Abstract
Background and Aim:
Extended-spectrum β-lactamase (ESBL)-producing Escherichia coli are gradually increasing worldwide and carry a serious public threat. This study aimed to determine the antimicrobial resistance pattern of ESBL-producing E. coli isolated from fecal samples of piglets and pig farm workers.
Materials and Methods:
Fecal samples from <3-month-old piglets (n=156) and farm workers (n=21) were processed for the isolation of ESBL-producing E. coli in MacConkey agar added with 1 µg/mL of cefotaxime. E. coli (piglets=124; farm workers=21) were tested for ESBL production by combined disk method and ESBL E-strip test. Each of the ESBL-positive isolate was subjected to antibiotic susceptibility testing. The ESBL-producing E. coli were further processed for genotypic confirmation to CTX-M gene.
Results:
A total of 55 (44.4%, 55/124) and nine (42.9%, 9/21) ESBL-producing E. coli were isolated from piglets and farm workers, respectively. Antibiotic susceptibility testing of the ESBL-positive E. coli isolates from piglets and farm workers showed 100% resistance to ceftazidime, cefotaxime, cefotaxime/clavulanic acid, ceftazidime/clavulanic acid, and cefpodoxime. A proportion of 100% (55/55) and 88.9% (8/9) ESBL-positive E. coli were multidrug resistance (MDR) in piglets and farm workers, respectively. On genotypic screening of the ESBL E. coli isolated from piglets (n=55), 15 were positive for the blaCTX-M gene and of the nine ESBL E. coli from farm workers, none were positive for the blaCTX-M gene.
Conclusion:
Although there was no significant difference in isolation of ESBL-producing E. coli between piglets and farm workers, the ESBL-positive E. coli from piglets showed relatively higher MDR than farm workers.
Keywords: CTX-M gene, India, multidrug resistance, organized farm, piglets, workers
Introduction
Treating infections associated with extended-spectrum β-lactamase (ESBL)-producing Escherichia coli are becoming difficult since they are capable of hydrolyzing penicillin, broad-spectrum cephalosporins, and monobactams and are often resistant to other antimicrobial classes such as fluoroquinolones, aminoglycosides, and trimethoprim-sulfamethoxazole [1]. The general trend of antibiotic resistance around the world indicates that an overall decline in the total stock of antibiotic effectiveness [2].
In the livestock sector, antibiotics are not only used for the treatment of infections but also for disease prevention and growth promotion [3]. India is one of the largest consumers of antibiotics in the world with 13 billion standard units in 2010 [2]. In India, human and animals live in close proximity which increases the risk of contamination and resistance between them and 95% of adults in India shows β-lactam antimicrobials resistance in Gram-negative bacteria [4,5]. Reports suggest the possibility of transferring ESBL-producing E. coli from farm workers to food animal and vice versa [6-10]. Multidrug-resistant E. coli are reported in food animals and pet animals of India [11-15]. Besides, food animals may act as a reservoir for ESBL-producing strains and foods as a vehicle for the transfer of β-lactam-producing bacteria [16].
This study was conducted to determine the antimicrobial resistance (AMR) pattern of ESBL-producing E. coli isolated from fecal samples of piglets and pig farm workers.
Materials and Methods
Ethical approval
Ethical approval was not required for this study.
Sample collection
A cross-sectional study was conducted between August 2016 and May 2017 to sample five government organized pig farms covering three states, namely, Uttar Pradesh, Karnataka, and Tamil Nadu. The sample was collected from five different organized pig farms, namely, Aligarh (41), Bareilly (30), Chennai farm 1 (28), Chennai farm 2(21), and Hassan (36). The fecal samples were aseptically collected with the help of fecal swab (HiMedia, Mumbai, India) directly from piglet’s rectum and in case of piglet farm workers, swabs were distributed to the farmers which were collected in the next morning (n=21) and transported to the laboratory under cold chain for the isolation of ESBL-producing E. coli.
