Abstract
Background and Aim:
Salmonella spp. are an important group of pathogens responsible for human and animal diseases. This study aimed to estimate the prevalence and identify and characterize of Salmonella spp. isolated from broiler farms of Gazipur, Tangail, and Dhaka districts of Bangladesh. This study also evaluated the difference of Salmonella positivity status between two groups of farms, good practices adapted in broiler rearing at the project intervened farms, and non-project intervened traditional farms.
Materials and Methods:
A total of 352 samples including 128 cloacal swabs, 32 whole carcasses, 64 feed, 64 water, and 64 attendants’ hand rinses were collected through convenient sampling technique from 16 poultry food safety project of Food and Agricultural Organization of United Nations Bangladesh intervened farms and other 16 non-project intervened farms in the same location. Various cultural based techniques and biochemical methods were employed for the estimation of prevalence, isolation, and identification of Salmonella spp. which was further evaluated by polymerase chain reaction. Antimicrobial susceptibility test using disk diffusion methods and serogrouping by slide agglutination test was accomplished for additional characterization.
Results:
Among the samples, an overall prevalence of Salmonella spp. was 31.25% (110/352) (95% confidence interval [CI]=26.44-36.38%). However, the prevalence of Salmonella spp. was 24.43% (43/176) (95% CI=18.28-31.47) in project intervened farms and 38.07% (67/176) (95% CI=30.87-45.68%) in non-intervened farms. Among the 110 isolates, 31.82% (35/110) were fitted under serogroup B, and the rest of the isolates 75 (68.18%) under serogroup D. Of 110 isolates, 82.72%, 77.27%, 81.82%, and 79.09% were susceptible to ciprofloxacin, gentamycin, norfloxacin, and streptomycin, respectively. In addition, 81.82% and 80% isolates were resistant to erythromycin and tetracycline, respectively. Isolated Salmonella spp. presented moderate resistance to both amoxicillin and azithromycin. Alarmingly, 80.91% (89/110) isolates were shown to be multidrug-resistant Salmonella spp.
Conclusion:
The study has presented a significant variation of the prevalence of Salmonella spp. between project intervened and non-project intervened farms, and this indicates project intervened farms are comparatively safer than the non-intervened farms considering public health and food safety grounds. This research outcome also has highlighted a substantial proportion of poultry origin multidrug resistance Salmonella spp. is a potential source of public health hazards. In this regard, proper awareness creation and motivational activities on good agriculture practices in poultry rearing and maintaining good personal hygiene at the farmers’ level are warranted through participatory training.
Keywords: good agriculture practices, hygienic practices, multidrug resistance, poultry, Salmonella spp
Introduction
Salmonella spp. are commonly responsible for various pathogenic processes in human and animal, including poultry [1]. Among the foodborne diseases caused by bacterial pathogens, Salmonella is one of the most important zoonotic pathogens which have more than 2600 serotypes can prompt of human and animal gastrointestinal infection such as gastroenteritis, typhoid fever, paratyphoid fever, and can cause of serious ailments for younger and aged people, and even result of death [2-4]. Human consumed different types of food such as food-producing animals including poultry especially broiler and layer chicken meat, eggs, seafood, beef, pork, vegetables, and contaminated water are the main source of foodborne illness in human [5,6]. It causes endemic salmonellosis worldwide and reasons a colossal economic loss in livestock and poultry industry in Bangladesh [7]. Among the bacterial diseases, Salmonella infection is one of the major problems for poultry farming in Bangladesh, which is considered a key threat of the poultry industry [8]. In Bangladesh, the occurrence of Salmonella infection is about 21-30% in layer and about 15% in broiler which is measured as the highest prevalence among different types of poultry disease [9,10], among which a variety of acute and chronic diseases in poultry are included [11]. Chicks can be infected with Salmonella spp. by vertical transmission through infected parents or by horizontal transmission through hatcheries, sexing in contaminated hatcheries, cloacal infection, and transportation of equipment and feed [12]. Motile Salmonella (paratyphoid group) infection causes salmonellosis in chickens with zoonotic significance [13].
It is very common of broiler farming with low or no biosecurity practices in Bangladesh where most of the broiler farms have been developed near the dwellings or close proximate to the human habitats is a significant hazard for public health at present time [14]. In addition, poultry feces are used in the agricultural field and/or as fish feed without proper treatment is deemed to be potentially risky practices for the public health view point. Showing antimicrobials’ resistance by pathogenic bacteria is a universal public health concern throughout the world especially in developing countries [3,14]. The results of imprudent use of antimicrobial agents to minimize bacterial infection or as a growth promoter in poultry production are the major determinants for the emergence of multidrug-resistant pathogenic bacteria [3,15]. Because of the phenomenon of developing multidrug-resistant Salmonella isolates, the management of Salmonella infection using regular drugs is very difficult [16]. Considering the urgency of the above, the survey of Salmonella in food animal production together with surveillance on antimicrobial resistance pattern was very essential [17].
Many previous Salmonella studies in country and abroad have used poultry, poultry products and environmental samples for isolation and identification of the organism [7,18-22]. Since, lack of study to evaluating the Salmonella spp. from broiler farms with the comparison between two groups of the farm, namely, project-intervened farms with best practices versus non-intervened farm with traditional practices. The farmers of the project-intervened farms were trained on poultry farming in compliance with good practices of biosecurity measures such as provision of perimeter fencing, netting of the farm, footwear clean entry in the farm, all in all-out, and cleaning and sanitation, and judicial use antibiotics through the Food and Agricultural Organization (FAO)-Food Safety Program project intervention to be appropriate for safer poultry production considering public health hazard.
