Abstract
Tea plant (Camellia sinensis), an important economic crop, is seriously affected by various abiotic stresses, including salt stress, which severely diminishes its widespread planting. However, little is known about the roles of long non-coding RNAs (lncRNAs) in transcriptional regulation under salt stress. In this study, high-throughput sequencing of tea shoots under salt-stress and control conditions was performed. Through sequencing analysis, 16,452 unique lncRNAs were identified, including 172 differentially expressed lncRNAs (DE-lncRNAs). The results of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of their cis- and trans-target genes showed that these DE-lncRNAs play important roles in many pathways such as the galactinol synthase (GOLS), calcium signaling pathway, and interact with transcription factors (TFs) under salt stress. The data from the gene-specific antisense oligodeoxynucleotide-mediated reduction in the lncRNA MSTRG.139242.1 and its predicted interacting gene, TEA027212.1 (Ca2+-ATPase 13), in tea leaves revealed that MSTRG.139242.1 may function in the response of tea plants to high salinity. In addition, 12 lncRNAs were predicted to be target mimics of 17 known mature miRNAs, such as miR156, that are related to the salt-stress response in C. sinensis. Our results provide new insights into lncRNAs as ubiquitous regulators in response to salt stress in tea plants.
Keywords: Camellia sinensis, long non-coding RNA, salt stress, antisense oligodeoxynucleotide suppression, endogenous target mimic
Introduction
Tea plant [Camellia sinensis (L.) O. Kuntze] is an important economic crop in many countries, such as China, Japan, and India. Tea made from tea plant leaves is one of the most consumed drinks worldwide. However, tea plants are seriously affected by many abiotic stresses, such as cold, salt, and drought stresses, due to their specific environmental requirements, which severely diminish the widespread planting of tea plants (Upadhyaya and Panda, 2013). Therefore, it is important for the tea industry to breed tea plant species that exhibit strong resistance and adaptability. Salt stress is a form of abiotic stress that severely affects the growth of tea plants and the quality of tea products (Upadhyaya and Panda, 2013). In our previous report, we performed an Illumina RNA sequencing (RNA-Seq) to compare the transcriptomes of tea plants treated with and without NaCl, and many differentially expressed mRNAs involved in signal transduction pathways, transcription factors (TFs), and other functional genes under salt stress were identified (Wan et al., 2018). Previous reports have also shown that many mRNAs and proteins in tea plants, such as CsSnRK2 (Zhang et al., 2018), CsAQP (Yue et al., 2014), and CsVQ (Guo et al., 2018), respond to salt stress. Long non-coding RNAs (lncRNAs) are important regulatory factors that respond to various abiotic stresses in plants; however, their functions in responding to salt stress have not been studied in tea plants. Therefore, the identification of important lncRNAs that respond to salt stress in tea plants is necessary.
LncRNAs are non-coding RNAs that are longer than 200 nt and usually have low protein-coding potential (Mercer et al., 2009). According to their genomic origins, lncRNAs are broadly divided into four types: intronic lncRNAs, intergenic lncRNAs (lincRNAs), sense lncRNAs, and antisense lncRNAs (Liu et al., 2012). LncRNAs are considered “transcriptional noise” because of their poor conservation and the limited evidence for their functions (Ponjavic et al., 2007). However, increasing research has revealed that lncRNAs function in transcriptional regulation and epigenetic gene regulation, and some lncRNAs function as cis- or trans-regulators in various biological processes (Ponting et al., 2009). In mammals, many lncRNAs have been proven to be related to a wide range of diseases, especially cancers and neurodegenerative diseases (Ma et al., 2012). In recent years, a large number of lncRNAs that are important in plant growth and development (Heo and Sung, 2011; Ding et al., 2012), biotic stress responses (Cui et al., 2017), and abiotic stress responses (Swiezewski et al., 2009; Qin et al., 2017) have been identified. For example, the reduced expression of the rice lncRNA LDMAR can lead to male sterility under long-day conditions in rice (Ding et al., 2012). In Arabidopsis, the antisense lncRNA FLORE can regulate the circadian clock to photoperiodic flowering (Henriques et al., 2017). A nucleus-localized lncRNA, DRIR, can enhance drought- and salt-stress tolerance in Arabidopsis (Qin et al., 2017). Currently, with the development of deep-sequencing technology, genome-wide lncRNA analyses have been performed on many species, such as Arabidopsis (Liu et al., 2012), rice (Zhang Y.C. et al., 2014), Zea mays (Boerner and McGinnis, 2012), wheat (Shumayla et al., 2017), and other species (Zhang and Chen, 2013). However, in many non-model plants, studies on lncRNAs are rather limited. Recently, Varshney et al. (2019) discovered 33,400 putative lncRNAs in different tissues of tea plants, which was the first report of lncRNAs in tea plants.
In this study, to explore the early response of lncRNA under salt stress in tea plants, we reanalyzed the transcriptomes of tea plants treated with and without NaCl based on the latest tea tree genome (Wei et al., 2018). LncRNAs were identified and classified systematically. The functions of these lncRNAs were predicted, and differentially expressed lncRNAs (DE-lncRNAs) that responded to salt stress were identified. In addition, the cis-targeting and trans-targeting relationship between lncRNAs and mRNAs and the possible interactions between lncRNAs and abiotic stress-related miRNAs were found to be involved in salt stress in tea plants. Notably, in our previous report, the Ca2+-transporting ATPase was shown to be involved in the salt-stress response in tea plants (Wan et al., 2018), and in this study, the differentially expressed Ca2+-transporting ATPase 13 (TEA027212.1) was proven to be co-expressed with the lncRNA MSTRG.139242.1, which indicates that this lncRNA may participate in Ca2+ signal transduction in response to salt stress. In addition, six DE-lncRNAs were randomly selected for quantitative real-time PCR (qRT-PCR) validation to confirm the reliability of the expression levels obtained from the RNA-Seq transcriptome. Overall, the results obtained in our study provide a valuable resource for studying lncRNAs involved in salt stress in tea plants and will enhance our understanding of the putative regulatory functions of lncRNAs in plants.
Materials and Methods
Plant Materials and NaCl Treatments
One-year-old “Pingyangtezao” tea plant cutting seedlings with consistent growth were pre-incubated in nutrient solution under standard growth conditions (temperature: 25 ± 3°C, air relative humidity: 60–70%, photoperiod: 12 h light/12 h dark) for 2 months. These tea plants were used for stress assays under the same growth conditions. In order to analyze the early response of tea plant to high salinity and evaluate the early salt responsive genes in the tea plant leaves, salt treatment was conducted by soaking the roots in nutrient solution that contained 250 mM NaCl (salt treatment) or standard nutrient solution (control), and fresh leaf samples (first and second leaves) were randomly collected at 4 h when tea plants began to show the symptoms of salt stress and wilt slightly after treatment. Control and NaCl treatment were both repeated three times (Control1, Control2, and Control3 and Salt1, Salt2, and Salt3). The samples were immediately immersed in liquid nitrogen and stored at −80°C until RNA-Seq, qRT-PCR validation, and further analysis.
Analysis of Transcriptomic Data Based on the Tea Tree Reference Genome
In our previous report, we sequenced six tea plant samples using RNA-Seq technology. A total of 79.40 Gb of clean data was obtained, and the Q30 base percentage of each sample was greater than 92.93% (Wan et al., 2018). The identification of lncRNAs was executed according to the pipeline shown in Figure 1A. All RNA-Seq datasets were aligned to the reference genome of C. sinensis var. sinensis (Wei et al., 2018) using the HISAT2 system1 (Kim et al., 2015) to reconstruct the transcriptome. After the alignment, the StringTie software2 (Pertea et al., 2016) was used to assemble reads into transcripts and for quantification. The assembled transcripts were annotated using the gffcompare program,3 and the unknown transcripts were used to screen for putative lncRNAs.
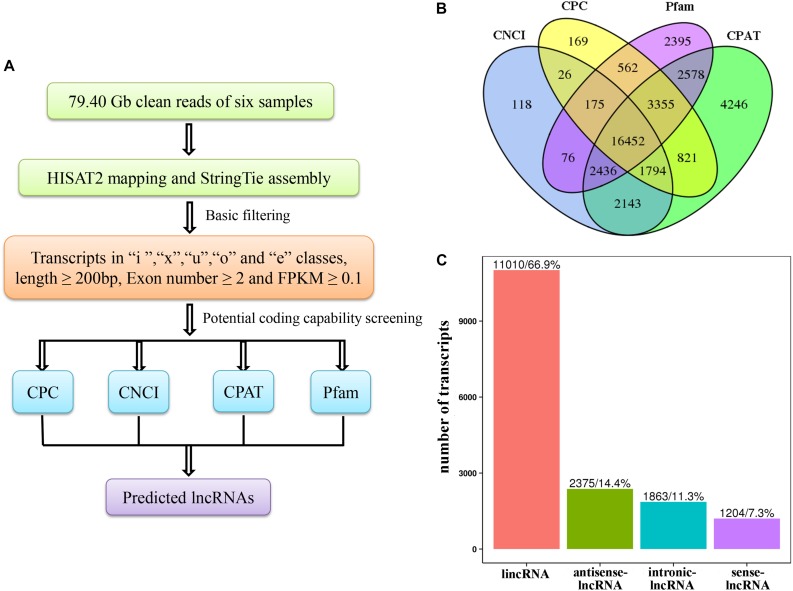
FIGURE 1.
