Abstract
Simultaneous, parental RNA interference (pRNAi) mediated knockdown of Hedgehog and Decapentaplegic (Dpp) signaling components, Pt-patched (Pt-ptc) and Pt-dpp, respectively, exhibited serious defects in the formation of the major embryonic axes in the model spider Parasteatoda tepidariorum. This paper describes a dataset of a custom oligonucleotide two-color microarray analysis that was carried out to compare the transcript expression levels between untreated (normal) and Pt-ptc + Pt-dpp double pRNAi embryos at late stage 5. Array spots that showed the intensity ratio of [Pt-ptc + Pt-dpp double pRNAi]/[normal] <0.6 were categorized as positive. The expressions of most, not all, of the transcripts related to the positive array spots were examined in embryos by whole-mount in situ hybridization. Some of the stained embryos showed distinct patterns of gene expression. The data presented may be useful for characterizing the mechanisms of embryonic patterning in spider embryos.
Keywords: Axis formation, Pattern formation, Embryology, Arthropod, Emerging model organism, RNA interference, Microarray, Signaling pathway
Specifications Table
| Subject | Developmental Biology |
| Specific subject area | Axis formation in animal embryos |
| Type of data | Table Image |
| How data were acquired | Custom oligonucleotide, two-color microarray; whole-mount in situ hybridization |
| Data format | Raw Analyzed |
| Parameters for data collection | No biological or technical replicates, with positive and negative controls |
| Description of data collection | Total RNAs that were extracted from late stage 5 embryos produced by the same parents before and after, respectively, Pt-ptc + Pt-dpp double pRNAi treatment were used for microarray analysis. |
| Data source location | Osaka, Japan |
| Data accessibility | For the microarray data, Repository name: Gene Expression Omnibus (GEO) at NCBI Data identification number: GSE112435 Direct URL to data: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE112435 For the WISH images, Repository name: Mendeley Data Data identification number: c7cfhyd2p3 Direct URL to data: https://data.mendeley.com/datasets/c7cfhyd2p3/3 |
Value of the Data
|
1. Data
We obtained embryos that showed serious defects in axis formation and extra-embryonic differentiation caused by simultaneous, parental RNA interference (pRNAi) mediated knockdown of Pt-patched (Pt-ptc) and Pt-decapentaplegic (Pt-dpp) (Movie S1), as was predictable from results of our previous experiments [1,2]. In Pt-ptc + Pt-dpp double pRNAi embryos, the migration of cumulus mesenchymal cells was impaired as observed in Pt-ptc single pRNAi embryos but no ectopic extra-embryonic differentiation occurred unlike in the Pt-ptc single pRNAi embryos [2]. This was presumably due to the simultaneous knockdown of Pt-dpp, which has been shown to be involved in the induction of extra-embryonic differentiation [1]. Using Combimatrix custom microarrays previously described [3], we compared the levels of the transcript expressions between untreated (normal) and Pt-ptc + Pt-dpp double pRNAi embryos at late stage 5. The microarray dataset deposited in the GEO Database at NCBI (GSE112435) consists of a data table showing the details of probe sequences for array spots (Platform: GPL11390 and GPL11391) and one showing the normalized signal intensity ratio of [Pt-ptc + Pt-dpp double pRNAi]/[normal] for each array spot (Sample: GSM3070092 and GSM3070093). Values of the [Pt-ptc + Pt-dpp double pRNAi]/[normal] intensity ratio from control probes are shown in Table 1. Array spots that showed the intensity ratio of [Pt-ptc + Pt-dpp double pRNAi]/[normal] < 0.6 were categorized as positive, and are listed in Table 2. Additional information about the control and positive array spots, including probe sequences, gene models, gene accessions, and notes based on the previously described developmental transcriptomes [4], is available in Supplementary Tables 1 and 2 (Tables S1 and S2), respectively. The expressions of most, not all, of the transcripts related to the positive array spots were examined in embryos by whole-mount in situ hybridization (Table S2). Some of the stained embryos showed distinct patterns of gene expression, which were photographed and are displayed in Fig. 1. The original images are available in the Mendeley data repository [5] and in the searchable databases of the Biohistory Research Hall (BRH) Data Resources (https://www.brh2.jp).
