Reports of human-pathogenic C. braakii strains, especially of strains showing resistance to carbapenems, are rare. To the best of our knowledge, our results represent the first detection of carbapenemase gene blaKPC-2 in C. braakii strains. In addition, we have studied detailed genetic characteristics of the novel IncR/IncP6 hybrid plasmid pCRE3-KPC, which was isolated from a clinical multidrug-resistant Citrobacter braakii CRE3 strain. Our results may provide further insight into the horizontal transfer of multidrug resistance genes in bacteria and into the genomic diversity and molecular evolution of plasmids.
KEYWORDS: Citrobacter braakii, blaKPC-2, IncR, IncP6, plasmid, transposon
ABSTRACT
Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae have become widespread in hospitals and the environment. Here, we describe a blaKPC-2-carrying plasmid called pCRE3-KPC, which was recovered from a clinical multidrug-resistant Citrobacter braakii CRE3 strain in China. The complete nucleotide sequence of pCRE3-KPC was determined by combining MiSeq and MinION sequencing and then compared with those of three related plasmids. Plasmid conjugal transfer and electroporation tests, modified carbapenem inactivation method, and bacterial antimicrobial susceptibility test were carried out. We compared this plasmid with three related plasmids to verify that the backbone of pCRE3-KPC was composed of the backbones of the IncR plasmid and IncP6 plasmid. Further bioinformatics analysis showed that pCRE3-KPC carried two resistance-related regions (the blaKPC-2 gene cluster and the aacC2-tmrB-related region). The aacC2-tmrB-related region included two novel insertion sequences (ISCfr28 and ISCfr16).
IMPORTANCE Reports of human-pathogenic C. braakii strains, especially of strains showing resistance to carbapenems, are rare. To the best of our knowledge, our results represent the first detection of carbapenemase gene blaKPC-2 in C. braakii strains. In addition, we have studied detailed genetic characteristics of the novel IncR/IncP6 hybrid plasmid pCRE3-KPC, which was isolated from a clinical multidrug-resistant Citrobacter braakii CRE3 strain. Our results may provide further insight into the horizontal transfer of multidrug resistance genes in bacteria and into the genomic diversity and molecular evolution of plasmids.
INTRODUCTION
Klebsiella pneumoniae strains that produce K. pneumoniae carbapenemase (KPC) were initially identified in the United States in 2001 (1). Citrobacter braakii, as a member of the Citrobacter freundii complex, was identified in 1993 (2) and has rarely been reported as a human pathogen (3–6). The blaKPC-2 gene, as a subtype of KPC genes, has widely spread in Enterobacteriaceae, such as K. pneumoniae (1), Citrobacter freundii (7), C. portucalensis (8), and Escherichia coli (9) strains. However, the blaKPC-2 gene had not previously appeared in C. braakii strains. Moreover, it has been found to be carried on several plasmids to date, namely, IncR, IncP, IncFII, IncL/M, IncN, IncA, IncC, and IncX plasmids (10–12). As of 22 May 2019, 54 plasmids containing both the IncR replicon and the blaKPC-2 gene and 16 plasmids containing both the IncP6 replicon and the blaKPC-2 gene had been documented in the GenBank database, and there was no documented instance of an IncR/IncP6 hybrid plasmid (see Table S1 and S2 in the supplemental material).
Prevalence statistics of plasmids containing both the IncR replicon and the blaKPC-2 gene. Data represent statistics of plasmids containing both the IncR replicon and the blaKPC-2 gene documented as of 22 May 2019. Download Table S1, DOCX file, 0.02 MB (26.9KB, docx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prevalence statistics of plasmids containing both the IncP6 replicon and the blaKPC-2 gene. Data represent statistics of plasmids containing both the IncP6 replicon and the blaKPC-2 gene documented as of 22 May 2019. Download Table S2, DOCX file, 0.02 MB (19.5KB, docx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The IncR replicon was first described in 2009 (13); since then, IncR plasmids have been increasingly reported in Enterobacteriaceae isolates (14). IncR replicons have also been found either as single replicons or as parts of multireplicon plasmids, which includes associations with IncA/C, IncF, IncFIIk, or nontypeable backbones (15). On the basis of prevalence statistics of plasmids containing both the IncR replicon and the blaKPC-2 gene (Table S1), we found that these plasmids usually contain multiple replicons. The blaKPC-2-carrying plasmid unnamed3 (GenBank accession no. CP027150) contains one IncR replicon from the K. pneumoniae AR_0363 strain, which was that initially reported.
IncP6 plasmids have a broad host range (16), and to date the blaKPC-2-carrying IncP6 plasmids have been found in Pseudomonas aeruginosa (16), K. oxytoca (GenBank accession no. KY913901), Enterobacter cloacae (GenBank accession no. CP018968), and C. freundii (17). Both blaKPC-2-carrying IncP6 plasmid pCOL-1 (GenBank accession no. KC609323) (18) and p10265-KPC (GenBank accession no. KU578314) (16) were recovered from P. aeruginosa strains.