(A) Pipeline for the identification of lncRNAs in Camellia sinensis. (B) Filtering of the candidate long non-coding RNAs (lncRNAs). Venn diagrams of coding potential analysis using four methods [Coding Potential Calculator (CPC), Coding-Non-Coding Index (CNCI), Coding Potential Assessment Tool (CPAT), and Protein family (Pfam)]. Those simultaneously shared by the four methods were predicted as candidate lncRNAs and used in subsequent analyses. (C) Classification of lncRNAs.
Identification of LncRNAs in Tea Plants
The prediction of lncRNAs consisted of two parts: basic filtering and potential coding capability screening. For basic filtering, transcripts with a class code of “i,” “x,” “u,” “o,” and “e” (Lv et al., 2013), a length ≥ 200 bp, an exon number ≥ 2, and a fragments per kilobase of transcript per million mapped reads (FPKM) value ≥ 0.1 (Kelley and Rinn, 2012) were selected. Subsequently, four methods, including Coding Potential Calculator (CPC)4 (Kong et al., 2007), Coding-Non-Coding Index (CNCI)5 (Sun L. et al., 2013), Coding Potential Assessment Tool (CPAT)6 (Wang et al., 2013), and Protein family (Pfam)7 (Finn et al., 2014) protein domain analysis were used to evaluate the coding capabilities of the transcripts, and potential lncRNAs were filtered out by combining the four results. Next, the identified lncRNAs were classified into four categories: lincRNA, antisense-lncRNA, intronic-lncRNA, and sense-lncRNA using cuffcompare.8 In addition, lncRNAs and mRNAs were comparatively analyzed according to their transcript length and exon number.
Prediction of Potential Target Genes of LncRNAs
The prediction of potential target genes of lncRNAs was based on the two interaction modes between lncRNAs and mRNAs. First, because lncRNA can regulate the expression of nearby genes, we searched coding genes 100 kb upstream and downstream of lncRNAs for cis-target genes. The other interaction mode of lncRNA and mRNA is due to the complementary pairing of bases. The LncTar (Li et al., 2015) tool was used for target gene prediction of lncRNAs. The free energy and standard free energy of paired sites were calculated, and the target genes with standard free energy threshold < −0.1 were considered trans-target genes of lncRNAs.
Identification and Functional Analysis of DE-LncRNAs
The expression levels of lncRNAs were quantified using FPKM values with the StringTie software. Differential expression analysis of the NaCl and Control groups was performed using the DESeq R package (1.10.1).9 The resulting P-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. LncRNAs with an adjusted value of P < 0.01 and an absolute value of log2 (fold change) > 1 found by DESeq were considered as differentially expressed.
Functional analyses of DE-lncRNAs were performed by annotation and classification of their cis- and trans-target genes. BLASTX alignment (value of E < 10–5) (Altschul et al., 1997) between target genes and public databases was performed, which included the Clusters of Orthologous Groups (COG) of proteins (Tatusov et al., 2000) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (Kanehisa et al., 2004). Gene Ontology (GO) classifications were conducted using Blast2GO and WEGO software (Ashburner et al., 2000).
Prediction of LncRNAs as Endogenous Target Mimics for miRNAs Under Salt Stress
The 172 DE-lncRNAs and 539 known conserved mature miRNAs of C. sinensis (Zhu and Luo, 2013; Zhang Y. et al., 2014; Jeyaraj et al., 2017a, b; Sun et al., 2018) were submitted to psRNATarget10 using a maximum expectation of 2.0 (Wu et al., 2013; Dai et al., 2018) for miRNA–lncRNA interaction prediction. Less than three mismatches and G/U pairs were allowed within the lncRNA and miRNA pairing regions. The co-expression network was established using Cytoscape,11 based on the DE-lncRNAs, target mRNAs of DE-lncRNAs, and miRNAs interacting with DE-lncRNAs.
LncRNA and Target mRNA Suppression in Tea Plants Using Antisense Oligonucleotides (AsODNs)
A gene-suppression assay in tea plants using antisense oligonucleotides (AsODNs) was conducted according to Liu et al. (2018). ODN sequences were selected using the Soligo software.12 The sequences of input and selected ODNs are shown in Supplementary Data Sheet S1: Table S1. To increase the efficiency of gene suppression, two or three independent ODNs were synthesized of each gene and mixed in equal proportions during treatment. The ODNs were purified by HPLC, and the concentration was diluted to 10 μM. Fresh tea shoots with a bud and two leaves were excised and inserted into centrifuge tubes containing 1 ml of mixed ODNs for treatment. All leaves from the shoots were harvested after 24 h of treatment, and each treatment had three biological repeats. At the same time, a random non-sense control was used (Supplementary Data Sheet S1: Table S1). A nucleotide BLAST search of the tea tree genome CDS and lncRNA sequences with this sequence was used to verify that the sequences did not overlap with other sequences.
qRT-PCR
qRT-PCR was performed to validate the expression patterns of salt-responsive lncRNAs. Total RNA was isolated from the six tea samples (Salt1, Salt2, Salt3, Control1, Control2, and Control3) using TRNzol reagent (TIANGEN, Beijing, China). First-strand cDNA was synthesized using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Dalian, China), after which qRT-PCR detection was completed using the EvaGreen qPCR MasterMix-No Dye kit (ABM, Richmond, BC, Canada) on a StepOne Plus PCR instrument (ABI, United States) following the manufacturers’ protocols. The tea plant Csβ-actin gene was chosen as the reference gene in accordance with previous methods (Wan et al., 2018), and the primers used in this assay are shown in Supplementary Data sheet S1: Table S2. The relative expression levels of the genes were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001), and the data are presented as the mean ± SD from three independent biological replicates. Group differences were tested using one-way ANOVA and Duncan’s test, and significant differences among various treatment groups are represented by different letters (P < 0.05). The SPSS 20.0 software was used to determine significant differences between the Control and 250 mM NaCl treatment data.
Results
Identification of LncRNAs in C. sinensis
To identify salt-responsive RNAs in C. sinensis, six cDNA libraries from three controls (Control1, Control2, Control3) and three salt-treated (Salt1, Salt2, Salt3) leaf samples that were treated with 250 mM NaCl for 4 h were constructed and sequenced using an Illumina HiSeq2500 platform in our previous report (Wan et al., 2018). In this report, the RNA-Seq results were aligned to the latest tea tree genome, and the alignment results are shown in Table 1. The mapped ratio of each sample ranged from 88.99 to 91.10%. After the data were filtered with CPC, CNCI, Pfam, and CPAT, 16,452 candidate lncRNAs were obtained from the six samples (Figure 1B). Then, the identified lncRNAs were classified into four categories: 11,010 lincRNAs (66.9%), 2,375 antisense-lncRNAs (14.4%), 1,863 intronic-lncRNAs (11.3%), and 1,204 sense-lncRNAs (7.3%) (Figure 1C).
TABLE 1.
Overview of the genome alignment result.