Isolation and identification of ESBL-producing E. coli
Fecal swabs were pre-enriched in buffered peptone water and incubated at 37°C for 4-6 h. Initial screening was done using MacConkey agar supplemented with cefotaxime at 1 mg/L and plates were then incubated at 37°C for 12 h. The purified cultures of presumptive E. coli were stored in nutrient agar for identification by biochemical tests (IMViC pattern). For phenotypic identification of ESBL producers, the combination disk method by cefotaxime and ceftazidime with and without clavulanic acid was used [17] and the isolates were further confirmed by Triple ESBL detection Ezy minimum inhibitory concentration (MIC) Strip (HiMedia, India). The isolates were also tested for antibiotic susceptibility pattern with ceftazidime/clavulanic acid (20/10 μg), chloramphenicol (30 μg), cefpodoxime (10 μg), ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), cefixime (5 μg), cefoxitin (30 μg), piperacillin-tazobactam (100/10 μg), cefotaxime/clavulanic acid (30/10 μg), tetracycline (30 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg), amikacin (30 μg), and ciprofloxacin (5 μg) by disk diffusion method. The MIC of cefotaxime, cefepime, and ceftazidime was determined by E strips (HiMedia, India). Antimicrobial susceptibility results were interpreted by the following criteria established by the Clinical and Laboratory Standards Institute [18]. E. coli showing resistance to at least three classes of antibiotics were categorized as multidrug-resistant strains.
Genotypic detection of blaCTX-M gene
The genomic DNA was extracted from ESBL-positive isolates using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) and the polymerase chain reaction (PCR) for blaCTX-M (forward: CAATGTGCAGCACCAAGTAA; reverse: CGCGA TATCGTTGGTGGTG) was carried out in a final reaction volume of 25 μL [11]. The amplified PCR product was visualized by a gel documentation system (UVP, UK) after electrophoresis in 1.5% (w/v) agarose gel containing ethidium bromide (0.5 µg/mL, Loba Chemie, India). The positive PCR amplicons were sent to commercial sequencing services (Eurofins Ltd., Bangalore) for further purification and sequencing by Sanger method. The homology searches were made using the BLAST algorithm available at http://blast.ncbi.nlm.ni.gov/Blast.cgi, and the representative sequences were submitted to GenBank for accession number.
Statistical analysis
The association of the isolation of ESBL-producing E. coli between piglets and farm workers was tested by Chi-square test/Fisher’s exact (two-tailed) using SPSS version 20.0 statistical software (IBM Corp., Armonk, NY, USA).
Results
A total of 124 and 21 E. coli were isolated from piglets and human fecal samples, respectively, on MacConkey agar supplemented with cefotaxime. There was no significant difference in isolation of ESBL-producing E. coli between piglets and farm workers (p>0.05). Of the 124 isolates from piglets and 21 from farm workers, 55 (44.4%) and 9 (42.9%) isolates were identified as ESBL producers, respectively, by combined disk method. The ESBL-producing isolates from piglets and farm workers had the MIC of cefotaxime >128 µg/mL, cefepime > 32 µg/mL, and ceftazidime> 64 µg/mL. Antibiotic susceptibility testing of the ESBL-positive E. coli isolates from piglets (n=55) showed 100% resistance to ceftazidime, cefotaxime, cefotaxime/clavulanic acid, ceftazidime/clavulanic acid, and cefpodoxime. In addition, they were also resistant to cefepime (53, 96.3%), piperacillin (52, 94.5%), amikacin (42, 76.4%) ciprofloxacin (16, 29.1%), chloramphenicol (18, 32.7%), tetracycline (34, 61.8%), and sulfamethoxazole/trimethoprim (28, 50.9%). The ESBL E. coli from farm workers showed 100% resistant to tetracycline, ceftazidime, cefotaxime, cefotaxime/clavulanic acid, ceftazidime/clavulanic acid, and cefpodoxime. Further, E. coli from farm workers were also resistant to cefepime (3, 33.3%) and piperacillin (4, 44%), amikacin (8, 88.8%), chloramphenicol (5, 55%), sulfamethoxazole/trimethoprim (7, 77%), and ciprofloxacin (6, 66.6%) (Figure-1). A proportion of 100% (55/55) and 88.9% (8/9) ESBL-positive E. coli was resistant to more than three classes of antibiotics in piglets and farm workers, respectively, and classified as multidrug resistance (MDR). On genotypic screening of ESBL E. coli (n=55), 15 isolates were positive for blaCTX-M gene and of the nine ESBL E. c oli from farm workers, none were positive for the blaCTX-M gene. The representative blaCTX-M gene sequences were submitted to GenBank which were assigned with the accession numbers (MF177899, MF 177900).