In this work, we have reported the first isolation of a blaKPC-2-positive C. braakii strain. In addition, we determined the whole genomic sequence of a blaKPC-2-carrying plasmid that we have named pCRE3-KPC, which was isolated from a clinical multidrug-resistant C. braakii CRE3 strain. We compared this plasmid with the following three related plasmids: plasmid unnamed3 (GenBank accession no. CP027150), p10265-KPC (GenBank accession no. KU578314), and pCOL-1 (GenBank accession no. KC609323). Interestingly, we found that plasmid pCRE3-KPC contains both an IncR replicon and an IncP6 replicon belonging to a novel IncR/IncP6 hybrid plasmid. To the best of our knowledge, this is the first report of an IncR/IncP6 hybrid plasmid. Our results may offer insight into the horizontal transfer of resistance genes and provide an overview of plasmid diversity and evolution.
RESULTS AND DISCUSSION
Characterization of C. braakii CRE3.
PCR screening revealed that the multiple antimicrobial resistance genes present in C. braakii CRE3 include blaKPC-2, blaTEM-1B, blaOXA-1, blaCMY-83, qnrB10, and aacC2. Plasmid pCRE3-KPC failed to transfer to E. coli EC600 through conjugation experiments but was successfully transferred to E. coli DH5α by electroporation to generate the blaKPC-2-positive electroporant CRE3-KPC-DH5α. This result illustrates that pCRE3-KPC is a nonconjugative but mobilizable plasmid. The antimicrobial susceptibility tests showed that both the C. braakii CRE3 and E. coli electroporant CRE3-KPC-DH5α strains were highly resistant to ampicillin, piperacillin, cefuroxime, ceftriaxone, aztreonam, imipenem, meropenem, and gentamicin (Table 1). Moreover, carbapenemase was produced in both of the strains mentioned above, as revealed by the modified carbapenem inactivation method (mCIM) (19).
TABLE 1.
Antimicrobial susceptibility profiles
| Antibiotic | MIC (mg/liter)/antimicrobial susceptibilitya
|
||
|---|---|---|---|
| C. braakii CRE3 | Electroporant CRE3-KPC-DH5α | E. coli DH5α | |
| Ampicillin | ≥32/R | ≥32/R | ≤2/R |
| Piperacillin | ≥128/R | ≥128/R | ≤4/S |
| Cefuroxime | ≥64/R | ≥64/R | 4/S |
| Ceftriaxone | ≥64/R | ≥64/R | ≤1/S |
| Ceftazidime | ≥64/R | 4/S | ≤1/S |
| Cefepime | ≥64/R | ≤1/S | ≤1/S |
| Aztreonam | ≥64/R | ≥64/R | ≤1/S |
| Imipenem | ≥16/R | ≥16/R | ≤1/S |
| Meropenem | ≥16/R | ≥16/R | ≤0.25/S |
| Amikacin | 32/I | ≤2/S | ≤2/S |
| Gentamicin | ≥16/R | ≥16/R | ≤1/S |
| Tobramycin | ≥16/R | 2/S | ≤2/S |
| Ciprofloxacin | ≥4/R | ≤0.25/S | ≤0.25/S |
| Levofloxacin | 4/I | ≤0.25/S | ≤0.25/S |
| Nitrofurantoin | 128/R | ≤16/S | ≤16/S |
The interpretation is derived from the Clinical and Laboratory Standards Institute guidelines (CLSI, 2018) (S, sensitive; R, resistant; I, intermediately resistant).
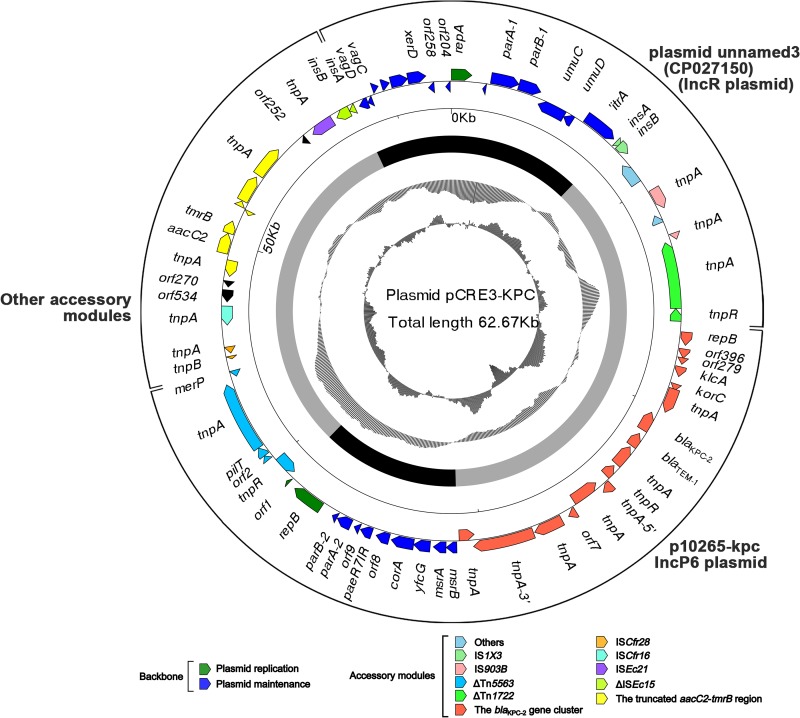
Overview of plasmid pCRE3-KPC.
The circular DNA sequence of pCRE3-KPC is 62,673 bp in length, with mean G+C content of 56%. Furthermore, it contains 71 predicted open reading frames (ORFs) and two distinct replicons (IncR replicon repA and IncP6 replicon repB) (Table 2) (Fig. 1).
