Abstract
Staphylococcus aureus lipase (SAL), a triacylglycerol esterase, is an important virulence factor and may be a therapeutic target for infectious diseases. Herein, we determined the 3D structure of native SAL, the mutated S116A inactive form, and the inhibitor complex using the anti-obesity drug orlistat to aid in drug development. The determined crystal structures showed a typical α/β hydrolase motif with a dimeric form. Fatty acids bound near the active site in native SAL and inactive S116A mutant structures. We found that orlistat potently inhibits SAL activity, and it covalently bound to the catalytic Ser116 residue. This is the first report detailing orlistat–lipase binding. It provides structure-based information on the production of potent anti-SAL drugs and lipase inhibitors. These results also indicated that orlistat can be repositioned to treat bacterial diseases.
Subject terms: X-ray crystallography, Biochemistry, Structural biology, Biophysics, Molecular biophysics, Molecular conformation
Introduction
Staphylococcus aureus (SA) can cause skin, throat, and digestive tract infections. SA is a native bacterium that resides in the nasal cavity, respiratory tract, and skin, and infection typically results in a localized collection of pus1,2. SA is related to several diseases and produces various types of pathogenic toxins3. Methicillin-resistant SA (MRSA) is one of the most well-known “super bugs,” which has developed resistance to almost all current antibiotics4,5. It is a common nosocomial infection that presents a serious risk to those with weak immune systems such as children and the elderly6. Identifying novel, effective drugs is of great importance for treating MRSA-related diseases.
SA lipase (SAL, also known as glycerol ester hydrolase), a triacylglycerol esterase (Fig. 1A), shows strong cytopathic activity in host cells7. SAL is a potential target of anti-SA drugs in skin diseases8. SAL degrades lipids, which attack pathogenic bacteria9,10. By reducing the effect of immune-responsive lipids, SA colonization increases on the skin surface or inside the body. It has also been reported that some inhibitors such as farnesol decrease SA production11. Recently, Chen et al. observed that SAL released by SA inhibits the activation of innate immune cells and interferes with the host immune system to affect innate immune recognition in the microbe12.
Figure 1.
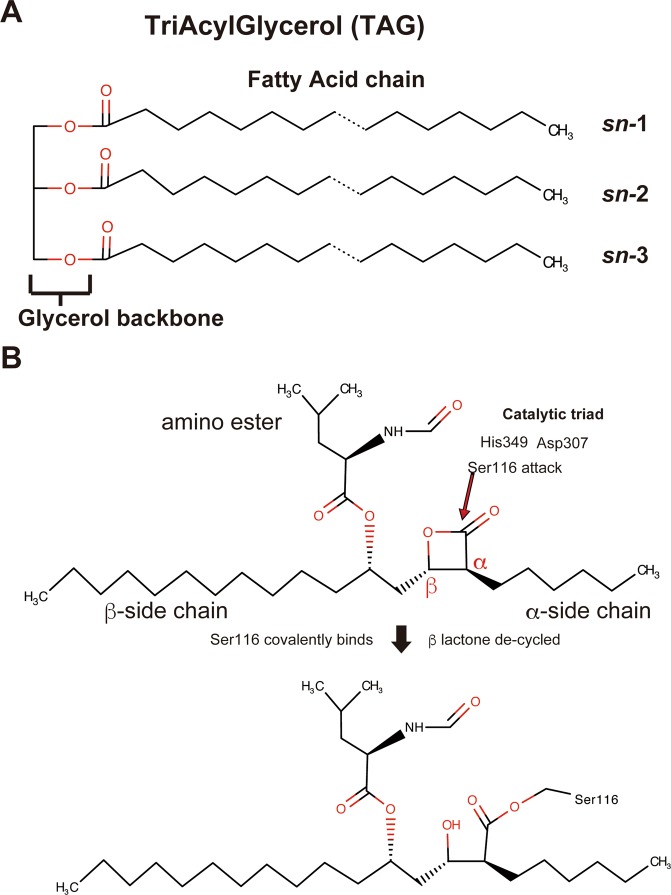
Chemical structures of triacylglycerol and orlistat. Triacylglycerol structure comprises one glycerol molecule and three fatty acids. (B) Proposed mechanism of orlistat binding. The catalytic triad His349, Asp307, and Ser116 attack orlistat. β-Lactone is hydrolyzed by Ser116 of SAL. The oxygen atom of Ser116 is covalently bound to the carbonyl carbon atom.
SAL, an ester bond-hydrolyzing enzyme, is secreted in prepolypeptide form containing 690 amino acid residues and is processed into its mature form containing 394 amino acid residues. Its active site is composed of a typical catalytic triad including Ser116, Asp 307, and His349 residues, similar to other serine hydrolases.
Very few specific SAL-related bacterial lipase inhibitors have been identified to date10,11; tetracycline is a known inhibitor of Staphylococcus epidermidis lipase13. An example of a SAL inhibitor is farnesol, which is a competitive inhibitor11. However, this monoterpenoid weakly inhibits SAL (IC50 value, 0.57 mM). These inhibitors were not so potent against SAL-related bacterial lipases. We screened various chemical libraries and discovered that the anti-obesity drug orlistat is a potent SAL inhibitor with an IC50 value of 2.4 μM.
Orlistat belongs to the lipstatin family, derived from the gram-positive bacterium Streptomyces toxytricini14. The IUPAC name of orlistat is (2S)-1-[(2S,3S)-3-hexyl-4-oxooxetan-2-yl]tridecan-2-yl (2S)-4-methyl-2-(2-oxoethyl)pentanoate (Fig. 1B). Its Y-shaped chemical structure is divided into three fragments: amino ester moiety (N-formyl- L-leucine substituent extending off the C5 carbon atom), α-heptyl alkyl tail, and long β-tetradecanyl alkyl chain (Fig. 1B)14,15. Orlistat is a potent inhibitor of human gastric and pancreatic lipases that play an important role in dietary fat digestion. It is currently available in the market as an anti-obesity drug (Xenical or Alli), which acts locally to block gastric lipases that are crucial for the digestion of long-chain triglycerides15. However, there is no detailed structural information on how orlistat molecules bind to these lipases.
We discovered that orlistat is a potent SAL inhibitor at µM concentrations, and we succeeded in obtaining its crystal structure complex. Herein, we also reported 3D structures of native SAL, the inactive S116A mutant, and SAL–orlistat complex. Based on this binding of SAL with orlistat, we predicted the binding manner of orlistat with mammalian gastric lipase. These structures will help in the development of anti-SAL drugs.