Figure-1.
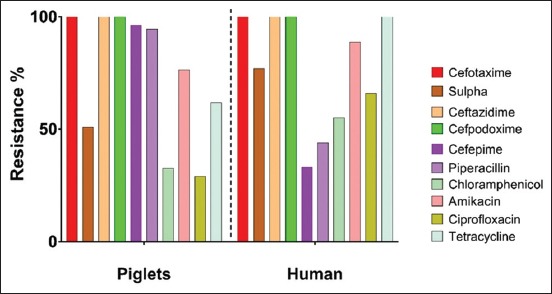
Antimicrobial resistance pattern of extended-spectrum β-lactamase-producing Escherichia coli isolates of piglets (n=55) and farm workers (n=9).
Discussion
The emergence and spread of ESBLs producing E. coli in food-producing animals is a major public health issue worldwide [11-15]. Piglets and farm workers harbored ESBL E. coli, the percentage of cefotaxime and ceftazidime resistance is higher among isolates as compared to ciprofloxacin and chloramphenicol indicates the abundant use of beta-lactam and cephalosporin antimicrobials in piglets and human [5,15]. A recent study documented that the occurrence of ESBL-positive E. coli in pigs was not related to total antimicrobial use, but associated with the presence or absence of cephalosporin use at the farm [10]. The current investigations showed that once the bacteria are on the farm or in the farm environment may be widely spread among animals (including insects and rodents) and can reach the environment through manure [19,20]. In this study, the ESBL-producing E. coli showed resistance to clinically important antibiotics such as cephalosporins, fluoroquinolones, and aminoglycosides and jeopardize the effective prevention and treatment of various bacterial infections [21]. The present study revealed that all the ESBL-producing E. coli were 100% resistant to cefotaxime and ceftazidime and were concordance with others [11-13]. This study reported that the entire ESBL-positive E. coli from piglets as MDR, similarly, in a study from China reported MDR phenotype in total ESBL-producing E. coli isolated from pigs [22]. AMR rate depends on the factors around the farm, human behavior, improper sanitation, and hygiene [23]; the improper antibiotic use with route of administration is important for multidrug-resistant bacteria. This study revealed lower resistance for ciprofloxacin, tetracycline, amikacin, and chloramphenicol compared with other antibiotics. The difference might be linked with the overall decline in the use of these antibiotics in India since 2000 [24]. It is also observed that ESBL producers also show resistance to non-β-lactam antimicrobials, such as fluoroquinolones, aminoglycosides, and sulfonamides [22]. The study noticed the CTX-M genotype in ESBL-positive isolates, similar findings were reported in ESBL-producing enterobacterials which might indicate rapid dissemination of blaCTX-M genes [25-27]. In ESBL E. coli, the most commonly identified enzymes are CTX-M family [28].
Conclusion
The study highlights the AMR pattern of ESBL-producing E. coli isolated from fecal samples of piglets and farm workers. The ESBL-positive E. coli from piglets showed relatively higher MDR than farm workers. Hence, necessary steps are needed to reduce the use of antimicrobials in pig farming to decrease the AMR.
