Abstract
IS1548, a 1316-bp element of the ISAs1 family affects the expression of several genes of the opportunistic pathogen Streptococcus agalactiae. Furthermore, certain lineages of S. agalactiae are more frequently associated to particular diseases than other [1, 2]. We took advantage of the release of the genome sequences of a huge number of epidemiologically unrelated S. agalactiae strains of various origin to analyze the prevalence of IS1548 among S. agalactiae strains. To this end, S. agalactiae genome available at the National Center for Biotechnology Information (NCBI) database were blasted with IS1548 DNA sequences. A sequence type (ST), based on the allelic profile of seven housekeeping genes, was assigned to each strain possessing IS1548. These strains were then grouped into clonal complexes (CCs). The data obtained will give the opportunity to compare the sequenced genomes of S. agalactiae based on their lineage and/or possession of IS1548, and to select the corresponding strains for comparative experimental studies. The data is related to the research article « Dual and divergent transcriptional impact of IS1548 insertion upstream of the peptidoglycan biosynthesis murB gene of Streptococcus agalactiae” [2].
Keywords: Mobile genetic element, ISAs1 family, Multi locus sequence typing, Sequence type, Clonal complex, Population structure
Specifications Table
| Subject | Microbiology |
| Specific subject area | Bacterial population structure and dynamics |
| Type of data | Raw: Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9 Raw: Table 1 |
| How data were acquired | BlastN program (https://blast.ncbi.nlm.nih.gov/) MLST program (http://pubmlst.org/sagalactiae/) goeBURST software (http://www.phyloviz.net/goeburst/) |
| Data format | Raw |
| Parameters for data collection | All the Streptococcus agalactiae genome sequences available as whole genome contigs or as complete genome sequences at the National Center for Biotechnology Information database on January 19, 2018 were analyzed |
| Description of data collection | Bioinformatic analyses of the above cited genomes by the BlastN program, the MLST program and the goeBURST software |
| Data source location | Institution: University of Tours City/Town/Region: Tours Country: France |
| Data accessibility | With the article |
| Related research article | Sarah Khazaal, Rim Al Safadi, Dani Osman, Aurelia Hiron, Philippe Gilot, Dual and divergent transcriptional impact of IS1548 insertion upstream of the peptidoglycan biosynthesis murB gene of Streptococcus agalactiae, Gene, 720, https://doi.org/10.1016/j.gene.2019.144094 |
Value of the Data
|
1. Data description
One hundred and twenty-one S. agalactiae strains with IS1548 positive blastN similarity finding were identified among the 911 strains whose genomes where sequenced and available at the NCBI database on January 19, 2018 [2]. The dataset comprises the analysis of the Multi Locus Sequence type of these strains with the MLST program (Table 1) and their classification in clonal complex with the goeburst software (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9), respectively.
Table 1.
Raw data of the Multi Locus Sequence Typing profiles of Streptococcus agalactiae strains possessing IS1548, obtained by submitting their sequenced genomes to the MLST databasea.