| Sample | Total reads | Mapped reads | Uniq mapped reads | Multiple mapped reads | Reads map to “ + ” | Reads map to “−” |
| Control1 | 112,129,142 | 101,328,235 (90.37%) | 70,079,537 (62.50%) | 31,248,698 (27.87%) | 3,8880,996 (34.68%) | 3,8760,991 (34.57%) |
| Control2 | 130,302,020 | 118,699,443 (91.10%) | 76,309,916 (58.56%) | 42,389,527 (32.53%) | 43,511,636 (33.39%) | 43,518,412 (33.40%) |
| Control3 | 78,517,558 | 71,255,097 (90.75%) | 47,395,231 (60.36%) | 23,859,866 (30.39%) | 26,782,952 (34.11%) | 26,888,454 (34.25%) |
| Salt1 | 119,801,158 | 106,611,134 (88.99%) | 80,043,176 (66.81%) | 26,567,958 (22.18%) | 43,845,279 (36.60%) | 43,746,111 (36.52%) |
| Salt2 | 84,340,792 | 75,281,525 (89.26%) | 54,731,132 (64.89%) | 20,550,393 (24.37%) | 30,080,568 (35.67%) | 29,993,645 (35.56%) |
| Salt3 | 110,082,884 | 98,490,021 (89.47%) | 71,042,037 (64.54%) | 27,447,984 (24.93%) | 39,127,799 (35.54%) | 39,106,854 (35.52%) |
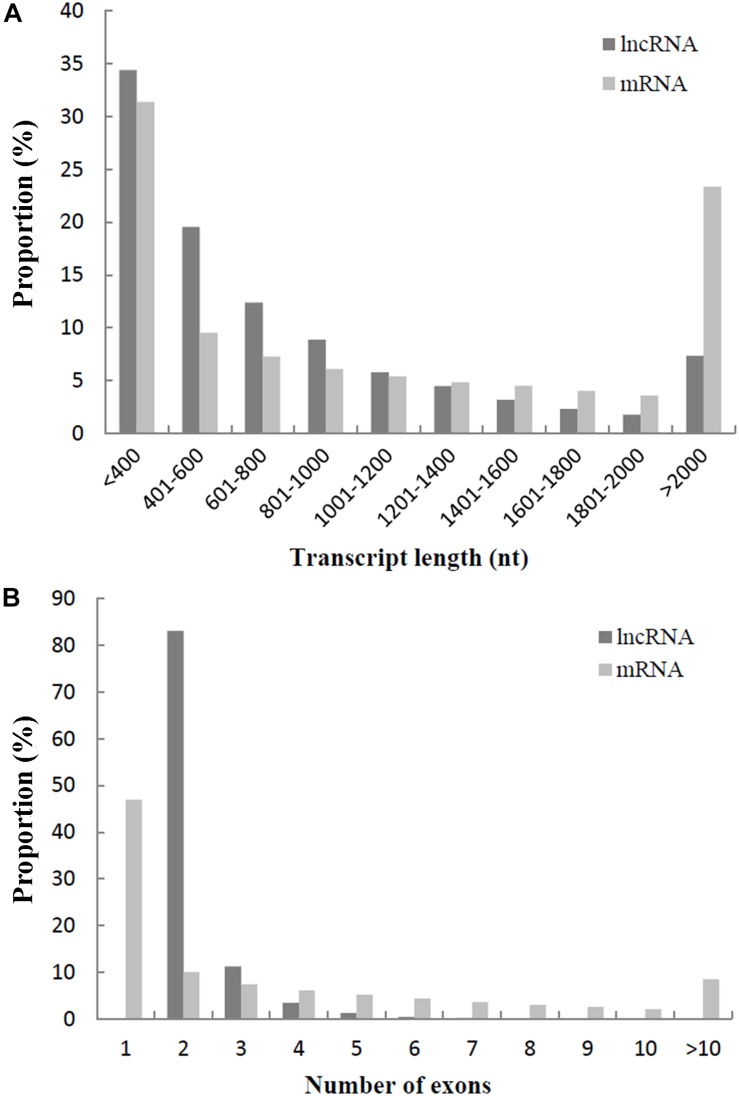
To understand the differences between lncRNAs and mRNAs in sequence and structure, lncRNAs were analyzed and compared with mRNAs, according to the transcript length and number of exons. The average length of lncRNAs was 822 nucleotides, and the average length of mRNAs was 1,348 nucleotides. As shown in Figure 2A, 75.19% of lncRNAs were < 1,000 nucleotides, while only 54.28% of mRNAs were < 1,000 nucleotides. In addition, 94.44% of lncRNAs had two (83.15%) or three (11.29%) exons, while 10.05% of mRNAs had two exons and 46.99% of mRNAs had only one exon (Figure 2B).
FIGURE 2.
(A) Comparison of transcript length between lncRNAs and mRNAs. (B) Comparison of exon number between lncRNAs and mRNAs.
DE-lncRNAs Involved in NaCl Stress
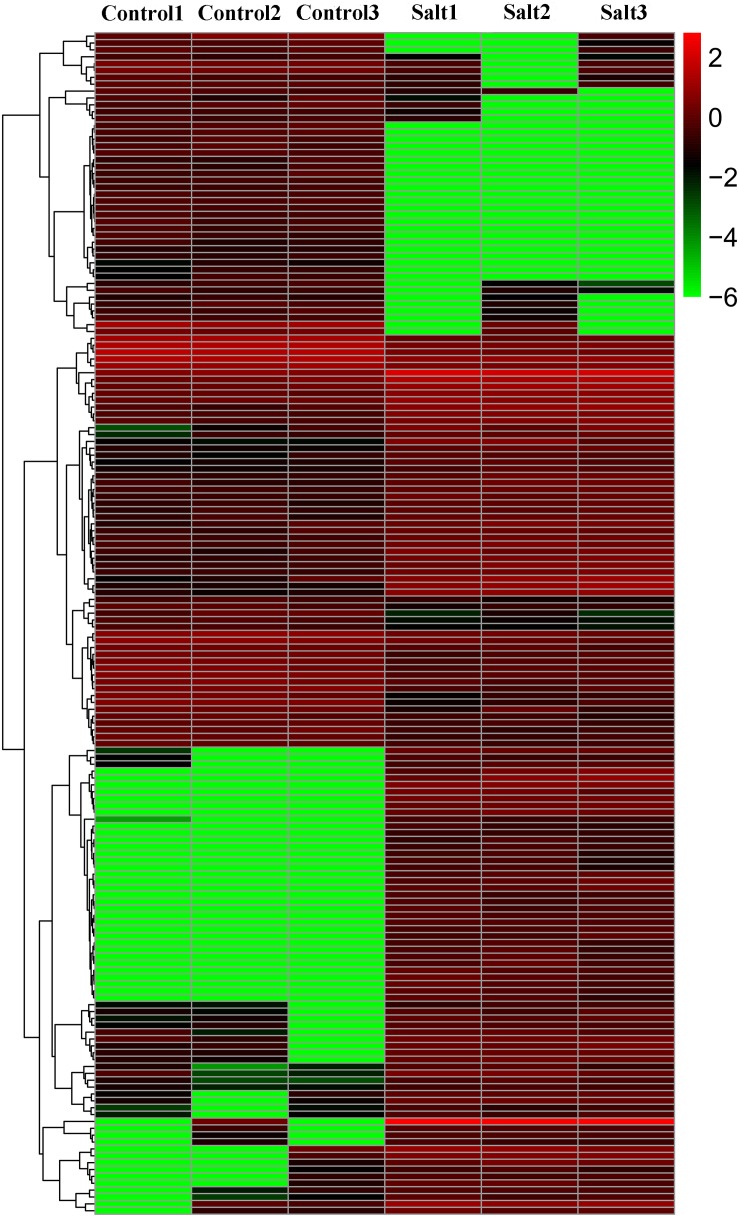
In this study, 172 DE-lncRNAs in tea plants were identified by DESeq using the FPKM value, including 101 upregulated and 71 downregulated lncRNAs (Figure 3 and Supplementary Table S1), suggesting that these lncRNAs may function in response to salt stress.
FIGURE 3.
Cluster heat map of differentially expressed lncRNAs (DE-lncRNAs) in control and NaCl-treated tea plants.
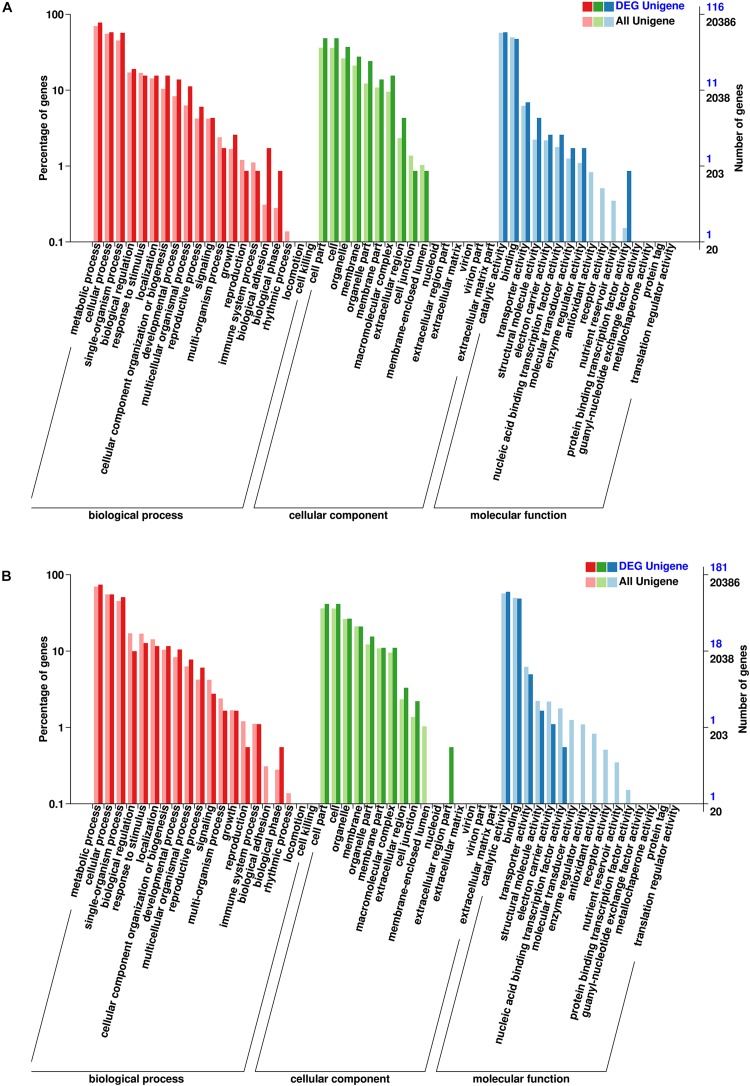
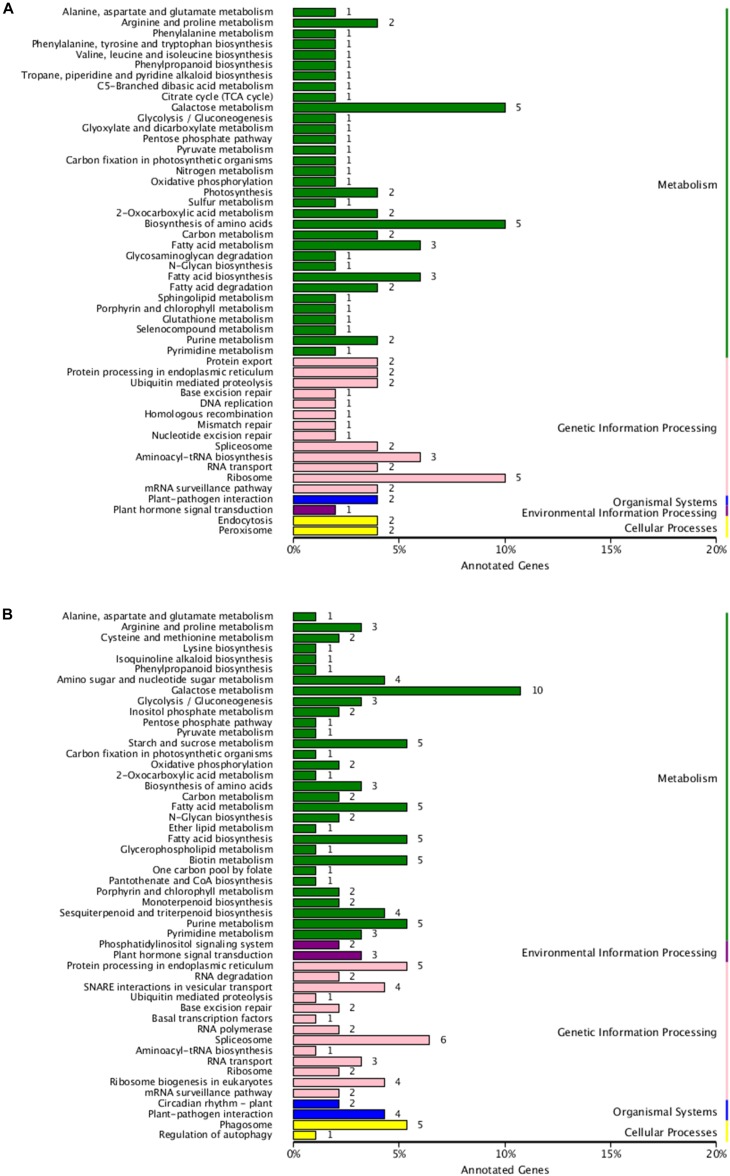
To analyze the potential functions of these lncRNAs, the predicted cis-target genes encoding genes 100 kb upstream or downstream of these DE-lncRNAs and trans-target genes predicted by complementary pairing of bases were searched and annotated. In total, 250 cis- and 421 trans-target genes of these DE-lncRNAs were predicted (Supplementary Tables S2, S3). Furthermore, GO and KEGG enrichment analyses were used to investigate the potential functions of the cis- and trans-target genes, respectively. As shown in Figures 4A,B, 116 cis-target and 181 trans-target genes were annotated in the GO database. For both cis- and trans-target genes, “Cell part” (GO: 0005623) was the largest subcategory in the cellular component category; “Catalytic activity” (GO: 0003824) in the molecular function category, and “Metabolic process” (GO: 0008152) in the biological process category were the most abundant terms. In addition, 50 cis- and 93 trans-target genes were assigned to 50 and 63 KEGG pathways, respectively (Figures 5A,B), which mainly included “Galactose metabolism” (ko00052), “Biosynthesis of amino acids” (ko01230), and “Ribosome” (ko03010). The results showed that these pathways may play important roles in the salt-stress response of tea plants.
FIGURE 4.
(A) Gene Ontology (GO) enrichment of cis-target genes of the DE-lncRNAs. (B) GO enrichment of trans-target genes of the DE-lncRNAs.