Table 1.
Values of the [Pt-ptc, Pt-dpp RNAi]/[normal] intensity ratios from control probes in the microarray analysis.
| Array No. | REF_ID | Ratio | EST clone ID | Sequence accession | RefSeq Gene ID | Description |
|---|---|---|---|---|---|---|
| 1 | 6978 | 0.998 | At_eW_003_D02 | FY217447 | LOC107439705 | catenin alpha |
| 1 | 7997 | 1.469 | At_eW_003_D02 | FY217447 | LOC107439705 | catenin alpha |
| 1 | 9278 | 0.867 | At_eW_003_D02 | FY217447 | LOC107439705 | catenin alpha |
| 1 | 11582 | 1.144 | At_eW_003_D02 | FY217447 | LOC107439705 | catenin alpha |
| 2 | 6342 | 0.988 | At_eW_003_D02 | FY217447 | LOC107439705 | catenin alpha |
| 2 | 11607 | 1.047 | At_eW_003_D02 | FY217447 | LOC107439705 | catenin alpha |
| 1 | 4354 | 1.054 | eS7_003_G08 | FY376809 | LOC107441347 | elongation factor 1-alpha |
| 1 | 10730 | 1.077 | eS7_003_G08 | FY376809 | LOC107441347 | elongation factor 1-alpha |
| 2 | 430 | 1.083 | eS7_003_G08 | FY376809 | LOC107441347 | elongation factor 1-alpha |
| 1 | 3623 | 1.006 | eS7_SB_037_C01 | FY380578 | LOC107447866 | histone H3.3 |
| 1 | 9723 | 0.927 | eS7_SB_037_C01 | FY380578 | LOC107447866 | histone H3.3 |
| 2 | 6417 | 0.928 | eS7_SB_037_C01 | FY380578 | LOC107447866 | histone H3.3 |
Table 2.
List of array spots that showed the intensity ratios [Pt-ptc + Pt-dpp double RNAi]/[normal] of <0.6.
| Array No. | REF_ID | Ratio | EST clone ID or gene namea | RefSeq Gene ID or GB_ACC | Description |
|---|---|---|---|---|---|
| 1 | 450 | 0.580 | At_eW_000_C16 | LOC107441590 | rap guanine nucleotide exchange factor 2-like |
| 1 | 1005 | 0.567 | At_eW_000_E06 | LOC107449884 | notch-regulated ankyrin repeat-containing protein-like |
| 1 | 6423 | 0.549 | At_eW_000_J22 | LOC107449884 | notch-regulated ankyrin repeat-containing protein-like |
| 2 | 8616 | 0.516 | eS7_SB_021_E05* | LOC107449884 | notch-regulated ankyrin repeat-containing protein-like |
| 2 | 10410 | 0.487 | S7_d1_18_A10 | LOC107449884 | notch-regulated ankyrin repeat-containing protein-like |
| 2 | 6880 | 0.521 | S7_d1_18_A10 | LOC107449884 | notch-regulated ankyrin repeat-containing protein-like |
| 1 | 6611 | 0.508 | At_eW_000_F24 | IABY01000175 | 18S ribosomal RNA gene |
| 1 | 11812 | 0.462 | At_eW_000_F24 | IABY01000175 | 18S ribosomal RNA gene |
| 1 | 7145 | 0.597 | At_eW_000_M09 | ||