| Strain | adhP | pheS | atr | glnA | sdhA | glcK | tkt | ST | |
|---|---|---|---|---|---|---|---|---|---|
| 351521 | 1 | 1 | 3 | 1 | 1 | 2 | 2 | 2 | |
| 418136 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| 446329 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| 0506A_35_952 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| 112469214_isolate1 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| 2603V/R | 1 | 1 | 3 | 2 | 2 | 2 | 9 | 110 | |
| 357_SAGA | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| 986_SAGA | 156 | 1 | 3 | 2 | 2 | 2 | 2 | 853 | |
| B37VS | 1 | 1 | 43 | 2 | 2 | 2 | 2 | 335 | |
| B41VS | 116 | 1 | 4 | 1 | 3 | 3 | 2 | 652 | |
| B81VD | 1 | 1 | 3 | 4 | 2 | 5 | 2 | 106 | |
| B848 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| B96V | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 17 | |
| BE-PW-051 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| BE-PW-095 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| BG-NI-012 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| BSU167 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| BSU174 | 10 | 1 | 12 | 1 | 3 | 2 | 2 | 41 | |
| BSU188 | 5 | 4 | 6 | 3 | 2 | 1 | 3 | 23 | |
| BSU442 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| BSU447 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| BV3L5 | 1 | 1 | 3 | 2 | 2 | 2 | 9 | 110 | |
| CCUG 19094 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| CCUG 24810 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| CCUG 37737 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| CCUG 37738 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| CCUG 37741 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| CCUG 37742 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| CCUG 44077 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | |
| CCUG 44104 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| CCUG 45061 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| CZ-NI-002 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| CZ-NI-003 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| DE-NI-007 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| DE-NI-041 | 1 | 1 | 3 | 4 | 2 | 5 | 2 | 106 | |
| DE-NI-042 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| DK-NI-011 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| DK-PW-060 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| DK-PW-066 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| DK-PW-162 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| DML120817 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| ES-PW-008 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| ES-PW-033 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| ES-PW-063 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| ES-PW-085 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| ES-PW-118 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| ES-PW-156 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| FSL F2-338 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| FSL S3-003 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| FSL S3-005 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| FSL S3-268 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| FSL S3-337 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB00092 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB00174 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| GB00264 | 9 | 1 | 4 | 1 | 3 | 3 | 2 | 10 | |
| GB00543 | 1 | 1 | 3 | 2 | 2 | 2 | 7 | 36 | |
| GB00561 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB00653 | 10 | 1 | 4 | 1 | 3 | 3 | 2 | 12 | |
| GB00663 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB00864 | 9 | 1 | 4 | 1 | 3 | 3 | 2 | 10 | |
| GB00865 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB00884 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB00904 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB00923 | 1 | 1 | 3 | 3 | 2 | 2 | 2 | 175 | |
| GB00929 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB00975 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| GB00984 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB01004 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| GB-NI-007 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB-NI-011 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB-NI-014 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| GB-NI-015 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB-PW-035 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB-PW-041 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB-PW-049 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB-PW-051 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB-PW-067 | 10 | 1 | 4 | 1 | 3 | 3 | 2 | 12 | |
| GB-PW-087 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GB-PW-088 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| GBS1-NY | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| GBS2-NM | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| GBS6 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| GBS11 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| Gottschalk 1003A | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| H002 | 1 | 1 | 110 | 2 | 2 | 2 | 2 | 928 | |
| IMMI_409 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| IT-NI-036 | 1 | 1 | 5 | 4 | 1 | 4 | 6 | 26 | |
| IT-NI-037 | 1 | 1 | 5 | 4 | 1 | 4 | 6 | 26 | |
| LMG 15089 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| LMG 15093 | 1 | 1 | 3 | 2 | 2 | 2 | 9 | 110 | |
| Madagascar-IP-47 | 9 | 1 | 4 | 1 | 3 | 3 | 2 | 10 | |
| MC627 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| MC628 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| MC632 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| MC633 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| MC634 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| ML41151 | 1 | 1 | 4 | 2 | 2 | 3 | 2 | 328 | |
| Mother12 | 1 | 1 | 3 | 4 | 2 | 2 | 2 | 27 | |
| MRI Z1-022 | 32 | 1 | 3 | 2 | 2 | 2 | 2 | 121 | |
| PSS_7568 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| PSS_7632a | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| RBH01 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| RBH03 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| RBH04 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| RBH06 | 13 | 3 | 1 | 3 | 1 | 1 | 1 | 22 | |
| RBH11 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| RBH12 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| S10-201 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| Sag158 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| SG-M25 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| SH0334 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| SH0655 | 10 | 1 | 4 | 2 | 3 | 3 | 2 | 286 | |
| SH1370 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | |
| SH3601 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 19 | |
| ST 610 | 13 | 40 | 6 | 65 | 3 | 52 | 50 | 610 | |
| ST 612 | 111 | 40 | 6 | 65 | 3 | 52 | 51 | 612 | |
| ST 613 | 111 | 40 | 67 | 65 | 3 | 52 | 51 | 613 | |
| ST 615 | 13 | 40 | 6 | 65 | 3 | 52 | 51 | 615 | |
| ST 617 | 13 | 40 | 68 | 65 | 3 | 52 | 51 | 617 | |
| ST 618 | 111 | 40 | 69 | 65 | 3 | 52 | 51 | 618 | |
| USS-107 | 1 | 1 | 3 | 2 | 18 | 2 | 2 | 182 | |
Sequence type (ST), alcohol dehydrogenase gene (adhP), phenylalanine tRNA synthetase gene (pheS), amino acid transporter gene (atr), glutamine synthetase gene (glnA), serine dehydratase gene (sdhA), glucose kinase gene (glcK) and transketolase gene (tkt). Multi Locus Sequence Typing (MLST) database at https://pubmlst.org/sagalactiae/.
