Abstract
Objective
The study aimed to detect and analyze long non-coding RNAs (lncRNAs) in plasma of children diagnosed with chronic gastritis, and to explore its biological functions and involved signaling pathways.
Methods
The plasma samples were collected from six children that were diagnosed with chronic gastritis by physical examination, gastroscopy, and pathological examination and six healthy children. The plasma samples were assayed for determining the expression profiles of lncRNA based upon the gen chip detection. The specific expression of lcnRNA in plasma of children with chronic gastritis was analyzed and its biological functions were speculated.
Results
Five lncRNAs (RP11-697M17.1, RP11-388M20.9, AFAP1-AS1, BC062758, and XLOC001406) were significantly up-regulated, and five lncRNAs (UNQ697, BX571672.5, CYP4F35P, ANKRD20A5P, and AL832737) were observed to be significantly down-regulated. The lncRNAs RP11-697M17.1, and UNQ697 were detected with the highest up-regulation and down-regulation, respectively. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed that the up-regulated lncRNAs were significantly enriched in 20 signaling pathways such as phosphoinositide-3-kinase–protein kinase B (PI3K-Akt) pathway, and the down-regulated lncRNAs target genes were significantly enriched in 20 signaling pathways such as the metabolic pathway.
Conclusion
The analysis of the lncRNA expression profiles in plasma of children with chronic gastritis revealed that the lncRNA RP11-697M17.1, and lncRNA UNQ697 may act as plasma markers for predicting chronic gastritis in children.
Keywords: RP11-697M17.1, UNQ697, Chronic gastritis, Plasma markers
Introduction
Chronic gastritis is an inflammation of the stomach lining that is caused by a combination of various factors, including the Helicobacter pylori (H. pylori) infection.1, 2, 3, 4, 5, 6, 7 It can lead to various conditions, such as hernia, nausea, pain etc. In the current scenario, the common methods for the treatment of chronic gastritis include diet therapy, symptomatic treatment, and eradication of H. pylori.8, 9, 10, 11, 12, 13 However, an effective approach for the treatment of chronic gastritis is the eradication of H. pylori that can markedly reduce its recurrence rate. The long non-coding RNAs (lncRNAs) are a group of non-coding transcripts that are greater than 200 nucleotides (nt) in length but lack the protein-coding capacity. Recently, studies have reported that lncRNAs are widely implicated in various physiological and pathological processes in the body, and also involved in regulation of the progression of various diseases.14, 15, 16, 17 Specific lncRNAs, such as H19 and HOX transcript antisense RNA (HOTAIR), have been shown to play important roles in digestive tract diseases.18 For example, H19 induces the production of microRNA-675 (miR-675) by the tumor suppressor runt-related transcription factor 1, and regulates the gastric cancer cell proliferation. Moreover, the overexpression of HOTAIR may be involved in tumor escape mechanisms. These observations strongly suggest that lncRNAs are a molecular etiology of digestive tract diseases. Notably, the circulating lncRNAs have been detected as novel biomarkers for the digestive tract diseases, which is promising for the monitoring of development and progression of digestive tract diseases, and screening of patients.19, 20, 21
The plasma RNA has been identified as a novel non-invasive diagnostic biomarker.22 It has been reported that the circulating RNA in the blood is encapsulated within the vesicles, such as exosomes enabling it to persist for a longer time. Furthermore, it is more heterogeneous than endoscopy for detection of the disease. Previously, the circulating biomarkers have been screened mainly in adults for the detection of digestive tract diseases. In the present study, we report the analysis of expression profile of lncRNAs and screening of specific lncRNA markers in the plasma of children that were diagnosed with chronic gastritis.
Methods
Ethical approval
The study was approved by the ethics committee of Shanghai Pudong New District Zhoupu Hospital. All clinical practices and observations were conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from each patient before the study was conducted.
Patients
The study included six children (8.0–12.0 years old) from Shanghai Pudong New District Zhoupu Hospital from December 2018 to August 2019. The patients were selected for the study on the basis of clinical symptoms, physical examination, gastroscopy, and pathological examination of chronic gastritis. Additionally, six blood samples were collected as control group from children that were not diagnosed with chronic gastritis by the same clinical examination.