| 1 | 8819 | 0.576 | At_eW_002_L19 | LOC107444999 | epidermal growth factor receptor kinase substrate 8-like protein 2 |
| 1 | 919 | 0.442 | At_eW_003_L15 | LOC107438525 | protein melted |
| 1 | 9418 | 0.509 | At_eW_003_L17 | ||
| 1 | 661 | 0.554 | At_eW_004_D03 | ||
| 1 | 12282 | 0.541 | At_eW_004_F24 | LOC107438715 | TBC1 domain family member 22B |
| 1 | 339 | 0.547 | At_eW_004_N23 | LOC107439340 | cilia- and flagella-associated protein 58 |
| 1 | 9968 | 0.592 | At_eW_005_C14 | ||
| 1 | 12481 | 0.536 | At_eW_005_D02 | ||
| 1 | 3856 | 0.582 | At_eW_005_D07 | ||
| 1 | 3745 | 0.583 | At_eW_005_P05 | LOC107451405 | U2 small nuclear ribonucleoprotein A′ |
| 1 | 10076 | 0.598 | At_eW_005_P06 | ||
| 1 | 3583 | 0.544 | At_eW_005_P09 | LOC110282483 | uncharacterized LOC110282483 |
| 1 | 2714 | 0.534 | At_eW_007_J22 | ||
| 1 | 2093 | 0.512 | At_eW_007_M22 | LOC107443747 | protein SHQ1 homolog |
| 1 | 2326 | 0.592 | At_eW_008_I02 | LOC107452247 | uncharacterized LOC107452247 |
| 1 | 11478 | 0.563 | At_eW_009_O12 | ||
| 1 | 7942 | 0.589 | At_eW_010_D11 | ||
| 1 | 1482 | 0.567 | At_eW_010_H20 | ||
| 1 | 150 | 0.577 | At_eW_010_L11 | LOC107444253 | transcription factor HES-1-A |
| 1 | 6462 | 0.591 | At_eW_011_C15 | LOC107436693 | transmembrane protein 165-like |
| 1 | 9029 | 0.595 | At_eW_011_D17 | ||
| 1 | 11467 | 0.528 | At_eW_012_N12 | LOC107446959 | uncharacterized LOC107446959 |
| 1 | 5534 | 0.572 | At_eW_013_F08 | ||
| 1 | 1284 | 0.463 | At_eW_013_I14 | ||
| 1 | 1922 | 0.570 | At_eW_014_K24 | LOC107448046 | anaphase-promoting complex subunit 1 |
| 1 | 4791 | 0.527 | At_eW_016_G24 | ||
| 1 | 8895 | 0.573 | At_eW_016_H18 | LOC107445612 | neurobeachin |
| 1 | 8551 | 0.583 | At_eW_016_K20 | LOC107454643 | fasciclin-2 |
| 1 | 9159 | 0.578 | At_eW_016_L03 | ||
| 1 | 10700 | 0.540 | At_eW_016_N10* | LOC107452890 | uncharacterized LOC107452890 |
| 1 | 9831 | 0.543 | At_eW_017_A06 | ||
| 1 | 6001 | 0.598 | At_eW_017_H11 | ||
| 1 | 12023 | 0.543 | At_eW_017_H20* | IABY01007316 | notch-regulated ankyrin repeat-containing protein-like |
| 1 | 12159 | 0.506 | At_eW_017_N01 | ||
| 1 | 436 | 0.587 | At_eW_017_P04* | ||
| 1 | 7805 | 0.595 | At_eW_018_F16 | ||
| 1 | 2925 | 0.565 | At_eW_018_K04 | LOC107447180 | protein Wnt-5b-like |
| 2 | 10650 | 0.561 | Pt-wnt5 | LOC107447180 | protein Wnt-5b-like |
