Abstract
Background and Aim:
According to the previous study, the circulating canine parvovirus (CPV) in Thailand is 2a and 2b. Nowadays, CPV mutants, including CPV-2c, have been identified in many parts of the world. This study aimed to investigate the genetic diversity of the circulating CPV in Thailand.
Materials and Methods:
Eighty-five CPV-positive fecal samples were obtained from dogs with either acute hemorrhagic diarrhea or diarrhea. The complete VP2 gene of these samples was amplified using VP2 specific primers and polymerase chain reaction (PCR). The obtained full-length VP2 sequences were analyzed and a phylogenetic tree was constructed.
Results:
Sixty and 25 CPV-positive fecal samples were collected in 2010 and 2018, respectively. Thirty-four samples were new CPV-2a and 31 samples were new CPV-2b due to amino acids substitution at position 297 (Ser-Ala). In 2018, 5 new CPV-2a, 19 CPV-2c, and 1 feline panleukopenia virus (FPV) were found, but no new CPV-2b was detected. Moreover, most of the CPV in this study had amino acids mutations at positions 324 and 440. The phylogenetic construction demonstrated the close relationship between the current new CPV-2a with the previous CPV-2a reported from Thailand, China, Uruguay, Vietnam, Singapore, and India. Interestingly, the current new CPV-2b in this study was not closely related to the previous CPV-2b reported in Thailand. The CPV-2c in this study was closer to Asian CPV-2c and further from either European or South America CPV-2c. Interestingly, FPV was identified in a diarrhea dog.
Conclusion:
The evolution of CPV in Thailand is very dynamic. Thus, it is important to monitor for CPV mutants and especially the clinical signs relating to these mutants to conduct surveillance for the emergence of new highly pathogenic CPV in the future.
Keywords: canine parvoviruses, diversity, Thailand, VP2 gene
Introduction
Canine parvovirus (CPV) is one of the most common viruses in domestic dogs. It causes acute hemorrhagic gastroenteritis, leukopenia, nausea, diarrhea, and sometimes fatal myocarditis in young puppies [1]. CPV belongs to the family Parvoviridae, subfamily Parvovirinae, and genus Parvovirus. It is a non-enveloped, icosahedral, linearized, and single-stranded DNA virus. The genome of CPV is approximately 5.2 kb in length. The virus encodes two nonstructural proteins (NS1 and NS2) and three structural proteins (VP1, VP2, and VP3). The VP2 capsid protein is the main capsid protein and plays an important role in the determination of the antigenicity and host range of CPV [2]. CPV type 2 (CPV-2) was first identified in the USA in 1978 and was found to have spread worldwide in domestic and wild canid populations [3]. CPV is genetically close to feline panleukopenia virus (FPV); however, CPV has at least seven amino acid differences from FPV which determine the canine or feline host range, such as amino acid positions 80 (Lys-Arg), 93 (Lys-Asn), 103 (Val-Ala), 232 (Val-Ile), 323 (Asp-Asn), 564 (Asn-Ser), and 568 (Ala-Gly) [4].
The original CPV-2 was replaced worldwide by CPV-2a and CPV-2b in 1985. CPV-2a and CPV-2b have some nucleotide changes at the VP2 gene compared to the original CPV-2. There are six amino acid differences at residues 87 (Met-Leu), 101 (Ile-Thr), 300 (Ala-Gly), 305 (Asp-Tyr), 375 (Asn-Asp), and 555 (Val-Ile) between CPV-2 and CPV-2a and six amino acid differences at residues 87 (Met-Leu), 101 (Ile-Thr), 300 (Ala-Gly), 305 (Asp-Tyr), 375 (Asn-Asp), and 426 (Asn-Asp) between CPV-2 and CPV-2b. The differences between CPV-2a and CPV-2b are the substitution of two amino acids in the VP2 capsid protein, namely, Asn-426 in 2a (Asp-426 in 2b) and Ile-555 in 2a (Val-555 in 2b). Recently, the emergence of new CPV-2a and CPV-2b has been reported having an amino acid mutation at position 297 (Ser-Ala). Moreover, the new CPV-2a has a mutation at amino acid position 555 that changes isoleucine back to valine [4-8]. CPV-2c is a new CPV strain that has a glutamate substitution at the 426 residues of the VP2 protein [9-11]. Recently, CPV-2c has been detected in Argentina [12], Australia [13], Italy [10], Laos [14], Spain [6], Taiwan [15], and Uruguay [16]. According to the information above, CPV has a high mutation rate and has been dynamically evolving in many parts of the world. There is a growing concern about the severity and effectiveness of vaccine regarding the new mutant or genotype of CPV.
Molecular surveillance may be used as a tool for the detection of the new mutant, prediction of disease severity and providing the important data for the development of the better vaccine or the better diagnostic test in the future. Moreover, the knowledge of the current genotype of the CPV in Thailand is limited. This study aimed to investigate the current genotype of CPV circulating in Thailand and to determine the existence of CPV-2c and other CPV strains.
Materials and Methods
Ethical approval
This study was approved by the Animal Ethics Committee of the Faculty of Veterinary Medicine, Kasetsart University, Thailand (ACKU62-VET-007).
Samples
Eighty-five fecal samples were used in this study based on a positive result for CPV according to routine polymerase chain reaction (PCR) testing. In 2010 and 2018, 60 and 25 positive samples, respectively, were collected (Table-1). These fecal samples were collected from dogs that displayed either acute hemorrhagic diarrhea or diarrhea and nausea at the Veterinary Teaching Hospital, Kasetsart University, Rattanatibeth Referral Animal Hospital, Bangkok, Thailand, and the Amphawa Pet Hospital, Samut Songkhram, Thailand. These dogs were either vaccinated or unvaccinated and were aged from 1 month to 5 years. The fecal samples were stored at −80°C until used for DNA extraction.
Table-1.