FIGURE 5.
(A) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of cis-target genes of the DE-lncRNAs. (B) KEGG enrichment of trans-target genes of the DE-lncRNAs.
Interactions of DE-lncRNAs With mRNAs
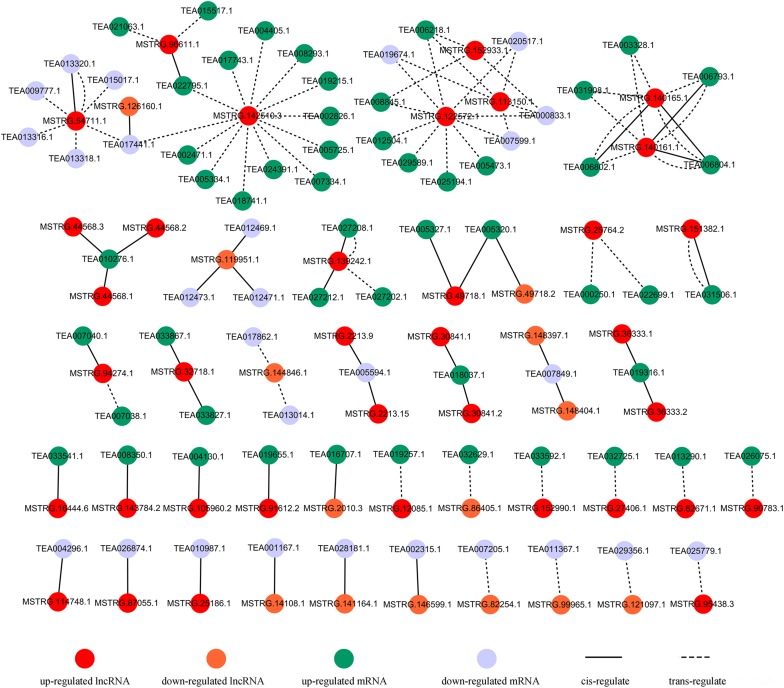
Recent studies in plants have found that lncRNAs may co-express with nearby coding genes, thereby regulating the downstream targets of the coding genes and exerting biological functions (Cui et al., 2017, 2018). Interestingly, in this study, 42 differentially expressed coding genes spaced less than 100 kb away from 35 DE-lncRNAs were identified (Figure 6 and Supplementary Data Sheet S1: Table S3). These coding genes were annotated as auxin-responsive protein (TEA005327.1), Ca2+-transporting ATPase 13 (TEA027212.1), vacuole membrane protein KMS1-like (TEA033827.1), etc. In addition, 67 differentially expressed coding genes were predicted to be trans-target genes of 23 DE-lncRNAs (Figure 6 and Supplementary Data Sheet S1: Table S4). These trans-target genes were annotated as galactinol synthase (GOLS) 2 (TEA006804.1), WRKY TF 31 isoform X2 (TEA005334.1), ethylene-responsive TF ABR1 (TEA015017.1), MYC2 TF (TEA000833.1), etc. These differentially expressed mRNAs may interact with corresponding lncRNAs and participate in the salt-stress response through various pathways.
FIGURE 6.
Predicted interaction networks among lncRNAs and mRNAs; details are shown in Supplementary Data Sheet S1: Tables S3, S4.
LncRNAs Participate in Galactinol Synthase, Calcium Signaling Pathway, and Interact With Transcription Factors Under Salt Stress
Synthesis of sugars and signal transduction pathways are crucial participants in regulatory networks of plant responses to salt stress. As presented in Table 2, eight cis/trans target genes GOLS 2 were identified to be upregulated under salt stress. Similarly, six cis/trans target genes related to the Ca2+ signaling pathway were identified, including genes encoding calcium-transporting ATPase, calmodulin-interacting protein, Ca2+-dependent protein kinases (CDPKs), and CBL-interacting protein kinases (CIPKs). Most of these genes were upregulated except CIPK 23 (TEA032544.1).
TABLE 2.
LncRNA target genes involved in galactinol synthase and calcium signaling pathway.
| Target gene | Regulation | Annotation | Interaction mode |
| TEA006791.1 | Up | Galactinol synthase 2 | Cis/trans |
| TEA006802.1 | Up | Galactinol synthase 2 | Cis/trans |
| TEA006793.1 | Up | Galactinol synthase 2 | Cis/trans |
| TEA006811.1 | Up | Galactinol synthase 2 | Cis/trans |
| TEA006804.1 | Up | Galactinol synthase 2 | Cis/trans |
| TEA011903.1 | Up | Galactinol synthase 2 | Trans |
| TEA031908.1 | Up | Galactinol synthase 2 | Trans |
| TEA003328.1 | Up | Galactinol synthase 2 | Trans |
| TEA027212.1 | Up | Calcium-transporting ATPase 13 | Cis |
| TEA020781.1 | Up | Calcium-dependent protein kinase 16 | Cis |
| TEA027208.1 | Up | Calcium-transporting ATPase 13 | Cis/trans |
| TEA023759.1 | Up | Calmodulin-interacting protein 111 | Trans |
| TEA019257.1 | Up | Calcium-dependent protein kinase 28 | Trans |
| TEA032544.1 | Down | CBL-interacting protein kinase 23 | Trans |
In addition, many TF families play vital roles in regulating plant resistance mechanisms under abiotic stress. In the present study, 23 DE-lncRNA target genes were identified, including HSF, GATA, MADS-box, bHLH, ERF, WRKY, ICE, LHY, and MYC TFs. Except all of the bHLH, LHY and MYC TFs and some of the TFs in GATA and MADS families were downregulated; the rest of the TFs were upregulated (Table 3).
TABLE 3.
