Abstract
Secretion systems are employed by bacteria to transport macromolecules across membranes without compromising their integrities. Processes including virulence, colonization, and motility are highly dependent on the secretion of effector molecules toward the immediate cellular environment, and in some cases, into the host cytoplasm. In Type II and Type III secretion systems, as well as in Type IV pili, homomultimeric complexes known as secretins form large pores in the outer bacterial membrane, and the localization and assembly of such 1 MDa molecules often relies on pilotins or accessory proteins. Significant progress has been made toward understanding details of interactions between secretins and their partner proteins using approaches ranging from bacterial genetics to cryo electron microscopy. This review provides an overview of the mode of action of pilotins and accessory proteins for T2SS, T3SS, and T4PS secretins, highlighting recent near‐atomic resolution cryo‐EM secretin complex structures and underlining the importance of these interactions for secretin functionality.
Keywords: bacterial virulence, protein–protein interactions, secretin, toxin secretion, Type IV pilus system, Types II and III secretion systems
1. INTRODUCTION
Bacteria depend on the transport of molecules, toxins, and macromolecules to (and from) the external environment in order to survive and proliferate. In Gram‐negative bacteria, such transport events require the crossing of the cytoplasmic membrane, the periplasmic space and the outer bilayer. In order to ensure substrate transport while still maintaining cell wall integrity, bacteria have developed secretion systems (T1SS–T9SS) and competence machineries that play key roles in competition with other microbes, virulence, surface attachment, and gene transfer processes. In addition, the various transported substrates include proteins (such as toxins and effectors), DNA, and protein‐DNA complexes.1, 2, 3, 4
One of the main challenges in the secretion process is the trespassing of the outer membrane without causing membrane damage and eventual cell rupture. Secretins, key members of the outer membrane complex in several systems, are essential for this stability. These homomultimeric complexes are key elements of the Type II and Type III secretion systems (T2SS and T3SS, respectively) as well as the Type IV pilus system (T4PS).5, 6, 7 Secretins are also present in bacteriophage extrusion systems8, 9 but these will not be discussed here. Other secretion systems employ different strategies to translocate molecules through the outer membrane, such as the formation of a pore by the secreted protein or a dedicated translocation partner in the T5SS,10 employment of the TolC oligomer by the T1SS,11 and transport through the 36‐stranded β‐barrel structures in the T8SS and the T9SS.12, 13, 14
Primary sequence analyses and electron microscopy studies of secretins from different transport systems and strains have revealed similarities in domain organization and overall structure. Secretin monomers display over 600 residues (Figure 1) and associate into oligomers of >1 MDa. The exact stoichiometry of secretin pores has been a matter of discussion. While assemblies of 12 protomers have been reported,18, 19, 20 more recent higher resolution cryo‐EM structures have indicated that the pentadecameric assembly is the most stable and predominant arrangement, at least for the T2SS and T3SS.15, 16, 21, 22, 23, 24, 25 Notably, structures of T4PS secretins have been reported to carry 12–14 subunits.17, 26, 27, 28, 29 The assembled secretin pores present two major domains: a C‐terminal, conserved core that folds into a 60‐stranded β‐barrel composed of inner and outer walls, and an N‐terminal region composed of a variable number of small α/β domains separated by flexible linkers (Figures 1 and 2). The latter are structurally similar to each other and may interact with the inner membrane (IM) platform, secreted substrates as well as internal structures.6, 25, 30, 31, 32 Secretins from the T2SS and T3SS also carry C‐terminal S‐domains, involved in localization, assembly and membrane stability15, 21, 25, 33, 34, 35, 36, 37, 38 (red in Figures 1 and 2). T4PS secretins, on the other hand, in some cases can present amidase N‐terminal (AMIN) domains, involved in peptidoglycan binding and secretin localization,27, 39, 40 or β‐domains that can act as a periplasmic gate.41
Figure 1.
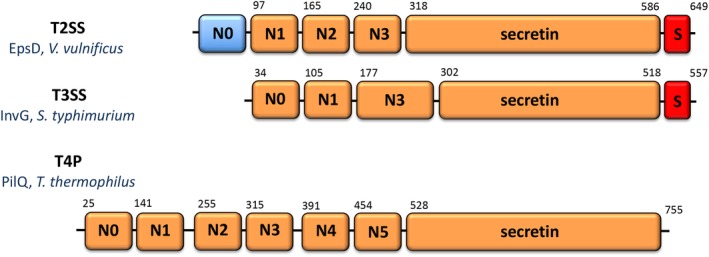
Schematic diagrams of secretin domains of the T2SS, T3SS, and T4PS. The number of N‐terminal domains can be variable, and T3SS do not carry N2 domains. N0 in EpsD from the T2SS of V. vulnificus was not traceable in the cryo‐EM map due to flexibility15 and is thus indicated in blue. Numbers indicate domain delimitations, as indicated in the publications describing the structures in Figure 2 15, 16, 17
Figure 2.
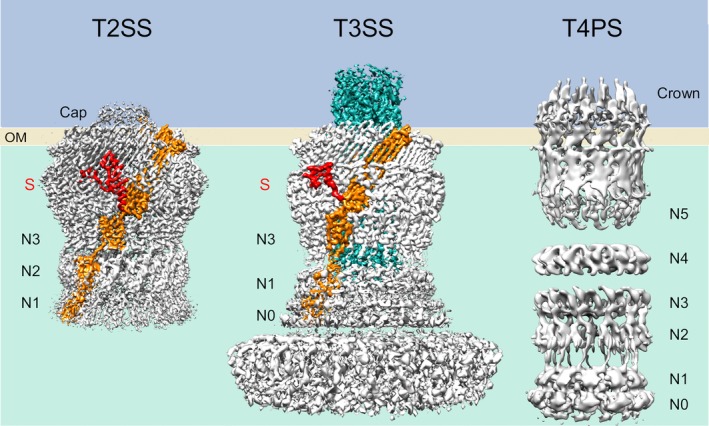
Cryo‐EM structures of secretins (T2SS, T4PS) and injectisome needle complex (T3SS). The resolution of the maps for the T2SS (EpsD from V. vulnificus, 3.4 Å15) and T3SS (InvG, PrgH, PrgK, PrgI from S. typhimurium; ~3.6 Å16) allowed for the positioning of the monomer (orange) including the S‐domain (red). The T3SS needle is indicated in cyan. The cryo‐EM structure of the T4PS secretin from T. thermophilus (right17) displays a large number of traceable N‐terminal domains, which is unusual due to their well‐documented flexibility in absence of the base components. OM, outer membrane; S, S‐domain. The figure was generated with PDB files and EM maps corresponding to: 6i1y and EMD‐0327 (T2SS); 6duz/6dwb/6dv3 and EMD‐8913/EMD‐8914/EMD‐8924 (T3SS); EMD‐3995/EMD‐3996/EMD‐3997/EMD‐3998 (T4PS)
Recent developments in cryo‐EM methods, associated to elegant genetic, biochemical, and microbiological studies have been essential not only for the understanding of secretin structures, but also of complete secretion machineries, as highlighted above. Importantly, mechanistic details of secretin targeting, assembly, and stability in the membrane, which are all essential for secretion system functionality, have also started to emerge. These events can be dependent on different classes of partner proteins or mechanisms (or a combination thereof): pilotins, ancillary molecules, and self‐piloting systems. This review will summarize the most recent evidence regarding such mechanisms for the T2SS, T3SS, and T4PS secretins, with particular emphasis on structural details regarding interactions between secretins and their partner molecules and their importance for substrate secretion and virulence.