Age of parvovirus-infected dogs, vaccination, year of sample collection, GenBank accession number, genotype of CPV, and amino acid at important position (NR=no report).
| No. | Sample | Amino acid position | CPV type | Previous vaccination | Age | GenBank accession number | Year of collection | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 80 | 87 | 93 | 103 | 232 | 267 | 297 | 300 | 305 | 323 | 324 | 370 | 426 | 440 | 555 | 564 | 568 | |||||||
| 1 | FPV-VT-2020 | K | M | N | V | V | F | S | A | D | D | Y | Q | N | T | V | N | A | FPV | NR | 1 years | MN270937 | 2018 |
| 2 | CPV-VT-1 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | NR | KP715658 | 2010 |
| 3 | CPV-VT-7 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 3 M | KP715659 | 2010 |
| 4 | CPV-VT-13 | R | L | N | A | I | F | A | G | Y | N | I | Q | N | T | V | S | G | New 2a | No | 3 M | KP715660 | 2010 |
| 5 | CPV-VT-14 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 2 M | KP715661 | 2010 |
| 6 | CPV-VT-30 | R | L | N | A | I | F | A | G | Y | N | I | Q | N | T | V | S | G | New 2a | Yes | 3 M | KP715662 | 2010 |
| 7 | CPV-VT-37 | R | L | N | A | I | F | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | Yes | 2 M | KP715663 | 2010 |
| 8 | CPV-VT-39 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 5 M | KP715664 | 2010 |
| 9 | CPV-VT-40 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 2 M | KP715665 | 2010 |
| 10 | CPV-VT-42 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 1 M | KP715666 | 2010 |
| 11 | CPV-VT-45 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | Yes | 2 M | KP715667 | 2010 |
| 12 | CPV-VT-51 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | Yes | 6 M | KP715668 | 2010 |
| 13 | CPV-VT-58 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 2 M | KP715669 | 2010 |
| 14 | CPV-VT-61 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 3 M | KP715670 | 2010 |
| 15 | CPV-VT-62 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | >3 M | KP715671 | 2010 |
| 16 | CPV-VT-71 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | Yes | 5 M | KP715672 | 2010 |
| 17 | CPV-VT-81 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 4 M | KP715673 | 2010 |
| 18 | CPV-VT-82 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 2 M | KP715674 | 2010 |
| 19 | CPV-VT-83 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 3 M | KP715675 | 2010 |
| 20 | CPV-VT-87 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 2 M | KP715676 | 2010 |
| 21 | CPV-VT-88 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | NR | 3 M | KP715677 | 2010 |
| 22 | CPV-VT-89 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 1.5 M | KP715678 | 2010 |
| 23 | CPV-VT-92 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 3 M | KP715679 | 2010 |
| 24 | CPV-VT-93 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 3 M | KP715680 | 2010 |
| 25 | CPV-VT-97 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | 3 M | KP715681 | 2010 |
| 26 | CPV-VT-103 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | No | >3 M | KP715682 | 2010 |
| 27 | CPV-VT-109 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | NR | NR | KP715683 | 2010 |
| 28 | CPV-VT-115 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | NR | NR | KP715684 | 2010 |
| 29 | CPV-VT-138 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | NR | 3 M | KP715685 | 2010 |
| 30 | CPV-VT-139 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | NR | 3.5 M | KP715686 | 2010 |
| 31 | CPV-VT-0561 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | NR | NR | MN270938 | 2018 |
| 32 | CPV-VT-1377 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | NR | NR | MN270939 | 2018 |
| 33 | CPV-VT-2097 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | NR | 2 M | MN270940 | 2018 |
| 34 | CPV-VT-2098 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | NR | 2 M | MN270941 | 2018 |
| 35 | CPV-VT-2961 | R | L | N | A | I | Y | A | G | Y | N | I | Q | N | A | V | S | G | New 2a | NR | 1 M | MN270942 | 2018 |
| 36 | CPV-VT-3 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 2 years | KP715687 | 2010 |
| 37 | CPV-VT-12 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | Yes | 7 M | KP715688 | 2010 |
| 38 | CPV-VT-18 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 2 M | KP715689 | 2010 |
| 39 | CPV-VT-28 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | Yes | 3 M | KP715690 | 2010 |
| 40 | CPV-VT-43 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | NR | 1 years | KP715691 | 2010 |
| 41 | CPV-VT-49 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | Yes | 4 M | KP715692 | 2010 |
| 42 | CPV-VT-53 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | A | V | S | G | New 2b | No | 2 M | KP715693 | 2010 |
| 43 | CPV-VT-54 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 2 M | KP715694 | 2010 |
| 44 | CPV-VT-56 | R | L | N | A | I | Y | A | G | Y | N | Y | Q | D | T | V | S | G | New 2b | No | 5 M | KP715695 | 2010 |
| 45 | CPV-VT-65 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | NR | KP715696 | 2010 |
| 46 | CPV-VT-66 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 2 years | KP715697 | 2010 |
| 47 | CPV-VT-68 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 4 M | KP715698 | 2010 |
| 48 | CPV-VT-74 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | NR | KP715699 | 2010 |
| 49 | CPV-VT-75 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 2 M | KP715700 | 2010 |
| 50 | CPV-VT-80 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 4 M | KP715701 | 2010 |
| 51 | CPV-VT-84 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 4 M | KP715702 | 2010 |
| 52 | CPV-VT-86 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 5 M | KP715703 | 2010 |