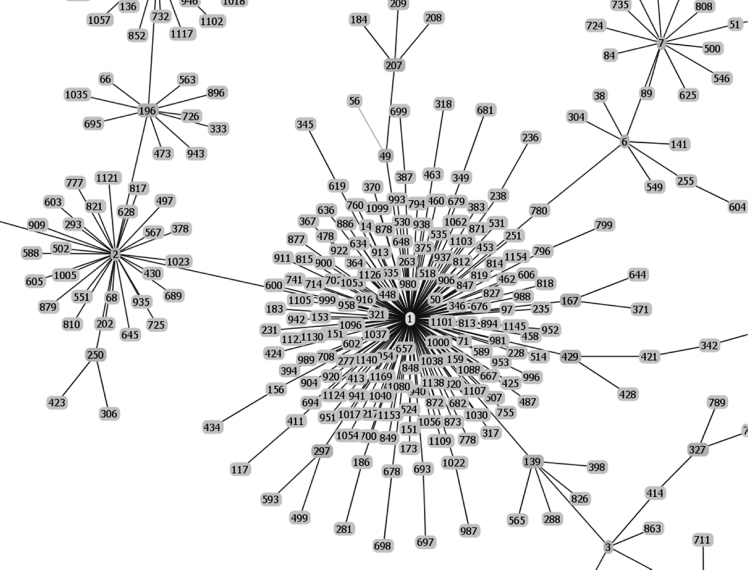
Fig. 1.
Raw data of the goeBURST analysis of the sequence types of strains possessing IS1548: Clonal Complex 1.
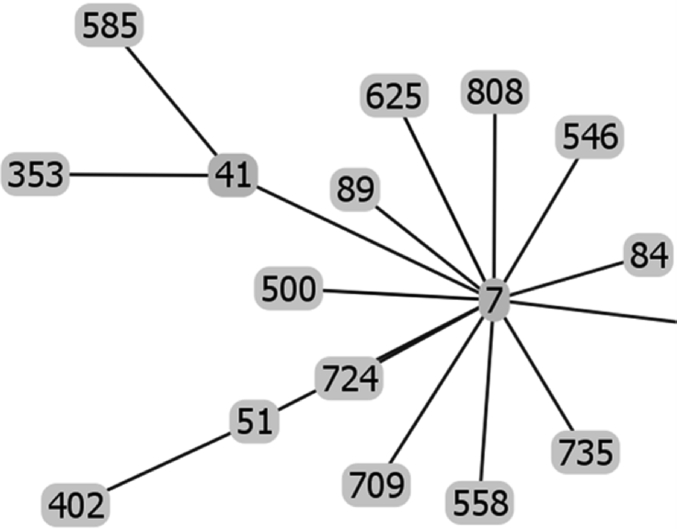
Fig. 2.
Raw data of the goeBURST analysis of the sequence types of strains possessing IS1548: Clonal Complex 7.