Results
Structure determination of native SAL, inactive S116A mutant, and SAL–orlistat complex
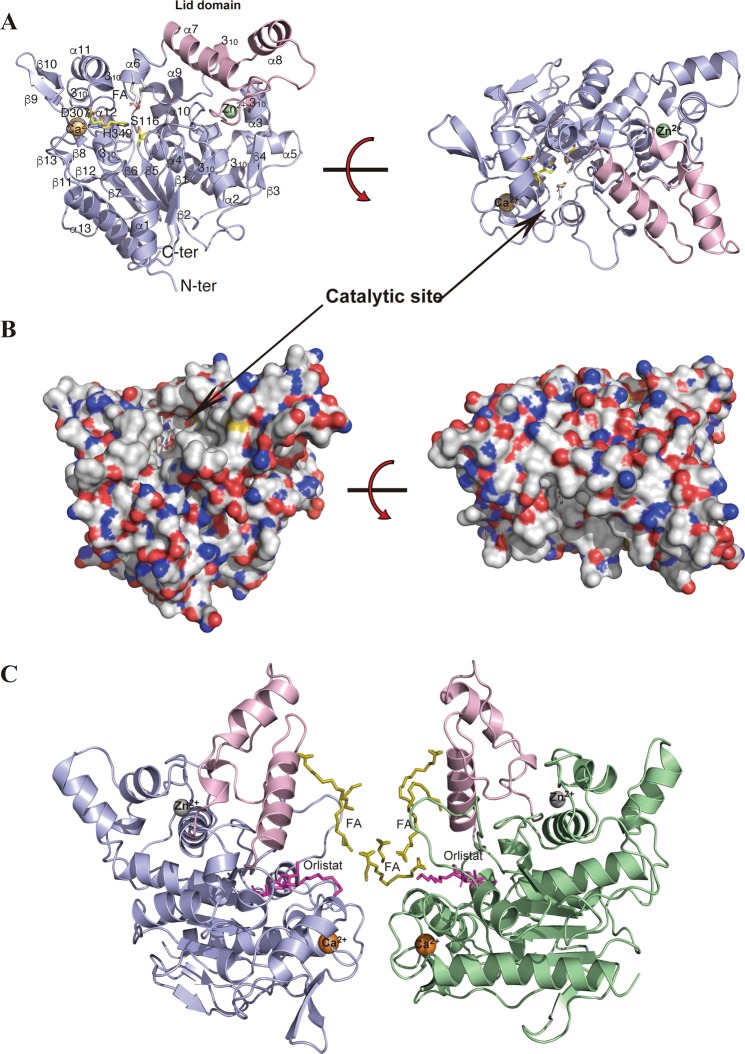
This report describes the crystal structures of native SAL, inactive S116A mutant, and SAL–orlistat complex at resolutions of 2.08, 2.6, and 2.23 Å, respectively (Table 1). The crystal structure of native SAL was determined using MR techniques. The Staphylococcus hyicus lipase (SHL) chain A of 2hih16, which shares 47.7% identity with that of SAL, was used as a search model using the CCP4 Phaser program17,18. The MR solution automatically allowed unequivocal tracing of the two defined chains A and B (each chain with amino acid residues 4–385 was unambiguously determined with the exception of the His-tagged linker peptides N-terminus 1–3 and C-terminus 386–394, which are not visible) and was subsequently refined at a resolution of 2.08 Å using REFMAC519. Five percent of the unique reflections were used to monitor the free R-factor. Final values of the general R-factor and free R-factor up to a resolution of 2.08 Å were 21.3% and 23.5%, respectively (Table 1). The dimeric refined model comprised 764 amino acids with 76 solvent molecules with 2 zinc and 2 calcium ions and 12 fatty acid fragments of various lengths (Fig. 2A–C).
Table 1.
Data collection, processing, and refinement.
| Native SAL | Inactive S116A mutant | SAL–orlistat complex | |
|---|---|---|---|
| PDB ID | 6KSI | 6KSL | 6KSM |
| Diffraction source | BL44XU/SPring-8 | BL44XU/SPring-8 | BL44XU/SPring-8 |
| Wavelength (Å) | 0.90 | 0.90 | 0.90 |
| Temperature (K) | 100 | 100 | 100 |
| Detector | Eiger X 16M | Eiger X 16M | Eiger X 16M |
| Crystal–detector distance (mm) | 300 | 380 | 300 |
| Rotation range per image (°) | 0.1 | 0.1 | 0.1 |
| Total rotation range (°) | 180 | 180 | 180 |
| Exposure time per image (s) | 0.1 | 0.1 | 0.1 |
| Space group | P4122 | P6122 | P4122 |
| a, b, c (Å) | 129.9, 129.9, 251.2 | 164.2, 164.2, 233.1 | 132.5, 132.5, 248.2 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 |
| Mosaicity (°) | 0.044 | 0.063 | 0.049 |
| Resolution range (Å) | 50–2.08 (2.21–2.08) | 50–2.59 (2.75–2.59) | 50–2.23 (2.37–2.23) |
| Total No. of reflections | 1761937 | 1155509 | 1467858 |
| No. of unique reflections | 246153 | 109044 | 205262 |
| Completeness (%) | 99.9 (99.3) | 99.8 (98.7) | 99.7 (98.5) |
| Redundancy | 7.2 (7.1) | 10.6 (10.4) | 7.2 (6.6) |
| 〈I/σ(I)〉 | 11.6 (0.7) | 16.8 (0.9) | 10.6 (0.7) |
| Rmeas | 0.089 (2.566) | 0.083 (2.138) | 0.125 (2.573) |
| CC1/2 | 0.999 (0.481) | 0.999 (0.523) | 0.999 (0.424) |
| Refinement | Refmac | Refmac | Refmac |
| Resolution range (Å) | 50-2.08 | 50-2.59 | 50-2.23 |
| R-factor/free R-factor (%) | 21.3/23.5 | 21.3/24.6 | 20.8/24.0 |
| No. of atoms | 6246 | 6110 | 6266 |
| No. of solvent atoms | 78 | 12 | 62 |
| No. of ligands | 16 | 9 | 13 |
| Ramachandran distribution (% favored, allowed, outlier) | 98.0, 2.0, 0 | 95.0, 5.0, 0 | 96.8, 3.2, 0 |
| RMS bonds (Å), angles (°) | 0.028, 2.91 | 0.020, 2.36 | 0.026, 2.69 |
| Average B value (Å2) | 47.1 | 65.0 | 45.8 |
Values for the outer shell are given in parentheses. Completeness for all reflections and for the highest-resolution shell in parentheses.
Figure 2.