TABLE 2.
Major features of plasmids in this work
| Plasmid | Accession no. or source | Species | Inc group | Country of origin | Total length (bp) | Total no. of ORFs | Mean G+C content (%) | Accessory module(s) (resistance genes harbored) |
|---|---|---|---|---|---|---|---|---|
| unnamed3 | CP027150 | K. pneumoniae | IncR | United States | 65,684 | 72 | 55 | MDR region, Tn4401a, aac(6′)-Ib-cr-related region |
| pCRE3-KPC | This study | C. braakii | IncR-P6 | China | 62,673 | 71 | 56 | blaKPC-2 gene cluster, aacC2-tmrB-related region |
| p10265-KPC | KU578314 | P. aeruginosa | IncP6 | China | 38,939 | 46 | 58 | blaKPC-2 gene cluster |
| pCOL-1 | KC609323 | P. aeruginosa | IncP6 | Colombia | 31,529 | 34 | 60 | Tn4401b |
FIG 1.
Schematic maps of plasmid pCRE3-KPC. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and in color, respectively. The innermost circle presents GC-skew [(G − C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle represents GC content.
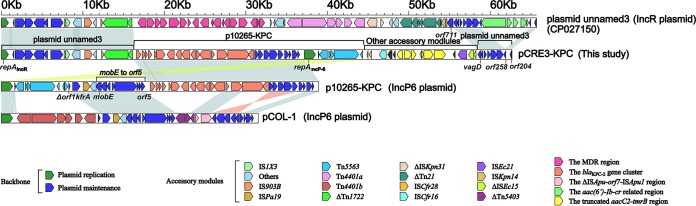
Linear comparisons of plasmid pCRE3-KPC with three related reference plasmids, namely, blaKPC-2-carrying IncR plasmid unnamed3 (GenBank accession no. CP027150), p10265-KPC (a blaKPC-2-carrying IncP6 plasmid first reported in China) (16), and pCOL-1 (a blaKPC-2-carrying IncP6 plasmid, initially identified in Colombia) (18), were conducted. The detailed comparisons revealed that the overall structure of plasmid pCRE3-KPC is highly mosaic and can be divided into the following three distinct modules (Fig. 1 and 2; see also Fig. S1 in the supplemental material): (i) a first module (∼20.5 kb) that is high homologous (>98.6% identity) to plasmid unnamed3 from the K. pneumoniae AR_0363 strain reported in the United States and extends from the resolution site (res) of ΔTn1722 to gene vagD (virulence-associated gene); (ii) a second module (∼27.8 kb) that shares >99.9% identity with plasmid p10265-KPC (16) from P. aeruginosa strain 10265 isolated in China and extends from the blaKPC-2 gene cluster to ΔTn5563; (iii) a third module comprising the other accessory modules (∼13.8 kb) with two novel insertion sequences (ISCfr28 and ISCfr16), the truncated aacC2-tmrB region, ISEc21, and ΔISEc15. On the basis of the study of the hybrid plasmids p675920-1 (20, 21) and pKP1034 (22), the majority of the backbone and accessory regions of unnamed3 and p10265-KPC were found to be present in pCRE3-KPC, so pCRE3-KPC may represent a combination resulting from plasmids like these. Compared to the backbone of unnamed3 and p10265-KPC, pCRE3-KPC lost part of its backbone genes (orf711 of unnamed3, Δorf1 and kfrA, and a fragment extending from mobE to orf5 of p10265-KPC) during the recombination process, suggesting that these genes may not be necessary in these plasmids. The gene functions of these plasmids are annotated in detail (see Data Set S1, S2, S3, and S4 in the supplemental material).
FIG 2.
Linear comparison of pCRE3-KPC with related plasmids. A linear comparison was carried out for the complete DNA sequences of plasmids unnamed3 (GenBank accession no. KC609323), pCRE3-KPC (this study), p10265-KPC (GenBank accession no. KU578314), and pCOL-1 (GenBank accession no. KC609323). Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on functional classification. Shading indicates regions of homology (>95% nucleotide identity). MDR, multidrug resistant.
Schematic maps of the related plasmids. The three plasmids unnamed3 (CP027150), p10265-KPC (KU578314), and pCOL-1 (KC609323) were included in the comparative analysis. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and in color, respectively. The innermost circle represents GC-skew [(G-C)/(G + C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle represents GC content. Download FIG S1, DOCX file, 1.2 MB (1.2MB, docx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Annotations of plasmid pCRE3-KPC. Download Data Set S1, XLSX file, 0.02 MB (23.7KB, xlsx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Annotations of plasmid unnamed3. Download Data Set S2, XLSX file, 0.02 MB (23.8KB, xlsx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Annotations of plasmid p10265-KPC. Download Data Set S3, XLSX file, 0.02 MB (18KB, xlsx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Annotations of plasmid pCOL-1. Download Data Set S4, XLSX file, 0.02 MB (16.1KB, xlsx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic comparison of the backbone regions from pCRE3-KPC and related plasmids.
The backbone of each plasmid was further divided into the replication genes and the plasmid maintenance genes, without the conjugal-transfer genes, such that the hybrid pCRE3-KPC plasmid comprised the IncR and IncP6 backbones. The resultant backbone includes two replication genes (IncR replicon repA and IncP6 replicon repB) and two sets of partitioning system parAB genes (Fig. 1).