Plasma sample collection and preservation
The plasma samples were collected from six children that were diagnosed with chronic gastritis, and six healthy children. The samples were collected in BD EDTA (Becton Dickinson [BD] Ethylenediaminetetraacetic acid) tubes. The plasma samples were initially centrifuged at a speed of 1500×g at 4 °C for 30 min, and then at 3000×g at 4 °C for 10 min. The supernatant was collected for each sample and stored in TRIzol® reagent (Qiagen, Inc. Valencia, California, USA) at −80 °C.
Total RNA extraction and reverse transcription
Total RNA was extracted from the plasma samples using the miRNeasy Serum/Plasma kit (REF 217184, Germany) according to the manufacturer's instructions. Total RNA concentration and purity were measured using the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific). Total RNA was then dissolved in RNase-free water and then immediately used for reverse transcription using the PrimeScript RT-PCR kit (REF RR036A, Japan) according to the manufacturer's instructions.
lncRNA gene chip detection
Twelve plasma samples were assayed for determining the expression profiles of lncRNA using the Agilent Human LncRNA Array v3.0 (8 × 60K, Arraystar, USA).
GO and KEGG analysis
Gene ontology was classified into three groups by Gene Ontology (GO) database: BP (biological process), CC (cell component), and MF (molecular function). KEGG pathway analysis was performed to determine the involvement of differentially expressed lncRNA genes in different biological pathways. The cut off P value (hypergeometric P value) was set at 0.05.
Statistical analysis
Numerical data were expressed as mean ± standard deviation (SD) and measured in triplicates. The comparison between the statistically normalized data sets was performed using the t-test or the Mann-Whitney U test. For statistical significance, P value was set at 0.05 and the confidence level was set at 95%.
Results
Differentially expressed lncRNAs in chronic gastritis