2. ASSEMBLY OF T2SS SECRETINS
The T2S apparatus is used by at least 32 genera of Proteobacteria to secrete folded proteins from the periplasm to the outer milieu or to the cell surface. Secreted substrates are involved in survival and growth in the environment and inside hosts, and are essential for virulence in the case of pathogens. The T2SS translocates folded substrates from the periplasm toward the outside of the cell, and well‐studied substrates secreted by the T2SS include the heat labile toxin of enterotoxigenic Escherichia coli,42 cholera toxin of Vibrio cholerae 43 and exotoxin A of Pseudomonas aeruginosa.44 The T2SS apparatus is composed of four major components: an IM platform which, together with the OM secretin channel, forms a passage that accommodates substrates to be secreted; the pseudopilus, which polymerizes and pushes substrates through the interior of the secretin pore; and an ATPase, which provides energy for pseudopilus polymerization.5, 25, 45
Most full‐length secretin structures published to date are from the T2SS, and include those from pathogens such as V. cholerae, E. coli,21, 23, 36 Vibrio vulnificus, Aeromonas hydrophila,15 Klebsiella pneumoniae,25 and Klebsiella oxytoca.20 In addition, the structural characterization of the periplasmic domains of T2SS secretins as well as inner T2SS components have been instrumental in providing insight toward a mechanistic understanding of the system as well as the development of targeted inhibitors.46, 47, 48
Several T2SS secretins depend on pilotins for their assembly in the membrane. Pilotins are small lipoproteins that target the secretin monomers to the inner leaflet of the outer membrane via the Lol pathway.49, 50 The pilotin‐dependent assembly mechanism was first described by Hardie and co‐workers for PulD from K. oxytoca,51 which was shown to be dependent on the outer membrane‐anchored protein PulS for assembly in the leaflet. Studies of the PulS‐PulD interaction showed that the role of the pilotin is to protect the secretin from degradation in the periplasm; in its absence, secretin assembly can occur within the IM, leading to the initiation of the phage shock response.52, 53 More recent secretin assembly studies have confirmed the role of pilotins in T2SS secretin stability in the outer membrane.15, 36, 38 It is of interest that T2SS secretins have been classified as Vibrio‐type, Klebsiella‐type or Pseudomonas‐type based not only on sequence homologies, but also on the identity of their cognate pilotins.38
Two families of T2SS pilotins have been identified and structurally characterized by X‐ray crystallography: the OutS‐PulS pilotins, that interact with Klebsiella‐type secretins, and the AspS‐GspSβ pilotins, present in strains expressing Vibrio‐type secretins.38, 54 Interestingly, representatives from the two pilotin families are distinct both at the sequence and the structural levels (Figure 3, Table 1). Pilotins from the OutS‐PulS family fold into a bundle of 4 α‐helices that assemble with a concave hydrophobic groove at the center.33, 61 In the case of interaction studies performed with PulS and the S‐domain of PulD, all four helices were shown to interact with a disordered segment of the S‐domain that undergoes a disorder‐to‐order transition and folds into a helix upon binding.33 However, pilotins from the AspS/GspSβ family display a completely different fold, consisting of a 5‐stranded β‐sheet flanked by 4 α‐helices.15, 36, 38 This fold is reminiscent of a cupped hand, where the central region of the β‐sheet, which is highly hydrophobic, could represent the palm (EpsS in Figure 3).
Figure 3.
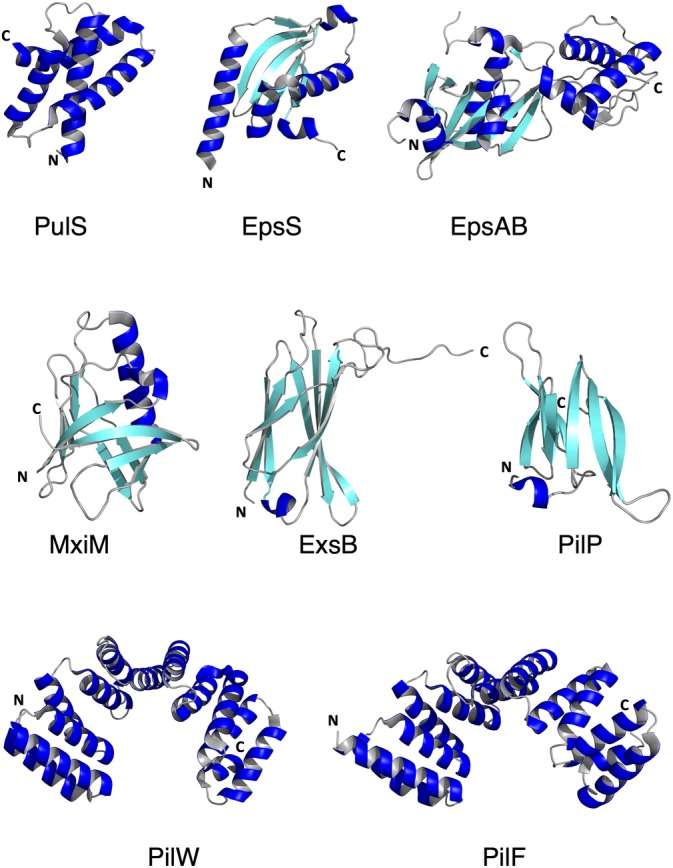
Gallery of X‐ray structures of pilotins and accessory molecules. The PDB codes for the different molecules include: T2SS: 4A56 (PulS33), 6I2V (EpsS15); 4G54 (EpsAB55); T3SS: 1Y9I (MxiM56), 2YJL (ExsB57); T4PS: 4AV2 (PilP58), 2VQ2 (PilW59), 2HO1 (PilF60)
Table 1.