| 1 | 3287 | 0.360 | eS6_d1_26_A11* | LOC107447180 | protein Wnt-5b-like |
| 1 | 11579 | 0.410 | At_eW_019_D19 | LOC107438410 | myosin regulatory light chain 2 |
| 1 | 7511 | 0.522 | At_eW_019_H22 | LOC107446659 | pituitary tumor-transforming gene 1 protein-interacting protein |
| 1 | 7367 | 0.528 | At_eW_019_L05 | IABY01019505 | beta-1,4-galactosyltransferase 7-like |
| 1 | 7525 | 0.597 | At_eW_019_M01 | ||
| 1 | 11212 | 0.454 | At_eW_019_O17 | LOC107440487 | heat shock 70 kDa protein cognate 4 |
| 1 | 2858 | 0.578 | At_eW_020_B15* | LOC107447678 | homeobox protein MSH-D-like, Msx1 |
| 1 | 10173 | 0.552 | At_eW_020_D06 | ||
| 1 | 4395 | 0.571 | At_eW_021_C05 | ||
| 1 | 9232 | 0.516 | At_eW_021_K24 | ||
| 1 | 11068 | 0.538 | At_eW_022_I21 | LOC107446292 | protein sel-1 homolog 1-like |
| 1 | 5400 | 0.590 | At_eW_023_A14 | LOC107453070 | uncharacterized LOC107453070 |
| 1 | 12508 | 0.501 | At_eW_023_I22* | LOC107438015 | growth arrest-specific protein 1 |
| 1 | 2821 | 0.493 | At_eW_023_I22* | LOC107438015 | growth arrest-specific protein 1 |
| 1 | 9995 | 0.548 | At_eW_023_J04 | ||
| 1 | 1123 | 0.593 | At_eW_023_M02 | LOC107441148 | lipopolysaccharide-induced tumor necrosis factor-alpha factor homolog |
| 1 | 12307 | 0.594 | At_eW_024_C09 | ||
| 1 | 5992 | 0.529 | At_eW_024_H11 | IABY01020283 | |
| 1 | 540 | 0.577 | At_eW_024_P15 | LOC107447475 | ubiquitin-protein ligase E3A-like |
| 1 | 8886 | 0.571 | At_eW_025_M12 | ||
| 1 | 3886 | 0.562 | At_eW_026_K05 | LOC107446429 | uncharacterized LOC107446429 |
| 1 | 9733 | 0.458 | At_eW_027_J20* | LOC107446595 | uncharacterized LOC107446595 |
| 1 | 11788 | 0.573 | At_eW_027_N08 | ||
| 1 | 612 | 0.389 | Pt-dpp | LOC107442925 | bone morphogenetic protein 4-like |
| 1 | 5343 | 0.546 | eS6_d1_12_H07 | LOC107442925 | bone morphogenetic protein 4-like |
| 1 | 6206 | 0.514 | Pt-cad | LOC107437910 | homeobox protein CDX-1-like |
| 1 | 8638 | 0.463 | Pt-gataC | LOC107448880 | endothelial transcription factor GATA-2-like |
| 1 | 9435 | 0.598 | eS6_d1_01_A08 | ||
| 1 | 9555 | 0.523 | eS6_d1_01_C03 | ||
| 1 | 12467 | 0.594 | eS6_d1_01_D11 | ||
| 1 | 6832 | 0.557 | eS6_d1_02_C06 | LOC107457141 | protein capicua homolog |
| 1 | 4902 | 0.566 | eS6_d1_02_G12* | LOC107439895 | cyclin-dependent kinase 6-like |