LncRNA target transcription factors.
| Target gene | Regulation | Interaction mode | Annotation |
| TEA022795.1 | Up | Cis/trans | HSF |
| TEA009409.1 | Down | Cis | GATA transcription factor 15 |
| TEA013465.1 | Down | Cis | GATA transcription factor 28 |
| TEA006583.1 | Up | Cis | MADS-box transcription factor |
| TEA011370.1 | Down | Cis | bHLH70 |
| TEA008966.1 | Up | Cis | MADS-box transcription factor 23 |
| TEA002032.1 | Up | Cis | GATA transcription factor 24 |
| TEA017704.1 | Up | Cis | ERF118 |
| TEA002471.1 | Up | Trans | WRKY7 |
| TEA013512.1 | Up | Trans | ICE1 |
| TEA011367.1 | Down | Trans | LHY |
| TEA007038.1 | Up | Trans | ERF4 |
| TEA024930.1 | Down | Trans | bHLH155 |
| TEA008962.1 | Down | Trans | MADS-box transcription factor 23 |
| TEA015017.1 | Up | Trans | ERF110 (ABR1) |
| TEA022018.1 | Down | Trans | bHLH74 |
| TEA007969.1 | Up | Trans | WRKY transcription factor 31 |
| TEA010590.1 | Down | Trans | bHLH transcription factor 1 |
| TEA010741.1 | Down | Trans | MYC1 |
| TEA005334.1 | Up | Trans | WRKY transcription factor 31 |
| TEA021401.1 | Up | Trans | ERF4 |
| TEA000833.1 | Up | Trans | MYC2 |
| TEA005142.1 | Up | Trans | WRKY6 |
Interactions of DE-lncRNAs With miRNAs
In total, 12 lncRNAs were predicted to be target mimics of 17 known mature miRNAs in C. sinensis (Table 4). Interestingly, four miRNAs, lja-miR7539, csn-miR156h, ath-miR156i, and csn-miR156h, were predicted to be targeted by more than one lncRNA. Similarly, five lncRNAs, MSTRG.56302.5, MSTRG.116911.1, MSTRG.36615.10, MSTRG.55773.2, and MSTRG.92784.12, were predicted to be targets of more than one miRNA. Thus, it can be inferred that these lncRNAs and miRNAs may form a complex regulatory network in response to salt stress.
TABLE 4.
Putative targets of lncRNAs for miRNAs.
| miRNA Acc. | Target Acc. | Expect | UPE | miRNA-aligned fragment | Target-aligned fragment | Inhibition |
| ath-miR156i | MSTRG.56302.5 | 1 | 11.063 | GAGAGAGAGAGAGAGAGCAG | UUUCUCUCUCUCUCUCUCUC | Cleavage |
| csn-miR156h | MSTRG.116911.1 | 1 | 4.055 | UGAGAGAGAGAGAGAGAGCAU | CCCCUCUCUCUCUCUCUCUCA | Cleavage |
| gma-miR1533c | MSTRG.118391.1 | 1 | 10.118 | AAAAUAAAAAUAAUAAUAA | UUAUUUUUAUUUUUAUUUU | Cleavage |
| lja-miR7539 | MSTRG.36615.10 | 1 | 19.202 | GAGAGAGAGAGAGCGAGAGG | CCUCUCUCUCUCUCUCUCUC | Cleavage |
| MSTRG.56302.5 | 1 | 10.734 | GAGAGAGAGAGAGCGAGAGG | UCUCUCUCUCUCUCUCUCUC | Cleavage | |
| MSTRG.55773.2 | 1 | 1.11 | GAGAGAGAGAGAGCGAGAGG | UCUCUCUCUCUCUCUCUCUC | Cleavage | |
| MSTRG.92784.12 | 1 | 6.259 | GAGAGAGAGAGAGCGAGAG | CUCUCUCUCUCUCUCUCUC | Cleavage | |
| ath-miR426 | MSTRG.106114.4 | 1.5 | 5.662 | AUUUGGAAAAGGAAAGAGAAAAG | UUUUUCUUUUUUCUUUUCUAAAU | Cleavage |
| ath-miR5021 | MSTRG.26810.3 | 1.5 | 6.423 | AGAGAAGAAGAAGAAGAAAA | UCUUCUUCUUCUUCUUCUUU | Cleavage |
| ath-miR5998b | MSTRG.143628.1 | 1.5 | 7.673 | UUAGUUUUUGUUUUGUUUUGU | AAAAAACAAAACAAAAACAAA | Cleavage |
| csn-miR156h | MSTRG.92784.12 | 1.5 | 6.259 | UGAGAGAGAGAGAGAGAGCA | CUCUCUCUCUCUCUCUCUCG | Cleavage |
| MSTRG.56302.5 | 1.5 | 6.06 | UGAGAGAGAGAGAGAGAGCAU | UCUCUCUCUCUCUCUCUCUCG | Cleavage | |
| lja-miR7539 | MSTRG.91236.3 | 1.5 | 7.404 | GAGAGAGAGAGAGCGAGAGG | ACUCUCUCUUUCUCUCUCUC | Cleavage |
| ppe-miR6281 | MSTRG.92784.12 | 1.5 | 6.259 | AUGAGAGAGAGAGAGAGUGAG | CUCUCUCUCUCUCUCUCUCGU | Cleavage |
| ath-miR156i | MSTRG.36615.10 | 2 | 17.653 | GAGAGAGAGAGAGAGAGCAG | GCCCUCUCUCUCUCUCUCUC | Cleavage |
| MSTRG.55773.2 | 2 | 1.11 | GAGAGAGAGAGAGAGAGCAG | CAUCUCUCUCUCUCUCUCUC | Cleavage | |
| MSTRG.116911.1 | 2 | 4.044 | GAGAGAGAGAGAGAGAGCAG | CCCCUCUCUCUCUCUCUCUC | Cleavage | |
| ath-miR5021 | MSTRG.88836.3 | 2 | 5.697 | AGAGAAGAAGAAGAAGAAAA | CUAUGUUCUUCUUCUUCUCU | Cleavage |
| csn-miR156h | MSTRG.36615.10 | 2 | 18.775 | UGAGAGAGAGAGAGAGAGCAU | GCCCUCUCUCUCUCUCUCUCU | Cleavage |
| MSTRG.55773.2 | 2 | 1.11 | UGAGAGAGAGAGAGAGAGCAU | CAUCUCUCUCUCUCUCUCUCU | Cleavage | |
| lja-miR7539 | MSTRG.116911.1 | 2 | 4.044 | GAGAGAGAGAGAGCGAGAGG | CCCCUCUCUCUCUCUCUCUC | Cleavage |
| mtr-miR2586a | MSTRG.123314.2 | 2 | 18.404 | AGAGGUGUCCGUGCUUCAU | AUGAAGCAUGGACACAUCU | Cleavage |
| ppe-miR6281 | MSTRG.116911.1 | 2 | 4.06 | AUGAGAGAGAGAGAGAGUGAG | CCCUCUCUCUCUCUCUCUCAC | Cleavage |
| ptc-miR6462a | MSTRG.56302.5 | 2 | 5.5 | UCUCUUUUGCAUUUUUGCUGCC | GAGAGCAAAGAUGAAGAAGAGA | Translation |
LncRNA MSTRG.139242.1 May Interact With Its Nearby mRNATEA027212.1
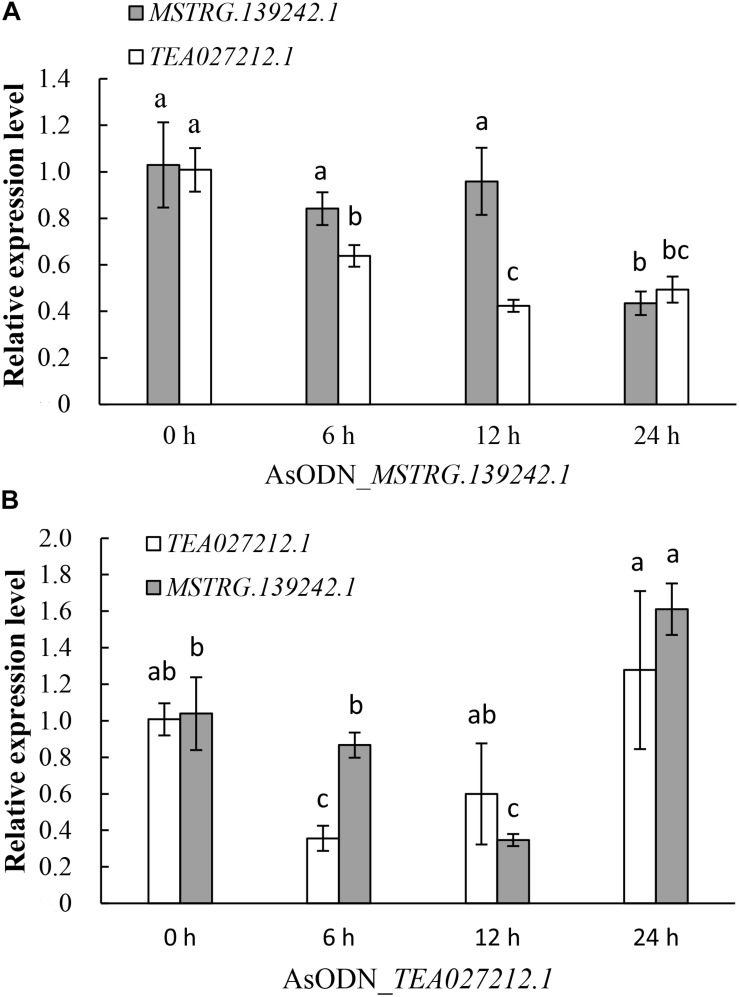
Among all of the target genes of the DE-lncRNAs, we found an upregulated coding gene, TEA027212.1 (Scaffold765 1270655–1272793), which was annotated as Ca2+-transporting ATPase 13, that was located only 5,911 bp adjacent to the upregulated lncRNA MSTRG.139242.1 (Scaffold765 1278704–1412939) according to the tea tree genome. To verify their expression relationship, a gene suppression assay in tea plants using AsODNs was conducted with MSTRG.139242.1 and TEA027212.1. qRT-PCR results showed that after gene suppression of lncRNA MSTRG.139242.1, its expression level was not significantly reduced until 12–24 h, but TEA027212.1expression level was significantly reduced from 0 to 12 h (Figure 7A). However, gene suppression of TEA027212.1 from 0 to 6 h significantly reduced MSTRG.139242.1 transcript levels from 6 to 12 h (Figure 7B). These results indicate that the lncRNA MSTRG.139242.1 may be regulated by its nearby coding gene, TEA027212.1, and may be involved in Ca2+ transport in response to salt stress in tea plants.