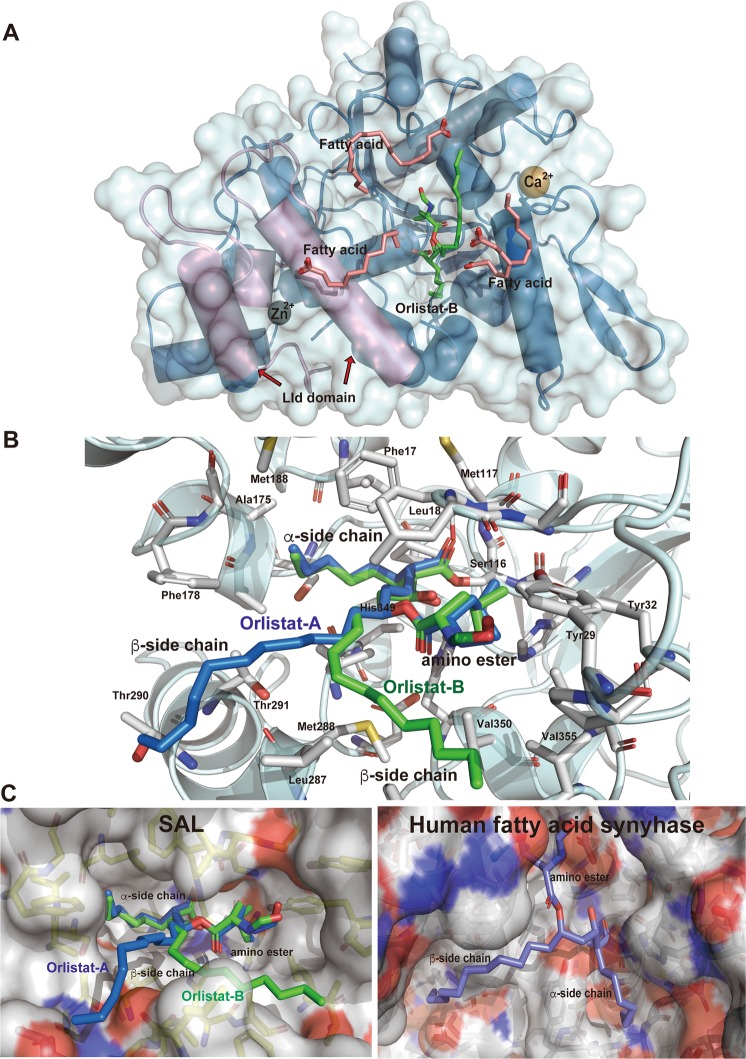
Crystal structure of native SAL and the SAL–orlistat complex. (A) Schematic of the SAL structure. α-Helices and β-strands are shown in blue; the lid domain is shown in pink. Fatty acid fragment molecules are shown as sticks. The calcium and zinc ions are shown in orange and green, respectively. (B) Molecular surface depiction of native SAL with positively charged areas shown in blue and negatively charged areas in red. (C) The dimeric form of the SAL complex with orlistat and fatty acids. Each monomer is shown in blue (chain A) and green (chain B), respectively. The lid domain is shown in pink. The orlistat and fatty acid fragment molecules are shown as sticks in magenta and yellow, respectively. All images of molecular structures were prepared using PyMOL (http://www.pymol.org/2).
Stereochemistry checks indicated that the refined models were in good agreement with the expectations with these models within this resolution range (Table 1). The asymmetric unit contained two SAL molecules (Fig. 2C).
Structures of Inactive S116A mutant and SAL–orlistat complex were also determined with the MR technique using the CCP4 Phaser program18. The SAL–orlistat complex, which was isomorphous with native SAL, showed strong positive electron density maps of orlistat molecules near the active sites of chains A and B. The crystallization condition of the S116A mutant crystal was different than the native SAL crystal, with the former displaying a hexagonal system and with two molecules in the asymmetric unit.
Overall structure
The SAL crystal structure exhibited a typical α/β-hydrolase fold comprising a central twisted β-sheet flanked by α-helices on both sides. The refined model comprised 13 β-strands, 13 α-helices, and 6 310 helices (Figs. 2A and 3A). The SAL crystal structure contained a conserved catalytic triad (Ser116, His349, and Asp307) (Fig. 3A), which was located at the concaved cleft (Fig. 2A). SAL had a unique shape like a cow face with two metal eyes and two ears formed by β-strands and α-helices with a deep central cavity as the catalytic site (Fig. 2A,B). The cavity within the substrate-binding domain contained two large hydrophobic alkyl chain-binding pockets and a shallow, more polar third pocket with a Ser116 catalytic center capable of binding a fatty acid or lactone ester (Fig. 2B).
Figure 3.
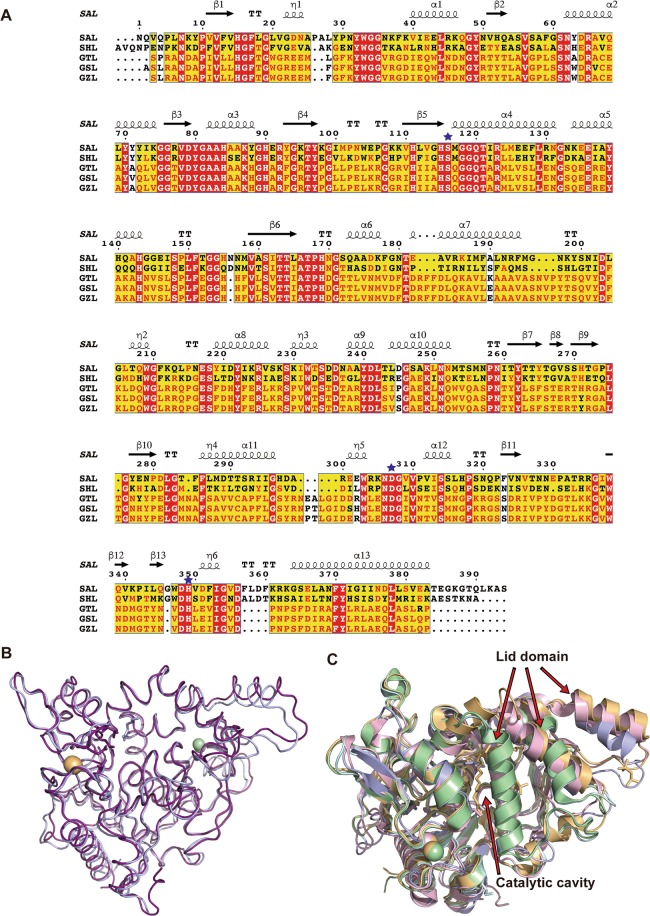
(A) Sequence alignments of SAL, SHL, GTL, GSL, and GZL. Multiple sequence alignment of SAL and these structural homologous lipases was performed with ClustalW, followed by the ESPRIT program. The secondary structure elements of SAL are given above their corresponding sequences. Arrows and springs above the sequences indicate β-strand and helical conformations, respectively. The catalytic triad residues are marked with an asterisk. (B) Superimposition of SAL and SHL structures are shown in blue and pink, respectively. (C) Structural comparison of SAL with other homologous lipases. SAL, SHL, and GTL are shown in their open-lid form in blue, pink, and orange, respectively. GSL and GZL are shown in the closed form in green and cyan, respectively. Fatty acid fragment molecules are shown as sticks. The lids of GSL and GZL overlap with the position where fatty acids are localized in SAL.