The IncR backbone from pCRE3-KPC was compared with plasmid unnamed3 (an IncR plasmid; GenBank accession no. CP027150), and their backbones were found to consist of the replication genes (IncR replicon and its iterons) as well as plasmid maintenance genes (parAB, umuCD, and vagDC). However, two differences in their backbones were identified as follows: (i) the orf711 gene (hypothetical protein) is deleted in pCRE3-KPC but complete in plasmid unnamed3 and (ii) the orf258 gene (hypothetical protein) is interrupted into two parts by the insertion of the aac(6′)-Ib-cr-related region in plasmid unnamed3 (Fig. 1 and 2; see also Fig. S1).
Furthermore, p10265-KPC (16) and pCOL-1 (18) can be assigned to the IncP6 incompatibility group, according to replicon-based schemes. The IncP6 backbone of pCRE3-KPC was compared with those of both of the plasmids named above, and the backbones were found to comprise the replication genes (IncP6 replicon and its iterons) and plasmid maintenance genes (kfrA, parABC, the mob gene cluster, the msrB-msrA-yfcG-corA-orf8 gene cluster, and paeR7IR). Three differences were notable among them (Fig. 1 and 2; see also Fig. S1): (i) pCRE3-KPC has lost genes (Δorf1 and kfrA) and a fragment extending from mobE (auxiliary protein) to orf5 (hypothetical protein); (ii) the numbers of copies of the 17-bp tandem repeat (GCGCCTGCCTTTGAGTA) within the iterons were 11 in pCRE3-KPC, 6 in p10265-KPC, and 12 in pCOL-1; and (iii) the Δorf8-corA-yfcG-msrA-msrB gene cluster was found to be inverted in pCOL-1.
Genomic comparison of the blaKPC-2 gene region from pCRE3-KPC with those from related plasmids.
The blaKPC-2 gene is associated with the core blaKPC platform (Tn3-ISKpn27-blaKPC-ΔISKpn6) in most Chinese Enterobacteriaceae strains (23–25). This core platform is integrated into a ΔISEc33-associated blaKPC-2 cluster, which was initially discovered in the p10265-KPC plasmid from a P. aeruginosa strain (16). In the blaKPC-2 gene cluster of p10265-KPC, the primary genetic structure, Tn3-ISKpn27-blaKPC-2-ΔISKpn6-korC-orf6-klcA-ΔrepB, may have undergone two evolutionary events (16): (i) insertion of a ΔblaTEM-1 gene between ISKpn27 and the Tn3 IRR (right inverted repeat) and (ii) disruption of the tnpA gene (transposase) from Tn3, resulting in its becoming two separate parts, an event caused by insertion of a composite transposon, ISApu1-orf7-ISApu2. The blaKPC-2-carrying pCRE3-KPC plasmid was detected in an inpatient at a tertiary care hospital in China, and the BLASTN analysis of it showed that the surrounding genetic environment of the blaKPC-2 gene in pCRE3-KPC is highly similar to that in p10265-KPC. The ΔISApu1-orf7-ISApu2 composite transposon is also present in pCOL-1, but it has not been inserted into Tn3 and occurs downstream of ΔTn5403. Furthermore, the blaKPC-2 gene cluster is located downstream of ΔTn1722. Tn1722, a Tn3-family transposon, consists of an IRL (left inverted repeat), tnpA, tnpR (resolvase), res, mcp (methyl-accepting chemotaxis protein), and an IRR (26). ΔTn1722 contains an IRR, tnpA, tnpR, and res in pCRE3-KPC, which is also present in plasmid unnamed3 (GenBank accession no. CP027150) (Fig. 3).
FIG 3.
The blaKPC-2 gene region from pCRE3-KPC and comparison with the related plasmids. Genes are denoted by arrows. Mobile elements, genes, and other features are colored based on functional classification. Numbers in parentheses denote GenBank numbers and the nucleotide positions within the corresponding plasmids. Shaded regions show shared DNA regions of homology (>95% nucleotide identity). For reference, the accession number of Tn1722 is X61367.
However, the Tn3-family Tn4401 transposon has contributed to the rapid dissemination of the blaKPC-2 gene in Europe and the Americas. A number of previously reported isoforms of Tn4401, which differ by a 100-to-200-bp sequence upstream of blaKPC-2, are currently known (27–29). For example, Tn4401b, which is a Tn4401 isoform, contains IRL, tnpR, tnpA, ISKpn7, blaKPC-2, ISKpn6, and IRR. Plasmid pCOL-1 (18) and plasmid unnamed3 (GenBank accession no. CP027150) originated from Colombia and the United States, respectively. The blaKPC-2 genes carried by plasmid pCOL-1 and plasmid unnamed3 are embedded in Tn4401b and Tn4401a, respectively. Compared with the complete Tn4401b, Tn4401a in plasmid unnamed3 (GenBank accession no. CP027150) has lost a 135-bp sequence upstream of blaKPC-2 (Fig. 3).
Genomic comparison of the aacC2-tmrB-related region from pCRE3-KPC with those from related plasmids.