Pilotins and accessory molecules of the T2SS, T3SS, and T4PS
| System | Pilotin | Accessory molecule | Cognate secretin | Bacterial species | Main references |
|---|---|---|---|---|---|
| T2SS | EpsS |
EpsA EpsB |
EpsD | Vibrio vulnificus |
Howard et al. 15 Strozen et al.55 |
| AspS | GspD | Escherichia coli (EPEC) |
Dunstan et al.38 Yin et al.36 |
||
| AspS | GspD | Vibrio cholerae | Dunstan et al.38 | ||
| OutS | OutD | Erwinia chrysanthemi | Rehman et al.61 | ||
| PulS | PulD | Klebsiella oxytoca |
Tosi et al.33 Nickerson et al.34 |
||
|
ExeA ExeB |
ExeD | Aeromonas hydrophila |
Li and Howard62 Ast et al.63 Vanderlinde et al.64 |
||
| T3SS | MxiM | MxiJ | MxiD | Shigella flexneri |
Schuch and Maurelli65 Lario et al.56 Okon et al.66 |
| ExsB | PscC | Pseudomonas aeruginosa |
Izoré et al.57 Perdu et al.67 |
||
| YscW | YscC | Yersinia enterocolitica |
Burghout et al.68 Ross and Plano132 |
||
| InvH | InvG | Salmonella enterica |
Daefler and Russell69 Craig and Koronakis70 |
||
| YsaP | YsaC | Yersinia enterocolitica | Rau and Darwin71 | ||
| T4aPS | PilW | PilP | PilQ | Neisseria meningitidis |
Trindade et al.59 Carbonnelle et al.72 Golovanov et al.58 |
| PilF | PilQ | Pseudomonas aeruginosa | Koo et al.60 | ||
| Tgl | PilQ | Myxococcus xanthus | Nudleman et al.73 | ||
| T4bPS | BfpG | BfpB | Escherichia coli | Lieberman et al.74 | |
| TcpQ | TcpC | Vibrio cholerae | Chang et al.75 | ||
| FimV | PilQ | Pseudomonas aeruginosa | Wehbi et al.76 | ||
| CpaE | CpaC | Caulobacter crescentus | Viollier et al.77 | ||
| TadD | RcpA | Aggregatibacter actinomycetemcomitans | Clock et al.78 |
Structural details showing the interaction between the latter family of pilotins and their cognate secretins have recently become available (E. coli GspD‐AspS, V. vulnificus EpsD‐EspS, and K. pneumoniae PulD‐PulS).15, 25, 36 A major player in the interaction is the S‐domain that forms a “belt” around the secretins and contains the two C‐terminal α‐helices (α11 and α12, in the case of the E. coli and V. vulnificus secretins) and the interconnecting loops. The cryo‐EM structure of the GspD‐AspS complex reveals that in order for the pilotin to bind, α12 must undergo an outward movement and place itself away from the body of the structure, after which recognition of AspS is possible through the hydrophobic platform formed by the central β‐sheet (Figure 4). In the structure of the GspD‐AspS complex, however, the lower resolution (approx. 5 Å) in the AspS binding region did not allow the visualization of details regarding the pilotin. Nevertheless, modeling of the interaction between V. vulnificus EpsD and its pilotin EpsS (a member of the AspS pilotin family) onto the GpsD‐AspS complex, where the structure of EpsS was at 1.7 Å resolution, shed more light on the issue. This analysis indicated that upon binding to α12, EpsS could undergo a minor conformational modification, as if “closing” its hand on the C‐terminal helix of EpsD (Figures 3 and 4). Mutational studies performed on E. coli and V. cholerae highlighted the importance of hydrophobic regions of both the S‐domain and the pilotins for secretin stability in the outer membrane and bacterial toxicity.15, 36
Figure 4.
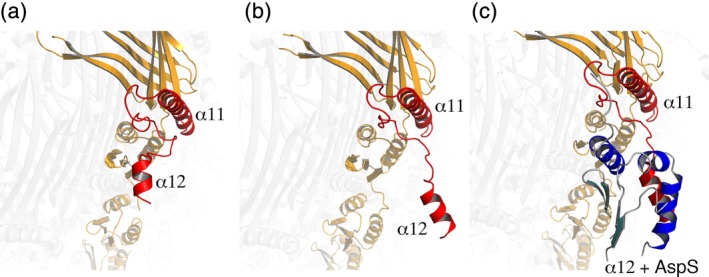
Pilotin recognition requires a conformational modification within the S‐domain of T2SS secretins. One monomer is indicated in orange, with the S domain in red, while the rest of the GspD secretin is indicated in light gray. (a) In the absence of the AspS pilotin, α12 is positioned close to the body of the secretin. (b) Pilotin recognition of α12 requires that it move away from the homo‐oligomer. (c) AspS (in blue) recognizes the “open” location of α1236
Some bacteria also present accessory proteins that may either substitute or work together with pilotins. ExeD from A. hydrophila, for example, requires the IM complex ExeA/ExeB for piloting and assembly. ExeA is an ATPase that can bind to peptidoglycan and form large multimers in the periplasm,62 while ExeB binds directly to the N0/N1 domains of ExeD.64 Notably, in the absence of the complex, ExeD is inserted in the IM,63 as in the case of PulS deletion mutants in K. oxytoca.53 It is of note that genomic analyses of A. hydrophila have not revealed the presence of a pilotin‐encoding gene, underlining the relevance of pilotin‐independent assembly mechanisms. Interestingly, ExeA/ExeB homologs have been identified as EpsA and EpsB in Vibrio spp, and are represented as a fusion protein in V. vulnificus 55 (Figure 3).
The T2SS secretin from P. aeruginosa, HxcQ, is an exception to all of the cases described above. Viarre and co‐workers showed that in addition to forming stable multimers within the outer membrane, HxcQ is also a lipoprotein, and carries a fatty acid at its extreme N‐terminus that plays a key role in its oligomerization in the membrane.79 Surprisingly, it was later identified that PA3611, a conserved T2SS protein of unknown function in P. aeruginosa, presents high structural similarity to AspS, but also has no lipoprotein signature sequence.38 This observation led authors to suggest that the function of PA3611 could involve recognition of the S‐domain of its cognate secretin HxcQ but could be limited to protecting it from proteolysis, which is plausible in the case of an autopiloting secretin like HxcQ. Interestingly, the T4PS also has examples of secretins capable of self‐piloting (below), indicating the importance of there being multiple mechanisms that guarantee the assembly of these important proteins.
3. ASSEMBLY OF T3SS SECRETINS
The T3SS, whose main structural element is the injectisome, is a complex machinery of more than 20 proteins that plays a key role in the secretion of substrates with the goal of modulating eukaryotic host cell function.80, 81 It is evolutionarily related to the flagellum assembly system and is widely used by bacteria to establish relations of mutualism or pathogenicity with eukaryotic organisms, including animals, plants, and fungi.81, 82, 83 A remarkable characteristic of this system is the needle complex that encompasses both inner and outer membrane rings and is completed by a protruding hollow needle. This apparatus allows the passage of substrates in semiunfolded form into the eukaryotic cytoplasm through a pore formed directly on the target cell membrane, thus forming a direct channel between the bacterial cytoplasm and that of the host.16, 24, 84, 85, 86, 87, 88, 89, 90
As is the case for the T2SS, T3SS secretin assembly can also be guided by partner proteins (Figure 3, Table 1). The first identified T3SS pilotin was YscW (formerly known as VirG) from Yersinia enterocolitica, a lipoprotein responsible for targeting the secretin YscC to the outer membrane, facilitating its oligomerization.68 Soon other pilotins were identified, such as InvH, responsible for localization and functionality of InvG in the outer membrane of Salmonella enterica,69, 70, 91 and MxiM, involved in stability, localization, and assembly of MxiD in Shigella flexneri.65 Other examples of T3SS pilotins include YsaP, involved in localization of the YsaC secretin in Y. enterocolitica,71 and ExsB, a YscW homologue shown to be critical for PscC targeting and assembly in P. aeruginosa 57, 67 (Figure 3, Table 1).