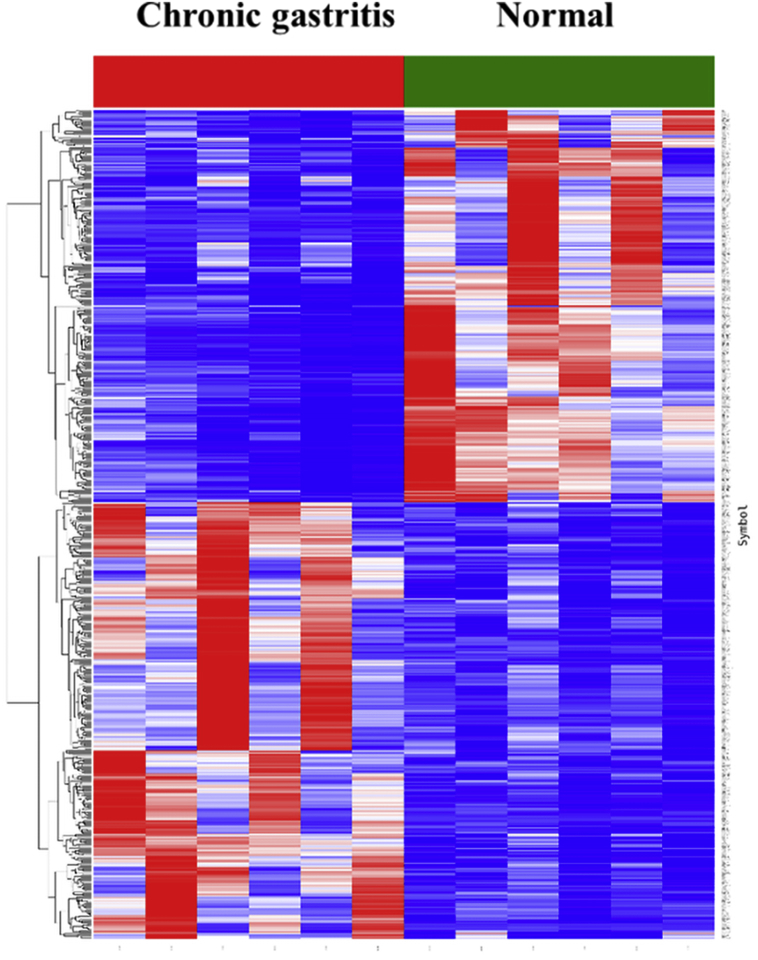
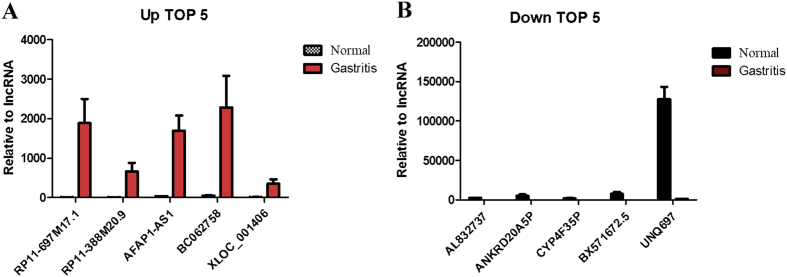
In gene chip detection, a specific lncRNA expression profile was obtained in children with chronic gastritis. A differential expression pattern of 2053 lncRNAs was observed in children with chronic gastritis (fold change ≥ 2.0, P ≤ 0.05) as compared to the healthy control group. Out of these, 934 and 1119 lncRNAs were significantly up-regulated and down-regulated, respectively in diseased children as compared to the healthy control group (Fig. 1). Among these, five lncRNAs (RP11-697M17.1, RP11-388M20.9, AFAP1-AS1, BC062758, and XLOC001406) were observed to be significantly up-regulated (Table 1), and five lncRNAs (UNQ697, BX571672.5, CYP4F35P, ANKRD20A5P, and AL832737) were observed to be significantly down-regulated (Table 2). The lncRNAs RP11-697M17.1 (Fig. 2A) and UNQ697 (Fig. 2B) were identified with the highest up-regulation and down-regulation, respectively.
Fig. 1.
Expression profiles of specific lncRNAs in children with chronic gastritis. 934 lncRNAs were significantly up-regulated and 1119 lncRNAs were significantly down-regulated.
Table 1.
Up-regulated lncRNAs in plasma of children with chronic gastritis.
| Symbols | Fold change | P | Threshold |
|---|---|---|---|
| RP11-697M17.1 | 188.73 | 0.029 | Up |
| RP11-388M20.9 | 76.68 | 0.032 | Up |
| AFAP1-AS1 | 50.07 | 0.0075 | Up |
| BC062758 | 43.63 | 0.039 | Up |
| XLOC_001406 | 34.32 | 0.026 | Up |
Table 2.
Down-regulated lncRNAs in children with chronic gastritis.
| Symbols | Fold change | P | Threshold |
|---|---|---|---|
| AL832737 | 0.035 | 3.8E-05 | Down |
| ANKRD20A5P | 0.027 | 0.04 | Down |
| CYP4F35P | 0.025 | 0.027 | Down |
| BX571672.5 | 0.017 | 0.025 | Down |
| UNQ697 | 0.0089 | 0.00049 | Down |
Fig. 2.
Five lncRNAs (RP11-697M17.1, RP11-388M20.9, AFAP1-AS1, BC062758, and XLOC001406) were significantly up-regulated (A), and 5 lncRNAs (UNQ697, BX571672.5, CYP4F35P, ANKRD20A5P, and AL832737) were significantly down-regulated (B).