| 53 | CPV-VT-90 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | Yes | 6 M | KP715704 | 2010 |
| 54 | CPV-VT-99 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 5 M | KP715705 | 2010 |
| 55 | CPV-VT-101 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 1 M | KP715706 | 2010 |
| 56 | CPV-VT-106 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | NR | NR | KP715707 | 2010 |
| 57 | CPV-VT-108 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | A | V | S | G | New 2b | NR | NR | KP715708 | 2010 |
| 58 | CPV-VT-114 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | NR | NR | KP715709 | 2010 |
| 59 | CPV-VT-120 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 3 M | KP715710 | 2010 |
| 60 | CPV-VT-121 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 2 M | KP715711 | 2010 |
| 61 | CPV-VT-123 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 8 M | KP715712 | 2010 |
| 62 | CPV-VT-129 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 1.5 years | KP715713 | 2010 |
| 63 | CPV-VT-135 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | NR | KP715714 | 2010 |
| 64 | CPV-VT-142 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | NR | 1 M | KP715715 | 2010 |
| 65 | CPV-VT-143 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | T | V | S | G | New 2b | No | 4 M | KP715716 | 2010 |
| 66 | CPV-VT-148 | R | L | N | A | I | Y | A | G | Y | N | I | Q | D | A | V | S | G | New 2b | NR | 2 M | KP715717 | 2010 |
| 67 | CPV-VT-0861 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 1 M | MN270943 | 2018 |
| 68 | CPV-VT-0937 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | NR | MN270944 | 2018 |
| 69 | CPV-VT-1161 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 11 M | MN270945 | 2018 |
| 70 | CPV-VT-1261 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 6 M | MN270946 | 2018 |
| 71 | CPV-VT-1361 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 4 M | MN270947 | 2018 |
| 72 | CPV-VT-1373 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | NR | MN270948 | 2018 |
| 73 | CPV-VT-1374 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | NR | MN270949 | 2018 |
| 74 | CPV-VT-1375 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | NR | MN270950 | 2018 |
| 75 | CPV-VT-1383 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | NR | MN270951 | 2018 |
| 76 | CPV-VT-1661 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 1 years | MN270952 | 2018 |
| 77 | CPV-VT-2016 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 2 M | MN270953 | 2018 |
| 78 | CPV-VT-2018 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 3 M | MN270954 | 2018 |
| 79 | CPV-VT-2019 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 3 years | MN270955 | 2018 |
| 80 | CPV-VT-2021 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 7 M | MN270956 | 2018 |
| 81 | CPV-VT-2388 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 3 M | MN270957 | 2018 |
| 82 | CPV-VT-2389 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 5 M | MN270958 | 2018 |
| 83 | CPV-VT-2391 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 2.5 years | MN270959 | 2018 |
| 84 | CPV-VT-2461 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 3 M | MN270960 | 2018 |
| 85 | CPV-VT-3761 | R | L | N | A | I | Y | A | G | Y | N | I | R | E | T | V | S | G | 2c | NR | 21 days | MN270961 | 2018 |
CPV=Canine parvovirus, FPV=Feline panleukopenia virus
DNA extraction and PCR
DNA was extracted from the fecal samples using the acid guanidinium thiocyanate-phenol-chloroform extraction method. A set of primers was designed to amplify the whole VP2 gene: F (5’-ATG AGT GAT GGA GCA GTT CA) and R (5’-TTA ATA TAA TTT TCT AGG TGC TAG TTG). The PCR mixture (25 µl) was composed of 1× buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl2), 0.2 mM dNTPs, 2.5 mM MgCl2, 100 pmol of each of the forward and reverse primers, 1 unit Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), and 2.5 µl of DNA template to give a total volume of 25 µl. After an initial denaturing at 94°C for 5 min, the amplification was performed using 35 cycles at 94°C for 40 s, annealing at 50°C for 40 s, and extension at 72°C for 90 s, and a final extension at 72°C for 10 min. The expected PCR products were 1755 bps in size. The PCR products were analyzed using 1% agarose gel electrophoresis at 100 V for 30 min and visualized under ultraviolet illumination. The PCR products were purified using an UltraClean®15 DNA Purification Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA) and cloned into plasmid pGEM-T Easy (Promega Corporation, Madison, WI, USA). The sequences of the cloned full-length VP2 were determined at First BASE Laboratories Sdn Bhd, Selangor, Malaysia.
Analysis and phylogenetic construction of full-length VP2 gene of CPVs
The nucleotide sequences were translated into amino acid and multiple alignments of the amino acid sequences using the Bioedit biological sequence alignment editor computer package (version 7.1.3; Ibis Biosciences, Carlsbad, CA, USA). The phylogenetic analysis was constructed from the amino acid sequences of all 85 samples in this study and other full-length VP2 sequences obtained from the GenBank database (Table-2) using the MEGA program (version 7.0, The Biodesign Institute, Tempe, AZ, USA) with the neighbor-joining method and running 1000 replicates in the bootstrap. Bayesian phylogenetic analysis was also performed for more extensive amino acids analysis to analyze the selective pressures on certain amino acids using mixed model analysis. The phylogenetic tree was created by MrBayes version 3.2.6 (https://nbisweden.github.io/MrBayes/download.html) [17]. The tree was viewed using FigTree software version 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Table-2.
GenBank accession numbers of CPV used in phylogenetic tree construction.