This study aimed to estimate the prevalence of and identify and characterize Salmonella spp. isolated from broiler farms of Gazipur, Tangail, and Dhaka districts of Bangladesh. This study also evaluated the difference of Salmonella positivity status between two groups of farms, good practices adapted in broiler rearing at the project intervened farms, and non-project intervened traditional farms.
Materials and Methods
Ethical approval and informed consents
The farms were selected after consultation with the sub-district (Upazila) livestock office of each study site taking into consideration of willingness of the farmers. No ethical approval was required; however, during the collection of samples; verbal consent was taken from each of the farm owner/managers.
Study area and study period
The study was conducted in three different districts (Dhaka, Gazipur, and Tangail) of Bangladesh under this study from May 2017 to December 2017 (Figure-1). Dhaka district is located in between 23°22’30” and 24°22’20” north latitudes and in between 89°41’6” and 90°59’23” east longitudes. Gazipur district is located in between 23°53’ and 24°20’24” north latitudes and in between 90°04’ and 90°49’ east longitudes. Tangail district is located in between 23°59’50” and 24°48’51” north latitudes and in between 89°48’50” and 90°51’25” east longitudes.
Figure-1.
Location of the broiler farms, equal number farms (n=16) both in project intervened and non-intervened category were included from three districts of Bangladesh (as the coordinates of some farms are same, all farms are not visualized in the map).
Farm selection, sample collection, and processing
Sixteen (16) project intervened farm with an inclusion criterion of a minimum flock size of ≥2000 that comprised 12 from Gazipur district, two from Dhaka, and two from Tangail district were included under this survey with good biosecurity and farm practices from May 2017 to December 2017. A similar number of farms (n=16) were included randomly from the same study sites to match the project intervened farm for comparing the best farm practices among the two groups.
Three hundred and fifty-two (352) different samples were randomly collected through convenient sampling technique from 32 broiler farms in three different districts, of which, 75% (n=264) samples (96 cloacal swab, 24 whole carcasses, 48 feed, 48 water, and 48 attendant hand rinse) were collected from 24 farms (n=12 project intervened, and n=12 project non-intervened) of Gazipur district, 12.5% (n=44) samples (16 cloacal swab, four whole carcasses, eight feed, eight water, and eight attendant hand rinse) were collected from four farms (n=2 project intervened, and n=2 project non-intervened) of Tangail district, and remaining 12.5% (n=44) samples (16 cloacal swab, four whole carcasses, eight feed, eight water, and eight attendant hand rinse) were collected from four farms (n=2 project intervened, and n=2 project non-intervened) of Dhaka district. Normal saline (0.85% NaCl) was used for the collection of cloacal swabs, 0.1% peptone water was used for the collection of attendants’ hand rinse water. After collection, samples were shifted to the Bacteriology and Molecular Microbiology Laboratory of the Department of Microbiology and Hygiene, Bangladesh Agricultural University maintaining proper cool chain using icebox. All collected samples were processed and cultures within 5-6 h of its collection time. The samples of 10 g of whole carcasses and 10 g of feed samples were performed homogenate using mortar and pestle and dissolved in 90 ml of 0.1% peptone water, respectively, for culture and further testing.
Isolation and identification of Salmonella spp.
Isolation and identification of Salmonella spp. were carried out according to the methods described by Akbar and Anal and ISO 6579:2002(E) [3,23] with a little modification. Separately, the processed sample of 50 µl was taken and poured on Xylose Lysine Deoxycholate agar (XLD) (HiMedia, India) and spread using glass spreader, and then incubated at 37°C for 24 h. After incubation, cultural characteristics were observed, on XLD agar, Salmonella presented pink colonies having a black center. The suspected colonies were then subcultured on XLD agar and incubated again at 37°C for 24 h for obtaining pure colonies. For identification of suspected colonies, Gram’s stain, motility test, and different biochemical tests including sugar fermentation (dextrose, sucrose, lactose, maltose, and mannitol), methyl red, Voges–Proskauer, indole, citrate, and urease tests were accomplished. The isolated colonies were then subjected to molecular confirmation through polymerase chain reaction (PCR), antimicrobial susceptibility test, and serogrouping.
Molecular detection of Salmonella spp.
For the molecular assay, the DNA template was equipped with the boiling method as described by Queipo-Ortuño et al. [24]. The 16S rRNA gene-based PCR was performed for the confirmation of the genus Salmonella. Primers were used for the amplification of the 16S rRNA gene according to the procedure described by Lin and Tsen [25] and shown in Table-1. The reaction mixture (20 µl) was prepared by mixing 10 µl master mixtures (Promega, USA), 1 µl forward primer (10 pmol), 1 µl reverse primer (10 pmol) (BioServe Biotechnologies Ltd., USA), 3 µl DNA template, and 5 µl deionized water. The PCR reactions were carried out using a thermocycler (Astec, Japan) with the following program: Initial denaturation with one cycle for 5 min at 94°C, 30 cycles each consist of denaturation with 30 s at 94°C, annealing with 30 s at 50°C, extension with 30 s at 72°C, and a final extension step of 5 min at 72°C. PCR products were analyzed by 2% agarose (Invitrogen, USA) gel electrophoresis and the bands were visualized with ultraviolet (UV) light after staining with ethidium bromide (0.5 µg/ml) for 10 min in a dark place. Bands were visualized and images were captured on a UV transilluminator (Biometra, Germany).