Structures of MxiM56, 66 and ExsB57 reveal folds that are different from T2SS pilotins. Both proteins are predominantly β‐stranded, but while the seven β‐strands of ExsB form an antiparallel sandwich with a short α‐helix between β4 and β5, MxiM presents eight β‐strands that form a pseudo‐barrel that is interrupted by a long α‐helix (Figure 3). NMR and ITC studies of the interaction of MxiM and InvH with the S‐domains of their cognate secretins revealed that central residues of these domains become structured upon binding of the pilotins, as was observed in the PulS‐PulD system described above.33, 66 Interestingly, in the case of the MxiM‐MxiD interaction, the cavity within the S‐domain of the secretin was shown to bind either lipid or the pilotin, suggesting a mechanism in which the pilotin only becomes lipid‐free in the presence of the secretin.66
Lastly, the presence of other accessory proteins that participate in secretin localization and assembly is not exclusive of T2S systems. In S. flexneri, one of the IM ring‐forming proteins, MxiJ, works synergistically with the pilotin MxiM, and the presence of either protein can stabilize the secretin.65 In enteropathogenic E. coli (EPEC), the IM ring protein EscD assists the oligomerization of EscC by interacting with its C‐terminus.92
4. ASSEMBLY OF T4PS SECRETINS
Many bacterial strains display surface pili, which are long (1–5 μM), fiber‐like appendages. In Gram‐negative organisms, these include the Type IV and chaperone‐usher pili.93 Type IV pili (T4P) are highly dynamic, and possess the remarkable ability to quickly extend and retract repeatedly. This ability is crucial for multiple functions, including adherence, motility, DNA uptake, and protein secretion.94, 95, 96, 97, 98, 99, 100
T4PS assembly is coordinated by a complex machinery that encompasses both inner and outer bacterial membranes, and the pilus itself is formed by polymerized pilin subunits arranged helically.101 The core T4PS machinery includes four main components: an ATPase, located at the base of the system and which provides energy for pilus extension; an IM platform; the pilus filament itself; and an outer membrane secretin.93 T4PS pilins are synthesized as prepilins in the cytoplasm and translocated toward the periplasmic side of the IM by the Sec system, as is the case of T2SS pseudopilins.102, 103 Prepilins are then processed by dedicated peptidases into mature pilins.104 Energy transduced from the cytoplasmic ATPase through an IM platform guides polymerization and depolymerization of the filament105, 106 that emerges in the outer membrane through the secretin channel.4, 107, 108, 109 Interestingly, Gram‐positive bacteria110 and archaea111 also present homologous T4P systems, though the secretin is mostly absent.
The most widely accepted classification of Type IV pili was generated based on sequence similarities of the pilin‐coding genes.112 Type IVa pili (T4aP) include those of Pseudomonas, Neisseria, Dichelobacter, Thermus, Myxococcus, and Deinococcus spp, among others, while Type IVb pili (T4bP) are represented in enteropathogenic, enterohemorrhagic, and enterotoxigenic E. coli, S. enterica, Caulobacter crescentus, and V. cholerae. In what relates to secretin assembly, outer‐membrane pilotin‐like accessory lipoproteins have been identified for T4aP,59, 60, 72, 73, 113 but T4bP secretins require other stability factors (Table 1) and often undergo auto‐assembly.74, 114, 115
4.1. T4aP
Stability and assembly of Neisseria spp, P. aeruginosa and Myxococcus xanthus T4aP secretins (PilQ) are dependent on the presence of the lipoproteins PilW, PilF, and Tgl, respectively, since in their absence only the monomeric form of PilQ can be detected.72, 73, 116 However, outer membrane targeting of these secretins seems to depend on different pathways: while in N. meningitidis deletion of pilW does not affect localization72 and PilQ is probably inserted by the BAM system,117 P. aeruginosa PilF pilots PilQ to the outer membrane in a Lol‐dependent manner.60, 118 It is interesting to note that Tgl can be transferred from tgl+ cells to tgl− mutants through a contact‐dependent mechanism, inducing the formation of PilQ multimers.73
Crystal structures of PilW and PilF reveal similar superhelical folds, with 13 anti‐parallel α‐helices that fold into six TPR (tetratricopeptide repeat) motifs.59, 60, 119 A similar number of TPRs is predicted for Tgl.73 TPRs are thermostable motifs that mediate protein–protein interactions and are often critical parts of large complexes.120 The TPR superhelix fold of PilW and PilF differs clearly from the α/β folds of the classical pilot proteins described above, indicating that their function could be distinct from the “piloting” of secretin monomers. Rather, their highly charged convex region could interact with other T4PS proteins or with the negatively charged outer membrane, while the concave groove could be involved in partner protein recognition.59, 120 Interestingly, TPR motifs found in chaperones involved in T3SS needle assembly do employ their conserved concave interface to allow partner binding.121, 122, 123 In PilW, a disulfide bond interconnects the two parts of the TPR superhelix, and plays a role in its functionality.59, 113
Other factors that may be involved in PilQ assembly and stabilization are the accessory factors FimV, PilP, and TsaP. FimV is a peptidoglycan‐binding factor found in P. aeruginosa that participates in IM subcomplex formation and ensures efficient multimerization of the secretin, in a similar manner to ExeAB in the T2SS of A. hydrophila, although the two proteins do not present sequence similarities.76, 124 In fact, FimV has multiple binding partners and possibly different functions, including a role in Type II secretion and regulation of cAMP production,125, 126 functions in which its TPR domains could play a key role.127
PilP is an inner‐membrane anchored lipoprotein present in N. meningitidis, N. gonorrhoeae, and P. aeruginosa. It binds directly to PilQ and is essential for T4PS formation.72, 128 Following the N‐terminal lipid attachment site, PilP presents an unstructured region and a C‐terminal globular domain that folds as a 7‐stranded β‐sandwich58 and presents structural homology with the T2SS IM protein GspC.41 Structural studies of the PilQ:PilP interaction involving cryo‐EM, NMR, and modeling indicate that PilP interacts at the interface between the central and peripheral rings of PilQ, with a potential role in stabilizing PilQ during the secretion process, in order to prevent channel disruption.116, 129
In the pathogen N. gonorrhoeae, absence of TsaP (T4PS secretin‐associated protein) results in formation of multiple pili in membrane protrusions instead of on the surface of the cell, indicating its function in extrusion of pili from the periplasm. This protein is in fact part of the peripheral ring observed in cryo‐EM maps of M. xanthus T4aP systems, and also plays roles in peptidoglycan attachment and T4PS localization to cell poles.28, 29
4.2. T4bP
Many T4bP secretins are lipoproteins.74, 115 In the EPEC bundle‐forming pilus (BFP), BfpB has been shown to be recognized by the Lol pathway,74 which can be involved in its outer membrane targeting, as is the case for HxcQ in the T2SS.79 In addition, the accessory protein BfpG is critical for assembly of a functional BfpB in the outer membrane, but only at the multimerization step.114 Likewise, in the toxin‐coregulated pilus (TCP) of V. cholerae, despite the fact that the TcpC secretin presents a lipidation signal, it also requires the periplasmic protein TcpQ for outer membrane localization and stability, and in its absence TcpC is degraded.115 Recent electron cryotomography studies reveal that TcpC appears as a ring around the periplasmic domain of TcpQ.75 Insight into the fold of TcpC was obtained by homology modeling using the Type 4 secretion system (T4SS) protein VirB7, which displays a compact globular structure that is reminiscent of the N0 domain of secretins.75
The Flp pilus or Tad pilus is a distinct subclass of T4bP130 and is sometimes called the Type IVc pilus.97 Unlike other T4bPs, its secretins (CpaC from C. crescentus and RcpA from Aggregatibacter actinomycetemcomitans) are not lipidated and in fact are very similar to those of the T2SS.78 CpaC requires CpaE for correct polar localization and for multimer assembly,77 and outer membrane insertion may be assisted by the BAM machinery.131 Meanwhile, in A. actinomycetemcomitans, the TPR‐containing TadD lipoprotein appears to be involved in RcpA stabilization, assembly, protection from proteolysis and targeting to the outer membrane,78 performing a role that is similar to that of pilotins.