| Order | Origin | GenBank accession number | Year of collection | Genetic type |
|---|---|---|---|---|
| 1 | China | MF467224 | 2015 | 2a |
| 2 | China | FJ435343 | 2008 | 2a |
| 3 | China | FJ435345 | 2008 | 2a |
| 4 | China | GU380304 | 2009 | 2a |
| 5 | China | GU569936 | 2008 | 2a |
| 6 | France | DQ026002 | - | 2a |
| 7 | India | KX469433 | 2015 | 2a |
| 8 | Italy | FJ005259 | 2008 | 2a |
| 9 | Singapore | KY083098 | 2014 | 2a |
| 10 | Thailand | GQ379047 | 2009 | 2a |
| 11 | Thailand | GQ379045 | 2009 | 2a |
| 12 | Thailand | GQ379046 | 2009 | 2a |
| 13 | Uruguay | KM457139 | 2011 | 2a |
| 14 | Uruguay | JF906788 | 2010 | 2a |
| 15 | Vietnam | LC214970 | 2013 | 2a |
| 16 | China | GU569937 | 2002 | 2b |
| 17 | China | KF482468 | 2009 | 2b |
| 18 | China | GU569938 | 2002 | 2b |
| 19 | China | GU569944 | 2002 | 2b |
| 20 | China | JQ743891 | 2010 | 2b |
| 21 | Italy | FJ005263 | 2005 | 2b |
| 22 | Thailand | FJ869123 | 2008 | 2b |
| 23 | Thailand | FJ869124 | 2008 | 2b |
| 24 | Vietnam | AB120724 | 2013 | 2b |
| 25 | USA | JX475261 | 2010 | 2b |
| 26 | Argentina | KM236569 | 2013 | 2c |
| 27 | China | KT162005 | 2014 | 2c |
| 28 | China | KT162016 | 2014 | 2c |
| 29 | China | KP260509 | 2014 | 2c |
| 30 | China | KY937641 | 2016 | 2c |
| 31 | Croatia | KP859576 | 2014 | 2c |
| 32 | Ecuador | KF149984 | 2012 | 2c |
| 33 | Germany | FJ005202 | 1997 | 2c |
| 34 | Germany | FJ005204 | 1999 | 2c |
| 35 | Greece | GQ865518 | 2008 | 2c |
| 36 | Indonesia | LC216905 | 2013 | 2c |
| 37 | Italy | HQ025913 | 2010 | 2c |
| 38 | Singapore | KY083092 | 2014 | 2c |
| 39 | Uruguay | KM457112 | 2008 | 2c |
| 40 | - | M74849 | - | 2b |
| 41 | - | M24003 | - | 2a |
| 42 | - | M38245 | - | 2 |
| 43 | - | FJ405225 | - | FPV |
CPV=Canine parvovirus, FPV=Feline panleukopenia virus
Results
Thirty-nine and nine samples out of 85 positive samples were from non-vaccinated and vaccinated dogs, respectively. The other 37 positive samples did not have a history of vaccination. The youngest CPV-infected dog was aged 21 days, and the oldest was 3 years. The youngest age of positive vaccinated dogs was 2 months; however, the individual histories of booster vaccination for these positive, vaccinated dogs were not available (Table-1).
For the 2010 data, the amino acid sequences analysis revealed that 29 samples were new CPV-2a and 31 samples were new CPV-2b due to amino acids substitution at position 297 (Ser-Ala) (Table-1). The number of positive samples for new CPV-2a and new CPV-2b was nearly equal in 2010. For 2018, 5 new CPV-2a, 19 CPV-2c, and 1 FPV were found, but CPV-2b was not detected (Table-3). In this study, most of the CPV had amino acid substitution at positions 324 and 440. Eighty-three CPV had 324 (Tyr-Ile) substitution due to a T-to-A transversion at nucleotide 970 and an A-to-T transversion at nucleotide 971 with the exception of 1 sample of new CPV-2b in 2010 (CPV-VT 56) (Figure-1). Thirty-two out of 34 new CPV-2a and three out of 31 new CPV-2b (CPV-VT 53, CPV-VT 108, and CPV-VT 148) had 440 (Thr-Ala) substitution due to an A-to-G transition at nucleotide 1318 (Table-1). The new CPV-2a in this study was closely related to CPV-2a from Thailand in 2009 (GQ379045, GQ379046, and GQ379047), and from Uruguay in 2010 and 2011 (JF906788 and KM457139), China in 2015 (MF467224), India in 2015 (KX469433), Singapore in 2014 (KY083098), and Vietnam in 2013 (LC214970) (Figures-2 and 3) due to similar amino acids at positions 267 (Tyr), 324 (Ile), and 440 (Ala). However, two new CPV-2a (CPV-VT 13 and 30) were closely related to new CPV-2a from China in 2008 (FJ435343, FJ435345, GU380304, and GU569936) (Figures-2 and 3) due to similar amino acids at positions 267 (Phe), 324 (Ile), and 440 (Thr). All CPVs in this study had 267 (Phe-Tyr) due to an A-to-T transversion at nucleotide 800 substitution except for the two new CPV-2a samples (CPV-VT 13 and 30) that did not have 440 (Thr-Ala) substitution and one new CPV-2a (CPV-VT 37) that had substitution at position 440. All CPV-2a samples had valine at position 555.
Table-3.
Number of parvoviruses found in each year.
| Year | Type of parvoviruses | ||||
|---|---|---|---|---|---|
| FPV | CPV2a | CPV2b | CPV2c | Total | |
| 2010 | 0 | 29 | 31 | 0 | 60 |
| 2018 | 1 | 5 | 0 | 19 | 25 |
| Total | 1 | 34 | 31 | 19 | 85 |
CPV=Canine parvovirus, FPV=Feline panleukopenia virus
Figure-1.