FIGURE 7.
Quantitative real-time PCR (qRT-PCR) analysis of the lncRNA MSTRG.139242.1 and its target mRNATEA027212.1, using tea leaves treated with AsODN_MSTRG.139242.1 and AsODN_TEA027212.1 for 0, 6, 12, and 24 h, respectively. Different letters in the columns represent a significant difference in the qRT-PCR results between the control and antisense oligodeoxynucleotide (AsODN)-treated individuals (P < 0.05).
qRT-PCR Validation
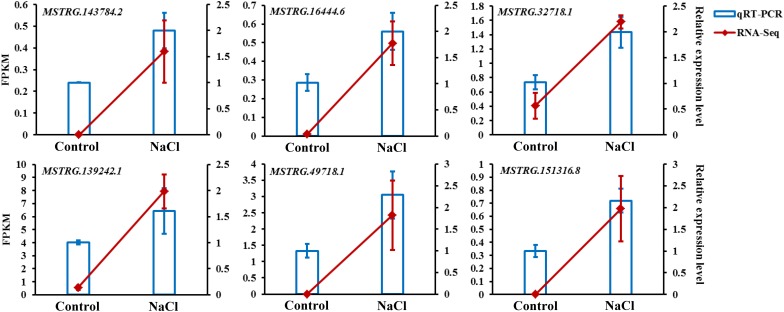
To confirm the reliability of the expression levels of lncRNAs obtained from the RNA-Seq transcriptome, six DE-lncRNAs, MSTRG.143784.2, MSTRG.16444.6, MSTRG.32718.1, MSTRG.139242.1, MSTRG.49718.1, and MSTRG.151316.8, were randomly selected for qRT-PCR validation. As shown in Figure 8, the expression levels of these lncRNAs closely corresponded to the transcript level estimated from the sequence data, which indicates the reproducibility and accuracy of the RNA-Seq results.
FIGURE 8.
qRT-PCR validation of six DE-lncRNAs selected from the RNA-Seq transcriptome. The expression levels of the RNA-Seq results were calculated using the FPKM value presented on the left axis; qRT-PCR relative expression levels were calculated using the 2– ΔΔCt method presented on the right axis; the values shown are the means ± standard deviation of n = 3 replicates.
Data Deposition
The sequencing data were deposited in the NCBI Sequence Read Archive (SRA) database13 under accession number SRP107589. LncRNA sequences identified are attached in Supplementary Table S4.
Discussion
Salt stress is an important factor that affects the growth of plants, including tea plants, under natural conditions, and the mechanism of the response to salt stress and salt stress-resistance breeding have been studied for a long period. Over the past decade, a number of genes involved in salt stress have been identified and verified, in addition to their regulatory pathways that mediate the transduction of stress signals and the process of the salt-stress response, such as the Ca2+ signaling pathway, ABA pathway, and MAPK cascade (Boudsocq and Sheen, 2009; Cheong and Kim, 2010; Yoshida et al., 2014; Abbasi et al., 2016; Zhu, 2016). In tea plants, many coding genes, such as AQP, VQ, and SnRK2, participate in the salt-stress response (Yue et al., 2014; Guo et al., 2018; Zhang et al., 2018). However, a few studies have been performed on the identification and mechanism of salt stress-related non-coding RNAs, especially lncRNAs, although lncRNAs have been proven to be involved in many biological processes in plants (Ponjavic et al., 2007; Ponting et al., 2009). In this study, we systematically identified tea plant lncRNAs based on the latest tea tree genome to find lncRNAs associated with salt stress and preliminarily analyzed their possible interactions with mRNAs or miRNAs. A number of lncRNAs were identified for the first time to be specifically expressed under high concentrations of NaCl and involved in the stress response.
A total of 16,452 candidate lncRNAs from six tea leaf samples were identified in this study. Similar to the lncRNAs identified in other species (Liu et al., 2012; Shuai et al., 2014; Zhu et al., 2015; Tian et al., 2016; Ma et al., 2018), the average length of lncRNAs in tea plants was much shorter than that of mRNAs, and most lncRNAs (66.9%) were lincRNAs. In some plants, functional lncRNAs that were differentially expressed under specific conditions were identified using transcriptome sequencing, and the functions were validated. For example, Zhu et al. (2015) identified 490 lncRNAs that were significantly upregulated in tomato ripening mutants using RNA-Seq, and two novel lncRNAs, lncRNA1459 and lncRNA1840, were proven to function in the delay of fruit ripening by gene silencing and mutagenesis (Li et al., 2018). Ding et al. (2014) conducted RNA-seq of salt-treated Arabidopsis seedlings, and Qin et al. (2017) obtained a putative lncRNA named DRIR that was later proven to increase tolerance to drought and salt stress. Similarly, 172 lncRNAs with various functions were found to be differentially expressed under NaCl stress in our results. Thus, these lncRNAs are involved in the salt-stress response of tea plants, and the results can provide candidate genes for salt tolerance studies in tea plants.
Previous studies have reported that lncRNAs play important roles in a variety of biological processes in response to stress by acting directly or indirectly on functional genes in plants (Wu et al., 2013; Fatica and Bozzoni, 2014; Wang et al., 2017). Currently, the interactions between lncRNAs, mRNAs, and miRNAs are the main research hotspots. Thus, in this study, the interactions between lncRNAs and mRNAs, and between lncRNAs and miRNAs were predicted.
To the best of our knowledge, lncRNAs can negatively or positively regulate the expression of protein-coding genes by acting in cis or in trans; lncRNAs work in cis when their target genes are on the same chromosome and within a close distance, and lncRNAs work in trans when they affect genes on other chromosomes (Kornienko et al., 2013). In addition, recent studies found that some cis-acting lncRNAs also have the ability to act in trans (Martianov et al., 2007; Schmitz et al., 2010). In this study, possible cis- and trans-target genes were predicted and annotated to explore the possible functions of the DE-lncRNAs. In total, 250 cis- and 421 trans-target genes of these DE-lncRNAs were predicted. GO and KEGG analyses indicated that these lncRNAs participated in various pathways, such as catalytic activity, galactose metabolism, and biosynthesis of amino acids in response to salt stress. Notably, 42 cis- and 67 trans-target genes were differentially expressed under salt stress. Among these target genes, Ca2+-transporting ATPase (TEA027212.1) was reported to be an important gene in Ca2+ signaling under abiotic stress (Wilkins et al., 2016), which suggests that its corresponding lncRNA, MSTRG.139242.1, may interact with Ca2+-transporting ATPase and may be involved in this process. Thus, a gene suppression assay of MSTRG.139242.1 and TEA027212.1 (Ca2+-transporting ATPase 13) using AsODNs was conducted and suggested that lncRNA MSTRG.139242.1 may be regulated by its nearby coding gene, TEA027212.1. In a previous study, the tomato lncRNA lncRNA33732 activated by WRKY1 induces RBOH expression and conferred resistance to Phytophthora infestans infection (Cui et al., 2018). In this study, we can speculate that lncRNA MSTRG.139242.1 may interact with its nearby mRNA TEA027212.1 in response to salt stress. In addition, an auxin-responsive protein (TEA005327.1) and ethylene-responsive TF ABR1 (TEA015017.1), the target genes of the lncRNAs MSTRG.49718.1 and MSTRG.54711.1, respectively, have been reported to be involved in the auxin, ethylene, and ABA signal transduction pathways in response to abiotic stresses in other plants (Pandey et al., 2005; Jain and Khurana, 2009; Feng et al., 2015), indicating that these two lncRNAs may participate in hormone signal transduction in response to salt stress in tea plants. We speculate that the expression of these target mRNAs may be regulated by their upstream lncRNAs in response to salt stress.