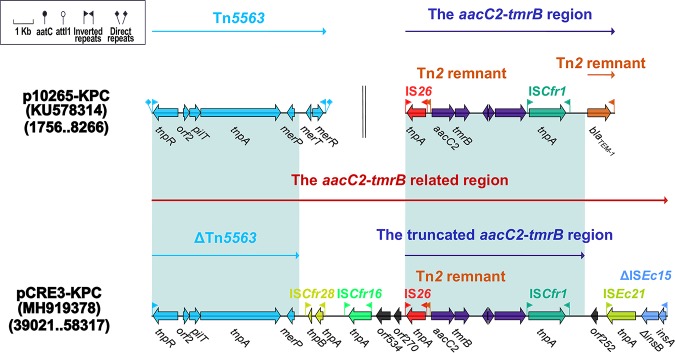
The aacC2-tmrB-related region from pCRE3-KPC is composed of ΔTn5563, two novel insertion sequences (ISCfr28 and ISCfr16), the truncated aacC2-tmrB region, ISEc21, and ΔISEc15. The Tn5563 element is organized sequentially with an IRL, tnpR, orf2 (hypothetical protein), pilT (PilT domain-containing protein), tnpA, merP (mercuric transport protein periplasmic component), merT (mercuric transport protein), merR (mercuric resistance operon regulatory protein), and an IRR. In p10265-KPC (16), Tn5563, which is located upstream of two consecutive backbone genes (Δorf1 and kfrA), differs from the prototype Tn5563 from pRA2 (30) with a 286-bp insertion occurring between merP (mercuric transport protein periplasmic component) and merT (mercuric transport protein). However, ΔTn5563 has undergone the deletion of a fragment extending from merT to the IRR in pCRE3-KPC (Fig. 2 and 3).
In addition, two novel insertion sequences (ISCfr28 and ISCfr16) are inserted downstream of ΔTn5563. ISCfr28, containing two transposase genes, tnpA and tnpB, and a Tn3 family element, is bordered by 13-bp IRs (IRL, GTCAGCCAAGAAG; IRR, CTTCTTGGCTGAC) (Fig. 4). The 1,025-bp ISCfr16 insertion sequence, a Tn3 family element, is made up of a transposase gene (tnpA) and 13-bp IRs (IRL, TAAGCTGCGAGCG; IRR, CGCTCGCAGCTAA). The aacC2 (aminoglycoside resistance)-tmrB (tunicamycin resistance) region is derived from transposon Tn2, and Tn2 has undergone the following molecular evolutionary changes (31, 32): (i) the tnpR-res-tnpA segment of Tn2 has been replaced by the aacC2-tmrB-orf192-orf228-orf1182-ISCfr1 module and (ii) the IS26 insertion sequence has been inserted at the right-hand end of Tn2. The complete aacC2-tmrB region was discovered in pEl1573 from E. cloacae (33), and its truncated forms have been integrated into transposon Tn6411 from the chromosome of P. aeruginosa 12939 (34). Because ISEc21 had inserted upstream of ISEc15, this may have led to the truncation of ISEc15 (Fig. 4).
FIG 4.
The aacC2-tmrB-related region from pCRE3-KPC and comparison with related plasmids. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on functional classification. Numbers in parentheses denote GenBank numbers and the nucleotide positions within the corresponding plasmids. Shaded regions indicate shared DNA regions of homology (>95% nucleotide identity). For reference, the accession number of the aacC2-tmrB region is JX101693.
MATERIALS AND METHODS
Bacterial isolates and identification.
The clinical C. braakii CRE3 strain was isolated from a drainage sample from a patient at a tertiary care hospital in China on 5 May 2018. Bacterial identification was carried out using a Vitek compact-2 automated system (bioMérieux, France) and was confirmed by 16S rRNA sequencing (35). The genes encoding extended-spectrum β-lactamase (36), carbapenemase (37), fluoroquinolone (38), and aminoglycoside (39) were detected by PCR. All the PCR amplicons were sequenced on an ABI 3730 platform (Applied Biosystems, USA).
Plasmid conjugal transfer.
The pCRE3-KPC plasmid was recovered from a clinical multidrug-resistant C. braakii CRE3 isolate. Conjugation experiments were carried out with cells of rifampin-resistant Escherichia coli strain EC600 as the recipient cells, and the transformation experiments were conducted using cells of E. coli DH5α Electro-Cells (TaKaRa, China) as the recipient cells for the plasmid electroporation. Plasmid pCRE3-KPC was extracted from the cells using a Qiagen Plasmid Midi kit (Qiagen, Germany). The plasmid conjugal transfer and electroporation tests were performed as described previously (40, 41).
Antimicrobial susceptibility and carbapenemase activity detection.
Antimicrobial susceptibility testing was conducted using a Vitek compact-2 automated system (bioMérieux, France). The results were interpreted according to the CLSI (Clinical and Laboratory Standards Institute) 2018 performance standards (42). Carbapenemase activities were detected using mCIM (19).
Sequencing and sequence assembly.