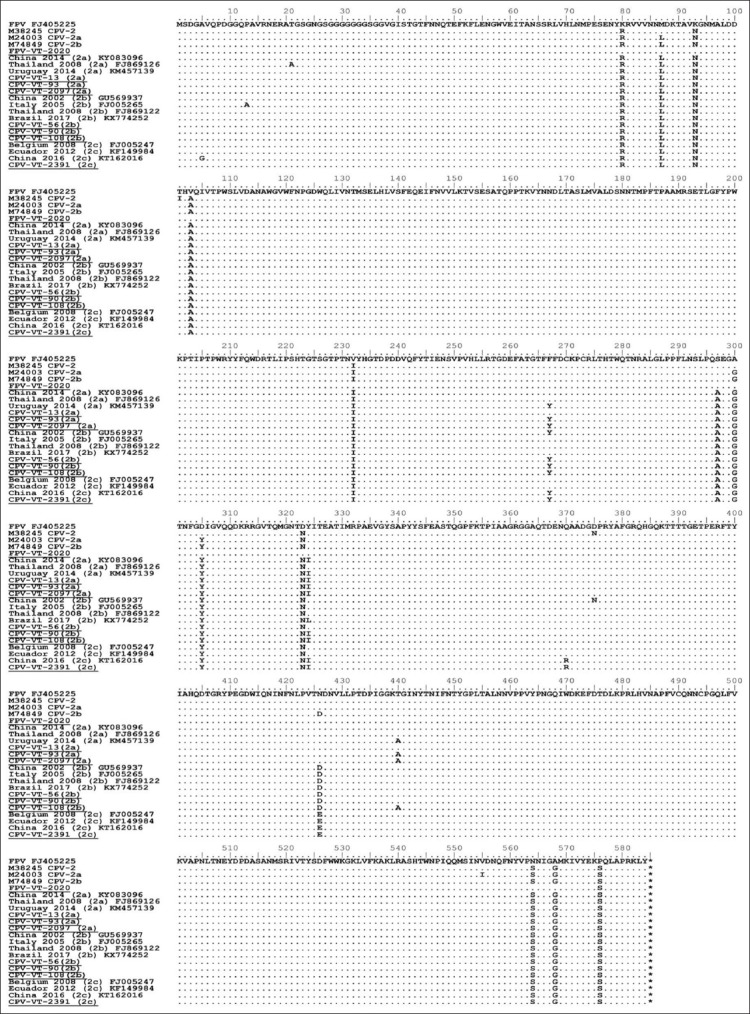
Amino acid comparison of VP2 gene of CPV-2a: CPV-VT13 (T440), CPV-VT93 (A440), CPV-VT2097, CPV-2b: CPV-VT56 (324Y), CPV-VT90 (440T), and CPV-VT108 (440A), CPV-2c: CPV-VT2391, FPV: FPV-VT2020 in this study, with reference strains FPV (FJ405225), CPV-2 (M38245), CPV-2a (M24003) and CPV-2b (M74849) and other isolates from other parts of the world. CPV=Canine parvovirus, FPV=Feline panleukopenia virus.
Figure-2.

Phylogenetic tree constructed from 80 amino acid sequences of the VP2 gene of canine parvovirus (CPV) and feline panleukopenia virus (FPV) in this study and other CPV and FPV sequences obtained from GenBank database using the neighbor-joining method and bootstrap analysis performed with 1000 trials. Drawn using MEGA version. ■=Samples collected in 2018 and ▲=Samples collected in 2010.
Figure-3.

Phylogenetic tree constructed from 80 amino acids of the VP2 gene of canine parvovirus (CPV) and feline panleukopenia virus (FPV) in this study and other CPV and FPV sequences obtained from GenBank database using MrBayes version 3.2.6 with Markov chain Monte Carlo, with 5 million generations. Node values (in percentages) indicate posterior clade probabilities. Vertical bars indicate clades of CPV in each group, A=CPV-2c in Europe and South America, B=new CPV-2b in this study, C=CPV-2c in this study, and Asia, D=new CPV-2b that has alanine at amino acid position 440, F=new CPV-2a in this study.
In contrast to new CPV-2a in this study, all new CPV-2b in this study was distanced from CPV-2b reported previously in Thailand in 2008 (FJ869123 and FJ869124) and Europe in 2005 (FJ005263) (Figures-2 and 3) because of the amino substitution at position 324 (Tyr-Ile). However, three new CPV-2b (CPV-VT 53, CPV-VT 108, and CPV-VT 148) were closely related to the new CPV-2b from China in 2010 (JQ743891) and were also closely related to a cluster of new CPV-2a. These three CPV-2b distanced from the other new CPV-2b in this study (Figures-2 and 3) due to amino substitution at positions 324 (Tyr-Ile) and 440 (Thr-Ala). Nineteen CPV-2c in this study had amino acids substitution at positions 267 (Phe-Tyr), 324 (Tyr-Ile), and 370 (Gln-Arg) due to an A-to-G transition at nucleotide 1109 and a G-to-T transversion at nucleotide 1110. CPV-2c in this study was closely related to CPV-2c from Asia including China in 2014 and 2016 (KP260509, KT162005, KT162016, and KY937641), Indonesia in 2013 (LC216905), and Singapore in 2014 (KY083092) due to these three amino acids substitution (Figures-2 and 3), but distanced from the CPV-2c in Europe and South America. Interestingly, one dog with clinical signs of diarrhea was positive for FPV. The six amino acids (at 80, 103, 232, 323, 564, and 568) out of the seven amino acids which determine the canine or feline host range were similar to the reference FPV; however, there was an amino acid substitution at position 93 (Lys-Asn). According to this amino acid substitution, FPV in this study was in the different clades compared to the reference FPV (Figures-2 and 3).
Discussion
The number of samples positive for new CPV-2a and 2b circulating in central Thailand in 2010 was approximately equal. This result contrasted with a study in Thailand (data collected between 2003 and 2008) in which the predominant genotype in central Thailand was CPV-2a [11]. Interestingly, the majority CPV genotype in 2018 was CPV-2c, but new CPV-2b was not found. This study showed that CPV-2c had become the predominant genotype during the year 2018 in Thailand. Besides of t he diminishing of CPV-2b, there was only one amino acid difference at position 426 between CPV-2b and CPV-2c but there were two amino acids differences at positions 426 and 440 between CPV-2a and CPV-2c. Thus, the emergence of CPV-2c in Thailand might have derived from the new CPV-2b (2010) that had a mutation at position 426 (Asp-Glu).
From the previous study in Thailand (2003-2009), both CPV-2a and CPV-2b had alanine at amino acid position 297 [11]. According to this finding, CPV-2a and CPV-2b (2003-2009) were new CPV-2a and new CPV-2b. However, most of the CPV in this study had additional amino acid substitution at position 324 (Tyr-Ile), except for 1 CPV-2b (CPV-VT 56). Thus, the new CPV-2b (2010) was different from the new CPV-2b in Thailand (2003-2009) and Europe (2005). The amino acid substitution at residue 324 (Tyr-Ile) was also reported in Brazil [18], China [19], Hungary [20], India [21,22], Italy [23], Nigeria [24], South Korea [25], Taiwan [1,15], and Uruguay [26]. According to the above, amino acid position 324 has been shown to undergo a strong positive selection in the parvovirus of all carnivores [27]. Amino acid position 324 was also adjacent to the critical amino acid position 323 that has been known to control canine cell infection. The changes in the region of the capsid surface around VP2 residue 300, within a raised region on the shoulder of the three-fold spike of the capsid, have been shown to influence the binding of the virus to the canine TfR [28-30]. This change may influence the interactions with their various host receptors and may be expected to result in an increased host range.