GO annotation and KEGG enrichment of differential genes
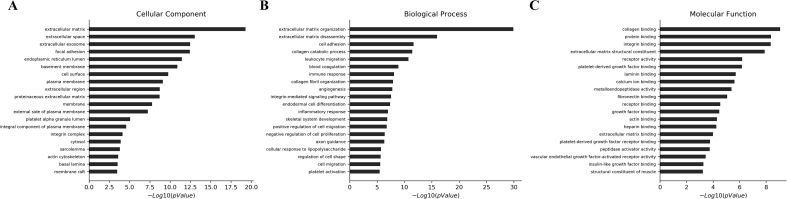
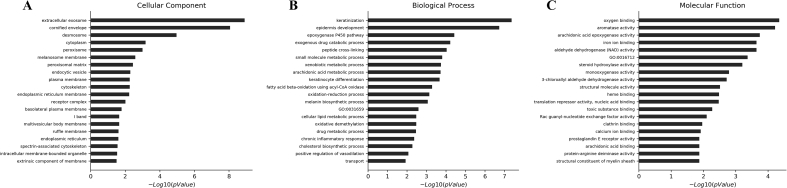
The information about GO functions was used to annotate 934 up-regulated and 1119 down-regulated genes, and each GO group was enriched separately. The results showed that 2683 up-regulated lncRNAs were enriched in BP, 435 were enriched in CC, and 675 were enriched in MF (P < 0.05). The GO analysis showed that 1418 down-regulated genes were enriched in BP, 261 were enriched in CC, and 428 were enriched in MF (P < 0.05). The functions of top 20 up-regulated and down-regulated genes are described in Fig. 3, Fig. 4, respectively.
Fig. 3.
The top 20 gene functions enriched in up-regulated lncRNAs in BP (biological processes), CC (cell components) and MF (molecular function) (P < 0.05).
Fig. 4.
The top 20 gene functions enriched in down-regulated lncRNAs in BP (biological processes), CC (cell components) and MF (molecular function) (P < 0.05).
KEGG signal pathway analysis
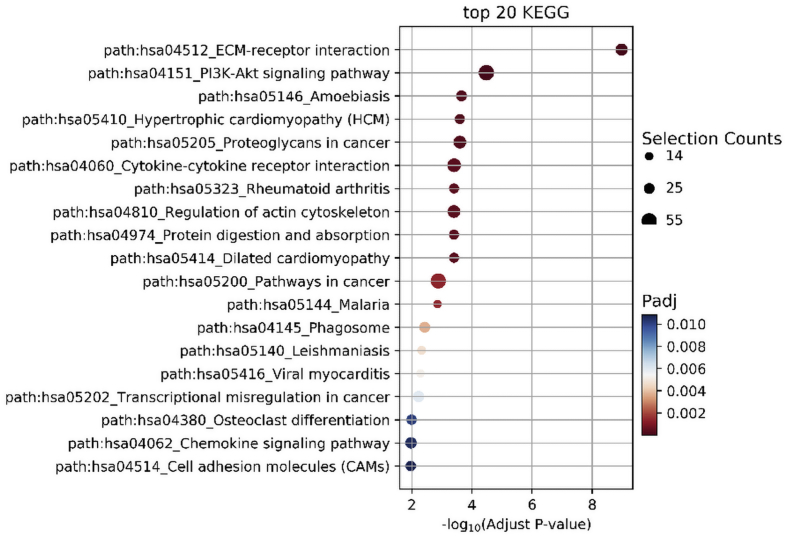
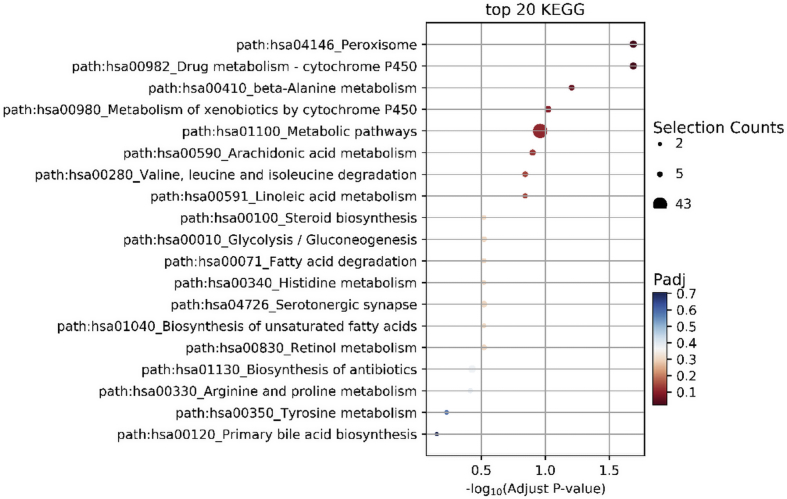
Based on the classification of GO annotations, KEGG pathway enrichment analysis was done for all identified 934 up-regulated and 1119 down-regulated genes. The up-regulated lncRNAs were mainly enriched in 20 signaling pathways, such as PI3K-Akt (Fig. 5) and the down-regulated lncRNAs target genes are significantly enriched in 20 signaling pathways, such as metabolic pathways (Fig. 6).