Table-1.
The list of primers used for the identification of Salmonella spp.
| Primer | Sequence (5’- 3’) | Target | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| Sal 16S rRNA F | TGTTGTGGTTAATAACCGCA | Salmonella 16S rRNA gene | 574 | [25] |
| Sal 16S rRNA R | CACAAATCCATCTCTGGA |
Serogrouping of Salmonella by O-antigen test
Serogrouping of isolated Salmonella spp. was done by slide agglutination test using commercial Salmonella-specific polyvalent O (A-I) antisera, Salmonella O Group B (Factor O: 4, 5, and 27) antisera, and Salmonella O Group D (Factor O: 9, 46) antisera kits (S & A Reagents Lab Ltd., Bangkok, Thailand) following the procedure described by Dhakal et al. [11].
Antimicrobial susceptibility test
All isolated Salmonella spp. were confirmed on antimicrobial susceptibility test by disk diffusion method to determine antimicrobial profile following the method described by Bauer et al. [26] and Clinical and Laboratory Standards Institute (CLSI) [27]. The following eight commercially available antimicrobial disks (HiMedia, India) were used at indicated concentration (µg/disk): Amoxicillin (AMX, 30 µg), azithromycin (AZM, 30 µg), ciprofloxacin (CIP, 5 µg), erythromycin (E, 30 µg), gentamicin (GEN, 10 µg), norfloxacin (NOR, 10 µg), streptomycin (S, 10 µg), and tetracycline (TE, 30 µg) to determine the antimicrobial susceptibility patterns. After preparing 0.5 McFarland standards bacterial suspension using normal saline, a sterile cotton bud was dipped into the bacterial suspension. The excess fluid of a swab was removed by pressing firmly against the inside of the tube just above the fluid level. The bud was streaked over the entire surface of Mueller-Hinton agar (HiMedia, India) medium 3 times, rotating the plate approximately 60 degrees after each application to ensure an even distribution of the inoculums. The antimicrobial disks were placed individually using sterile forceps and then gently press down onto the agar. The plates were inverted and incubated at 37°C overnight. After incubation, the zone of growth inhibition (diameter) of each antimicrobial agent was measured according to the guidelines of CLSI [27].
Data management and statistical analysis
The data were captured and recorded in Microsoft Excel® worksheet and imported into Epi Info 7 program [28] for statistical analysis. A univariate logistic regression model was used to calculate the odds ratio (OR) for evaluating the association of best farm practices among two groups of farms (project-intervened and non-project-intervened) with p=0.05 were used to determine statistical significance. Proportion, percentage, and 95% confidence interval (CI) were calculated using an excel data analysis tool pack for estimating prevalence status in various parameters of two groups of farms.
Results
Prevalence estimation and isolation of Salmonella spp.
A total of 352 samples were collected from 32 broiler farms of three different districts where 50% (n=176) samples were collected from project intervened farms and rest 50% (n=176) samples were collected from non-project intervened farms. Of 352 samples, overall prevalence of Salmonella spp. was estimated at 31.25% (110/352) (95% CI=26.44%-36.38%), a prevalence of 24.43% (43/176) (95% CI=18.28-31.47) was estimated in the project intervened farms and 38.07% (67/176) (95% CI=30.87-45.68) prevalence in non-project intervened farms (Table-2).
Table-2.
Prevalence of Salmonella spp. in broiler farms of three districts of Bangladesh (project intervened farms, n=176, and project non-intervened farms, n=176).
| District | Category of farms | Number of sample (n) | Number of isolates (positive) | Prevalence | 95% CI |
|---|---|---|---|---|---|
| Gazipur | Project intervened | 132 | 33 | 25 | 17.88-33.28 |
| Non-project intervened | 132 | 48 | 36.36 | 28.17-45.18 | |
| Overall Gazipur | 264 | 81 | 30.68 | 25.17-36.62 | |
| Tangail | Project intervened | 22 | 7 | 31.82 | 13.86-54.87 |
| Non-project intervened | 22 | 9 | 40.91 | 20.71-63.65 | |
| Overall Tangail | 44 | 16 | 36.36 | 22.41-52.23 | |
| Dhaka | Project intervened | 22 | 3 | 13.64 | 2.91-34.91 |
| Non-project intervened | 22 | 10 | 45.45 | 24.39-67.79 | |
| Overall Dhaka | 44 | 13 | 29.55 | 16.76-45.20 | |
| Three districts (Gazipur, Tangail, and Dhaka) | Project intervened | 176 | 43 | 24.43 | 18.28-31.47 |
| Non-project intervened | 176 | 67 | 38.07 | 30.87-45.68 | |
| Overall (three districts) | 352 | 110 | 31.25 | 26.44-36.38 |
CI=Confidence interval
Of 128 cloacal swab samples, 46.09% (n=59) samples were found positive for Salmonella spp. (Figure-2). Similarly, of 64 feed samples, 64 water samples, 18.75% (n=12), and 17.19% (n=11) were found positive, respectively, for Salmonella spp. A total of 64 water samples, 64 farm attendant’s hand rinse water sample, 32 whole carcasses samples, 17.19% (n=11), 23.44% (n=15), and 40.63% (n=13), were shown positive for Salmonella spp. (Figure-2).