5. CONCLUDING REMARKS
The combination of structural biology, biochemistry, and microbiology approaches have been instrumental in the comprehension of the assembly and functionality of secretins and of the systems in which they are involved. Nevertheless, there are still multiple questions that should be addressed in the future. One of the most interesting points involves the precise orchestration of secretin regulatory steps, as well as their potential interaction with the peptidoglycan and secreted substrates. Advances in single particle electron microscopy, cryo‐electron tomography, and the application of high‐resolution fluorescence microscopy strategies to the study of secretins and their complexes will undoubtedly shed light on these questions both from in vitro and in situ perspectives.
ACKNOWLEDGMENTS
Work in the Dessen lab on secretion systems and virulence factors is supported by grant 2017/12436‐9 from FAPESP (São Paulo Research Foundation) and the Laboratoire Intenational Associé (LIA) BACWALL (CNRS). We acknowledge the platforms of the Grenoble Instruct ERIC center (ISBG; UMS 3518 CNRS‐CEA‐UGA‐EMBL) within the Grenoble Partnership for Structural Biology (PSB). The IBS acknowledges integration into the Interdisciplinary Research Institute of Grenoble (IRIG, CEA). YROS was supported by grant 2018/04344‐0 from FAPESP.
Silva YRO, Contreras‐Martel C, Macheboeuf P, Dessen A. Bacterial secretins: Mechanisms of assembly and membrane targeting. Protein Science. 2020;29:893–904. 10.1002/pro.3835
Funding information Fundação de Amparo à Pesquisa do Estado de São Paulo, Grant/Award Numbers: 2017/12436‐9, 2018/04344‐0
REFERENCES
- 1. Costa TR, Felisberto‐Rodrigues C, Meir A, et al. Secretion systems in gram‐negative bacteria: Structural and mechanistic insights. Nat Rev Microbiol. 2015;13:343–359. [DOI] [PubMed] [Google Scholar]
- 2. Gaytán MO, Martínez‐Santos VI, Soto E, González‐Pedrajo B. Type three secretion system in attaching and effacing pathogens. Front Cell Infect Microbiol. 2016;6:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Christie P. The rich tapestry of bacterial protein translocation systems. Protein J. 2019;38:389–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denise R, Abby SS, Rocha EPC. Diversification of the type IV filament superfamily into machines for adhesion, protein secretion, DNA uptake, and motility. PLoS Biol. 2019;17:e3000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thomassin J‐L, Moreno JS, Guilvout I, Tran Van Nhieu G, Francetic O. The trans‐envelope architecture and function of the type 2 secretion system: New insights raising new questions. Mol Microbiol. 2017;105:211–226. [DOI] [PubMed] [Google Scholar]
- 6. Majewski DD, Worrall LJ, Strynadka NJC. Secretins revealed: Structural insights into the giant gated outer membrane portals of bacteria. Curr Opin Struct Biol. 2018;51:61–72. [DOI] [PubMed] [Google Scholar]
- 7. Hospenthal MK, Costa TRD, Waksman G. A comprehensive guide to pilus biogenesis in gram‐negative bacteria. Nat Rev Microbiol. 2017;15:365–379. [DOI] [PubMed] [Google Scholar]
- 8. Marciano DK, Russel M, Simon SM. Assembling filamentous phage occlude pIV channels. Proc Natl Acad Sci U S A. 2001;98:9359–9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Opalka N, Beckmann R, Boisset N, Simon MN, Russel M, Darst SA. Structure of the filamentous phage pIV multimer by cryo‐electron microscopy. J Mol Biol. 2003;325:461–470. [DOI] [PubMed] [Google Scholar]
- 10. Leo JC, Grin I, Linke D. Type V secretion: Mechanism(s) of autotransport through the bacterial outer membrane. Phil Trans R Soc Lond B. 2012;367:1088–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thomas S, Holland IB, Schmitt L. The Type 1 secretion pathway—The hemolysin system and beyond. Biochim Biophys Acta. 2013;1843:1629–1641. [DOI] [PubMed] [Google Scholar]
- 12. Lauber F, Deme JC, Lea SM, Berks BC. Type 9 secretion system structures reveal a new protein transport mechanism. Nature. 2018;564:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goyal P, Krasteva PV, Van Gerven N, et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature. 2014;516:250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cao B, Zhao Y, Kou Y, Ni D, Zhang XC, Huang Y. Structure of the nonameric bacterial amyloid secretion channel. Proc Natl Acad Sci U S A. 2014;111:E5439–E5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Howard SP, Estrozi LF, Bertrand Q, et al. Structure and assembly of pilotin‐dependent and ‐independent secretins of the Type II secretion system. PLoS Pathog. 2019;15:e1007731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu J, Worrall LJ, Hong C, et al. Cryo‐EM analysis of the T3S injectisome reveals the structure of the needle and open secretin. Nat Comm. 2018;9:3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. D'Imprima E, Salzer R, Bhaskara RM, et al. Cryo‐EM structure of the bifunctional secretin complex of Thermus thermophilus . Elife. 2017;6:e30483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reichow SL, Korotkov KV, Hol WGJ, Gonen T. Structure of the cholera toxin secretion channel in its closed state. Nat Struct Mol Biol. 2010;17:1226–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kowal J, Chami M, Ringler P, et al. Structure of the dodecameric Yersinia enterocolitica secretin YscC and its trypsin‐resistant core. Structure. 2013;21:2152–2161. [DOI] [PubMed] [Google Scholar]
- 20. Tosi T, Estrozi LF, Job V, et al. Structural similarity of secretins from type II and type III secretion systems. Structure. 2014;22:1348–1355. [DOI] [PubMed] [Google Scholar]
- 21. Hay ID, Belousoff MJ, Dunstan RA, Bamert RS, Lithgow T. Structure and membrane topography of the Vibrio‐type secretin complex from the type 2 secretion system of enteropathogenic Escherichia coli . Bacteriol. 2018;200:e00521–e00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hay ID, Belousoff MJ, Lithgow T. Structural basis of type 2 secretion system engagement between the inner and outer bacterial membranes. MBio. 2017;8:e01344–e01317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yan Z, Yin M, Xu D, Zhu Y, Li X. Structural insights into the secretin translocation channel in the type II secretion system. Nat Struct Mol Biol. 2017;24:177–183. [DOI] [PubMed] [Google Scholar]
- 24. Worrall LJ, Hong C, Vuckovic M, et al. Near‐atomic‐resolution cryo‐EM analysis of the salmonella T3S injectisome basal body. Nature. 2016;540:597–601. [DOI] [PubMed] [Google Scholar]
- 25. Chernyatina AA, Low HH. Core architecture of a bacterial type II secretion system. Nat Comm. 2019;10:5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koo J, Lamers RP, Rubinstein JL, Burrows LL, Howell PL. Structure of the Pseudomonas aeruginosa type IVa pilus secretin at 7.4 Å. Structure. 2016;24:1778–1787. [DOI] [PubMed] [Google Scholar]