Fig. 5.
Up-regulated lncRNAs are significantly enriched to the PI3K-Akt signaling pathway.
Fig. 6.
Down-regulated lncRNAs target genes are significantly enriched into Metabolic signaling pathway.
Discussion
So far, the gold standard for the diagnosis of chronic gastritis still depends upon the endoscopy and histo-pathological evaluation.23 Various studies have shown that RNA in the blood plays an important role in the processes of cell-to-cell communication i.e. the inflammatory reactions, and tissue regeneration.24, 25, 26, 27 Circulating RNA in the blood may act as a potential biomarker for various diseases, which can be more heterogeneous than endoscopy in detecting various diseases.28, 29, 30, 31, 32, 33, 34
In the present study, we have determined the specific expression profiles of lncRNAs in plasma of children with chronic gastritis and normal healthy children. The results showed that 2053 lncRNAs were specifically expressed in children with chronic gastritis. It was determined for the first time that the lncRNA RP11-697M17.1, BC062758, XLOC001406, lncRNAs UNQ697, BX571672.5, CYP4F35P, ANKRD20A5P and AL832737. In addition, lncRNA RP11-388M20.9 and lncRNA AFAP1-AS1 have been reported. It found that lncRNA RP11-388M20.9 were significantly up-regulated with increasing concentrations of Cr(VI) in human bronchial epithelial cells. In gastric cancer, overexpression of lncRNA AFAP1-AS1 promotes cell proliferation and invasion.
Studies have found that PI3K/Akt is the main signaling pathway in gastrointestinal diseases gastrointestinal disorders. Helicobacter pylori (HP) mediated PI3K/AKT/GSK3β signal pathways in the occurrence of gastric cancer. In addition H. pylori causes a significant increase of PI3K/Akt in gastritis. We know that metabolic activities are also very rich in gastritis. So bioinformatics predicts that metabolic pathways are enriched more, which is consistent with previous reports. In the present study, it has been detected that lncRNA RP11-697M17.1, and lncRNA UNQ697 were significantly expressed in gastritis patients as compared to the healthy control group. Moreover, that the bioinformatics analysis revealed that the lncRNA RP11-697M17.1, and lncRNA UNQ697 might participate in PI3K-Akt and metabolic pathways. Further, it was speculated in our study that the lncRNA RP11-697M17.1 and lncRNA UNQ697, may be involved in regulation rather than protein-coding.
One main limitation of the present study is the small sample size, where only six samples of children with chronic gastritis were included for the analysis. Further validation experiments should be performed using the large patient population to explore the role of circulating lncRNAs in gastritis disease.
Conclusively, the plasma lncRNA RP11-697M17.1, and lncRNA UNQ697 are the most promising biomarkers for the diagnosis of chronic gastritis in children.
Conflict of interest
None.
Funding
This work was supported by a grant from Science and Technology Development Fund of Shanghai Pudong New Area (PKJ2016-Y12).
Edtied by Yan-Gang Ren and Yi Cui
Footnotes
Peer review under responsibility of Chinese Medical Association.