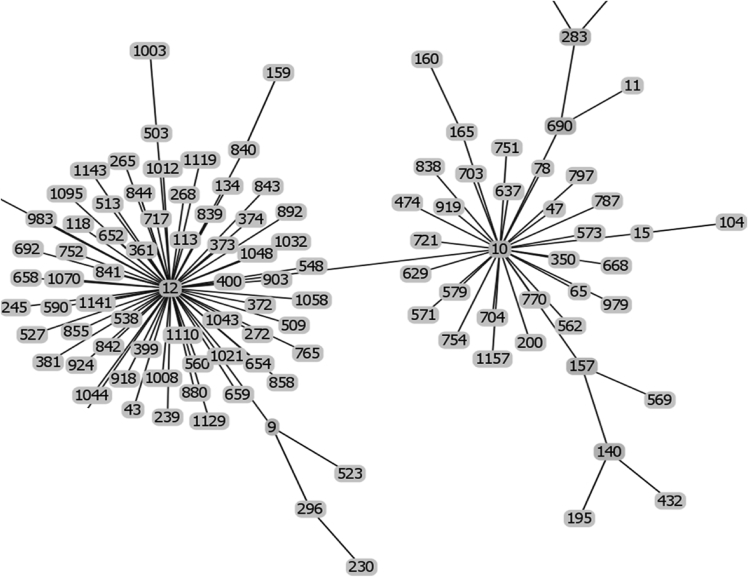
Fig. 3.
Raw data of the goeBURST analysis of the sequence types of strains possessing IS1548: Clonal Complex 10 and 12.
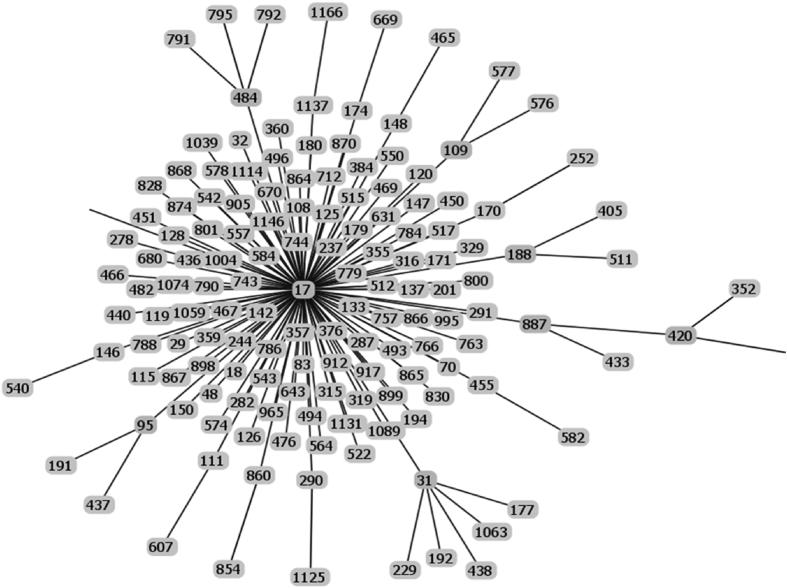
Fig. 4.
Raw data of the goeBURST analysis of the sequence types of strains possessing IS1548: Clonal Complex 17.
Fig. 5.