Moreover, most of the current new CPV-2a had alanine at amino acid position 440; however, most of the new CPV-2b still had threonine. Amino acid substitution at position 440 (Thr-Ala) was also reported in the previous studies in China [19], India [21,22], Nigeria [24,31], Pakistan [32], South Africa [33], South Korea [25], and Uruguay [26]. Amino acid position 440 is also important because it is located at the top of the three-fold spike, the main antigenic site of the virus [34,35]. Interestingly, this mutation was not detected in a previous study in Thailand [11]. In the current study, two new CPV-2a (CPV-VT13 and 30) had phenylalanine and threonine at amino acid positions 267 and 440 similar to CPV-2a in 2003-2004 as reported in the previous study in Thailand, but these two strains had isoleucine at amino acid position 324 as seen in CPV-2a in 2008-2009 and new CPV-2a in this study [11]. However, most of the current new CPV-2a in the current study had tyrosine and alanine at positions 267 and 440, as seen in CPV-2a in 2008-2009, respectively. A CPV-2a (CPV-VT37) had phenylalanine and isoleucine at positions 267 and 324 similar to CPV-2a in 2003-2004 but had alanine at amino acid position 440 as seen in CPV-2a in 2008-2009 and new CPV-2a in this study. CPV-VT13, 30, and 37 might represent the transition evolution of the original new CPV-2a to the current new CPV-2a. CPV-2b (CPV-VT56) had similar amino acids at amino acid positions 267, 324, 426, and 440 (phenylalanine, tyrosine, aspartic acid, and threonine, respectively), as reported in the previous study in Thailand [11]. However, new CPV-2b in this study had isoleucine at amino acid 324. Thus, CPV-VT56 might also represent the transition evolution of the original new CPV-2b in the current new CPV-2b.
The CPV-2c in this study was similar to CPV-2c in Asia, such as in China [36], Laos [14], and Taiwan [15,37] due to amino acid substitution at amino acid positions 267 (Tyr), 324 (Ile), and 370 (Arg). CPV-2c in this study differs from CPV-2c in Europe and South America at three amino acid positions (Phe267Tyr, Tyr324Ile, and Gln370Arg). The amino acid substitution at position 440 (Thr-Ala) has been found in Argentina, but this change was not found in this study [12]. Interestingly, FPV was found in a dog that had clinical signs of diarrhea. This FPV sample had an amino acid substitution at position 93 (Lys-Asn). The amino acid at position 93 is important because it is one of the amino acids that determine the canine host range [38]. FPV infection in dogs has also been reported in Pakistan [39]. These findings demonstrated that parvoviruses in Thailand have been dynamically evolving as those in the other part of the world [40,41]. This mutation rate is as high as seen in RNA viruses [41]. The rapid mutation of CPV has resulted in growing concern about the effectiveness of vaccines regarding the new mutant or genotype of CPV. Molecular surveillance of CPV is crucial for the prediction of disease severity and may be important for the development of more effective vaccines or diagnostic tests in the future.
Conclusion
Two genotypes of CPV (new CPV-2a and CPV-2c) are circulating in Central Thailand and the predominant circulating genotype of CPV has been changed from CPV-2a in the past to CPV-2c at present. Currently, CPV-2b has not been found in Central Thailand. The current new CPV-2a circulating in Thailand has amino acid substitutions at positions 324 (Tyr-Ile) and 440 (Thr-Ala). FPV was found in a dog that had acute diarrhea; however, the importance of this finding remains to be determined. Our results provided additional information on the dynamic evolution of CPV in Thailand, which is following the same evolutionary trend observed in the others part of the world.
Authors’ Contributions
NI and TS designed the experiment and made DNA extraction, PCR, multiple alignment, and phylogenetic study. NM, SK, KS, and TS were involved in scientific discussion and provided suggestions for the overall work. All authors read and approved the final manuscript.