References
- 1.Bacha D., Walha M., Ben Slama S. Chronic gastritis classifications. Tunis Med. 2018;96:405–410. [PubMed] [Google Scholar]
- 2.Zhang F., Wang F., Chen C. Prediction of progression of chronic atrophic gastritis with Helicobacter pylori and poor prognosis of gastric cancer by CYP3A4. J Gastroenterol Hepatol. 2019 doi: 10.1111/jgh.14844. [DOI] [PubMed] [Google Scholar]
- 3.Efficacy and safety of acupuncture therapy for chronic atrophic gastritis: a meta-analysis and trial sequential analysis protocol: Erratum. Medicine (Baltim) 2019;98 doi: 10.1097/MD.0000000000017514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castaneda C.A., Castillo M., Chavez I. Prevalence of Helicobacter pylori infection, its virulent genotypes, and Epstein-Barr virus in Peruvian patients with chronic gastritis and gastric cancer. J Glob Oncol. 2019;5:1–9. doi: 10.1200/JGO.19.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandra P., Nath S., Jain D. Pancreatitis, cholangitis, and gastritis: the triumvirate of immunoglobulin G4-related disease identified simultaneously on 18F-fluorodeoxyglucose positron emission tomography/computed tomography. Indian J Nucl Med. 2019;34:335–337. doi: 10.4103/ijnm.IJNM_110_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins K., Mnayer L., Shen P. Burkitt-like lymphoma with 11q aberration. Clin Case Rep. 2019;7:1823–1824. doi: 10.1002/ccr3.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dohi O., Majima A., Naito Y. Can image-enhanced endoscopy improve the diagnosis of Kyoto classification of gastritis in the clinical setting? Dig Endosc. 2019;32:191–203. doi: 10.1111/den.13540. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Castro K.I., Franceschi M., Noto A. Clinical manifestations of chronic atrophic gastritis. Acta Biomed. 2018;89:88–92. doi: 10.23750/abm.v89i8-S.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J.A., Tian Y.L., Cong L., Fan S., Duan L.W. Endoscopic submucosal dissection for treating gastritis cystica profunda. J Biol Regul Homeost Agents. 2019;33:1577–1580. doi: 10.23812/19-190-L. [DOI] [PubMed] [Google Scholar]
- 10.Liu X.Q., Wang L. Effect of modified Zhengqi Powder in treating chronic gastritis and on patients' life quality and inflammatory factors (in Chinese) China J Chin Mater Med. 2019;44:181–185. doi: 10.19540/j.cnki.cjcmm.2019.0004. [DOI] [PubMed] [Google Scholar]
- 11.Liu X.F., Li DG. Liu J.P., Du Y.R., Bai H.Y. Medicine-syndrome research and analysis of professor Li Dian-gui in treating chronic atrophic gastritis with intestinal metaplasia (in Chinese) China J Chin Mater Med. 2017;42:1792–1796. doi: 10.19540/j.cnki.cjcmm.20170224.013. [DOI] [PubMed] [Google Scholar]
- 12.Wei W., Yang Y., Shi H.X. Status, challenges, and prospects of treating chronic atrophic gastritis by Chinese medical diagnosis and treatment (in Chinese) Chin J Integr Tradit West Med. 2015;35:1424–1426. [PubMed] [Google Scholar]
- 13.Meltzer A.C., Winter L.E., Kulie P. Treating gastritis, peptic ulcer disease, and dyspepsia in the emergency department: the feasibility and patient-reported outcomes of testing and treating for Helicobacter pylori infection. Ann Emerg Med. 2015;66:131–139. doi: 10.1016/j.annemergmed.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Pyfrom S.C., Luo H., Payton J.E. PLAIDOH: a novel method for functional prediction of long non-coding RNAs identifies cancer-specific LncRNA activities. BMC Genomics. 2019;20:137. doi: 10.1186/s12864-019-5497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian W., Cai X., Qian Q. lncRNA ZEB1-AS1 promotes pulmonary fibrosis through ZEB1-mediated epithelial-mesenchymal transition by competitively binding miR-141-3p. Cell Death Dis. 2019;10:129. doi: 10.1038/s41419-019-1339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye Z.M., Yang S., Xia Y.P. LncRNA MIAT sponges miR-149-5p to inhibit efferocytosis in advanced atherosclerosis through CD47 upregulation. Cell Death Dis. 2019;10:138. doi: 10.1038/s41419-019-1409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L., Wang Y., Zhang L. ZBTB7A, a miR-663a target gene, protects osteosarcoma from endoplasmic reticulum stress-induced apoptosis by suppressing LncRNA GAS5 expression. Cancer Lett. 2019;448:105–116. doi: 10.1016/j.canlet.2019.01.046. [DOI] [PubMed] [Google Scholar]