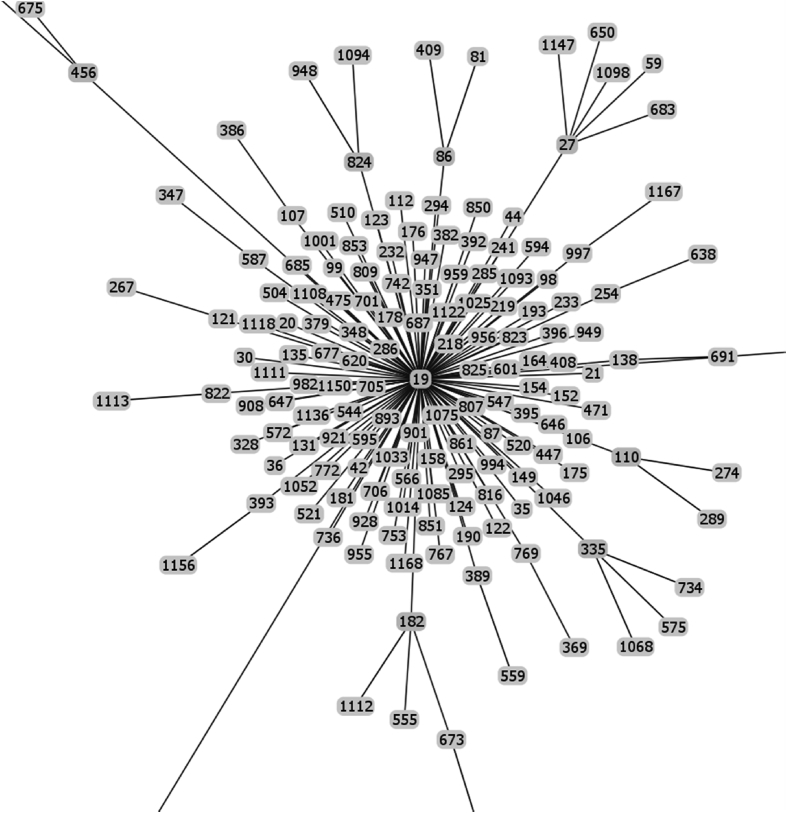
Raw data of the goeBURST analysis of the sequence types of strains possessing IS1548: Clonal Complex 19.
Fig. 6.
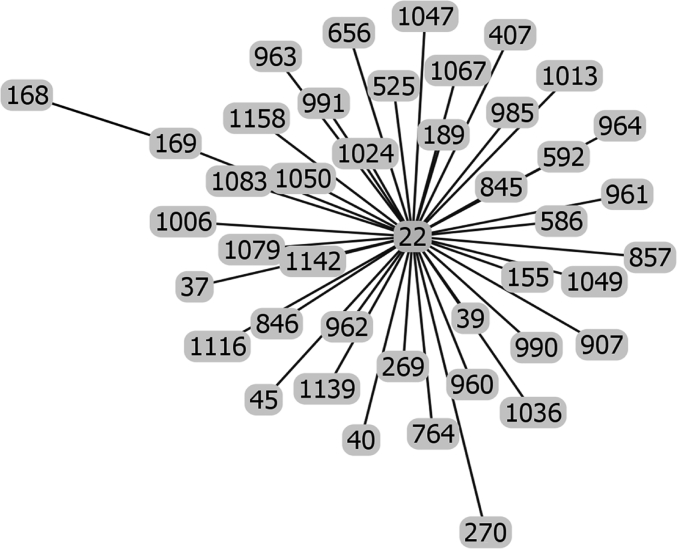
Raw data of the goeBURST analysis of the sequence types of strains possessing IS1548: Clonal Complex 22.
Fig. 7.
Raw data of the goeBURST analysis of the sequence types of strains possessing IS1548: Clonal Complex 23.
Fig. 8.
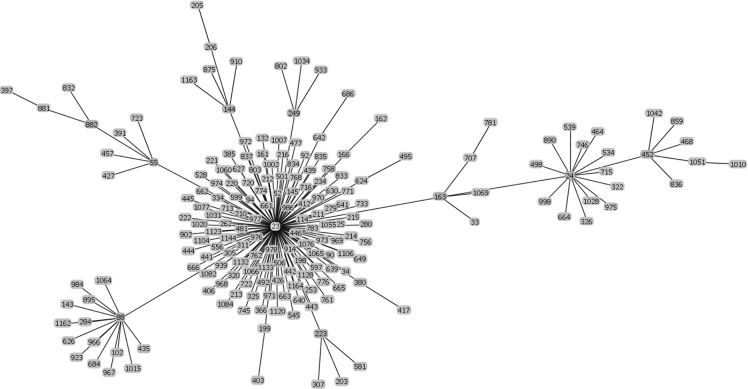
Raw data of the goeBURST analysis of the sequence types of strains possessing IS1548: Clonal Complex 27.