Catalytic triad
Ser116, a catalytic residue, is important for cleaving substrates and has the same catalytic mechanism as other serine hydrolases. The nucleophile Ser116 within the consensus GXSXG lipase motif was located at the “β-elbow” loop between β-strand 5 and α-helix 4 of the α/β hydrolase core20. The backbone nitrogen atoms Met117 and Phe17 formed an oxyanion hole that can stabilize the intermediate transition state during catalytic reactions. Ser116 was located at the bottom of an approximately 11-Å-deep and 23-Å-wide cavity. The entrance of this cavity was formed by hydrophobic clusters (Phe17, Leu18, Ty29, Pro30, Phe178, Met188, Leu287, Phe286, Val350, and Val355).
Metal-binding site
The zinc- and calcium-biding sites of lipases are highly conserved across many types of species16. The amino acid residues that recognize these metals are also highly conserved in various types of lipases. These two metal ions were located near the active site pocket opposite each other, possibly to stabilize the 3D SAL structure.
The SAL zinc ion was primarily surrounded by two histidine (His84 and His90) and two aspartate (Asp64 and Asp236) residues, showing same binding as SHL (Supplementary Fig. 1A). These four residues are highly conserved among various homologous lipases21. The two tyrosine residues (Tyr80 and Tyr88) surrounded the zinc ion with benzene rings (Supplementary Fig. 1A).
The SAL calcium ion was primarily surrounded by four aspartate residues (Asp348, Asp351, Asp356, and Asp359), similar to SHL (Supplementary Fig. 1B)16. The Gly283 carbonyl oxygen was also present at a resolution of 2.2 Å, which has the same form as SHL and other lipases16.
Dimerization complex
Similar to SHL, SAL was observed in dimeric form in the crystal structure, but the combination of dimer molecules was different than that in SHL16. In the native SAL crystal structure, dimer interactions were typically hydrophobic (Fig. 2C). There were 12 interacting residues, located near each calcium-binding site. Calcium ions were located at a distance of 16.5 Å from each dimer. Hydrophobic clusters were constructed at the loop regions surrounding calcium ions by Asp281, Leu282, Phe286, Phe357, Leu358, and Phe360 (Fig. 2C). The S116A mutant crystal had a different crystallization condition than the native crystal, as shown by the distinct dimeric form.
Remarkably, fatty acid branches were located between the dimer interface, which should stabilize the dimeric form of SAL. Fatty acids may have bound during the production of SAL in Escherichia coli cultivation and continued to bind to SAL even after the three-step chromatography. Crystal packing was stabilized by the insertion of fatty acids between molecules. Gel filtration showed that the dimeric form was stable in solution form at pH 7.5 in a physiological buffer (Supplementary Fig. 2).
Lid domain
Similar to other lipases, SAL had a “lid domain” to regulate the catalytic cavity between the open and closed forms22. The lid domain was formed from Thr181 to Thr233, and it was mainly constructed in a loop form by two α helices, i.e., helix 7 and helix 8, which were located on the surface of the SAL structure (Fig. 2A,C). The native SAL structure was observed as an open form (Fig. 2A). The SAL–orlistat complex also had an open form, with the lid region located as far as possible from the catalytic cavity (Fig. 2C). The overall structures of SAL and SHL were relatively similar, except the lid domain, which was due to the differences in crystal packing between SAL and SHL (Fig. 3B).
Figure 3C shows the overlay of various lipases in the open and closed forms23–25. In lipases with closed forms, the two lid helices shifted toward the catalytic cavity, and the catalytic cavity was completely occupied by long α-helix 7 (Fig. 3A,C)22. Remarkably, the lid of the opened form, the α-helix 8 of SAL, was approximately 13 Å outward from the closed form position, thereby allowing substrates or inhibitors to enter the catalytic cavity (Fig. 3C).
Characterization of SAL inhibition by orlistat
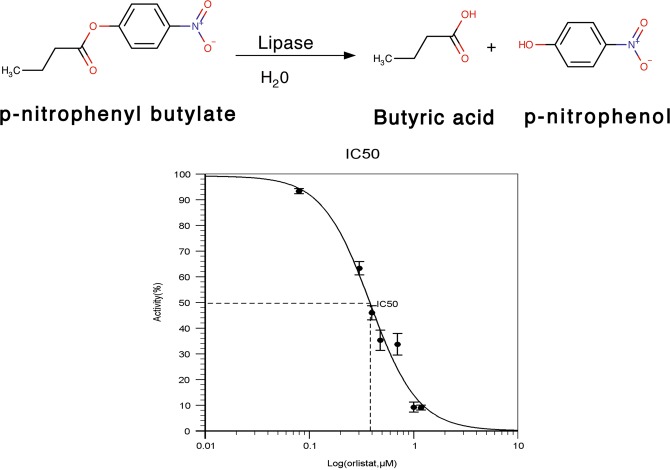
The potent inhibitors of SAL were searched via chemical libraries. We also tested commercially available lipase inhibitors. p-Nitrophenyl butyrate ester was used in an in vitro lipase assay, and the absorbance of liberated p-nitrophenol was monitored continuously at a wavelength of 405 nm. The assay revealed that lipase activity was inhibited in an orlistat dose-dependent manner, and the IC50 value was calculated as 2.4 µM (Fig. 4).
Figure 4.
Inhibition of SAL by orlistat. SAL activity toward p-nitro phenylbutyrate. Enzymatic activity was measured by monitoring p-nitrophenol production. Relative activities were measured at various inhibitor concentrations. Data are shown as mean ± s.d. (n = 3). IC50 value was determined using the IC50 calculator (https://www.aatbio.com/tools/ic50-calculator).
Inhibitor–fatty acid binding
The crystal structure of the SAL–orlistat complex showed an inhibitor-binding site and atomic-level details of binding interactions (Fig. 5A,B). Notably, the β-lactone rings of orlistat molecules were decyclized by their hydrolyzing ring cleavage reaction with SAL on chains A and B. Ser116 attached the carbonyl carbon atom of the orlistat tetracyclic rings at the active sites (Figs. 1 and 5B). Then, orlistat covalently bound to Ser116, with the oxygen atom of Ser116 and orlistat maintaining the bond. The same binding process has been shown for orlistat and the thioesterase domain of the human fatty acid synthase complex, wherein the β-lactone ring of orlistat was hydrolyzed and the carbonyl oxygen covalently bound to catalytic serine residues26. However, the mode of interaction of SAL with orlistat is different from that previously determined for human fatty acid synthase complexes (Fig. 5C).
Figure 5.