- 18.Lin Y., Xu L., Wei W., Zhang X., Ying R. Long noncoding RNA H19 in digestive system cancers: a meta-analysis of its association with pathological features. BioMed Res Int. 2016;2016:4863609. doi: 10.1155/2016/4863609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S., Du L., Wang L. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23:1396–1405. doi: 10.1111/jcmm.14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C., Yang X., Qi Q., Gao Y., Wei Q., Han S. lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 2018;21:651–659. doi: 10.3233/CBM-170727. [DOI] [PubMed] [Google Scholar]
- 21.Shen X., Zhang Y., Wu X. Upregulated lncRNA-PCAT1 is closely related to clinical diagnosis of multiple myeloma as a predictive biomarker in serum. Cancer Biomark. 2017;18:257–263. doi: 10.3233/CBM-160158. [DOI] [PubMed] [Google Scholar]
- 22.Cai Y., Yang Y., Chen X. Circulating 'lncRNA OTTHUMT00000387022' from monocytes as a novel biomarker for coronary artery disease. Cardiovasc Res. 2016;112:714–724. doi: 10.1093/cvr/cvw022. [DOI] [PubMed] [Google Scholar]
- 23.Cao Z.M., You W.Z., Chen H.B. A long-term follow-up study with endoscope in chronic gastritis (in Chinese) Chin J Intern Med. 1993;32:743–745. [PubMed] [Google Scholar]
- 24.Simonian M., Mosallayi M., Mirzaei H. Circulating miR-21 as novel biomarker in gastric cancer: diagnostic and prognostic biomarker. J Cancer Res Ther. 2018;14:475. doi: 10.4103/0973-1482.175428. [DOI] [PubMed] [Google Scholar]
- 25.Xie J.F., Xie N.S., Tang J.H., Gu Q.P., Luo Q.T. Circulating SHIP2 mRNA as a novel biomarker in the diagnosis and prognosis of gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21:5129–5134. doi: 10.26355/eurrev_201711_13829. [DOI] [PubMed] [Google Scholar]
- 26.Wu J., Li G., Wang Z. Circulating MicroRNA-21 is a potential diagnostic biomarker in gastric cancer. Dis Markers. 2015;2015:435656. doi: 10.1155/2015/435656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valladares-Ayerbes M., Reboredo M., Medina-Villaamil V. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med. 2012;10:186. doi: 10.1186/1479-5876-10-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penyige A., É Márton, Soltész B. Circulating miRNA profiling in plasma samples of ovarian cancer patients. Int J Mol Sci. 2019;20:4533. doi: 10.3390/ijms20184533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwee L.C., Neely M.L., Grass E. Associations of osteopontin and NT-proBNP with circulating miRNA levels in acute coronary syndrome. Physiol Genom. 2019;51:506–515. doi: 10.1152/physiolgenomics.00033.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan H.N., Ho S.L., He D., Li H.W. Direct and sensitive detection of circulating miRNA in human serum by ligase-mediated amplification. Talanta. 2020;206:120217. doi: 10.1016/j.talanta.2019.120217. [DOI] [PubMed] [Google Scholar]
- 31.Hao Q.Q., Wang Q.H., Xia W., Qian H.Z. Circulating miRNA expression profile and bioinformatics analysis in patients with occult hepatitis B virus infection. J Med Virol. 2020;92:191–200. doi: 10.1002/jmv.25594. [DOI] [PubMed] [Google Scholar]
- 32.Zhou X., Fang S., Wang M. Diagnostic value of circulating miRNA-122 for hepatitis B virus and/or hepatitis C virus-associated chronic viral hepatitis. Biosci Rep. 2019;39 doi: 10.1042/BSR20190900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swellam M., Ezz El Arab L., Al-Posttany A.S., B Said S. Clinical impact of circulating oncogenic MiRNA-221 and MiRNA-222 in glioblastoma multiform. J Neuro Oncol. 2019;144:545–551. doi: 10.1007/s11060-019-03256-2. [DOI] [PubMed] [Google Scholar]
- 34.Santos A.S., Cunha Neto E., Fukui R.T., Ferreira L., Silva M. Increased expression of circulating microRNA 101-3p in type 1 diabetes patients: New insights into miRNA-regulated pathophysiological pathways for type 1 diabetes. Front Immunol. 2019;10:1637. doi: 10.3389/fimmu.2019.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]