Fig. 9.
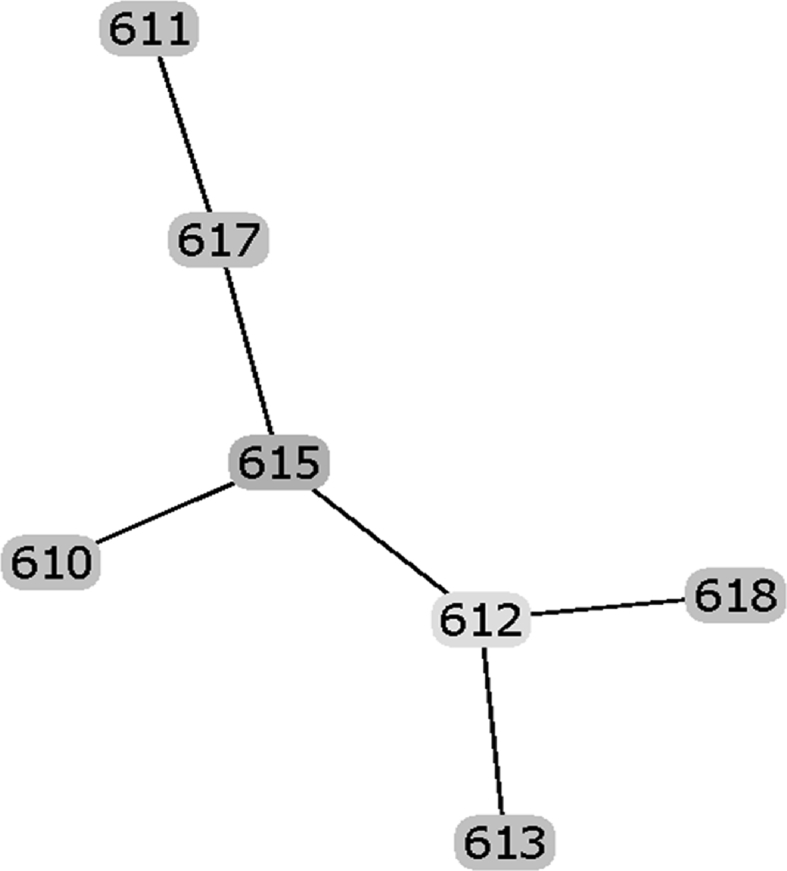
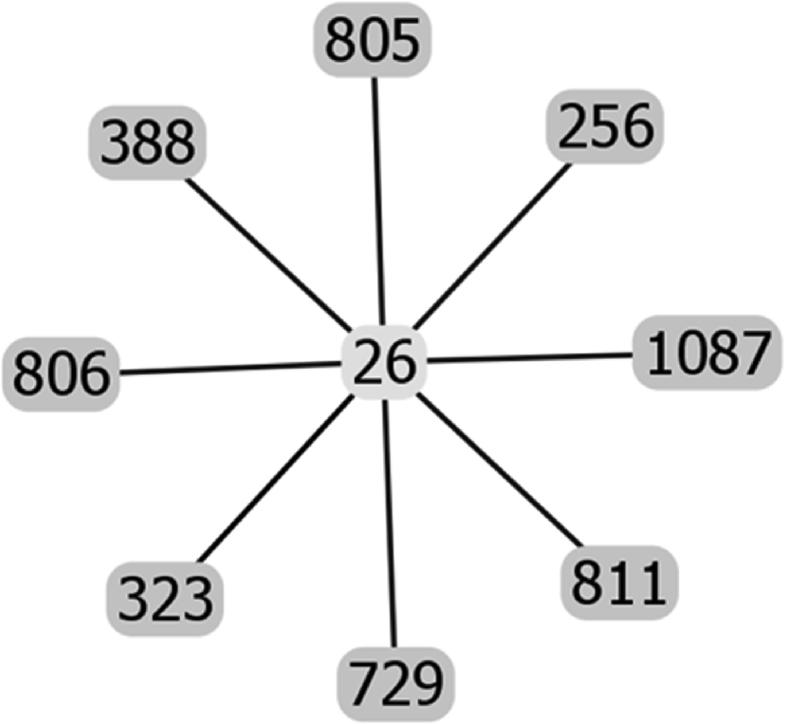
Raw data of the goeBURST analysis of the sequence types of strains possessing IS1548: Clonal Complex 615.