Binding interaction between SAL and orlistat. (A) Overall structure of SAL complex with orlistat and fatty acids. (B) Close-up view. The recognition mode of orlistat at the SAL catalytic site. Two orlistat molecules are overlapped in blue (chain A) and green (chain B) in the stick model. The residues, which indicate orlistat recognitions, are presented as stick models in white. (C) Orlistat-binding regions are highlighted as surface models. (left) Orlistat (blue and green sticks) and SAL complex; (right) Orlistat (blue stick) and the thioesterase domain of human fatty acid synthase complex. Positively charged areas are shown in blue, and negatively charged areas are shown in red.
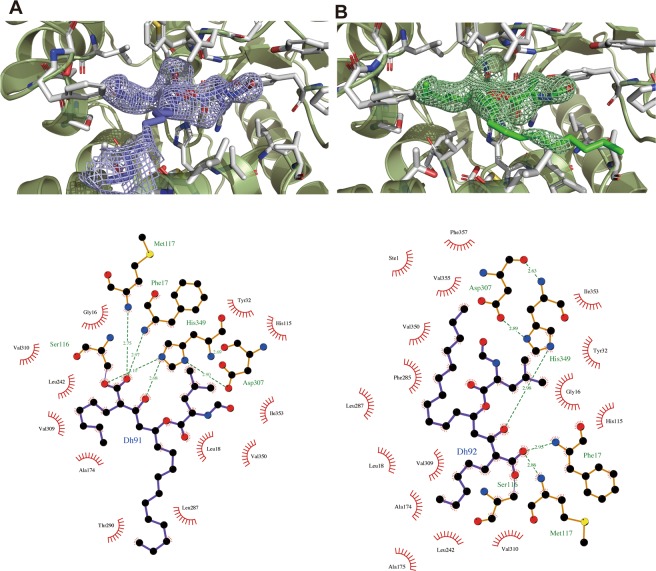
SAL monomer bound one orlistat molecule each in a similar manner. Both β-lactone rings were decyclized, but minor differences were observed in the long alkyl β-side-chain moieties (Figs. 5B, 6A,B). Seventeen data sets of the SAL–orlistat complex crystal were collected. All electron density maps for the alkyl β-side chain were not strong, indicating the flexibility of this portion. Two possible conformations of this moiety were observed (Fig. 6A,B). The Ser116-binding decyclized region and amino ester portions were tightly bound to the SAL active site, with a clear electron density map (Fig. 6A,B). The binding mode of orlistat to SAL active sites was divided into three main parts, which matched orlistat’s unique Y-shaped form (Fig. 1). Orlistat has a long β-chain and short α-chain, which are constructed by alkyl hydrophobic carbon atoms and are fitted to the hydrophobic grooves of the SAL structure (Fig. 5C). The decycled β-lactone presented as one hydroxy group and one carboxylate moiety with extended forms (Fig. 1B). The oxygen atom of the hydroxy group is hydrogen bound to the His349 side-chain nitrogen atom (2.67 Å) (Figs. 5B and 6). The Ser116 side-chain residue covalently bound to the carboxylate group and shared one oxygen atom with both SAL and orlistat (Figs. 1, 5B, and 6). The other oxygen of the decycled carboxylate group formed hydrogen bonds with two nitrogen atoms of the main chain Phe17 (2.97 Å) and Met117 (2.75 Å), which form the oxyanion hole (Fig. 6A).
Figure 6.
Omit maps of orlistat are shown for chains A and B. (A,B) The residues that indicate orlistat recognitions are presented as stick models in white. The key interactions of orlistat at the SAL active site are shown using a LIGPLOT + diagram39.
The amino ester moiety of orlistat was a modification of the leucine compound (Fig. 1B) and was located close to the enzyme in a configuration typically seen during hydrophobic contact. The leucyl moiety made hydrophobic contact with Tyr32, Il353, His115, and Val350. The terminal N-formyl carbonyl group of the amino ester moiety was located in the hydrophobic pocket formed by Leu18, Tyr29, Pro30, Tyr32, Val350, Ile353, and Val355 (Fig. 5B).
The α-side chain of orlistat was also primarily bound to a hydrophobic pocket, which was formed by Ala174, Ala176, Pro168, Leu242, Val309, Val310, and partially Phe17 at the bottom of its deep active site (Fig. 5B). Two possible conformations were observed on the β-side chain of orlistat. One was extended (in chain A), whereas the other was bent at an angle of 90° (in chain B) (Fig. 6A,B).
The β-chain of orlistat on chain A contained the hydrophobic residues Leu18, Phe178, Leu287, Thr290, Thr291, and Val309. Its terminal alkyl chain was outside the active site (Fig. 6A). The β-alkyl chain of orlistat on chain B, which was parallel to the leucyl moiety, demonstrated hydrophobic contact with Leu18, Phe285, Leu287, Val350, Val355, and Phe357 (Fig. 6B). The fatty acids of the native SAL crystal also bound at the same site. These formed two alkyl chain recognition sites and a leucyl moiety site (Fig. 6).
An inactive Ser116 mutant with Ala substitution, which lost PNPB-hydrolyzing activity, was also constructed. The S116A mutant crystal structure also bound to fatty acids near the Ser116 residue (Fig. 7A, Supplementary Fig. 3B). Wild-type S116A and S116A mutant differentially bound to fatty acids at the active site due to amino acid differences between Ser and Ala. The carboxylate moieties of fatty acids shifted closer to Ala116 in the mutant than in the wild type (Fig. 7A).
Figure 7.
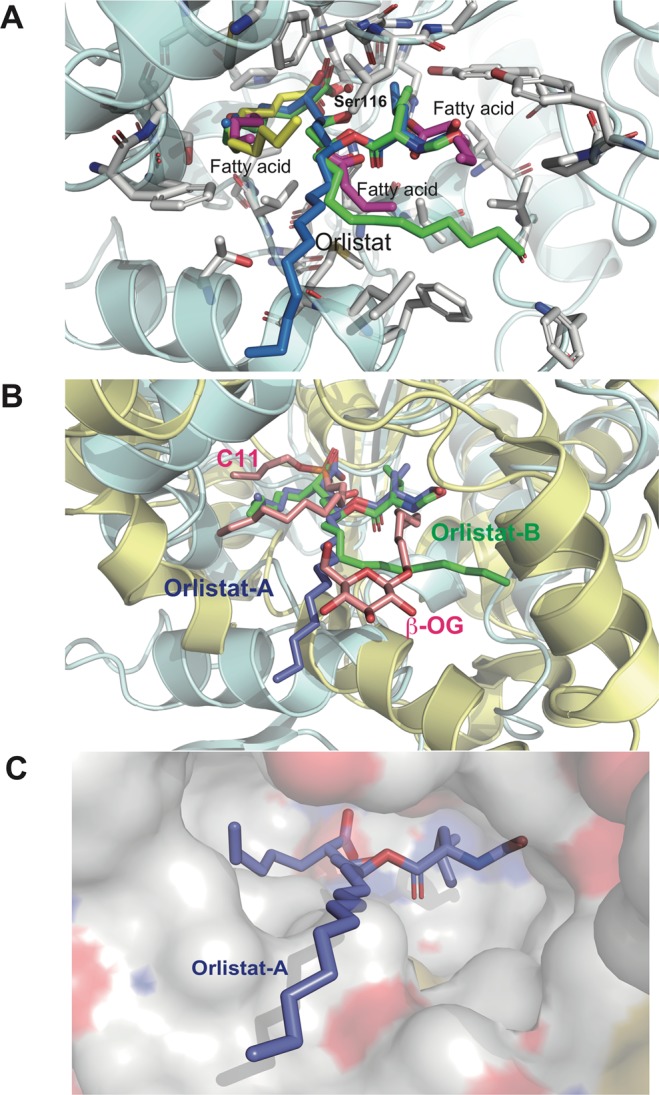
Superimposition of SAL-binding molecules. The orlistat complex (blue and green) and fatty acids fragments in native (magenta) SAL and S116A mutant (yellow). The key residue side chains are shown as sticks, and the protein backbone is shown schematically in light cyan. The catalytic triad residues are shown as sticks (separated structures shown in Supplementary Figure 3). (B) Superimposition of the orlistat complex of SAL (blue) and the phosphonate inhibitor C11 and β-octyl glycoside complex of DGL. SAL (light blue) and DGL (light yellow) overlap. (C) The predicted model of orlistat and DGL (surface model) binding mode. Orlistat molecules are shown as sticks in blue. Positively charged areas are shown in blue, and negatively charged areas are shown in red.