Figure-2.
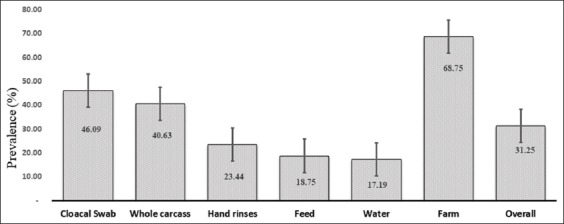
Frequency of prevalence of Salmonella spp. with a standard error of the mean at different parameters of broiler farming practices (farm=32, sample=352).
Of 32 farms, 68.75% (n=22) farms were found positive with Salmonella spp. of which 28.15% (n=9) farms were under project intervened category and remaining 40.63% (n=13) farms under non-project intervened category (Figure-2). The variation of prevalence among two groups of farms (project intervened and non-intervened) was observed among the three districts (Table-2). The non-intervened farms were found to be riskier than the project-intervened farms considering Salmonella positivity status (OR=1.9, 95% CI=1.20-3.00, p=0.005) and statistically found significant (Table-3).
Table-3.
Univariable logistic regression analysis for associating the best farm practices between two groups of broiler farms with the likelihood of Salmonella infection in different parameters.
| Parameter/sample type | Farm type | Positive | Negative | Odds ratio | 95% CI | p value |
|---|---|---|---|---|---|---|
| Farm | Project non-intervened | 13 | 3 | 3.37 | 0.71-16.06 | 0.12 |
| Project intervened | 9 | 7 | ||||
| Cloacal swab | Project non-intervened | 34 | 30 | 1.84 | 0.91-3.72 | 0.08 |
| Project intervened | 24 | 39 | ||||
| Feed | Project non-intervened | 4 | 24 | 1.17 | 0.26-5.17 | 0.83 |
| Project intervened | 4 | 28 | ||||
| Water | Project non-intervened | 7 | 25 | 1.96 | 0.52-7.39 | 0.32 |
| Project intervened | 4 | 28 | ||||
| Attendants’ hand rinse water | Project non-intervened | 22 | 10 | 0.41 | 0.12-1.34 | 0.14 |
| Project intervened | 27 | 5 | ||||
| Whole carcass | Project non-intervened | 8 | 8 | 2.2 | 0.53-9.20 | 0.28 |
| Project intervened | 5 | 11 | ||||
| Overall | Project non-intervened | 67 | 109 | 1.9 | 1.20-3.00 | 0.005 |
| Project intervened | 43 | 133 |
CI=Confidence interval
Molecular detection by PCR
Genus specific 16S rRNA gene-based PCR was performed for the confirmation of Salmonella isolates. All Salmonella isolates gave specific amplification (574 bp). The results of PCR are presented in Figure-3.
Figure-3.
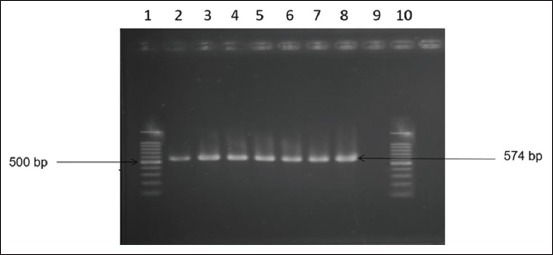
16S rRNA gene-based polymerase chain reaction of Salmonella spp. Lane 1 and 10: 100 bp DNA ladder; Lane 2-8: Tested samples were positive for the 16S rRNA gene, Lane 9: negative control without DNA.
Serogrouping of Salmonella spp.
Serogrouping of Salmonella isolates was performed by slide agglutination test using commercial Salmonella specific polyvalent O (A-I) antisera, Salmonella O Group B (Factor O: 4, 5, 27) antisera, and Salmonella O Group D (Factor O: 9, 46) antisera (S & A Reagent Lab). All isolates were positive to Salmonella Poly A-I antisera. Of 110 isolates, 39.1% (n=43) isolates were from project intervened farms, of which 30.23% (n=13) were classified under serogroup B (O:4,5,27) and 69.77% (n=30) under serogroup D (O:9,46). In other respects 60.9% (n=67) isolates were confirmed from non-project intervened farms, of which 32.84% (n=22) were classified under serogroup B (O:4,5,27) and 67.16% (n=45) under serogroup D(O:9,46). More than two-third (68.18%, 75/110) isolates of two categories of farms were classified under serogroup D (O:9,46) and rest of the isolates (31.82%, 35/110) were under the serogroup B (O:4,5,27)(Table-4).
Table-4.
Summary of Salmonella spp. serogrouping.
| Category | Isolates No. | Number (%) of Salmonella isolates | ||
|---|---|---|---|---|
| Poly A-I | Group B (O: 4,5,27) | Group D (O: 9,46) | ||
| Project intervened | 43 | 100 | 13 (30.23) | 30 (69.77) |
| Non-project intervened | 67 | 100 | 22 (32.84) | 45 (67.16) |
| Total | 110 | 100 | 35 (31.82) | 75 (68.18) |
Antimicrobial susceptibility of Salmonella spp.
Antimicrobial susceptibility test was carried out in 110 Salmonella isolates against eight selected antimicrobial agents. The results of susceptibility analysis showed that 42.73%, 82.72%, 77.27%, 81.82%, and 79.09% of Salmonella isolates were susceptible to amoxicillin, ciprofloxacin, gentamycin, norfloxacin, and streptomycin, respectively. The resistance analysis showed that 42.73%, 47.27%, 81.82%, and 80% of Salmonella isolates were resistant to amoxicillin, azithromycin, erythromycin, and tetracycline, respectively (Figure-4).