2. Experimental design, materials, and methods
The list of the S. agalactiae strains analyzed is available in Table S1 of reference [2]. Sequences of the genomes of these strains were available as whole genome contigs or as complete genome sequences at the NCBI database on January 19, 2018 (https://www.ncbi.nlm.nih.gov/genome/genomes/186?). Strains with an IS1548 genomic insertion were identified by blasting these genomes with the IS1548 DNA sequences described in Ref. [2]. To this end, the nucleotide collection (nr/nt) and the whole-genome shotgun contigs (wgs) databases of S. agalactiae (taxid:1311) were screened with the blastN program optimized for highly similar sequences (megablast). The algorithm parameters were defined as followed: maximum number of aligned sequences to display: 1000; parameters for short input sequences automatically adjusted; expect threshold: 10; word size: 28; maximum matches in a query range: 0; match/mismatch scores: 1,-2; gap costs: linear; filter for low complexity region activated; mask for lookup table only activated. S. agalactiae strains with an IS1548 positive blastN similarity finding were typed by Multi Locus Sequence Typing (MLST). Seven housekeeping genes were analyzed: alcohol dehydrogenase gene (adhP), phenylalanine tRNA synthetase gene (pheS), amino acid transporter gene (atr), glutamine synthetase gene (glnA), serine dehydratase gene (sdhA), glucose kinase gene (glcK) and transketolase gene (tkt). A sequence type, based on the allelic profile of these housekeeping genes, was assigned to each strain by submitting the complete genome sequence or all of the contigs sequences of each strain to the Streptococcus agalactiae MLST database (http://pubmlst.org/sagalactiae/, [3], Table 1). Identified MLST types were then assigned to clonal complexes defined by the goeBURST algorithm (http://www.phyloviz.net/goeburst/, [4], Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9) with the ST1 to ST1193 MLST profiles available at the MLST database (https://pubmlst.org/bigsdb?db=pubmlst_sagalactiae_seqdef&page=downloadProfiles&scheme_id=1). A goeBURST clonal complex was defined as all allelic profiles sharing six identical alleles with at least one other member of the group.
Acknowledgments
SK was supported by PhD fellowships of the Lebanese University and AZM & SAADÉ and of the Lebanese Association for Scientific Research (LASeR). This publication made use of the PubMLST website (https://pubmlst.org/) developed by Keith Jolley and sited at the University of Oxford. The development of that website was funded by the Wellcome Trust.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.105066.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Fléchard M., Gilot P. Physiological impact of transposable elements encoding DDE transposases in the environmental adaptation of Streptococcus agalactiae. Microbiology. 2014;160:1298–1315. doi: 10.1099/mic.0.077628-0. [DOI] [PubMed] [Google Scholar]
- 2.Khazaal S., Al Safadi R., Osman D., Hiron A., Gilot P. Dual and divergent transcriptional impact of IS1548 insertion upstream of the peptidoglycan biosynthesis murB gene of Streptococcus agalactiae. Gene. 2019;720:144094. doi: 10.1016/j.gene.2019.144094. [DOI] [PubMed] [Google Scholar]
- 3.Jolley K., Bray J., Maiden M. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francisco A.P., Bugalho M., Ramirez M., Carrico J.A. Global Optimal eBURST analysis of Multilocus typing data using a graphic matroid approach. BMC Bioinf. 2009;10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.