- 27. Zhao X, Schwartz CL, Pierson J, et al. Three‐dimensional structure of the ultraoligotrophic marine bacterium “Candidatus Pelagibacter ubique”. Appl Environ Microbiol. 2017;83:e02807–e02816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang Y‐W, Rettberg LA, Treuner‐Lange A, Iwasa J, Søgaard‐Andersen L, Jensen G. Architecture of the type Iva pilus machine. Science. 2016;351:aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siewering K, Jain S, Friedrich C, et al. Peptidoglycan‐binding protein TsaP functions in surface assembly of type IV pili. Proc Natl Acad Sci U S A. 2014;111:E953–E961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Korotkov KV, Pardon E, Steyaert J, Hol WGJ. Crystal structure of the N‐terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure. 2009;17:255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Korotkov KV, Delarosa JR, Hol WGJ. A dodecameric ring‐like structure of the N0 domain of the type II secretin from enterotoxigenic Escherichia coli . J Struct Biol. 2013;183:354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Douzi B, Trinh NTT, Michel‐Souzy S, et al. Unraveling the self‐assembly of the Pseudomonas aeruginosa XcpQ secretin periplasmic domain provides new molecular insights into type II secretion system secreton architecture and dynamics. MBio. 2017;8:e01185–e01117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tosi T, Nickerson NN, Mollica L, et al. Pilotin‐secretin recognition in the type II secretion system of Klebsiella oxytoca . Mol Microbiol. 2011;82:1422–1432. [DOI] [PubMed] [Google Scholar]
- 34. Nickerson NN, Tosi T, Dessen A, et al. Outer membrane targeting of secretin PulD protein relies on disordered domain recognition by a dedicated chaperone. J Biol Chem. 2011;286:38833–38843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spreter T, Yip CK, Sanowar S, et al. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat Struct Mol Biol. 2009;16(5):468–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yin M, Yan Z, Li X. Structural insight into the assembly of the type II secretion system pilotin‐secretin complex from enterotoxigenic Escherichia coli . Nat Microbiol. 2018;3:581–587. [DOI] [PubMed] [Google Scholar]
- 37. Gu S, Rehman S, Wang X, Shevchik VE, Pickersgill RW. Structural and functional insights into the pilotin‐secretin complex of the type II secretion system. PLoS Pathog. 2012;8:e1002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dunstan RA, Heinz E, Wijeyewickrema LC, et al. Assembly of the type II secretion system such as found in Vibrio cholerae depends on the novel pilotin AspS. PLoS Pathog. 2013;9:e1003117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carter T, Buensuceso RN, Tammam S, et al. The type IVa pilus machinery is recruited to sites of future cell division. MBio. 2017;8:e02103–e02116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tarry MJ, Jääskeläinen M, Paino A, Tuominen H, Ihalin R, Högbom M. The extra‐membranous domains of the competence protein HofQ show DNA binding, flexibility and a shared fold with type I KH domains. J Mol Biol. 2011;409:642–653. [DOI] [PubMed] [Google Scholar]
- 41. Korotkov KV, Johnson TL, Jobling MG, et al. Structural and functional studies on the interaction of GspC and GspD in the type II secretion system. PLoS Pathog. 2011;7:e1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tauschek M, Gorrell RJ, Strugnell RA, Robins‐Browne RM. Identification of a protein secretory pathway for the secretion of heat‐labile enterotoxin by an enterotoxigenic strain of Escherichia coli . Proc Natl Acad Sci U S A. 2002;99:7066–7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rivera‐Chávez F, Mekalanos JJ. Cholera toxin promotes pathogen acquisition of host‐derived nutrients. Nature. 2019;572:244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Voulhoux R, Taupiac MP, Czjzek M, Beaumelle B, Filloux A. Influence of deletions within domain II of exotoxin A on its extracellular secretion from Pseudomonas aeruginosa . J Bacteriol. 2000;182:4051–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kooger R, Szwedziak P, Böck D, Pilhofer M. CryoEM of bacterial secretion systems. Curr Opin Struct Biol. 2018;52:64–70. [DOI] [PubMed] [Google Scholar]
- 46. Fulara A, Vandenberghe I, Read RJ, Devreese B, Savvides SN. Structure and oligomerization of the periplasmic domain of GspL from the type II secretion system of Pseudomonas aeruginosa . Sci Rep. 2018;8:16760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Michel‐Souzy S, Douzi B, Cadoret F, et al. Direct interaction between the secreted effector and the T2SS components GspL and GspM reveal a new effector‐sensing step during type 2 secretion. J Biol Chem. 2018;293:19441–19450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y, Faucher F, Zhang W, et al. Structure‐guided disruption of the pseudopilus tip complex inhibits the type II secretion in Pseudomonas aeruginosa . PLoS Pathog. 2018;14:e1007343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Okuda S, Tokuda H. Lipoprotein sorting in bacteria. Annu Rev Microbiol. 2011;65:239–259. [DOI] [PubMed] [Google Scholar]
- 50. Collin S, Guilvout I, Nickerson NN, Pugsley AP. Sorting of an integral outer membrane protein via the lipoprotein‐specific Lol pathway and a dedicated lipoprotein pilotin. Mol Microbiol. 2011;80:655–665. [DOI] [PubMed] [Google Scholar]
- 51. Hardie KR, Lory S, Pugsley AP. Insertion of an outer membrane protein in Escherichia coli requires a chaperone‐like protein. EMBO J. 1996;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- 52. Collin S, Krehenbrink M, Guilvout I, Pugsley AP. The targeting, docking and anti‐proteolysis functions of the secretin chaperone PulS. Res Microbiol. 2013;164:390–396. [DOI] [PubMed] [Google Scholar]
- 53. Guilvout I, Chami M, Engel A, Pugsley AP, Bayan N. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 2006;25:5241–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Strozen TG, Li G, Howard SP. YghG (GspSβ) is a novel pilot protein required for localization of the GspSβ type II secretion system secretin of enterotoxigenic Escherichia coli . Infect Immun. 2012;80:2608–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Strozen TG, Stanley H, Gu Y, et al. Involvement of the GspAB complex in assembly of the type II secretion system secretin of Aeromonas and Vibrio species. J Bacteriol. 2011;193:2322–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lario PI, Pfuetzner RA, Frey EA, et al. Structure and biochemical analysis of a secretin pilot protein. EMBO J. 2005;24:1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Izoré T, Perdu C, Job V, Attree I, Faudry E, Dessen A. Structural characterization and membrane localization of ExsB from the type III secretion system (T3SS) of Pseudomonas aeruginosa . J Mol Biol. 2011;413:236–246. [DOI] [PubMed] [Google Scholar]
- 58. Golovanov AP, Balasingham S, Tzitzilonis C, et al. The solution structure of a domain from the Neisseria meningitidis lipoprotein PilP reveals a new β‐sandwich fold. J Mol Biol. 2006;364:186–195. [DOI] [PubMed] [Google Scholar]