Figure-4.
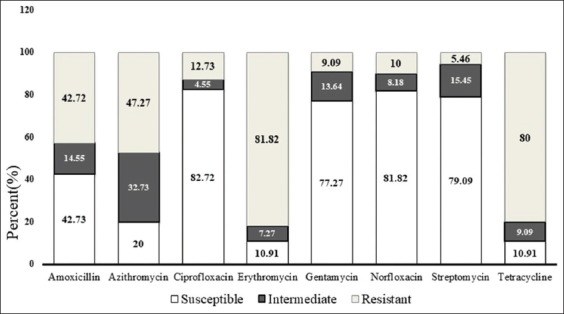
Proportion of antimicrobial susceptibility against eight selected antimicrobial agents is amoxicillin, azithromycin, ciprofloxacin, erythromycin, gentamycin, norfloxacin, streptomycin, and tetracycline presented in three categories (susceptible, intermediate, and resistant) of the pattern.
Antimicrobial resistance patterns of Salmonella spp.
The results of antimicrobial resistance patterns of Salmonella spp. are summarized in Table-5. Of 110 (n=110) Salmonella spp., 9.09% (n=10) isolates were resistant to one agent (TE), 6.63% (n=7) isolates were resistant to one agent (E), 3.64% (n=4) isolates were resistant to one agent (AMX), 1.82% (n=2) isolates were resistant against two agents (TE-AZM), 17.27% (n=19) isolates were resistant against three agents (TE-E-AMX), 30 27.27% (n=30) isolates were resistant against three agents (TE-E-AZM), 7.27% (n=8) isolates were resistant against three agents (TE-E-CIP), 4.55% (n=5) isolates were resistant against three agents (E-AMX-GEN), 14 12.73% (n=14) isolates were resistant against four agents (TE-E-AMX-AZM), 4.55 % (n=5) isolates were resistant against four agents (TE-AMX-GEN-NOR), and 5.45% (n=6) isolates were resistant against four agents (AMX-CIP-NOR-S) (Table-5).
Table-5.
Antimicrobial resistance pattern of Salmonella spp.
| Isolates | No. of agents | Antimicrobial resistance profile | No. (%) of isolates | No. (%) of multidrug resistant isolates |
|---|---|---|---|---|
| Salmonella spp. | No. resistance demonstrated | - | - | 89 (80.91) |
| 1 | TE | 10 (9.09) | ||
| 1 | E | 7 (6.63) | ||
| 1 | AMX | 4 (3.64) | ||
| 2 | TE-AZM | 2 (1.82) | ||
| 3 | TE-E-AMX | 19 (17.27) | ||
| 3 | TE-E-AZM | 30 (27.27) | ||
| 3 | TE-E-CIP | 8 (7.27) | ||
| 3 | E-AMX-GEN | 5 (4.55) | ||
| 4 | TE-E-AMX-AZM | 14 (12.73) | ||
| 4 | TE-AMX-GEN-NOR | 5 (4.55) | ||
| 4 | AMX-CIP-NOR-S | 6 (5.45) | ||
| Total resistant isolates | 110 (100) | |||
In this study, multidrug-resistant Salmonella spp. were identified and presented resistant against two or more antimicrobials, alarmingly, 80.91% (n=89) Salmonella isolates were established as multidrug-resistant in this survey (Table-5).
Discussion
Despite the importance of the poultry sector in the National Economy of Bangladesh, insufficient disease data brings bottlenecks toward understanding the disease burden like its true prevalence, spatial and temporal distribution, and economic impact [29]. Among the different types of bacterial and viral origin diseases in Bangladesh, Salmonella infection is cogitated to be one of the major problems nowadays [9,10]. In view of that, this survey was rational to determine the status of Salmonella spp. in broiler farming system that will pave the way for a baseline data depository in the National Disease Control Program.
The study estimated the overall prevalence of Salmonella spp. was 31.25% (95% CI=26.44%-36.38%), in broiler farms of poultry dense districts of Bangladesh. The prevalence was found to be lower in project intervened farms (24.43%, 95% CI=18.28-31.47) than the non-project intervened traditional farm (38.07%, 95% CI=30.87-45.68). Due to intervention of good practices of biosecurity measures such as provision of perimeter fencing, netting of the farm, footwear clean entry in the farm, all-in all-out practice, and cleaning and sanitation practices, the likelihood of bacterial contamination was lessen in project intervened farms than the non-project intervened farms [30]. The project intervened farms were found to be protective considering Salmonella infection in this study (OR=1.9, 95% CI=1.20-3.00, p=0.005). This finding has validated the impact of good agriculture practices (GAP) on poultry rearing at the farmers’ level.