Acknowledgments
This research was supported by grants from the Kasetsart University Research and Development Institute, Bangkok, Thailand (Grant no. 21.53), and the Center of Excellence on Agricultural Biotechnology, Science and Technology Postgraduate Education and Research Development Office, Office of Higher Education Commission, Ministry of Education, Science, Research and Innovation (AG-BIO/PERDO-CHE), Bangkok, Thailand (Grant no. AG-BIO/59-002-015). We would like to thank the Faculty of Veterinary Technology, Kasetsart University, for providing technical support, the Veterinary Teaching Hospital, Faculty of Veterinary Medicine, Kasetsart University, Rattanatibeth Referral Animal Hospital, Bangkok, Thailand, and the Amphawa Pet Hospital, Samut Songkhram, Thailand, for providing specimens.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Lin C.N, Chien C.H, Chiou M.T, Chueh L.L, Hung M.Y, Hsu H.S. Genetic characterization of type 2a canine parvoviruses from Taiwan reveals the emergence of an Ile324 mutation in VP2. Virol. J. 2014;11(1) doi: 10.1186/1743-422X-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hueffer K, Govindasamy L, Agbandje-McKenna M, Parrish C.R. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 2003;77(18):10099–10105. doi: 10.1128/JVI.77.18.10099-10105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appel M.J, Scott F.W, Carmichael L.E. Isolation and immunization studies of a canine parvo-like virus from dogs with hemorrhagic enteritis. Vet. Rec. 1979;105(8):156–159. doi: 10.1136/vr.105.8.156. [DOI] [PubMed] [Google Scholar]
- 4.Miranda C, Thompson G. Canine parvovirus:The worldwide occurrence of antigenic variants. J. Gen. Virol. 2016;97(9):2043–2057. doi: 10.1099/jgv.0.000540. [DOI] [PubMed] [Google Scholar]
- 5.Martella V, Decaro N, Elia G, Buonavoglia C. Surveillance activity for canine parvovirus in Italy. J. Vet. Med. B. Infect. Dis. Vet. Public Health. 2005;52(7-8):312–315. doi: 10.1111/j.1439-0450.2005.00875.x. [DOI] [PubMed] [Google Scholar]
- 6.Decaro N, Martella V, Desario C, Bellacicco A.L, Camero M, Manna L, d' Aloja D, Buonavoglia C. First detection of canine parvovirus type 2c in pups with haemorrhagic enteritis in Spain. J. Vet. Med. B. Infect. Dis. Vet. Public Health. 2006;53(10):468–472. doi: 10.1111/j.1439-0450.2006.00974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinchkar S.R, Subramanian B.M, Rao N.H, Rangarajan P.N, Thiagarajan D, Srinivasan V.A. Analysis of VP2 gene sequences of canine parvovirus isolates in India. Arch. Virol. 2006;151(9):1881–1887. doi: 10.1007/s00705-006-0753-8. [DOI] [PubMed] [Google Scholar]
- 8.Kang B.K, Song D.S, Lee C.S, Jung K.I, Park S.J, Kim E.M, Park B.K. Prevalence and genetic characterization of canine parvoviruses in Korea. Virus Genes. 2008;36(1):127–133. doi: 10.1007/s11262-007-0189-6. [DOI] [PubMed] [Google Scholar]
- 9.Buonavoglia C, Martella V, Pratelli A, Tempesta M, Cavalli A, Buonavoglia D, Bozzo G, Elia G, Decaro N, Carmichael L. Evidence for evolution of canine parvovirus type 2 in Italy. J. Gen. Virol. 2001;82(21):3021–3025. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- 10.Decaro N, Buonavoglia C. Canine parvovirus a review of epidemiological and diagnostic aspects, with emphasis on type 2c. Vet. Microbiol. 2012;155(1):1–12. doi: 10.1016/j.vetmic.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phromnoi S, Sirinarumitr K, Sirinarumitr T. Sequence analysis of VP2 gene of canine parvovirus isolates in Thailand. Virus Genes. 2010;41(1):23–29. doi: 10.1007/s11262-010-0475-6. [DOI] [PubMed] [Google Scholar]
- 12.Calderon M.G, Romanutti C, Wilda M, D'Antuono A, Keller L, Giacomodonato M.N, Mattion N, La Torre J. Resurgence of canine parvovirus 2a strain in the domestic dog population from Argentina. J. Virol. Methods. 2015;222:145–149. doi: 10.1016/j.jviromet.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Woolford L, Crocker P, Bobrowski H, Baker T, Hemmatzadeh F. Detection of the canine parvovirus 2c subtype in Australian dogs. Viral Immunol. 2017;30(5):371–376. doi: 10.1089/vim.2017.0019. [DOI] [PubMed] [Google Scholar]
- 14.Vannamahaxay S, Vongkhamchanh S, Intanon M, Tangtrongsup S, Tiwananthagorn S, Pringproa K, Chuammitri P. Molecular characterization of canine parvovirus in Vientiane, Laos. Arch. Virol. 2017;162(5):1355–1361. doi: 10.1007/s00705-016-3212-1. [DOI] [PubMed] [Google Scholar]
- 15.Lin Y.C, Chiang S.Y, Wu H.Y, Lin J.H, Chiou M.T, Liu H.F, Lin C.N. Phylodynamic and genetic diversity of canine parvovirus type 2c in Taiwan. Int. J. Mol. Sci. 2017;18(12):2703. doi: 10.3390/ijms18122703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pintos A.B, Larrama C.B.N, Baratta E.E.R, Barthe M.B.B, Rodonz J.R.A. Isolation and characterization of canine parvovirus type 2c circulating in Uruguay. Cienc. Rural. 2011;41(8):1436–1440. [Google Scholar]
- 17.Ronquist F, Huelsenbeck J.P. MrBayes 3:Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 18.Silva S.P, Silva L, Rodrigues E.D.L, Cardoso J.F, Tavares F.N, Souza W.M, Santos C.M.P, Martins F.M.S, Jesus I.S, Brito T.C, Moura T.P.C, Nunes M.R.T, Casseb L.M.N, Filho E.S, Casseb A.R. Full-length genomic and molecular characterization of canine parvovirus in dogs from North of Brazil. Genet. Mol Res. 2017;16(3):gmr16039719. doi: 10.4238/gmr16039719. [DOI] [PubMed] [Google Scholar]
- 19.Li C, Tang J, Chen Z, Li Q, Huang Z, Wang Q, Meng C, Wang Y, Liu G. Genetic characterization of the complete genome of a mutant canine parvovirus isolated in China. Arch. Virol. 2018;163(2):521–525. doi: 10.1007/s00705-017-3586-8. [DOI] [PubMed] [Google Scholar]
- 20.Cságola A, Varga S, Lorincz M, Tuboly T. Analysis of the full-length VP2 protein of canine parvoviruses circulating in Hungary. Arch. Virol. 2014;159(9):2441–2444. doi: 10.1007/s00705-014-2068-5. [DOI] [PubMed] [Google Scholar]