The bacterial genomic DNA extracted from the CRE3 isolate using a Wizard Genomic DNA purification kit (Promega, USA) was sequenced on the MiSeq (Illumina, USA) and the MinION (Oxford Nanopore, United Kingdom) platforms. The DNA library was constructed in accordance with a NEB Next Ultra II DNA Library Prep kit for Illumina, and the Illumina sequencing read length used was 300. The library preparations for the MinION platform were performed by the use of a rapid barcoding sequencing kit (SQK-RBK004) according to the protocol of the manufacturer (Oxford Nanopore Technologies), and the results were then loaded into the flow cell (FLO-MIN106D, Oxford Nanopore Technologies) for sequencing. Short Illumina reads were trimmed to remove poor-quality reads using Trimmomatic, and the contigs were assembled using Newbler3.0 (43). The long reads from MinION were combined with the short Illumina reads, which were subjected to hybrid assembly using SPAdesv3.11.1 (44). The hybrid assembly produced several scaffolds, and further bioinformatics analysis verified that the scaffold of the pCRE3-KPC plasmid was successfully cyclized by our in-house script. The correctness was then demonstrated by mapping the Illumina reads to the cyclized scaffold using CLC Genomics Workbench 9.0 (CLC Bio, Denmark), with an average level of read mapping coverage of 817×. The final consensus sequence obtained from CLC Genomics Workbench 9.0 was considered to represent the complete sequence of plasmid pCRE3-KPC.
Sequence annotation and genome comparisons.
Annotation of open reading frames (ORFs) and pseudogenes was performed using RAST2.0 (45) combined with BLASTP/BLASTN searches against the UniProtKB/Swiss-Prot (46) and RefSeq (47) databases. Resistance genes, mobile elements, and other features were predicted using ResFinder3.2 (48), INTEGRALL (49), ISfinder (50), and PlasmidFinder2.1 (51) online databases. Paired-sequence comparisons and multiple-sequence comparisons were carried out using BLASTN and MUSCLE 3.8.31 (52), respectively. Gene organization diagrams were drawn in Inkscape 0.48.1 (https://inkscape.org/en/).
Accession number(s).
The complete sequence of pCRE3-KPC was submitted to GenBank and deposited under accession number MH919378.
ACKNOWLEDGMENTS
This work was supported in part by the National Natural Science Foundation of China (grant no. 81802107), the National Science and Technology Major Project (grant no. 2018ZX10201001), the Science and Technology Plan of Yantai City (2019MSGY132), the Science and Technology Development Plan of Yeda Hospital of Yantai (201801), the Foundation of the Affiliated Hospital of Qingdao University (2019+X), and the Youth Foundation of the Affiliated Hospital of Qingdao University (2019).
REFERENCES
- 1.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner DJ, Grimont PA, Steigerwalt AG, Fanning GR, Ageron E, Riddle CF. 1993. Classification of citrobacteria by DNA hybridization: designation of Citrobacter farmeri sp. nov., Citrobacter youngae sp. nov., Citrobacter braakii sp. nov., Citrobacter werkmanii sp. nov., Citrobacter sedlakii sp. nov., and three unnamed Citrobacter genomospecies. Int J Syst Bacteriol 43:645–658. doi: 10.1099/00207713-43-4-645. [DOI] [PubMed] [Google Scholar]
- 3.Samonis G, Karageorgopoulos DE, Kofteridis DP, Matthaiou DK, Sidiropoulou V, Maraki S, Falagas ME. 2009. Citrobacter infections in a general hospital: characteristics and outcomes. Eur J Clin Microbiol Infect Dis 28:61–68. doi: 10.1007/s10096-008-0598-z. [DOI] [PubMed] [Google Scholar]
- 4.Lai CC, Tan CK, Lin SH, Liu WL, Liao CH, Huang YT, Hsueh PR. 2010. Bacteraemia caused by non-freundii, non-koseri Citrobacter species in Taiwan. J Hosp Infect 76:332–335. doi: 10.1016/j.jhin.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Carlini A, Mattei R, Mazzotta L, Lucarotti I, Pioli R, Bartelloni A, Antonelli A. 2005. Citrobacter braakii, an unusual organism as cause of acute peritonitis in PD patients. Perit Dial Int 25:405–406. doi: 10.1177/089686080502500417. [DOI] [PubMed] [Google Scholar]
- 6.Gupta R, Rauf SJ, Singh S, Smith J, Agraharkar ML. 2003. Sepsis in a renal transplant recipient due to Citrobacter braakii. South Med J 96:796–798. doi: 10.1097/01.SMJ.0000051068.52066.E2. [DOI] [PubMed] [Google Scholar]
- 7.Wu W, Espedido B, Feng Y, Zong Z. 2016. Citrobacter freundii carrying blaKPC-2 and blaNDM-1: characterization by whole genome sequencing. Sci Rep 6:30670. doi: 10.1038/srep30670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J, Ding H, Shi Y, Zhao Y, Hu X, Ren J, Huang G, Wu R, Zhao Z. 21 August 2018, posting date Further spread of a blaKPC-harboring untypeable plasmid in Enterobacteriaceae in China. Front Microbiol doi: 10.3389/fmicb.2018.01938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, Siu LK. 2014. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother 69:628–631. doi: 10.1093/jac/dkt409. [DOI] [PubMed] [Google Scholar]
- 10.Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob Agents Chemother 53:1998–2004. doi: 10.1128/AAC.01355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother 57:269–276. doi: 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother 54:4493–4496. doi: 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Fernandez A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 63:274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 14.Papagiannitsis CC, Miriagou V, Giakkoupi P, Tzouvelekis LS, Vatopoulos AC. 2013. Characterization of pKP1780, a novel IncR plasmid from the emerging Klebsiella pneumoniae ST147, encoding the VIM-1 metallo-beta-lactamase. J Antimicrob Chemother 68:2259–2262. doi: 10.1093/jac/dkt196. [DOI] [PubMed] [Google Scholar]