In this study, Salmonella spp. were isolated from hand rinses of farm attendants as the earliest research in Bangladesh. The presence of Salmonella in poultry attendants’ hand rinse water (23.44%, 15/64) could pose a serious impact on public health. This finding has partially dissenting with the studies of Paul et al. [7] and Akond et al. [31] as 50% and 6% poultry retailer hand rinse water were found positive, respectively. In this study, feed samples were found to be contaminated with Salmonella spp. (18.75%, 12/64). This finding was reasonably supported by several authors [32,33], as 28.56% Salmonella spp. described by Al-Mamun et al. [32]. A total of 64 water samples were tested, and 11 17.19% (n=11) samples were found positive. This finding is compatible with the findings of several researchers [32-34]. In this study, of 32 whole carcasses, 40.63% (n=13) samples were positive for Salmonella spp. This finding has similarities with the studies of a few investigators [14,32]; however, Karim et al. [8] showed only 20% Salmonella positivity in broiler meat. A total of 128 cloacal swab samples were collected, of which 46.09% (n=59) samples were found positive for Salmonella spp. This finding was in conformity with the studies by some researchers [14,31]; however, Paul et al. [7] showed a very high prevalence (80%) in the cloacal swab. The overall prevalence of Salmonella spp. in broiler farming system was 31.25% and the farm level prevalence was 68.75% (Figure-4). This finding was well-matched with the studies by several researchers [5,31,35]; nonetheless, Paul et al. [7] and Islam et al. [18] showed a relatively higher prevalence of Salmonella spp. from broiler farms as 53.33% and 66.67%, respectively.
In this study, isolation and identification of Salmonella spp. were done through culturing of sample on selective media, Gram’s stain, different biochemical tests, and finally confirmed by 16S rRNA gene-based PCR. This method also used by several researches [14,32,36-38].
Serogrouping of Salmonella isolates was performed by slide agglutination test using commercial Salmonella-specific polyvalent O (A-I) antisera, Salmonella O Group B (Factor O: 4, 5, 27) antisera, and Salmonella O Group D (Factor O: 9, 46) antisera. Among the 110 isolates, 31.82% (n=35) were under serogroup B and 68.18% (n=75) isolates were under to serogroup D. The most prevalent serogroup identified in this study was serogroup D. This conclusion was in agreement with the findings of Al-Mamun et al. [32]. About 30.56% (n=11) isolates were fitted to serogroup B and 69.44% (n=25) fitted to serogroup D.
The results of susceptibility test showed that isolated Salmonella spp. were highly resistant to erythromycin (81.72%) and tetracycline (80%), and moderately resistant to amoxicillin (42.73%) and azithromycin (47.27%). This finding is compatible with the studies by many researchers [5,14,31,32]; however, Islam et al. [18] showed that the isolated Salmonella spp. were highly sensitive and Ifeanyichukwu et al. [39] revealed as highly resistant to amoxicillin, and Akbar and Anal presented that the isolated Salmonella spp. from ready-to-eat poultry were highly susceptible to erythromycin [3]. On the other hand, most of the isolated Salmonella spp. were susceptible to ciprofloxacin (82.72%), gentamicin (77.27%), norfloxacin (81.82%), and streptomycin (79.09%) [7,14,18,31]. On the contrary, this finding is incompatible with the findings of several researchers, among them Ifeanyichukwu et al. [39] presented the data where isolated Salmonella spp. were found to be highly resistant to gentamycin, and Paul et al. [7] showed the data where ciprofloxacin was susceptible against merely 20% Salmonella isolates.
In this investigation, of 110 Salmonella isolates, 56.36% (n=62) and 22.73% (n=25) isolates showed resistance against at least three and four antimicrobial agents, respectively, and two or more antimicrobial agents 80.91% (n=89) isolates as multidrug-resistant. The latter finding is well-matched with the studies by several researchers [14,32]. The high proportion of multidrug resistance showed by the isolated Salmonella spp. may be the result of the unjudicial use of different types of antimicrobial agents in poultry production in Bangladesh with an aim to retard the bacterial infection [14]. In addition, some antibiotics are being used unscrupulously in poultry feed and water by the feed millers and farmers, respectively, and these are also be the another cause for showing multidrug resistance. The findings of this study suggested that multidrug-resistant Salmonella spp. isolated from broiler farms might be an important concern for public health.
Conclusion
The presence of Salmonella spp. was confirmed from a wide range of samples at the broiler farming system specifies severe public health importance. This finding streamlines to GAP in poultry farming relating to maintaining proper farm biosecurity, all-in all-out practices, withdrawal period, safe disposal of poultry waste and prudent use of antimicrobial agents along with maintaining personal hygiene are needed to minimize the likelihood of Salmonella infection in poultry and its antimicrobial-resistant, and further transmission in human as a consequence. To minimize these public health threats, awareness creation and motivational activity on good hygienic practices and GAP for poultry farmers through participatory training under the “One Health” platform are very much necessitated.
Authors’ Contributions
SMLK planned and designed the study. DM, MNU, BA, AHMTA, and SKSI assisted in data collection, laboratory work, data analysis, and drafting of the manuscript. SMLK, SKSI, MSI, and MSRK assisted in data analysis and drafting of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
We appreciate the kind facilitation and valuable suggestions from Technical Expert, Food Safety Program of FAO of United Nations (FAO-UN), Bangladesh, for conducting the study. We are indebted to sub-district (Upazila) and district livestock officers for their cooperation for farm selection under this study, and also grateful to poultry farmers/attendants for their assistant to sample collection.
This study was supported by FAO-UN under the project named “Improving Food Safety Program in Bangladesh (Grant no. GCP/BGD/047/NET) in collaboration with the Department of Livestock of Services under the Ministry of Fisheries and Livestock, Bangladesh.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published map and institutional affiliation.