In addition, it is worth noting that many lncRNA target genes were classified as GOLS, calcium signaling, and some crucial TFs. GOLS was reported to be a key enzyme in the synthesis of raffinose family oligosaccharides (RFOs) and catalyzes the condensation of UDP-galactose with myo-inositol to produce galactinol as the sole donor for the synthesis of RFOs. In addition, RFOs are used for the transport and storage of carbohydrates and as compatible solutes for protection against abiotic and biotic stresses (Peters and Keller, 2009; Philippe et al., 2010). Sun Z.B. et al. (2013) have reported that TsGOLS2 enhances tolerance to high salinity and osmotic stresses in Arabidopsis thaliana. Thus, in the present study, lncRNAs targeting the eight upregulated GOLS genes may participate in RFO synthesis in response to salt stress. The Ca2+ signaling pathway was also reported to mediate plant response to salt stress, which starts with Ca2+ transporters such as Ca2+-ATPases, Ca2+ sensors, and relay proteins such as CaM, CMLs, CDPKs, CBLs, and CIPKs (Wilkins et al., 2016). LncRNAs targeting calcium-transporting ATPase, calmodulin-interacting protein, CDPKs and CIPKs in this study were predicted to be involved in salt-stress response through the Ca2+ signaling pathway. TFs have long been an important topic in plant resistance research, and some TF families such as WRKY, bHLH, ERF, GATA, and MYC play significant roles in salt stress adaptation (Lindemose et al., 2013). Here, the results provide many candidate lncRNAs, which may interact with vital TFs and response to salt stress.
On the other hand, lncRNAs can also interact with miRNAs. Some miRNAs have been found to respond to different environmental stresses (Zhang, 2015). Some lncRNAs can bind to miRNAs, thus inhibiting the effect of miRNAs on downstream genes. This type of lncRNA is termed an endogenous target mimic (eTM) (Wu et al., 2013). Jiang et al. (2019) reported that the tomato lncRNA23468 functions as an eTM to modulate NBS-LRR genes by mimicking miR482b in the tomato–P. infestans interaction. In this study, 12 lncRNAs were predicted to be target mimics of 17 known mature miRNAs. Interestingly, five lncRNAs, MSTRG.56302.5, MSTRG.116911.1, MSTRG.92784.12, MSTRG.36615.10, and MSTRG.55773.2, were predicted to be eTMs of miR156. According to previous reports, miR156 is induced by drought, and salinity stresses in several different plant species specifically regulates downstream SPL TFs and affects plant morphology, phase change, and seed germination (Stief et al., 2014). Thus, the five lncRNAs may influence the effect of miRNA156 on SPL in response to salt stress. These results provide candidate lncRNAs for exploring the interactions between lncRNAs and miRNAs in response to salt stress.
Conclusion
In the present study, genome-wide identification of lncRNAs was performed using high-throughput RNA-Seq and bioinformatics analysis. lncRNAs (16,452) were identified in tea plants, among which, 172 lncRNAs were differentially expressed and responsive to salt stress by cis- or trans-interaction with important coding genes. These lncRNAs may participate in GOLS, calcium signaling pathway, and interact with TFs in response to salt stress. Notably, 35 DE-lncRNAs were predicted to interact with 42 differentially expressed coding genes, which may participate in pathways such as the auxin response, ABA, and Ca2+ signal transduction pathways under salt stress. AsODN suppression of the lncRNA MSTRG.139242.1 and its predicted interacting gene, TEA027212.1 (Ca2+-ATPase 13), indicated that MSTRG.139242.1 interacts with Ca2+-ATPase 13 in the Ca2+-transport pathway in response to high salinity in tea plants. In addition, 12 lncRNAs were predicted to be target mimics of 17 known mature miRNAs in C. sinensis, thereby affecting the expression of downstream functional genes. This study can provide a source of lncRNAs and benefit an in-depth understanding of the function and regulatory mechanisms in tea plant response to salt stress.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
YYu and YYa designed the experiments. SW and YZ performed the experiments and data analysis. SW and YZ wrote the manuscript. WW and QX provided valuable advice on the manuscript. LH and MD revised the manuscript. All authors discussed the results and contributed to the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the earmarked fund for Modern Agro-industry Technology Research System (CARS-19), the Agricultural Special Fund Project of Shaanxi Province, and the special fund for University-Supported Extension Model (XTG2019-04).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00218/full#supplementary-material
Statistics of DE-lncRNAs.
Statistics of cis-target genes of DE-lncRNAs.
Statistics of trans-target genes of DE-lncRNAs.
LncRNA sequences identificated in this study.
Sequences for AsODN assay, primers used in qRT-PCR and differentially expressed target genes.
References
- Abbasi H., Jamil M., Haq A., Ali S., Ahmad R., Malik Z., et al. (2016). Salt stress manifestation on plants, mechanism of salt tolerance and potassium role in alleviating it: a review. Zemdirbyste Agric. 103 229–238. 10.13080/z-a.2016.103.030 [DOI] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J. H., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerner S., McGinnis K. M. (2012). Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS One 7:e43047. 10.1371/journal.pone.0043047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Sheen J. (2009). “Stress signaling II: calcium sensing and signaling,” in Abiotic Stress Adaptation in Plants, eds Pareek A., Sopory S., Bohnert H. (Dordrecht: Springer; ), 10.1007/978-90-481-3112-9-4 [DOI] [Google Scholar]
- Cheong Y. H., Kim M. C. (2010). Functions of MAPK cascade pathways in plant defense signaling. Plant Pathol. J. 26 101–109. 10.5423/PPJ.2010.26.2.101 [DOI] [Google Scholar]
- Cui J., Jiang N., Meng J., Yang G., Liu W., Zhou X., et al. (2018). LncRNA33732-respiratory burst oxidase module associated with WRKY1 in tomato-Phytophthora infestans interactions. Plant J. 97 933–946. 10.1111/tpj.14173 [DOI] [PubMed] [Google Scholar]
- Cui J., Luan Y. S., Jiang N., Bao H., Meng J. (2017). Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 89 577–589. 10.1111/tpj.13408 [DOI] [PubMed] [Google Scholar]
- Dai X., Zhuang Z., Zhao P. X. (2018). psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46 W49–W54. 10.1093/nar/gky316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F., Cui P., Wang Z. Y., Zhang S. D., Ali S., Xiong L. M. (2014). Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genom. 15:431. 10.1186/1471-2164-15-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J. H., Lu Q., Ouyang Y. D., Mao H. L., Zhang P. B., Yao J. L., et al. (2012). A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. U.S.A. 109 2654–2659. 10.1073/pnas.1121374109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Bozzoni I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15 7–21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- Feng S., Yue R., Tao S., Yang Y., Zhang L., Xu M., et al. (2015). Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J. Integr. Plant Biol. 57 783–795. 10.1111/jipb.12327 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42 D222–D230. 10.1093/nar/gkh121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. H., Chen J. F., Yang J. K., Yu Y. B., Yang Y. J., Wang W. D. (2018). Identification, characterization and expression analysis of the VQ motif-containing gene family in tea plant (Camellia sinensis). BMC Genom. 19:710. 10.1186/s12864-018-5107-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R., Wang H., Liu J., Boix M., Huang L. F., Chua N. H. (2017). The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. New Phytol. 216 854–867. 10.1111/nph.14703 [DOI] [PubMed] [Google Scholar]
- Heo J. B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331 76–79. 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- Jain M., Khurana J. P. (2009). Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J. 276 3148–3162. 10.1111/j.1742-4658.2009.07033.x [DOI] [PubMed] [Google Scholar]
- Jeyaraj A., Liu S. R., Zhang X., Zhang R., Shangguan M. Z., Wei C. L. (2017a). Genome-wide identification of microRNAs responsive to Ectropis oblique feeding in tea plant (Camellia sinensis L.). Sci. Rep. 7:13634. 10.1038/s41598-017-13692-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaraj A., Zhang X., Hou Y., Shangguan M. Z., Gajjeraman P., Li Y. Y., et al. (2017b). Genome-wide identification of conserved and novel microRNAs in one bud and two tender leaves of tea plant (Camellia sinensis) by small RNA sequencing, microarray-based hybridization and genome survey scaffold sequences. BMC Plant Biol. 17:212. 10.1186/s12870-017-1169-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Cui J., Shi Y. S., Yang G. L., Zhou X. X., Hou X. X., et al. (2019). Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction. Horticul. Res. 6:28. 10.1038/s41438-018-0096-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32 D277–D280. 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D., Rinn J. (2012). Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genom. Biol. 13:R107. 10.1186/gb-2012-13-11-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Landmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 357–U121. 10.1038/NMETH.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Zhang Y., Ye Z. Q., Liu X. Q., Zhao S. Q., Wei L., et al. (2007). CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35 W345–W349. 10.1093/nar/gkm391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko A. E., Guenzl P. M., Barlow D. P., Pauler F. M. (2013). Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11:59. 10.1186/1741-7007-11-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. W., Ma W., Zeng P., Wang J. Y., Geng B., Yang J. C., et al. (2015). LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 16 806–812. 10.1093/bib/bbu048 [DOI] [PubMed] [Google Scholar]