- 15.Compain F, Frangeul L, Drieux L, Verdet C, Brisse S, Arlet G, Decre D. 2014. Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob Agents Chemother 58:4207–4210. doi: 10.1128/AAC.02773-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai X, Zhou D, Xiong W, Feng J, Luo W, Luo G, Wang H, Sun F, Zhou X. 10 March 2016, posting date The IncP-6 plasmid p10265-KPC from Pseudomonas aeruginosa carries a novel DeltaISEc33-associated bla KPC-2 gene cluster. Front Microbiol doi: 10.3389/fmicb.2016.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao Y, Lazaro-Perona F, Falgenhauer L, Valverde A, Imirzalioglu C, Dominguez L, Canton R, Mingorance J, Chakraborty T. 28 June 2017, posting date Insights into a novel blaKPC-2-encoding IncP-6 plasmid reveal carbapenem-resistance circulation in several Enterobacteriaceae species from wastewater and a hospital source in Spain Front Microbiol doi: 10.3389/fmicb.2017.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naas T, Bonnin RA, Cuzon G, Villegas MV, Nordmann P. 2013. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J Antimicrob Chemother 68:1757–1762. doi: 10.1093/jac/dkt094. [DOI] [PubMed] [Google Scholar]
- 19.Tamma PD, Opene BN, Gluck A, Chambers KK, Carroll KC, Simner PJ. 2017. Comparison of 11 phenotypic assays for accurate detection of carbapenemase-producing Enterobacteriaceae. J Clin Microbiol 55:1046–1055. doi: 10.1128/JCM.02338-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi L, Feng J, Zhan Z, Zhao Y, Zhou H, Mao H, Gao Y, Zhang Y, Yin Z, Gao B, Tong Y, Luo Y, Zhang D, Zhou D. 2018. Comparative analysis of bla KPC-2- and rmtB-carrying IncFII-family pKPC-LK30/pHN7A8 hybrid plasmids from Klebsiella pneumoniae CG258 strains disseminated among multiple Chinese hospitals. Infect Drug Resist 11:1783–1793. doi: 10.2147/IDR.S171953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Xie J, Yang L, Chen D, Peters BM, Xu Z, Shirtliff ME. 2018. Identification of the KPC plasmid pCT-KPC334: new insights on the evolutionary pathway of epidemic plasmids harboring fosA3-blaKPC-2 genes. Int J Antimicrob Agents 52:510–511. doi: 10.1016/j.ijantimicag.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Xiang D-R, Li J-J, Sheng Z-K, Yu H-Y, Deng M, Bi S, Hu F-S, Chen W, Xue X-W, Zhou Z-B, Doi Y, Sheng J-F, Li L-J. 2015. Complete sequence of a novel IncR-F33:A-:B- plasmid, pKP1034, harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an epidemic Klebsiella pneumoniae sequence type 11 strain in China. Antimicrob Agents Chemother 60:1343–1348. doi: 10.1128/AAC.01488-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. 2009. Novel genetic environment of the carbapenem-hydrolyzing β-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother 53:4333–4338. doi: 10.1128/AAC.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Hu H, Chavda KD, Zhao S, Liu R, Liang H, Zhang W, Wang X, Jacobs MR, Bonomo RA, Kreiswirth BN. 2014. Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob Agents Chemother 58:2422–2425. doi: 10.1128/AAC.02587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, Zhang Y, Bi D, Shen P, Ai F, Liu H, Tian Y, Ma Y, Wang B, Rajakumar K, Ou HY, Jiang X. 2015. First report of a clinical, multidrug-resistant Enterobacteriaceae isolate coharboring fosfomycin resistance gene fosA3 and carbapenemase gene blaKPC-2 on the same transposon, Tn1721. Antimicrob Agents Chemother 59:338–343. doi: 10.1128/AAC.03061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allmeier H, Cresnar B, Greck M, Schmitt R. 1992. Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene 111:11–20. doi: 10.1016/0378-1119(92)90597-I. [DOI] [PubMed] [Google Scholar]
- 27.Chmelnitsky I, Shklyar M, Leavitt A, Sadovsky E, Navon-Venezia S, Ben Dalak M, Edgar R, Carmeli Y. 2014. Mix and match of KPC-2 encoding plasmids in Enterobacteriaceae-comparative genomics. Diagn Microbiol Infect Dis 79:255–260. doi: 10.1016/j.diagmicrobio.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Cuzon G, Naas T, Nordmann P. 2011. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garbari L, Busetti M, Dolzani L, Petix V, Knezevich A, Bressan R, Gionechetti F, Tonin EA, Lagatolla C. 2015. pKBuS13, a KPC-2-encoding plasmid from Klebsiella pneumoniae sequence type 833, carrying Tn4401b inserted into an Xer site-specific recombination locus. Antimicrob Agents Chemother 59:5226–5231. doi: 10.1128/AAC.04543-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo CC, Tham JM, Kwong SM, Yiin S, Poh CL. 1998. Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pRA2 of Pseudomonas alcaligenes. FEMS Microbiol Lett 165:253–260. doi: 10.1111/j.1574-6968.1998.tb13154.x. [DOI] [PubMed] [Google Scholar]
- 31.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 35:820–855. doi: 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 32.Liang Q, Yin Z, Zhao Y, Liang L, Feng J, Zhan Z, Wang H, Song Y, Tong Y, Wu W, Chen W, Wang J, Jiang L, Zhou D. 2017. Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int J Antimicrob Agents 49:709–718. doi: 10.1016/j.ijantimicag.2017.01.021. [DOI] [PubMed] [Google Scholar]