Authors’ Contributions
ORVK, BRS, and DKS conceptualized and designed this research. The research was carried out by ST and BSP. ORVK analyzed the data and result. ORVK, ST, SVS, RK, RR, DKS, and BRS drafted, revised, and finalized the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors thank the staff of Division of Epidemiology, Indian Veterinary Research Institute for the help in sample processing. We also acknowledge the Director, Joint Director (A), and Joint Director (R) of Indian Veterinary Research Institute, Izatnagar, India for their kind support in providing necessary facilities for this study. The authors did not receive any funds for this study.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Onnberg A, Molling P, Zimmermann J, Soderquist B. Molecular and phenotypic characterization of Escherichia coli and Klebsiella pneumonia producing extended-spectrum β-lactamases with focus on CTX-M in a low-endemic area in Sweden. APMIS. 2011;119(4-5):287–295. doi: 10.1111/j.1600-0463.2011.02730.x. [DOI] [PubMed] [Google Scholar]
- 2.Centre for Disease Dynamics, Economics and Policy. Drug Resistance Index and Resistance Map. 2015. [Last accessed on 05-09-2019]. Available from: http://www.cddep.org.
- 3.Wise R, Hart T, Cars O, Sprenger M.J.W. Antimicrobial resistance. BMJ Clin. Res. 1998;317(7159):609–610. doi: 10.1136/bmj.317.7159.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burch D.G.S. Problems of antibiotics resistance in pigs in UK. Practice. 2005;27(27):37–43. [Google Scholar]
- 5.Walsh T.R, Weeks J, Livermore D.M, Toleman M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health:An environmental point prevalence study. Lancet Infect. Dis. 2011;11(5):355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 6.Carattoli A. Importance of integrons in the diffusion of resistance. Vet. Res. 2001;32(3-4):243–259. doi: 10.1051/vetres:2001122. [DOI] [PubMed] [Google Scholar]
- 7.Moodley A, Guardabassi L. Transmission of IncN plasmid carrying blaCTX-M-1 between commensal Escherichia coli in pigs and farm workers. Antimicrob. Agents Chemother. 2009;53(4):1709–1711. doi: 10.1128/AAC.01014-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abraham S, Wong H.S, Turnidge J, Johnson J.R, Trott D.J. Carbapenemase-producing bacteriain companion animals:A public health concern on the horizon. J. Antimicrob. Chemother. 2014;69(5):1155–1157. doi: 10.1093/jac/dkt518. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X.L, Wang F, Zhu D.M, Wu S, Wu P.C, Chen Y.D, Wang Y.Q, Zhou L. The carriage of Escherichia coli resistant to antibiotics in healthy populations in Shanghai. Biomed. Environ. Sci. 1998;11(4):314–320. [PubMed] [Google Scholar]
- 10.Dohmen W, Bonten M.J.M, Bos M.E.H. Carriage of extended-spectrum β-lactamases in pig farmers is associated with occurrence in pigs. Clin. Microbiol. Infect. 2017;21(10):917–923. doi: 10.1016/j.cmi.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 11.Pruthvishree B.S, Kumar O.R.V, Sinha D.K, Malik Y.P.S, Dubal Z.B, Desingu P.A, Shivakumar M, Krishnaswamy N, Singh B.R. Spatial molecular epidemiology of carbapenem-resistant and New Delhi metallo beta-lactamase (blaNDM) producing Escherichia coli in the piglets of organized farms in India. J. Appl. Microbiol. 2017;122(6):1537–1546. doi: 10.1111/jam.13455. [DOI] [PubMed] [Google Scholar]
- 12.Pruthvishree B.S, Kumar O.R.V, Sivakumar M, Tamta S, Sunitha R, Sinha D.K, Singh B.R. Molecular characterization of extensively drug-resistant (XDR), extended-spectrum beta-lactamases (ESBL) and New Delhi Metallo beta-lactamase-1 (blaNDM1) producing Escherichia coli isolated from a male dog-a case report. Vet. Arh. 2018;88(1):139–148. [Google Scholar]
- 13.Nirupama K.R, Kumar O.R.V, Pruthvishree B.S, Sinha D.K, Murugan M.S, Krishnaswamy N, Singh B.R. Molecular characterization of blaOXA-48 carbapenemase-, extended-spectrum β-lactamase-and Shiga toxin-producing Escherichia coli isolated from farm piglets in India. J. Glob. Antimicrob. Resist. 2018;13(1):201–205. doi: 10.1016/j.jgar.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Murugan S.M, Sinha D.K, Kumar O.R.V, Yadav A.K, Pruthvishree B.S, Vadhana P, Nirupama K.R, Baradwaj M, Singh B.R. Epidemiology of carbapenem-resistant Escherichia coli and first report of blaVIM carbapenemases gene in calves from India. Epidemiol. Infec. 2019;147(e159):e159. doi: 10.1017/S0950268819000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar O.R.V, Singh B.R, Sinha D.K, Pruthvishree B.S, Tamta S, Dubal Z.B, Karthikeyan R, Rupner R.N, Malik Y.S. Risk factor analysis, antimicrobial resistance and pathotyping of Escherichia coli associated with pre-and post-weaning piglet diarrhea in organized farms, India. Epidemiol. Infect. 2019;147(e174):e147. doi: 10.1017/S0950268819000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg. Infect. Dis. 2011;17(7):1216–1222. doi: 10.3201/eid1707.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrews J. Detection of extended-spectrum beta-lactamases (ESBLs) in Escherichia coli and Klebsiella species. J. Antimicrob. Chemother. 2012. [Last accessed on 18-01-2019]. Available from: http://www.bsac.org.uk/wp-content/uploads/2012/02/ecoliklebsiella.pdf.