| 1 | 12346 | 0.569 | eS6_d1_03_B05 | LOC107436245 | polypeptide N-acetylgalactosaminyltransferase 1-like |
| 1 | 9898 | 0.404 | eS6_d1_03_D06 | ||
| 1 | 6599 | 0.588 | eS6_d1_03_D09 | ||
| 1 | 9521 | 0.594 | eS6_d1_04_F03* | LOC107443591 | BMP and activin membrane-bound inhibitor homolog |
| 1 | 7264 | 0.563 | eS6_d1_05_E04 | ||
| 1 | 2563 | 0.565 | eS6_d1_09_B04 | IABY01006050 | |
| 1 | 10790 | 0.590 | eS6_d1_09_B09 | LOC107454942 | uncharacterized LOC107454942 |
| 1 | 6404 | 0.582 | eS6_d1_12_D08 | ||
| 1 | 1398 | 0.412 | eS6_d1_13_E07 | ||
| 1 | 8383 | 0.561 | eS6_d1_14_A02 | ||
| 1 | 2126 | 0.478 | eS6_d1_15_H06 | ||
| 1 | 4375 | 0.597 | eS6_d1_21_A11 | ||
| 1 | 11328 | 0.589 | eS6_d1_23_G03 | LOC107449017 | 1-acyl-sn-glycerol-3-phosphate acyltransferase beta |
| 1 | 3476 | 0.517 | eS6_d1_23_H04 | ||
| 1 | 2074 | 0.575 | eS6_d1_25_E09 | ||
| 1 | 107 | 0.592 | eS6_d1_26_H06 | ||
| 1 | 6730 | 0.588 | eS6_d1_27_C12* | LOC107437911 | serine protease 27 |
| 1 | 4870 | 0.595 | eS6_d1_28_E12 | ||
| 1 | 8615 | 0.585 | eS6_d1_29_A10 | ||
| 1 | 8116 | 0.483 | eS6_d1_30_H10 | IABY01009517 | |
| 1 | 7465 | 0.537 | eS6_d1_31_D10* | ||
| 1 | 8137 | 0.404 | eS6_d1_32_D12 | LOC107443710 | cadherin-related tumor suppressor-like |
| 1 | 8396 | 0.476 | eS6_d1_32_G05* | LOC107452006 | transcription factor AP-2-beta-like |
| 1 | 1224 | 0.517 | eS6_d1_33_C11 | LOC107448603 | uncharacterized LOC107448603 |
| 2 | 8704 | 0.530 | eS6_d1_34_D05* | LOC107447504 | homeobox protein Hox-B4a-like, ftz-B |
| 2 | 4565 | 0.433 | eS7_SB_035_D03 | LOC107447504 | homeobox protein Hox-B4a-like, ftz-B |
| 2 | 11882 | 0.391 | eS7_SB_035_D03 | LOC107447504 | homeobox protein Hox-B4a-like, ftz-B |
| 2 | 6178 | 0.306 | eS7_SB_037_E07 | LOC107447504 | homeobox protein Hox-B4a-like, ftz-B |
| 2 | 6529 | 0.571 | eS6_d1_35_F10 | LOC107453461 | argininosuccinate synthase-like |
| 2 | 10205 | 0.554 | eS6_d1_36_B07 | ||
| 2 | 3126 | 0.592 | eS6_d1_43_B11 | ||
| 2 | 5110 | 0.509 | eS6_d1_44_D10 | ||
| 2 | 9590 | 0.588 | eS6_d1_51_D07 | LOC107456383 | zinc finger protein 25 |
| 2 | 654 | 0.590 | eS6_d1_51_H02 | IABY01004033 | |
| 2 | 9647 | 0.587 | eS6_d1_52_C02* | LOC107447988 | homeobox protein engrailed-like ceh-16, Noto1 |
| 2 | 4037 | 0.597 | eS6_d1_57_F09 | LOC107456922 | segment polarity protein dishevelled homolog DVL-3 |