References
- 1.Freeman B.A. Burrows Textbook of Microbiology. 22nd ed. Philadelphia: W. B. Saunders Company; 1985. pp. 464–472. [Google Scholar]
- 2.Zhao X, Gao Y, Ye C, Yang L, Wang T, Chang W. Prevalence and characteristics of Salmonella isolated from free-range chickens in Shandong province, China. Biomed. Res. Int, 2016. 2016:8183931. doi: 10.1155/2016/8183931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akbar A, Anal A.K. Isolation of Salmonella from ready-to-eat poultry meat and evaluation of its survival at low temperature, microwaving and simulated gastric fluids. J. Food Sci. Technol. 2015;52(5):3051–3057. doi: 10.1007/s13197-014-1354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Xie X, Xu X, Wang X, Chang H, Wang C, Wang A, He Y, Yu H, Wang X, Zeng M. Nontyphoidal Salmonella infection in children with acute gastroenteritis:Prevalence, serotypes, and antimicrobial resistance in Shanghai, China. Foodborne Pathog. Dis. 2014;11(3):200–206. doi: 10.1089/fpd.2013.1629. [DOI] [PubMed] [Google Scholar]
- 5.Im M.C, Jeong S.J, Kwon Y.K, Jeong O.M, Kang M.S, Lee Y.J. Prevalence and characteristics of Salmonella spp. Isolated from commercial layer farms in Korea. Poult. Sci. 2015;94(4):1691–1698. doi: 10.3382/ps/pev137. [DOI] [PubMed] [Google Scholar]
- 6.Orji M.U, Onuigbo H.C, Mbata T.I. Isolation of Salmonella from poultry droppings and other environmental sources in Awka, Nigeria. Int. J. Infect. Dis. 2005;9(2):86–89. doi: 10.1016/j.ijid.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 7.Paul P, Akther S, Ali M.Z, Banu H, Khan M.S.R, Khatun M.M. Isolation, identification and antibiogram study of Salmonella spp. from poultry farm environment. Int. J. Anim. Biol. 2017;3(2):5–11. [Google Scholar]
- 8.Karim M.R, Giasuddin M, Samad M.A, Mahmud M.S, Islam M.R, Rahman M.H, Yousuf M.A. Prevalence of Salmonella spp. In poultry and poultry products in Dhaka, Bangladesh. Int. J. Anim. Biol. 2017;3(4):18–22. [Google Scholar]
- 9.Rahman M.A, Rahman M.M, Moonmoon M, Alam K.J, Islam M.Z. Prevalence of common diseases of broiler and layer at Gazipur district in Bangladesh. Asian J. Med. Biol. Res. 2017;3(2):290–293. [Google Scholar]
- 10.Al Mamun M, Islam K.M, Rahman M.M. Occurrence of poultry diseases at Kishoregonj district of Bangladesh. MOJ Proteomics Bioinform. 2019;8(1):7–12. [Google Scholar]
- 11.Dhakal L.B, Dhakal I.P, Yadav S.K, Ahaduzzaman M, Islam M.Z. Prevalence and antibiotic resistance profile of Salmonella from livestock and poultry raw meat, Nepal. Int. J. Mol. Vet. Res. 2016;6(1):1–22. [Google Scholar]
- 12.Opitz H.M, El-Begearmi M, Flegg P, Beane D. Effectiveness of five feed additives in chicks infected with Salmonella Enteritidis phage Type 13A. J. Appl. Poult. Res. 1993;2(2):147–153. [Google Scholar]
- 13.Kabir S.M.L. Avian colibacillosis and salmonellosis:A closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. Int. J. Environ. Res. Public Health. 2010;7(1):89–114. doi: 10.3390/ijerph7010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahman M.A, Rahman A.K.M, Islam M.A, Alam M.M. Detection of multi-drug resistant Salmonella from milk and meat in Bangladesh. Bangladesh J. Vet. Med. 2018;16(1):115–120. [Google Scholar]
- 15.Rahman M.M, Rahman M.M, Meher M.M, Khan M.S.I, Anower A.K.M. Isolation and antibiogram of Salmonella spp. from duck and pigeon in Dinajpur, Bangladesh. J. Adv. Vet. Anim. Res. 2016;3(4):386–391. [Google Scholar]
- 16.Mirza S.H, Beeching N.J, Hart C.A. Multi-drug resistant typhoid:A global problem. J. Med. Microbiol. 1996;44(1996):317–319. doi: 10.1099/00222615-44-5-317. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez A.E, Calleja A.C, Fernández G.C, Capita R. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from poultry in Spain:Comparison between 1993 and 2006. Int. J. Food Microbiol. 2012;153(3):281–287. doi: 10.1016/j.ijfoodmicro.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Islam M.J, Mahbub-E-Elahi A.T.M, Ahmed T, Hasan M.K. Isolation and identification of Salmonella spp. from broiler and their antibiogram study in Sylhet, Bangladesh. J. Appl. Biol. Biotech. 2016;4(3):46–51. [Google Scholar]
- 19.Al-Salauddin A.S, Hossain M.F, Dutta A, Mahmud S, Islam M.S, Saha S, Kabir S.M.L. Isolation, identification and antibiogram studies of Salmonella species and Escherichia coli from boiler meat in some selected areas of Bangladesh. Int. J. Basic Clin. Pharmacol. 2015;4(5):999–1003. [Google Scholar]