- 18.Clinical and Laboratory Standard Institute. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standard Institute; 2014. pp. pM100–S124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer J, Rodriguez I, Schmoger S. Salmonella enterica subsp. enterica producing VIM-1 carbapenemase isolated from livestock farms. J. Antimicrob. Chemother. 2013;68(2):478–480. doi: 10.1093/jac/dks393. [DOI] [PubMed] [Google Scholar]
- 20.Guerra B. An Emerging Problem for Public Health:Carbapenemase Producing Microorganisms are also Present in Food Producing Animals, their Environment and Wild Birds. Abstracts of the 5th Symposium on Antimicrobial Resistance in Animals and the Environment, Ghent, Belgium. 2013:8. [Google Scholar]
- 21.World Health Organization. Antimicrobial Resistance, Fact Sheet No 194. Geneva: World Health Organization; 2015. [Google Scholar]
- 22.Liu X, Liu H, Wang L, Peng Q, Li Y, Zhou H, Li Q. Molecular characterization of extended-spectrum β-lactamase-producing multidrug-resistant Escherichia coli from swine in Northwest China. Front. Microbiol. 2018;9(1):1756. doi: 10.3389/fmicb.2018.01756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burroughs T, Najafi M, Lemon S.M, Knobler S.L. The Resistance Phenomenon in Microbes and Infectious Disease Vectors:Implications for Human Health and Strategies for Containment:Workshop Summary. Washington, DC: National Academies Press; 2003. [PubMed] [Google Scholar]
- 24.Laxminarayan R, Chaudhury R.R. Antibiotic resistance in India:Drivers and opportunities for action. PLoS Med. 2016;13(3):1001974. doi: 10.1371/journal.pmed.1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parajuli N.P, Maharjan P, Joshi G, Khanal P.R. Emerging perils of extended-spectrum β-lactamase producing Enterobacteriaceae clinical isolates in a teaching hospital of Nepal. BioMed. Res. Int. 2016:Article ID 1782835. doi: 10.1155/2016/1782835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Upadhyay S, Hussain A, Mishra S, Maurya A.P, Bhattacharjee A, Joshi S.R. Genetic environment of plasmid-mediated CTX-M-15 extended-spectrum beta-lactamases from clinical and foodborne bacteria in North-Eastern India. PLoS One. 2015;10(9):e0138056. doi: 10.1371/journal.pone.0138056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz S.J. First characterization of CTX-M-15-producing Escherichia coli ST131 and ST405 clones causing community-onset infections in South America. J. Clin. Microbiol. 2011;49(5):1993–1996. doi: 10.1128/JCM.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adamus W, Baraniak A, Wawszczak M, Głuszek S, Gad B, Wróbel K, Bator P, Majchrzak M, Parniewski P. The genetic background of antibiotic resistance among clinical uropathogenic Escherichia coli strains. Mol. Biol. Rep. 2018;45(5):1055–1065. doi: 10.1007/s11033-018-4254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