| 2 | 7784 | 0.556 | eS7_005_F03 | LOC107456962 | probable basic-leucine zipper transcription factor J |
| 2 | 12252 | 0.591 | eS7_SB_009_G07 | IABY01005160 | |
| 2 | 2630 | 0.520 | eS7_SB_011_D07* | LOC107456088 | iroquois-class homeodomain protein IRX-6, mirr4 |
| 2 | 879 | 0.568 | eS7_SB_011_D07* | LOC107456088 | iroquois-class homeodomain protein IRX-6, mirr4 |
| 2 | 1216 | 0.581 | S7_d1_29_C06 | LOC107456088 | iroquois-class homeodomain protein IRX-6, mirr4 |
| 2 | 8972 | 0.515 | eS7_SB_018_F06 | LOC107455065 | zinc finger protein-like 1 homolog |
| 2 | 4626 | 0.179 | eS7_SB_028_C07* | LOC107445228 | protein gooseberry, Prd2 |
| 2 | 5659 | 0.593 | eS7_SB_030_B11 | ||
| 2 | 8303 | 0.462 | eS7_SB_035_C08 | LOC107452623 | small glutamine-rich tetratricopeptide repeat-containing protein beta-like |
| 2 | 2111 | 0.501 | eS7_SB_038_H11* | LOC107448645 | transcription factor Sp9 |
| 2 | 7540 | 0.370 | S7_d1_24_G01 | LOC107448645 | transcription factor Sp9 |
| 2 | 10234 | 0.465 | eS7_SB_042_D01 | LOC107454524 | phospholipase A-2-activating protein |
| 2 | 341 | 0.592 | eS7_SB_043_C05 | ||
| 2 | 6422 | 0.589 | eS7_SB_047_C01* | LOC107437200 | inosine-5′-monophosphate dehydrogenase 2 |
| 2 | 2691 | 0.440 | S7_d1_04_F07 | ||
| 2 | 2109 | 0.595 | S7_d1_06_C11 | LOC107457213 | alpha-(1,3)-fucosyltransferase C |
| 2 | 10975 | 0.504 | S7_d1_08_C05 | LOC107454396 | bone morphogenetic protein receptor type-2 |
| 2 | 47 | 0.561 | S7_d1_18_F06 | ||
| 2 | 10933 | 0.545 | S7_d1_19_D06 | ||
| 2 | 3648 | 0.576 | S7_d1_20_H05* | IABY01019901 | |
| 2 | 5931 | 0.583 | S7_d1_21_H03 | LOC107436591 | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial |
| 2 | 1902 | 0.597 | S7_d1_29_B03 | LOC107437456 | cytochrome P450 302a1, mitochondrial |
| 2 | 6935 | 0.480 | S7_d1_30_G04 | ||
| 2 | 11999 | 0.528 | S7_d1_33_A05 | LOC107437124 | lipoyltransferase 1, mitochondrial-like |
| 2 | 9564 | 0.540 | S7_d1_35_G02* | LOC107449043 | toll-like receptor Tollo |
| 2 | 9079 | 0.569 | S7_d1_39_A09* | ||
| 2 | 1615 | 0.515 | S7_d1_40_C07 | LOC107441637 | DNA replication licensing factor mcm4-A |
| 2 | 2079 | 0.474 | S7_d1_40_G11 |
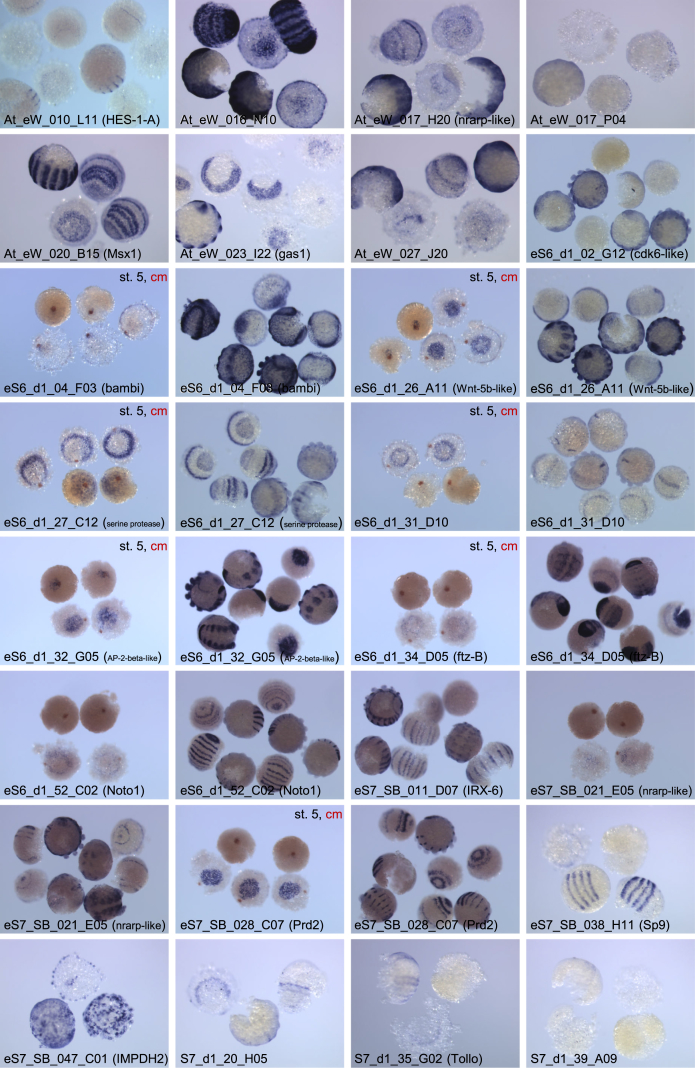
Expression of the transcripts related to the EST clones indicated by asterisks (*) was examined by whole-mount in situ hybridization (see Fig. 1).