- 20.Sultana M, Bilkis R, Diba F, Hossain M.A. Predominance of multidrug-resistant zoonotic Salmonella Enteritidis genotypes in poultry of Bangladesh. J. Poult. Sci. 2014;51(4):424–434. [Google Scholar]
- 21.Jahan F, Kabir S.M.L, Amin M.M. Identification and antimicrobial resistance profiles of Salmonella isolated from the broiler dressing plants associated with their environments. Adv. Res. J. Microbiol. 2013;1(1):1–9. [Google Scholar]
- 22.Lu Y, Wu C.M, Wu G.J, Zhao H.Y, He T, Cao X.Y, Dai L, Xia L.N, Qin S.S, Shen J.Z. Prevalence of antimicrobial resistance among Salmonella isolates from chicken in China. Foodborne Pathog. Dis. 2011;8(1):45–53. doi: 10.1089/fpd.2010.0605. [DOI] [PubMed] [Google Scholar]
- 23.ISO. Microbiology of Food and Animal Feeding Stuffs-horizontal Method for the Detection of Salmonella spp. 4th ed. Geneva, Switzerland: International Standard Organization; [Google Scholar]
- 24.Queipo-Ortuño M.I, De Colmeneroi Dios J, Macias M, Bravo M.J, Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin. Vaccine Immunol. 2008;15(2):293–296. doi: 10.1128/CVI.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin C.K, Tsen H.Y. Use of two 16S DNA targeted oligonucleotides as PCR primers for the specific detection of Salmonella in foods. J. Appl. Bacteriol. 1996;80(6):659–666. doi: 10.1111/j.1365-2672.1996.tb03271.x. [DOI] [PubMed] [Google Scholar]
- 26.Bauer A.W, Kirby W.M.M, Sheris J.C, Truck M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966;45(4):493–496. [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. Wayne, PA: Clinical and Laboratory Standards Institute supplement M100S; 2016. pp. 1–256. [Google Scholar]
- 28.Centers for Disease Control and Prevention. Epi Info™7. User Guide. [Last accessed on 09-12-2019]. Available on https://www.cdc.gov/epiinfo/support/userguide.html .
- 29.Hamid M.A, Rahman M.A, Ahmed S, Hossain K.M. Status of poultry industry in Bangladesh and the role of private sector for its development. Asian J. Poult. Sci. 2017;11(1):1–13. [Google Scholar]
- 30.Ratananakorn L, Wilson D. Zoning and compartmentalisation as risk mitigation measures:An example from poultry production. Rev. Sci. Tech. 2011;30(1):297. doi: 10.20506/rst.30.1.2029. [DOI] [PubMed] [Google Scholar]
- 31.Akond M, Shirin M, Alam S, Hassan S, Rahman M.M, Hoq M. Frequency of drug-resistant Salmonella spp. isolated from poultry samples in Bangladesh. Stamford J. Microbiol. 2013;2(1):15–19. [Google Scholar]
- 32.Al-Mamun M.A, Kabir S.M.L, Islam M.M, Lubna M, Islam S.S, Akhter A.T, Hossain M.M. Molecular identification and characterization of Salmonella species isolated from poultry value chains of Gazipur and Tangail districts of Bangladesh. Afr. J. Microbiol. Res. 2017;11(11):474–481. [Google Scholar]
- 33.Islam M.M, Islam M.N, Sharifuzzaman Fakhruzzaman M. Isolation and identification of Escherichia coli and Salmonella from poultry litter and feed. Int. J. Nat. Soc. Sci. 2014;1(2014):1–7. [Google Scholar]
- 34.Saha A.K, Sufian M.A, Hossain M.I, Hossain M.M. Salmonellosis in layer chickens:Pathological features and isolation of bacteria from ovaries and inner content of laid eggs. J. Bangladesh Agric. Univ. 2012;10(1):61–67. [Google Scholar]
- 35.Mahmud M.S, Bari M. L, Hossain M.A. Prevalence of Salmonella Serovars and antimicrobial resistance profiles in poultry of Savar area, Bangladesh. Foodborne Pathog. Dis. 2011;8(10):1111–1118. doi: 10.1089/fpd.2011.0917. [DOI] [PubMed] [Google Scholar]
- 36.Hyeon J.Y, Chon J.W, Hwang I.G, Kwak H.S, Kim M.S, Kim S.K, Choi I.S, Song C.S, Park C, Seo K.H. Prevalence, antibiotic resistance, and molecular characterization of Salmonella Serovars in retail meat products. J. Food Prot. 2011;74(1):161–166. doi: 10.4315/0362-028X.JFP-10-327. [DOI] [PubMed] [Google Scholar]
- 37.Muktaruzzaman M, Haider M.G, Ahmed A.K.M, Alam K.J, Rahman M.M, Khatun M.B, Rahman M.H, Hossain M.M. Validation and refinement of Salmonella Pullorum (SP) colored antigen for diagnosis of Salmonella infections in the field. Int. J. Poult. Sci. 2010;9(8):801–808. [Google Scholar]
- 38.Buxton A, Fraser G. Animal Microbiology. Oxford, London, Edinburg, Melbourne: Blackwell Scientific Publications; 1977. pp. 93–197. [Google Scholar]
- 39.Ifeanyichukwu I, Chika E, Ogonna A, Chidinma I, Monique A, Ikechukwu M, Stanley E, Emmanuel N, Ngozi A, Agabus N. Prevalence and antibiogram of Salmonella species isolated from poultry products. J. Adv. Vet. Anim. Res. 2016;3(4):353–359. [Google Scholar]