Fatty acids were observed in the native SAL, S116A mutant, and SAL–orlistat complex structures (Fig. 7A, Supplementary Fig. 3). In the native SAL crystal, three fatty acids were observed near the active site, whereas only one fragment was bound to S116A mutant. SAL specifically hydrolyzes triacylglycerol (Fig. 1A), and these three fatty acids may reflect a triacylglycerol position for hydrolysis (Fig. 7A, Supplementary Fig. 3).
In some cases, when using PEG solution for crystallization, multiple rounds of analysis were required to detect PEG fragments in structures. The native and S116A mutant crystals did not use PEG under the crystallization condition or during cultivation or purification. Therefore, the observation that SAL binds to fatty acids is interesting because they may have been derived from culture broths of E. coli. Fatty acids were present at the same position on the long β-side and short α-side chains. The fatty acid bound in the S116A mutant was located on the short α-side chain and was almost 12 carbon atoms in length (Fig. 7A, Supplementary Fig. 3). These results strongly supported that SAL has a high affinity for fatty acids and hydrolyzes them to reduce the effect of immune-responsive lipids9,12.
Discussion
DALI showed that several bacterial lipase structures match the 3D SAL structure well27, with the best-matched lipase being SHL (PDB ID: 2hih; RMSD, 1.3 Å), followed by Geobacillus thermocatenulatus lipase (GTL; PDB ID: 5ce5; RMSD, 1.8 Å), G. stearothermophilus lipase (GSL; PDB ID: 1ji3; RMSD of 2.3 Å), and G. zalihae lipase (GZL; PDB ID: 3umj; RMSD 2.3 Å)16,23–25. Multiple sequence alignment of SAL and structural homologous lipases was performed using ClustalW, and Fig. 3A was constructed using the ESPIRIT program28. The 3D structures of these lipases were also determined. They had no similarity in terms of amino acid sequences with human gastric lipase (HGL; data not shown).
In these alignments, SAL, SHL, and GTL crystal structures had an open-lid conformation (Fig. 3C). Among these, GSL and GZL crystal structures had a closed conformation24,25. It has been observed that structures with open-lid forms may bind ligands in the lid domain, possibly inhibiting lid closure. Although fatty acids bound to SAL, no small molecules were observed bound to SHL due to low-resolution analysis (approximately 2.8 Å). PEG and MPD molecules bound to GTL. Further, open forms of lipases have PEG or fatty acid molecules inserted within the active site cleft. These molecules prevented lid movement at α-helix 7, resulting in an open-lid conformation in crystal structures (Fig. 3C).
The native SAL crystal structure has the same fold as the SHL crystal structure, which has phospholipase activity16. This shape may be suitable for accessing membrane phospholipids. When SAL attaches to the membrane, it can hydrolyze the proximal lipid chains of the membrane.
Various fatty acids bind to the active and nonactive sites of the SAL crystal structure in its native form (12 fatty acids) and the SAL–orlistat complex (7 fatty acids). The other four long fatty acids were also found bound far from active sites on chains A and B of the SAL structure, particularly at the lid of α-helix 7 and helix 8. This also blocked the lid domain from moving forward to the active site, thereby maintaining the lid in its open form. These surplus strong findings of electron density maps of fatty acids were also observed for the SAL–orlistat complex structure covering the α-helix 7 lid, which seemed to be preventing its movement.
SAL shares 54% identity and 67% similarity with the SHL primary structure, respectively. These two lipases possessed the same whole structures and have almost identical secondary folds. The difference lies in helix 8, which is located at the lid domain (Fig. 3B) near the zinc-binding site, and is shifted by approximately 6 Å between each other. The lid domain is not close to the catalytic pocket in the SAL structure, whereas the SHL structure is externally opened with α-helix 10. It was observed that the structural differences in these loop regions occurred due to crystal packing. The packing combinations of SAL and SHL were also different from each other.
This study succeeded in determining the 3D structure of the SAL–orlistat complex. However, it remains unclear how orlistat binds to human lipases based on the current SAL–orlistat complex. No similarity was found with HGL in terms of amino acid sequence or 3D structure. The 3D alignment of SAL to HGL (PDB ID: 1hlg) showed that the small α-helix of HGL overlapped to interfere with the SAL active sites and disturbed the binding mode of orlistat, which led HGL to have a closed form29.
Although HGL has been reported as a closed form, the dog gastric lipase (DGL; PBD ID: 1k8q) complex has been reported as an open form in the presence of the phosphonate inhibitor C1130. The current study compared the DGL–C11 complex with the SAL–orlistat complex. Using the SSM module of the Coot program31, the DGL complex structure was fitted to the SAL–orlistat complex structure (Fig. 7B). The overlapped orlistat molecule showed a similar fit between the active site of DGL and the SAL-binding mode (Fig. 7B). In the DGL– C11 complex, β-octylglucoside also bound to the active site30 and was well-aligned with the β-chain moiety of the orlistat-binding site (Fig. 7B). C11 covalently bound to catalytic Ser153 of DGL, which also overlapped with the α-chain moiety in orlistat. The docking study also showed that the binding interaction between orlistat and DGL is similar to that between orlistat and SAL (Fig. 7C). Therefore, the interactions between orlistat and HGL are possibly similar to that between orlistat and DGL.