Fig. 1.
Staining of stage 5−8 embryos for selected transcripts by WISH. The identity of EST clones that were used for the synthesis of RNA probes is indicated in each panel. Some panels show stage 5 embryos additionally stained in red for a cumulus cell marker (cm).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.dib.2019.105088.
The following is the supplementary data related to this article:
Movie S1. Time-lapse observation of Pt-ptc + Pt-dpp double pRNAi embryos. These embryos were from the same egg sac that was used for RNA extraction in the microarray experiment. Time (day: h: min) after the start of recording (late stage 4) is indicated. The time point when Pt-ptc + Pt-dpp double pRNAi embryos were lysed for the RNA extraction was about 00:07:20. The time-lapse recording lasted more than two days, which should have covered the stages of germ band formation and elongation and limb bud formation. Apparently, the embryos observed failed to develop the orthogonal body axes and extra-embryonic tissues. The related phenotypes have been described in our previous work [1,2].
2. Experimental design, materials, and methods
2.1. Parental RNA interference (pRNAi)
The general procedure for pRNAi-mediated gene knockdown in P. tepidariorum was previously described [1]. Specifically, a mated female was injected with approximately 2.0 μl of Pt-ptc and Pt-dpp dsRNA mixture (0.6–1.0 μg/μl each) 5 times at the intervals of 2–3 days. The 709-bp (nt 1–709) region of Pt-ptc cDNA (GB_ACC: AB433900.1) and the 736-bp region (nt 1005–1740) of Pt-dpp cDNA (GB_ACC: AB096072.1) were used for the synthesis of the Pt-ptc and Pt-dpp dsRNAs, whose specific knockdown effects were previously described [1,2]. Embryos derived from an egg sac produced by the female two days before (normal) and 24 days after (Pt-ptc + Pt-dpp double pRNAi) the first injection of the dsRNA were used for RNA extraction. The morphological phenotype of the Pt-ptc + Pt-dpp double pRNAi embryos from the same egg sac that was used for the RNA extraction was recorded by time-lapse microscopy (Movie S1).