- 59. Trindade MB, Job V, Contreras‐Martel C, Pelicic V, Dessen A. Structure of a widely conserved type IV pilus biogenesis factor which affects the stability of secretin multimers. J Mol Biol. 2008;378:1031–1039. [DOI] [PubMed] [Google Scholar]
- 60. Koo J, Tammam S, Ku S‐Y, Sampaleanu LM, Burrows LL, Howell PL. PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa type IV pilus secretin. J Bacteriol. 2008;190:6961–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rehman S, Gu S, Shevchik VE, Pickersgill RW. Anatomy of secretin binding to the Dickeya dadantii type II secretion system pilotin. Acta Cryst. 2013;D69:1381–1386. [DOI] [PubMed] [Google Scholar]
- 62. Li G, Howard SP. ExeA binds to peptidoglycan and forms a multimer for assembly of the type II secretion apparatus in Aeromonas hydrophila . Mol Microbiol. 2010;76:772–781. [DOI] [PubMed] [Google Scholar]
- 63. Ast VM, Schoenhofen IC, Langen GR, Stratilo CW, Chamberlain MD, Howard SP. Expression of the ExeAB complex of Aeromonas hydrophila is required for the localization and assembly of the ExeD secretion port multimer. Mol Microbiol. 2002;44:217–231. [DOI] [PubMed] [Google Scholar]
- 64. Vanderlinde EM, Zhong S, Li G, Martynowski D, Grochulski P, Howard SP. Assembly of the type two secretion system in Aeromonas hydrophila involves direct interaction between the periplasmic domains of the assembly factor ExeB and the secretin ExeD. PLoS One. 2014;9:e102038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schuch R, Maurelli AT. MxiM and MxiJ, base elements of the Mxi‐Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J Bacteriol. 2001;183:6991–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Okon M, Moraes TF, Lario PI, et al. Structural characterization of the type III pilot‐secretin complex from Shigella flexneri . Structure. 2008;16:1544–1554. [DOI] [PubMed] [Google Scholar]
- 67. Perdu C, Huber P, Bouillot S, et al. ExsB is required for correct assembly of the Pseudomonas aeruginosa type III secretion apparatus in the bacterial membrane and full virulence in vivo . Infect Immun. 2015;83:1789–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Burghout P, Beckers F, de Wit E, et al. Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica . J Bacteriol. 2004;186:5366–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Daefler S, Russel M. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol Microbiol. 1998;28:1367–1380. [DOI] [PubMed] [Google Scholar]
- 70. Craig AM, Koronakis V. Salmonella InvG forms a ring‐like multimer that requires the InvH lipoprotein for outer membrane localization. Mol Microbiol. 1998;30:47–56. [DOI] [PubMed] [Google Scholar]
- 71. Rau R, Darwin AJ. Identification of YsaP, the pilotin of the Yersinia enterocolitica Ysa type III secretion system. J Bacteriol. 2015;197:2770–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Carbonnelle E, Helaine S, Prouvensier L, Nassif X, Pelicic V. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol Microbiol. 2005;55:54–64. [DOI] [PubMed] [Google Scholar]
- 73. Nudleman E, Wall D, Kaiser D. Polar assembly of the type IV pilus secretin in Myxococcus xanthus . Mol Microbiol. 2006;60:16–29. [DOI] [PubMed] [Google Scholar]
- 74. Lieberman JA, Frost NA, Hoppert M, et al. Outer membrane targeting, ultrastructure, and single molecule localization of the enteropathogenic Escherichia coli type IV pilus secretin BfpB. J Bacteriol. 2012;194:1646–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chang YW, Kjær A, Ortega DR, et al. Architecture of the Vibrio cholerae toxin‐coregulated pilus machine revealed by electron cryotomography. Nat Microbiol. 2017;2:16269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wehbi H, Portillo E, Harvey H, et al. The peptidoglycan‐binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin. J Bacteriol. 2011;193:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Viollier PH, Sternheim N, Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 2002;21:4420–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Clock SA, Planet PJ, Perez BA, Figurski DH. Outer membrane components of the tad (tight adherence) secreton of Aggregatibacter actinomycetemcomitans . J Bacteriol. 2008;190:980–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Viarre V, Cascales E, Ball G, Michel GPF, Filloux A, Voulhoux R. HxcQ liposecretin is self‐piloted to the outer membrane by its N‐terminal lipid anchor. J Biol Chem. 2009;284:33815–33823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wagner S, Grin I, Malmsheimer S, Singh N, Torres‐Vargas CW, Westerhauser S. Bacterial type III secretion systems: a complex device for the delivery of bacterial effector proteins into eukaryotic host cells. FEMS Microbiol Lett. 2018;365:fny201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Deng W, Marshall NC, Rowland JL, et al. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. 2017;15:323–337. [DOI] [PubMed] [Google Scholar]
- 82. Abby SS, Rocha EPC. The non‐flagellar type III secretion system evolved from the bacterial flagellum and diversified into host‐cell adapted systems. PLoS Genet. 2012;8:e1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Diepold A, Amstutz M, Abel S, Sorg I, Jenal U, Cornelis GR. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 2010;29:1928–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Park D, Lara‐Tejero M, Waxham MN, et al. Visualization of the type III secretion mediated Salmonella‐host cell interface using cryo‐electron tomography. Elife. 2018;7:e39514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Matteï P‐J, Faudry E, Job V, Izoré T, Attree I, Dessen A. Membrane targeting and pore formation by the type III secretion system translocon. FEBS J. 2011;278:414–426. [DOI] [PubMed] [Google Scholar]
- 86. Dortet L, Lombardi C, Cretin F, Dessen A, Filloux A. Pore‐forming activity of the Pseudomonas aeruginosa type III secretion system translocon alters the host epigenome. Nat Microbiol. 2018;3:378–386. [DOI] [PubMed] [Google Scholar]
- 87. Nauth T, Huschka F, Schweizer M, et al. Visualization of translocons in Yersinia type III protein secretion machineries during host infection. PLoS Pathog. 2018;14:e1007527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nans A, Kudryashev M, Saibil H, Hayward RD. Structure of a bacterial type III secretion system in contact with a host membrane in situ . Nat Commun. 2015;6:10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Schoehn G, Di Guilmi AM, Lemaire D, Attree I, Weissenhorn W, Dessen A. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas . EMBO J. 2003;22:4957–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Schraidt O, Lefebre MD, Brunner MJ, et al. Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 2010;6:e1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pati NB, Vishwakarma V, Jaiswal S, Periaswamy B, Hardt WD, Suar M. Deletion of invH gene in Salmonella enterica serovar Typhimurium limits the secretion of Sip effector proteins. Microbes Infect. 2013;15:66–73. [DOI] [PubMed] [Google Scholar]