Although it has not yet been determined how orlistat binds to human lipases, this study showed how orlistat bound to a bacterial lipase derived from SA, which causes human skin diseases and MRSA proliferation. These results suggest that orlistat is repositioned in dermatosis or MRSA-related diseases.
In summary, we found that orlistat is a potent SAL inhibitor, provided a novel 3D structure of SAL, and characterized the detailed molecular interactions between orlistat and SAL. Biochemical analyses and crystallographic studies of SAL and its inactive mutant allowed for the understanding of the catalytic mechanism of SAL. Using this structural information of SAL, highly potent structure-based drugs can be designed and developed for the treatment of SA skin disease and MRSA-related disease.
Materials and methods
Cloning and expression of SAL
The SAL gene was cloned into a pCold II vector (Takara Bio) and amplified by PCR with the KOD FX DNA polymerase (Toyobo) using chromosomal DNA as previously reported32. The plasmid for the expression of the Ser116Ala mutant was constructed using site-directed mutagenesis with 5′-CTTGTAGGGCATgcTATGGGTGG-3′ as the mutagenesis primer. The PCR reaction mixture included 0.25 U Tks Gflex DNA polymerase (Takara Bio), 0.2 mM deoxynucleotide triphosphate, polymerase buffer, 0.2 µM primer, and 20 ng pCold II-mSAL plasmid DNA solution as the template DNA in a total volume of 50 µL. The samples were preheated at 94 °C for 30 s, followed by 25 cycles of amplification under the following conditions: denaturation at 94 °C for 30 s, annealing at 55 °C for 1 min, and elongation at 72 °C for 6 min using a thermal cycler (GeneAtlas). After PCR, the products were digested by 10 U DpnI at 37 °C for 1 h. The DpnI-treated PCR products were introduced to E. coli DH5α cells using chemical transformation. The consequent plasmid was extracted from the transformant, and the mutation in the plasmid was confirmed using DNA sequencing. Finally, the constructed plasmid, termed pCold II–S116A mSAL, was introduced into the E. coli BL21(DE3) strain (Novagen).
Protein expression and purification
Recombinant SAL and S116A-SAL were expressed in E. coli and purified as described previously32 with three-step chromatography. The eluted protein containing SAL was further purified using size-exclusion chromatography to analyze its molecular size in solution (Supplementary Fig. 2). The purified SAL (10 mg/mL) was injected at 20 °C into the GE Superdex 200 increase 10/300 GL column. Chromatography was performed with 50 mM Tris-HCl (pH 7.5) containing 0.2 M NaCl at a flow rate of 0.5 ml/min, using BSA as a standard protein for calibration. The protein was concentrated to 30 mg mL−1 using the VIVASPIN 15 turbo concentrator (Sartorius, Germany) and sterilized by filtration through a 0.1-µm Ultrafree-MC device (Merck Millipore, Germany). The homogeneity of the purified preparation was confirmed by 12% SDS-PAGE.
Determination of specific activity
Enzymatic activities were determined using p-nitrophenylbutyrate (pNPB) ester substrate as previously reported32,33. Enzyme concentration was adjusted to 0.002 mM and pNPB concentration to 0.08 mM. Each assay was performed at 25 °C with 50 mM HEPES (pH 7.5) containing 10% dimethylformamide in 1-ml cuvettes. Various concentrations of orlistat (0–0.01 mM) were prepared to determine IC50 values. Before starting the SAL/pNPB reaction, SAL was incubated with orlistat for 15 min. Reaction curves were obtained following the addition of substrate at 25 °C. The absorbance of liberated p-nitrophenol was monitored continuously at a wavelength of 405 nm, and the initial linear velocity was used to calculate the specific activity of SAL. An experimentally determined absorption coefficient of 18.5 mM−1 cm−1 was used for all calculations33. Determination of the initial rate was performed at least thrice. IC50 values were determined with data averaged at each concentration using an IC50 calculator (https://www.aatbio.com/tools/ic50-calculator). The various orlistat concentrations inhibited SAL activities by more or less than 50% (Fig. 4).
Cocrystallization
For crystallization experiments, a 30 mg mL−1 solution of the recombinant SAL was prepared in 0.2 M NaCl and 10 mM Tris-HCl at pH 8.0. The inhibitor concentration was adjusted to 5–10-fold molar equivalents. Concentrated SAL was incubated overnight with 5–10-fold molar excess of the inhibitor dissolved in DMSO.
Crystallization trials were set up as sitting-drop vapor-diffusion experiments at 295 K on Cryschem crystallization plates. Initial screening was performed using the sparse matrix method34 with commercial crystal screening kits (Hampton Research, Wizard). Several trials were performed at varying protein concentrations, precipitant concentrations, and pH values to optimize experimental conditions.
Data collection and processing
X-ray diffraction data of SAL crystals were collected at 100 K in a flowing nitrogen stream. The mother liquor solution was supplemented with 30% glycerol as a cryoprotectant. X-ray data sets were collected from a single crystal using the beamline BL41XU and BL44XU at SPring-8. The X-ray wavelength was tuned to 0.9 Å. Further, the oscillation angle was 0.2°, and the crystal-to-detector distance was approximately 300 mm. Diffraction images were integrated and scaled using the XDS program package35 with the KAMO automatic program at SPring-836. The resolution limits were defined as CC1/2 of approximately 0.537.
Structure determination, model building, and refinement
Crystal structures were determined by molecular replacement using Phaser20. Model building and inspection were based on Coot31 and refined by Refmac5 on CCP419. All images of molecular structures were prepared using PyMOL (http://www.pymol.org/2).
Molecular Docking of orlistat in DGL
Structure refinement and energy minimization of the DGL–orlistat complex (Fig. 7B) was conducted using the YASARA Energy Minimization Server38.
Accession codes
The atomic coordinates and structure factors for native SAL, inactive S116A mutant, and SAL–orlistat complex have been deposited in the Protein Data Bank with accession codes 6KSI, 6KSL, and 6KSM, respectively.
Supplementary information
Acknowledgements
We are grateful to all members of beamline BL41XU, BL44XU, and BL45XU at SPring-8 for their help in data collection. Use of the synchrotron beamline BL44XU at SPring-8 was possible through the Cooperative Research Program of the Institute for Protein Research, Osaka University (proposal Nos. 2019A6912, 2018A6813, 2018B6813, 2017A6716, 2017B67165, 2016A6615, and 2016B6615). This research was partially supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED (BINDS support numbers 0370, 1175, and 1285). This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Science, and Technology of Japan.
Author contributions
K.K. and S.K. designed research. K.K., M.T., T.H., Y.O., M.Y. and S.K. performed research. K.K., M.T., T.H., Y.O. and M.Y. collect and analyzed the X-ray data at Spring-8. M.T., and Y.O. measured and analyzed the enzymatic activity. K.K. and S.K. wrote the main manuscript text and K.K. prepared all figures. M.T. prepared figure 4. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-62427-8.