- 21.Mittal M, Chakravarti S, Mohapatra J.K, Chug P.K, Dubey R, Upmanuyu V, Narwal P.S, Kumar A, Churamani C.P, Kanwar N.S. Molecular typing of canine parvovirus strains circulating from 2008 to 2012 in an organized kennel in India reveals the possibility of vaccination failure. Infect. Genet. Evol. 2014;23:1–6. doi: 10.1016/j.meegid.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Nookala M, Mukhopadhyay H.K, Sivaprakasam A, Balasubramanian B, Antony P.X, Thanislass J, Srinivas M.V, Pillai RM. Full-length VP2 gene analysis of canine parvovirus reveals the emergence of newer variants in India. Acta. Microbiol. Immunol. Hung. 2016;63(4):411–426. doi: 10.1556/030.63.2016.010. [DOI] [PubMed] [Google Scholar]
- 23.Mira F, Dowgier G, Purpari G, Vicari D, Di Bella S, Macaluso G, Gucciardi F, Randazzo V, Decaro N, Guercio A. Molecular typing of a novel canine parvovirus type 2a mutant circulating in Italy. Infect. Genet. Evol. 2018;61:67–73. doi: 10.1016/j.meegid.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apaa T.T, Daly J.M, Tarlinton R.E. Canine parvovirus (CPV-2) variants circulating in Nigerian dogs. Vet. Rec. Open. 2016;3(1):e000198. doi: 10.1136/vetreco-2016-000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon S.H, Jeong W, Kim H.J, An D.J. Molecular insights into the phylogeny of canine parvovirus 2 (CPV-2) with emphasis on Korean isolates:A bayesian approach. Arch. Virol. 2009;154(8):1353–1360. doi: 10.1007/s00705-009-0444-3. [DOI] [PubMed] [Google Scholar]
- 26.Perez R, Bianchi P, Calleros L, Francia L, Hernandez M, Maya L, Panzera Y, Sosa K, Zoller S. Recent spreading of a divergent canine parvovirus type 2a (CPV-2a) strain in a CPV-2c homogenous population. Vet. Microbiol. 2012;155(2-4):214–219. doi: 10.1016/j.vetmic.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Hoelzer K, Shackelton L.A, Holmes E.C, Parrish C.R. Within-host genetic diversity of endemic and emerging parvoviruses of dogs and cats. J. Virol. 2008;82(22):11096–11105. doi: 10.1128/JVI.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodman L.B, Lyi S.M, Johnson N.C, Cifuente J.O, Hafenstein S.L, Parrish C.R. Binding site on the transferrin receptor for the parvovirus capsid and effects of altered affinity on cell uptake and infection. J. Virol. 2010;84(10):4969–4978. doi: 10.1128/JVI.02623-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palermo L.M, Hueffer K, Parrish C.R. Residues in the apical domain of the feline and canine transferrin receptors control host-specific binding and cell infection of canine and feline parvoviruses. J. Virol. 2003;77(16):8915–8923. doi: 10.1128/JVI.77.16.8915-8923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palermo L.M, Hafenstein S.L, Parrish C.R. Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J Virol. 2006;80(17):8482–8492. doi: 10.1128/JVI.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fagbohun O.A, Omobowale T.O. Sequence and phylogenetic analysis of canine parvovirus-2 isolates in dogs revealed the circulation of three subtypes in Nigeria. Virus Dis. 2018;29(3):411–415. doi: 10.1007/s13337-018-0475-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shabbir M.Z, Sohail M.U, Chaudhary U.N, Yaqub W, Rashid I, Saleem M.H, Munir M. Genetic characterization of canine parvovirus from dogs in Pakistan. Acta Virol. 2017;61(2):175–182. doi: 10.4149/av_2017_02_06. [DOI] [PubMed] [Google Scholar]
- 33.Dogonyaro B.B, Bosman A.M, Sibeko K.P, Venter E.H, van Vuuren M. Genetic analysis of the VP2-encoding gene of canine parvovirus strains from Africa. Vet. Microbiol. 2013;165(3-4):460–465. doi: 10.1016/j.vetmic.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 34.Tsao J, Chapman M.S, Agbandje M, Keller W, Smith K, Wu H, Luo M, Smith T.J, Rossmann M.G, Compans R.W. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251(5000):1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- 35.Hueffer K, Parker J.S, Weichert W.S, Geisel R.E, Sgro J.Y, Parrish C.R. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 2003;77(3):1718–1726. doi: 10.1128/JVI.77.3.1718-1726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H, Wang J, Jiang Y, Cheng Y, Lin P, Zhu H, Han G, Yi L, Zhang S, Guo L, Cheng S. Typing of canine parvovirus strains circulating in North-East China. Transbound. Emerg. Dis. 2017;64(2):495–503. doi: 10.1111/tbed.12390. [DOI] [PubMed] [Google Scholar]
- 37.Chiang S.Y, Wu H.Y, Chiou M.T, Chang M.C, Lin C.N. Identification of a novel canine parvovirus type 2c in Taiwan. J. Virol. 2016;13(1) doi: 10.1186/s12985-016-0620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An D.J, Jeong W, Jeoung H.Y, Yoon S.H, Kim H.J, Park J.Y, Park B.K. Phylogenetic analysis of feline panleukopenia virus (FPLV) strains in Korean cats. Res. Vet. Sci. 2011;90(1):163–167. doi: 10.1016/j.rvsc.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Ahmed N, Riaz A, Zubair Z, Saqib M, Ijaz S, Nawaz-Ul-Rehman M.S, Al-Qahtani A, Mubin M. Molecular analysis of partial VP-2 gene amplified from rectal swab samples of diarrheic dogs in Pakistan confirms the circulation of canine parvovirus genetic variant CPV-2a and detects sequences of feline panleukopenia virus (FPV) Virol. J. 2018;15(1):45. doi: 10.1186/s12985-018-0958-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castro T.X, Costa E.M, Leite J.P, Labarthe N.V, Cubel Garcia R.C. Monitoring of canine parvovirus (CPV) strains detected in vaccinated puppies in Brazil. Res. Vet. Sci. 2011;90(2):336–340. doi: 10.1016/j.rvsc.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Shackelton L.A, Parrish C.R, Truyen U, Holmes E.C. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. U. S. A. 2005;102(2):379–384. doi: 10.1073/pnas.0406765102. [DOI] [PMC free article] [PubMed] [Google Scholar]