2.2. Microarray analysis
40-mer oligonucleotide probes designed were embedded in custom microarrays (CombiMatrix CustomArray 12K×2, CustomArray, Inc.). The same microarray design was used in our previous work [3]. The details of the custom microarray design including the probe sequences are available from the GEO database (GPL11390 and GPL11391). The total RNAs used for microarray analysis were extracted from approximately 250 embryos at late stage 5 using MagExtractor (Toyobo). The time point when Pt-ptc + Pt-dpp double pRNAi embryos were lysed for the RNA extraction was about 00:07:20 (day: h: min) in Movie S1. The RNA integrity was examined with an Agilent Bioanalyzer 2100. The cRNA labeled with Cy3 or Cy5 was prepared from 2 μg of total RNA using RNA Transcript SureLABEL Core Kit (Takara). The cRNA probes were hybridized to microarrays using Hybridization buffer (5× SSC, 0.1% SDS, 10% formamide) at 42 °C for 16–20 h. The microarray slides were scanned using a GenePix 4000B Scanner (Molecular Devices). There were no biological replicates. The obtained images were analyzed using an Array-Pro Analyzer ver. 4.5 (Media Cybernetics, Inc.). The quantitative data were subjected to Loess normalization. The ratio of the normalized intensity values ([Pt-ptc + Pt-dpp double pRNAi]/[normal]) for each array spot was calculated. The array spots for alpha-catenin (GB_ACC: AB433907; GI: LOC107439705), elongation factor 1-alpha (GB_ACC: AB433908; GI: LOC107441347), and histone H3 (GB_ACC: AB433909; GI, LOC107447866) served as negative controls (Table 1), while some of the array spots for Pt-dpp (GB_ACC: AB096072; GI: LOC107442925) and Pt-cad (GB_ACC: AB096075; GI: LOC107437910) were detected as positive, as expected from previous work [1,2]. The values from these positive and negative array spots validated the microarray experiment.
2.3. Embryo staining by whole-mount in situ hybridization (WISH)
Since most EST clones that were associated with positive array spots were instantly available, they were used for the synthesis of Digoxigenin-labeled RNA probes for WISH. The EST clone At_eW_022_P10 was used for the synthesis of fluorescein-labeled RNA probe, which marked the cumulus mesenchymal cells in stage 5 embryos [2]. Single- and double-staining of embryos at stages 5–8 by WISH were performed as described [1]. The stained embryos were photographed using a stereomicroscope (SZX12, Olympus) equipped with a color CCD camera (C7780-10, Hamamatsu Photonics).
Acknowledgments
We thank Akiko Noda for technical assistance. This work was supported in part by the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (KAKENHI) awards to HO (15K07139) and YA (26440130).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.105088.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Akiyama-Oda Y., Oda H. Axis specification in the spider embryo: dpp is required for radial-to-axial symmetry transformation and sog for ventral patterning. Development. 2006;133:2347–2357. doi: 10.1242/dev.02400. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama-Oda Y., Oda H. Cell migration that orients the dorsoventral axis is coordinated with anteroposterior patterning mediated by Hedgehog signaling in the early spider embryo. Development. 2010;137:1263–1273. doi: 10.1242/dev.045625. [DOI] [PubMed] [Google Scholar]
- 3.Kanayama M., Akiyama-Oda Y., Nishimura O., Tarui H., Agata K., Oda H. Travelling and splitting of a wave of hedgehog expression involved in spider-head segmentation. Nat. Commun. 2011;2:500. doi: 10.1038/ncomms1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwasaki-Yokozawa S., Akiyama-Oda Y., Oda H. Genome-scale embryonic developmental profile of gene expression in the common house spider Parasteatoda tepidariorum. Data Brief. 2018;19:865–867. doi: 10.1016/j.dib.2018.05.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oda H., Akiyama-Oda Y. Data for: Dataset on gene expressions affected by simultaneous knockdown of Hedgehog and Dpp signaling components in embryos of the spider Parasteatoda tepidariorum Version 3. Mendeley Data. 2019 doi: 10.1016/j.dib.2019.105088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Time-lapse observation of Pt-ptc + Pt-dpp double pRNAi embryos. These embryos were from the same egg sac that was used for RNA extraction in the microarray experiment. Time (day: h: min) after the start of recording (late stage 4) is indicated. The time point when Pt-ptc + Pt-dpp double pRNAi embryos were lysed for the RNA extraction was about 00:07:20. The time-lapse recording lasted more than two days, which should have covered the stages of germ band formation and elongation and limb bud formation. Apparently, the embryos observed failed to develop the orthogonal body axes and extra-embryonic tissues. The related phenotypes have been described in our previous work [1,2].