References
- 1.Lowy FD. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Findley K, Grice EA. The skin microbiome: a focus on pathogens and their association with skin disease. PLoS Pathog. 2014;10:e1004436. doi: 10.1371/journal.ppat.1004436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clinic. Microbio. Rev. 2000;13:16–34. doi: 10.1128/CMR.13.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindsay JA, Holden MT. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 2004;12:378–385. doi: 10.1016/j.tim.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Lakhundi S, Zhang K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018;31:e00020–18. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zetola N, Francis JS, Nuermberger EL, Bishai WR. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect. Dis. 2005;5:275–286. doi: 10.1016/S1473-3099(05)70112-2. [DOI] [PubMed] [Google Scholar]
- 7.Kamitani S, et al. Induction of Neutral Lipid-Containing Granules by Staphylococcal Lipase from Clinical Isolates. SOJ | Microbiology Infectious Diseases. 2015;2:1–7. [Google Scholar]
- 8.Ünlü A, Tanriseven A, Sezen IY, Çelik A. A new lipase as a pharmaceutical target for battling infections caused by Staphylococcus aureus: Gene cloning and biochemical characterization. Biotechnol. Appl. Biochem. 2015;62:642–651. doi: 10.1002/bab.1316. [DOI] [PubMed] [Google Scholar]
- 9.Delekta PC, Shook JC, Lydic TA, Mulks MH, Hammer ND. Staphylococcus aureus Utilizes Host-Derived Lipoprotein Particles as Sources of Fatty Acids. J. Bacteriol. 2018;200:e00728–17. doi: 10.1128/JB.00728-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu C, Xiong N, Zhang Y, Rayner S, Chen S. Functional characterization of lipase in the pathogenesis of Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2012;419:617–620. doi: 10.1016/j.bbrc.2012.02.057. [DOI] [PubMed] [Google Scholar]
- 11.Kuroda M, Nagasaki S, Ito R, Ohta T. Sesquiterpene farnesol as a competitive inhibitor of lipase activity of Staphylococcus aureus. FEMS Microbiol. Lett. 2007;273:28–34. doi: 10.1111/j.1574-6968.2007.00772.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Alonzo F., III Bacterial lipolysis of immune-activating ligands promotes evasion of innate defenses. Proc. Natl Acad. Sci. USA. 2019;116:3764–3773. doi: 10.1073/pnas.1817248116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dougherty JM, et al. The role of tetracycline in chronic blepharitis. Inhibition of lipase production in staphylococci. Invest. Ophthalmol. Vis. Sci. 1991;32:2970–2975. [PubMed] [Google Scholar]
- 14.Bai T, et al. Operon for biosynthesis of lipstatin, the beta-lactone inhibitor of human pancreatic lipase. Appl. Environ. Microbiology. 2014;80:7473–7483. doi: 10.1128/AEM.01765-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballinger A, Peikin SR. Orlistat: its current status as an anti-obesity drug. Eur. J. Pharmacol. 2002;440:109–117. doi: 10.1016/S0014-2999(02)01422-X. [DOI] [PubMed] [Google Scholar]
- 16.Tiesinga JJ, van Pouderoyen G, Nardini M, Ransac S, Dijkstra BW. Structural basis of phospholipase activity of Staphylococcus hyicus lipase. J. Mol. Biol. 2007;371:447–456. doi: 10.1016/j.jmb.2007.05.041. [DOI] [PubMed] [Google Scholar]
- 17.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta. Crystallogr. D. Biol. Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nardini MI, Dijkstra BW. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 1999;9:732–737. doi: 10.1016/S0959-440X(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 21.Jeong ST, et al. Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase. J. Biol. Chem. 2002;277:17041–17047. doi: 10.1074/jbc.M200640200. [DOI] [PubMed] [Google Scholar]
- 22.Khan FI, Al., et al. The Lid Domain in Lipases: Structural and Functional Determinant of Enzymatic Properties. Front. Bioeng. Biotechnol. 2017;5:16. doi: 10.3389/fbioe.2017.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timucin E, Cousido-Siah A, Mitschler A, Podjarny A, Sezerman OU. Probing the roles of two tryptophans surrounding the unique zinc coordination site in lipase family I.5. Proteins. 2016;84:129–142. doi: 10.1002/prot.24961. [DOI] [PubMed] [Google Scholar]
- 24.Ruslan R, et al. Improvement of Thermal Stability via Outer-Loop Ion Pair Interaction of Mutated T1 Lipase from Geobacillus zalihae Strain T1. Int. J. Mol. Sci. 2012;13:943–960. doi: 10.3390/ijms13010943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyndall JDA, Sinchaikul S, Fothergill-Gilmore LA, Taylor P, Walkinshaw MD. Crystal Structure of a Thermostable Lipase from Bacillus stearothermophilus P1. J.Mol.Biol. 2002;323:859–869. doi: 10.1016/S0022-2836(02)01004-5. [DOI] [PubMed] [Google Scholar]
- 26.Pemble CW, 4th, Johnson LC, Kridel SJ, Lowther WT. Crystal structure of the thioesterase domain of human fatty acid synthase inhibited by Orlistat. Nat. Struct. Mol. Biol. 2007;14:704–709. doi: 10.1038/nsmb1265. [DOI] [PubMed] [Google Scholar]
- 27.Holm L, Laakso LM. Dali server update. Nucleic acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roussel A, et al. Crystal structure of human gastric lipase. J. Biol. Chem. 1999;274:16995–17002. doi: 10.1074/jbc.274.24.16995. [DOI] [PubMed] [Google Scholar]
- 30.Roussel A, et al. Crystal structure of the open form of dog gastric lipase in complex with a phosphonate inhibitor. J. Biol. Chem. 2002;277:2266–74. doi: 10.1074/jbc.M109484200. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka M, Kamitani S, Kitadokoro K. Staphylococcus aureus lipase: purification, kinetic characterization, crystallization and crystallographic study. Acta Crystallogr. F. Struct. Biol. Commun. 2018;74:567–570. doi: 10.1107/S2053230X18010506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowers GN, Jr, McComb RB, Christensen RG, Schaffer R. High-purity 4-nitrophenol: purification, characterization, and specifications for use as a spectrophotometric reference material. Clin. Chem. 1980;26:724–729. doi: 10.1093/clinchem/26.6.724. [DOI] [PubMed] [Google Scholar]
- 34.Jancarik J, Kim SH. Sparse matrix sampling: a screening method for crystallization of proteins. J. Appl. Cryst. 1991;24:409–411. doi: 10.1107/S0021889891004430. [DOI] [Google Scholar]
- 35.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashita K, Hirata K, Yamamoto M. KAMO: towards automated data processing for microcrystals. Acta Crystallogr. D. Biol. Crystallogr. 2018;74:441–449. doi: 10.1107/S2059798318004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krieger E, et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins. 2009;77:114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.