- 33.Partridge SR, Ginn AN, Paulsen IT, Iredell JR. 2012. pEl1573 carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob Agents Chemother 56:6029–6032. doi: 10.1128/AAC.01189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhan Z, Hu L, Jiang X, Zeng L, Feng J, Wu W, Chen W, Yang H, Yang W, Gao B, Yin Z, Zhou D. 2018. Plasmid and chromosomal integration of four novel blaIMP-carrying transposons from Pseudomonas aeruginosa, Klebsiella pneumoniae and an Enterobacter sp. J Antimicrob Chemother 73:3005–3015. doi: 10.1093/jac/dky288. [DOI] [PubMed] [Google Scholar]
- 35.Dubois D, Leyssene D, Chacornac JP, Kostrzewa M, Schmit PO, Talon R, Bonnet R, Delmas J. 2010. Identification of a variety of Staphylococcus species by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 48:941–945. doi: 10.1128/JCM.00413-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galas M, Decousser J-W, Breton N, Godard T, Allouch PY, Pina P. 2008. Nationwide study of the prevalence, characteristics, and molecular epidemiology of extended-spectrum-β-lactamase-producing Enterobacteriaceae in France. Antimicrob Agents Chemother 52:786–789. doi: 10.1128/AAC.00906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Shao H, Zhou H, Zhang T, Zhao X, Jiang Z, Wang Q. 18 December 2018, posting date Preparation of molecularly imprinted hybrid monoliths for the selective detection of fluoroquinolones in infant formula powders. J Chromatogr A doi: 10.1016/j.chroma.2018.12.038. [DOI] [PubMed] [Google Scholar]
- 39.Zurfluh K, Tasara T, Stephan R. 2016. Full-genome sequence of Escherichia coli K-15KW01, a uropathogenic E. coli B2 sequence type 127 isolate harboring a chromosomally carried blaCTX-M-15 gene. Genome Announc 4:e00927-16. doi: 10.1128/genomeA.00927-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srijan A, Margulieux KR, Ruekit S, Snesrud E, Maybank R, Serichantalergs O, Kormanee R, Sukhchat P, Sriyabhaya J, Hinkle M, Crawford JM, McGann P, Swierczewski BE. 2018. Genomic characterization of nonclonal mcr-1-positive multidrug-resistant Klebsiella pneumoniae from clinical samples in Thailand. Microb Drug Resist 24:403–410. doi: 10.1089/mdr.2017.0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuzon G, Naas T, Villegas M-V, Correa A, Quinn JP, Nordmann P. 2011. Wide dissemination of Pseudomonas aeruginosa producing β-lactamase blaKPC-2 gene in Colombia. Antimicrob Agents Chemother 55:5350–5353. doi: 10.1128/AAC.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CLSI. 2018. Performance standards for antimicrobial susceptibility testing: twenty-fifth informational supplement M100-S28. CLSI, Wayne, PA. [Google Scholar]
- 43.Nederbragt AJ. 2014. On the middle ground between open source and commercial software - the case of the Newbler program. Genome Biol 15:113. doi: 10.1186/gb4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, et al. . 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:8. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bansal P, Bridge AJ, Poux S, Bougueleret L, Xenarios I. 2016. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol Biol 1374:23–54. doi: 10.1007/978-1-4939-3167-5_2. [DOI] [PubMed] [Google Scholar]
- 48.Kleinheinz KA, Joensen KG, Larsen MV. 2014. Applying the ResFinder and VirulenceFinder Web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 4:e27943. doi: 10.4161/bact.27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moura A, Soares M, Pereira C, Leitao N, Henriques I, Correia A. 2009. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 50.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 1 January 2006, posting date ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prevalence statistics of plasmids containing both the IncR replicon and the blaKPC-2 gene. Data represent statistics of plasmids containing both the IncR replicon and the blaKPC-2 gene documented as of 22 May 2019. Download Table S1, DOCX file, 0.02 MB (26.9KB, docx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prevalence statistics of plasmids containing both the IncP6 replicon and the blaKPC-2 gene. Data represent statistics of plasmids containing both the IncP6 replicon and the blaKPC-2 gene documented as of 22 May 2019. Download Table S2, DOCX file, 0.02 MB (19.5KB, docx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Schematic maps of the related plasmids. The three plasmids unnamed3 (CP027150), p10265-KPC (KU578314), and pCOL-1 (KC609323) were included in the comparative analysis. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and in color, respectively. The innermost circle represents GC-skew [(G-C)/(G + C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle represents GC content. Download FIG S1, DOCX file, 1.2 MB (1.2MB, docx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Annotations of plasmid pCRE3-KPC. Download Data Set S1, XLSX file, 0.02 MB (23.7KB, xlsx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Annotations of plasmid unnamed3. Download Data Set S2, XLSX file, 0.02 MB (23.8KB, xlsx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Annotations of plasmid p10265-KPC. Download Data Set S3, XLSX file, 0.02 MB (18KB, xlsx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Annotations of plasmid pCOL-1. Download Data Set S4, XLSX file, 0.02 MB (16.1KB, xlsx) .
Copyright © 2020 Dong et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.