- Li R., Fu D. Q., Zhu B. Z., Luo Y. B., Zhu H. L. (2018). CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 94 513–524. 10.1111/tpj.13872 [DOI] [PubMed] [Google Scholar]
- Lindemose S., O’Shea C., Jensen M. K., Skriver K. (2013). Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 14 5842–5878. 10.3390/ijms14035842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G. F., Liu J. J., He Z. R., Wang F. M., Yang H., Yan Y. F., et al. (2018). Implementation of CsLIS/NES in linalool biosynthesis involves transcript splicing regulation in Camellia sinensis. Plant Cell Environ. 41 176–186. 10.1111/pce.13080 [DOI] [PubMed] [Google Scholar]
- Liu J., Jung C., Xu J., Wang H., Deng S. L., Bernad L., et al. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24 4333–4345. 10.1105/tpc.112.102855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lv J., Cui W., Liu H. B., He H. J., Xiu Y. C., Guo J., et al. (2013). Identification and characterization of long non-coding RNAs related to mouse embryonic brain development from available transcriptomic data. PLoS One 8:e71152. 10.1371/journal.pone.0071152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. D., Hao Y., Dong X. R., Gong Q. T., Chen J. Q., Zhang J. F., et al. (2012). Molecular mechanisms and function prediction of long noncoding RNA. Sci. World J. 2012:541786. 10.1100/2012/541786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K. S., Shi W. S., Xu M. Y., Liu J. X., Zhang F. X. (2018). Genome-wide identification and characterization of long non-coding RNA in wheat roots in response to Ca2+channel blocker. Front. Plant Sci. 9:244. 10.3389/fpls.2018.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I., Ramadass A., Barros A. S., Chow N., Akoulitchev A. (2007). Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445 666–670. 10.1038/nature05519 [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Mattick J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10 155–159. 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- Pandey G. K., Grant J. J., Cheong Y. H., Kim B. G., Li L., Luan S. (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 139 1185–1193. 10.1104/pp.105.066324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11 1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S., Keller F. (2009). Frost tolerance in excised leaves of the common bugle (Ajuga reptans L.) correlates positively with the concentrations of raffinose family oligosaccharides (RFOs). Plant Cell Environ. 32 1099–1107. 10.1111/j.1365-3040.2009.01991.x [DOI] [PubMed] [Google Scholar]
- Philippe R. N., Ralph S. G., Mansfield S. D., Bohlmann J. (2010). Transcriptome profiles of hybrid poplar (Populus trichocarpa × deltoides) reveal rapid changes in undamaged, systemic sink leaves after simulated feeding by forest tent caterpillar (Malacosoma disstria). New Phytol. 188 787–802. 10.1111/j.1469-8137.2010.03392.x [DOI] [PubMed] [Google Scholar]
- Ponjavic J., Ponting C. P., Lunter G. (2007). Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 17 556–565. 10.1101/gr.6036807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Oliver P. L., Reik W. (2009). Evolution and Functions of Long Noncoding RNAs. Cell 136 629–641. 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- Qin T., Zhao H. Y., Cui P., Albesher N., Xiong L. M. (2017). A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 175 1321–1336. 10.1104/pp.17.00574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz K. M., Mayer C., Postepska A., Grummt I. (2010). Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24 2264–2269. 10.1101/gad.590910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai P., Liang D., Tang S., Zhang Z. J., Ye C. Y., Su Y. Y., et al. (2014). Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 65 4975–4983. 10.1093/jxb/eru256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumayla, Sharma S., Taneja M., Tyagi S., Singh K., Upadhyay S. K. (2017). Survey of high throughput RNA-Seq data reveals potential roles for lncRNAs during development and stress response in bread wheat. Front. Plant Sci. 8:1019. 10.3389/fpls.2017.01019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief A., Altmann S., Hoffmann K., Pant B. D., Scheible W. R., Baurle I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26 1792–1807. 10.1105/tpc.114.123851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Luo H. T., Bu D. C., Zhao G. G., Yu K. T., Zhang C. H., et al. (2013). Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 41:e166. 10.1093/nar/gkt646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. B., Qi X. Y., Wang Z. L., Li P. H., Wu C. X., Zhang H., et al. (2013). Overexpression of TsGOLS2, a galactinol synthase, in Arabidopsis thaliana enhances tolerance to high salinity and osmotic stresses. Plant Physiol. Biochem. 69 82–89. 10.1016/j.plaphy.2013.04.009 [DOI] [PubMed] [Google Scholar]
- Sun P., Zhang Z. L., Zhu Q. F., Zhang G. Y., Xiang P., Lin Y. L., et al. (2018). Identification of miRNAs and target genes regulating catechin biosynthesis in tea (Camellia sinensis). J. Integr. Agricult. 17 1154–1164. 10.1016/S2095-3119(17)61654-X [DOI] [Google Scholar]
- Swiezewski S., Liu F. Q., Magusin A., Dean C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 462 799–U122. 10.1038/nature08618 [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28 33–36. 10.1093/Nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J. X., Song Y. P., Du Q., Yang X. H., Ci D., Chen J. H., et al. (2016). Population genomic analysis of gibberellin-responsive long non-coding RNAs in Populus. J. Exp. Bot. 67 2467–2482. 10.1093/jxb/erw057 [DOI] [PubMed] [Google Scholar]
- Upadhyaya H., Panda S. K. (2013). Abiotic stress responses in tea [Camellia sinensis L (O) Kuntze]: an overview. Rev. Agricult. Sci. 1 1–10. 10.7831/ras.1.1 26308611 [DOI] [Google Scholar]
- Varshney D., Rawal H. C., Dubey H., Bandyopadhyay T., Bera B., Kumar P. M., et al. (2019). Tissue specific long non-coding RNAs are involved in aroma formation of black tea. Indust. Crops Products 133 79–89. 10.1016/j.indcrop.2019.03.020 [DOI] [Google Scholar]
- Wan S. Q., Wang W. D., Zhou T. S., Zhang Y. H., Chen J. F., Xiao B., et al. (2018). Transcriptomic analysis reveals the molecular mechanisms of Camellia sinensis in response to salt stress. Plant Growth Regul. 84 481–492. 10.1007/s10725-017-0354-4 [DOI] [Google Scholar]
- Wang J. J., Meng X. W., Dobrovolskaya O. B., Orlov Y. L., Chen M. (2017). Non-coding RNAs and their roles in stress response in plants. Genom. Proteom. Bioinform. 15 301–312. 10.1016/j.gpb.2017.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Park H. J., Dasari S., Wang S. Q., Kocher J. P., Li W. (2013). CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 41:6. 10.1093/nar/gkt006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Yang H., Wang S., Zhao J., Liu C., Gao L., et al. (2018). Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc Natl Acad Sci U.S.A. 115 E4151–E4158. 10.1073/pnas.1719622115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins K. A., Matthus E., Swarbreck S. M., Davies J. M. (2016). Calcium-mediated abiotic stress signaling in roots. Front. Plant Sci. 7:1296. 10.3389/fpls.2016.01296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. J., Wang Z. M., Wang M., Wang X. J. (2013). Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 161 1875–1884. 10.1104/pp.113.215962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Mogami J., Yamaguchi-Shinozaki K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21 133–139. 10.1016/j.pbi.2014.07.009 [DOI] [PubMed] [Google Scholar]
- Yue C., Cao H. L., Wang L., Zhou Y. H., Hao X. Y., Zeng J. M., et al. (2014). Molecular cloning and expression analysis of tea plant aquaporin (AQP) gene family. Plant Physiol. Biochem. 83 65–76. 10.1016/j.plaphy.2014.07.011 [DOI] [PubMed] [Google Scholar]
- Zhang B. H. (2015). MicroRNA: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 66 1749–1761. 10.1093/jxb/erv013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhu X. J., Chen X., Song C. N. A., Zou Z. W., Wang Y. H., et al. (2014). Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol. 14:271. 10.1186/S12870-014-0271-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. C., Liao J. Y., Li Z. Y., Yu Y., Zhang J. P., Li Q. F., et al. (2014). Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genom. Biol. 15:512. 10.1186/s13059-014-0512-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. C., Chen Y. Q. (2013). Long noncoding RNAs: new regulators in plant development. Biochem. Biophys. Res. Commun. 436 111–114. 10.1016/j.bbrc.2013.05.086 [DOI] [PubMed] [Google Scholar]
- Zhang Y. H., Wan S. Q., Wang W. D., Chen J. F., Huang L. L., Duan M. S., et al. (2018). Genome-wide identification and characterization of the CsSnRK2 family in Camellia sinensis. Plant Physiol. Biochem. 132 287–296. 10.1016/j.plaphy.2018.09.021 [DOI] [PubMed] [Google Scholar]
- Zhu B. Z., Yang Y. F., Li R., Fu D. Q., Wen L. W., Luo Y. B., et al. (2015). RNA sequencing and functional analysis implicate the regulatory role of long non-coding RNAs in tomato fruit ripening. J. Exp. Bot. 66 4483–4495. 10.1093/jxb/erv203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167 313–324. 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q. W., Luo Y. P. (2013). Identification of miRNAs and their targets in tea (Camellia sinensis). J. Zhejiang Univ. Sci. B 14 916–923. 10.1631/jzus.B1300006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistics of DE-lncRNAs.
Statistics of cis-target genes of DE-lncRNAs.
Statistics of trans-target genes of DE-lncRNAs.
LncRNA sequences identificated in this study.
Sequences for AsODN assay, primers used in qRT-PCR and differentially expressed target genes.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.