SJ; Grady EF; Cirino G; Gerard N; Basbaum AI; Andrade-Gordon P; Hollenberg MD; Wallace JL, Proteinase-Activated Receptor-2 and Hyperalgesia: A Novel Pain Pathway. Nat. Med. (N. Y., NY, U. S.) 2001, 7, 821–826. [DOI] [PubMed] [Google Scholar]
- (48).Goodman AM; Hogan NJ; Gottheim S; Li C; Clare SE; Halas NJ, Understanding Resonant Light-Triggered DNA Release from Plasmonic Nanoparticles. ACS Nano 2017, 11, 171–179. [DOI] [PubMed] [Google Scholar]
- (49).Davis AA; Farrar MJ; Nishimura N; Jin MM; Schaffer CB, Optoporation and Genetic Manipulation of Cells Using Femtosecond Laser Pulses. Biophys. J 2013, 105, 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Xiong R; Raemdonck K; Peynshaert K; Lentacker I; De Cock I; Demeester J; De Smedt SC; Skirtach AG; Braeckmans K, Comparison of Gold Nanoparticle Mediated Photoporation: Vapor Nanobubbles Outperform Direct Heating for Delivering Macromolecules in Live Cells. ACS Nano 2014, 8, 6288–6296. [DOI] [PubMed] [Google Scholar]
- (51).Yao C; Rudnitzki F; Huttmann G; Zhang Z; Rahmanzadeh R, Important Factors for Cell-Membrane Permeabilization by Gold Nanoparticles Activated by Nanosecond-Laser Irradiation. Int. J. Nanomed 2017, 12, 5659–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Chen X; Munjiza A; Zhang K; Wen D, Molecular Dynamics Simulation of Heat Transfer from a Gold Nanoparticle to a Water Pool. J. Phys. Chem. C 2014, 118, 1285–1293. [Google Scholar]
- (53).Sarkar D; Kang P; Nielsen SO; Qin Z, Non-Arrhenius Reaction-Diffusion Kinetics for Protein Inactivation over a Large Temperature Range. ACS Nano 2019, 13, 8669–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Takeda Y; Kondow T; Mafune F, Degradation of Protein in Nanoplasma Generated Around Gold Nanoparticles in Solution by Laser Irradiation. J. Phys. Chem. B 2006, 110, 2393–2397. [DOI] [PubMed] [Google Scholar]
- (55).Takeda Y; Mafuné F; Kondow T, Selective Degradation of Proteins by Laser Irradiation onto Gold Nanoparticles in Solution. J. Phys. Chem. C 2009, 113, 5027–5030. [Google Scholar]
- (56).Rahmanzadeh R; Rundnitzki F; Huttmann G, Two Ways to Inactivate the Ki‐67 Protein—Fragmentation by Nanoparticles, Crosslinking with Fluorescent Dyes. J. Biophotonics 2019, 12, 12:e201800460. [DOI] [PubMed] [Google Scholar]
- (57).Mayor U; Guydosh NR; Johnson CM; Grossmann JG; Sato S; Jas GS; Freund SM; Alonso DO; Daggett V; Fersht AR, The Complete Folding Pathway of a Protein from Nanoseconds to Microseconds. Nature 2003, 421, 863–867. [DOI] [PubMed] [Google Scholar]
- (58).Huang YC; Lei KF; Liaw JW; Tsai SW, The Influence of Laser Intensity Activated Plasmonic Gold Nanoparticle-Generated Photothermal Effects on Cellular Morphology and Viability: a Real-Time, Long-Term Tracking and Monitoring System. Photochem. Photobiol. Sci 2019, 18, 1419–1429. [DOI] [PubMed] [Google Scholar]
- (59).Arita Y; Ploschner M; Antkowiak M; Gunn-Moore F; Dholakia K, Laser-Induced Breakdown of an Optically Trapped Gold Nanoparticle for Single Cell Transfection. Opt. Lett 2013, 38, 3402–3405. [DOI] [PubMed] [Google Scholar]
- (60).Johannsmeier S; Heeger P; Terakawa M; Kalies S; Heisterkamp A; Ripken T; Heinemann D, Gold Nanoparticle-Mediated Laser Stimulation Induces a Complex Stress Response in Neuronal Cells. Sci. Rep 2018, 8, 6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Kalies S; Antonopoulos GC; Rakoski MS; Heinemann D; Schomaker M; Ripken T; Meyer H, Investigation of Biophysical Mechanisms in Gold Nanoparticle Mediated Laser Manipulation of Cells Using a Multimodal Holographic and Fluorescence Imaging Setup. PLoS One 2015, 10, e0124052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Burrows ND; Harvey S; Idesis FA; Murphy CJ, Understanding the Seed-Mediated Growth of Gold Nanorods through a Fractional Factorial Design of Experiments. Langmuir 2017, 33, 1891–1907. [DOI] [PubMed] [Google Scholar]
- (63).Wu Y; Ali MRK; Dong B; Han T; Chen K; Chen J; Tang Y; Fang N; Wang F; El-Sayed MA, Gold Nanorod Photothermal Therapy Alters Cell Junctions and Actin Network in Inhibiting Cancer Cell Collective Migration. ACS Nano 2018, 12, 9279–9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Scarabelli L; Sanchez-Iglesias A; Perez-Juste J; Liz-Marzan LM, A “Tips and Tricks” Practical Guide to the Synthesis of Gold Nanorods. J. Phys. Chem. Lett 2015, 6, 4270–4279. [DOI] [PubMed] [Google Scholar]
- (65).Hirsch LR; Stafford RJ; Bankson JA; Sershen SR; Rivera B; Price RE; Hazle JD; Halas NJ; West JL, Nanoshell-Mediated Near-Infrared Thermal Therapy of Tumors Under Magnetic Resonance Guidance. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 13549–13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Brink CB; Harvey BH; Bodenstein J; Venter DP; Oliver DW, Recent Advances in Drug Action and Therapeutics: Relevance of Novel Concepts in G-Protein-Coupled Receptor and Signal Transduction Pharmacology. Br. J. Clin. Pharmacol 2004, 57, 373–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Bale SS; Kwon SJ; Shah DA; Banerjee A; Dordick JS; Kane RS, Nanoparticle-Mediated Cytoplasmic Delivery of Proteins to Target Cellular Machinery. ACS Nano 2010, 4, 1493–1500. [DOI] [PubMed] [Google Scholar]
- (68).Miersch S; Sidhu SS, Intracellular Targeting with Engineered Proteins. F1000Research 2016, 5, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Gopinath SC; Lakshmipriya T; Chen Y; Arshad MK; Kerishnan JP; Ruslinda AR; Al-Douri Y; Voon CH; Hashim U, Cell-Targeting Aptamers Act as Intracellular Delivery Vehicles. Appl. Microbiol. Biotechnol 2016, 100, 6955–6969. [DOI] [PubMed] [Google Scholar]
- (70).Herce HD; Schumacher D; Schneider AFL; Ludwig AK; Mann FA; Fillies M; Kasper MA; Reinke S; Krause E; Leonhardt H; Cardoso MC; Hackenberger CPR, Cell-Permeable Nanobodies for Targeted Immunolabelling and Antigen Manipulation in Living Cells. Nature Chem. 2017, 9, 762–771. [DOI] [PubMed] [Google Scholar]
- (71).Mandal S; Debnath K; Jana NR; Jana NR, Trehalose-Functionalized Gold Nanoparticle for Inhibiting Intracellular Protein Aggregation. Langmuir 2017, 33, 13996–14003. [DOI] [PubMed] [Google Scholar]
- (72).Kimling J; Maier M; Okenve B; Kotaidis V; Ballot H; Plech A, Turkevich Method for Gold Nanoparticle Synthesis Revisited. J. Phys. Chem. B 2006, 110, 15700–15707. [DOI] [PubMed] [Google Scholar]
- (73).Chattopadhyay N; Cai Z; Pignol JP; Keller B; Lechtman E; Bendayan R; Reilly RM, Design and Characterization of HER-2-Targeted Gold Nanoparticles for Enhanced X-Radiation Treatment of Locally Advanced Breast Cancer. Mol. Pharmaceutics 2010, 7, 2194–2206. [DOI] [PubMed] [Google Scholar]
- (74).Moy JK; Khoutorsky A; Asiedu MN; Black BJ; Kuhn JL; Barragan-Iglesias P; Megat S; Burton MD; Burgos-Vega CC; Melemedjian OK; Boitano S; Vagner J; Gkogkas CG; Pancrazio JJ; Mogil JS; Dussor G; Sonenberg N; Price TJ, The MNK-eIF4E Signaling Axis Contributes to Injury-Induced Nociceptive Plasticity and the Development of Chronic Pain. J. Neurosci 2017, 37, 7481–7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Schindelin J; Arganda-Carreras I; Frise E; Kaynig V; Longair M; Pietzsch T; Preibisch S; Rueden C; Saalfeld S; Schmid B; Tinevez JY; White DJ; Hartenstein V; Eliceiri K; Tomancak P; Cardona A, Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Plech A; Kotaidis V; Gresillon S; Dahmen C; von Plessen G, Laser-Induced Heating and Melting of Gold Nanoparticles Studied by Time-Resolved X-Ray Scattering. Phys. Rev. B 2004, 70, 195423. [Google Scholar]
- (77).Daw MS; Baskes MI, Embedded-Atom Method: Derivation and Application to Impurities, Surfaces, and Other Defects in Metals. Phys. Rev. B 1984, 29, 6443–6453. [Google Scholar]
- (78).MacKerell AD; Bashford D; Bellott M; Dunbrack RL; Evanseck JD; Field MJ; Fischer S; Gao J; Guo H; Ha S; Joseph-McCarthy D; Kuchnir L; Kuczera K; Lau FT; Mattos C; Michnick S; Ngo T; Nguyen DT; Prodhom B; Reiher WE; Roux B; Schlenkrich M; Smith JC; Stote R; Straub J; Watanabe M; Wiorkiewicz-Kuczera J; Yin D; Karplus M, All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- (79).Bohren CF; Huffman DR, Absorption and Scattering of Light by Small Particles. John Wiley & Sons, Inc: New York, 1983. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