- 92. Tseytin I, Dagan A, Oren S, Sal‐Man N. The role of EscD in supporting EscC polymerization in the type III secretion system of enteropathogenic Escherichia coli . Biochim Biophys Acta Biomembr. 2017;1860:384–395. [DOI] [PubMed] [Google Scholar]
- 93. Craig L, Forest KT, Maier B. Type IV pili: Dynamics, biophysics and functional consequences. Nat Rev Microbiol. 2019;17:429–440. [DOI] [PubMed] [Google Scholar]
- 94. Nieto V, Kroken AR, Grosser MR, et al. Type IV pili can mediate bacterial motility within epithelial cells. MBio. 2019;10:e02880–e02818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Higashi DL, Lee SW, Snyder A, Weyand NJ, Bakke A, So M. Dynamics of Neisseria gonorrhoeae attachment: Microcolony development, cortical plaque formation, and cytoprotection. Infect Immun. 2007;75:4743–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lee CK, de Anda J, Baker AE, et al. Multigenerational memory and adaptive adhesion in early bacterial biofilm communities. Proc Natl Acad Sci U S A. 2018;115:4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ellison CK, Dalia TN, Vidal Ceballos A, et al. Retraction of DNA‐bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae . Nat Microbiol. 2017;3:773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Seitz P, Blokesch M. DNA‐uptake machinery of naturally competent Vibrio cholerae . Proc Natl Acad Sci U S A. 2013;110:17987–17992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Han X, Kennan RM, Parker D, Davies JK, Rood JI. Type IV fimbrial biogenesis is required for protease secretion and natural transformation in Dichelobacter nodosus . J Bacteriol. 2007;189:5022–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yuen AS, Kolappan S, Ng D, Craig L. Structure and secretion of CofJ, a putative colonization factor of enterotoxigenic Escherichia coli . Mol Microbiol. 2013;90:898–918. [DOI] [PubMed] [Google Scholar]
- 101. Craig L, Taylor RK, Pique ME, et al. Type IV pilin structure and assembly: X‐ray and EM analyses of Vibrio cholerae toxin‐coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11:1139–1150. [DOI] [PubMed] [Google Scholar]
- 102. Francetic O, Buddelmeijer N, Lewenza S, Kumamoto CA, Pugsley AP. Signal recognition particle‐dependent inner membrane targeting of the PulG pseudopilin component of a type II secretion system. J Bacteriol. 2007;189:1783–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Arts J, van Boxtel R, Filloux A, Tommassen J, Koster M. Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle‐sec pathway. J Bacteriol. 2007;189:2069–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. de Bentzmann S, Aurouze M, Ball G, Filloux A. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J Bacteriol. 2006;188:4851–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Takhar HK, Kemp K, Kim M, Howell PL, Burrows LL. The platform protein is essential for type IV pilus biogenesis. J Biol Chem. 2013;288:9721–9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kruse K, Salzer R, Averhoff B. The traffic ATPase PilF interacts with the inner membrane platform of the DNA translocator and type IV pili from Thermus thermophilus . FEBS Open Bio. 2019;9:4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Collins RF, Frye SA, Balasingham S, Ford RC, Tonjum T, Derrick JP. Interaction with type IV pili induces structural changes in the bacterial outer membrane secretin PilQ. J Biol Chem. 2005;280:18923–18930. [DOI] [PubMed] [Google Scholar]
- 108. Goosens VJ, Busch A, Georgiadou M, et al. Reconstitution of a minimal machinery capable of assembling periplasmic type IV pili. Proc Natl Acad Sci U S A. 2017;114:E4978–E4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gold VA, Salzer R, Averhoff B, Kuhlbrandt W. Structure of a type IV pilus machinery in the open and closed state. Elife. 2015;4:e07380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Melville S, Craig L. Type IV pili in gram‐positive bacteria. Microbiol Mol Biol Rev. 2013;77:323–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Pohlschroder M, Esquivel RN. Archaeal type IV pili and their involvement in biofilm formation. Front Microbiol. 2015;6:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Strom MS, Lory S. Structure‐function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. [DOI] [PubMed] [Google Scholar]
- 113. Szeto TH, Dessen A, Pelicic V. Structure/function analysis of Neisseria meningitidis PilW, a conserved protein that plays multiple roles in type IV pilus biology. Infect Immun. 2011;79:3028–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Schmidt SA, Bieber D, Ramer SW, Hwang J, Wu CY, Schoolnik G. Structure‐function analysis of BfpB, a secretin‐like protein encoded by the bundle‐forming pilus operon of enteropathogenic Escherichia coli . J Bacteriol. 2001;183:4848–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Bose N, Taylor RK. Identification of a TcpC‐TcpQ outer membrane complex involved in the biogenesis of the toxin‐coregulated pilus of Vibrio cholerae . J Bacteriol. 2005;187:2225–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Jain S, Mościcka KB, Bos MP, et al. Structural characterization of outer membrane components of the type IV pili system in pathogenic Neisseria . PLoS One. 2011;6:e16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. [DOI] [PubMed] [Google Scholar]
- 118. Hoang HH, Nickerson NN, Lee VT, et al. Outer membrane targeting of Pseudomonas aeruginosa proteins shows variable dependence on the components of Bam and Lol machineries. MBio. 2011;2:e00246–e00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kim K, Oh J, Han D, Kim EE, Lee BC, Kim Y. Crystal structure of PilF: Functional implication in the type 4 pilus biogenesis in Pseudomonas aeruginosa . Biochem Biophys Res Commun. 2006;340:1028–1038. [DOI] [PubMed] [Google Scholar]
- 120. Perez‐Riba A, Itzhaki LS. The tetratricopeptide‐repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr Opin Struct Biol. 2019;54:43–49. [DOI] [PubMed] [Google Scholar]
- 121. Bröms JE, Edqvist PJ, Forsberg A, Francis MS. Tetratricopeptide repeats are essential for PcrH chaperone function in Pseudomonas aeruginosa type III secretion. FEMS Microbiol Lett. 2006;256:57–66. [DOI] [PubMed] [Google Scholar]
- 122. Edqvist PJ, Broms JE, Betts HJ, Forsberg A, Pallen MJ, Francis MS. Tetratricopeptide repeats in the type III secretion chaperone, LcrH: Their role in substrate binding and secretion. Mol Microbiol. 2006;59:31–44. [DOI] [PubMed] [Google Scholar]
- 123. Quinaud M, Ple S, Job V, et al. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc Natl Acad Sci U S A. 2007;104:7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Howard SP, Gebhart C, Langen GR, Li G, Strozen TG. Interactions between peptidoglycan and the ExeAB complex during assembly of the type II secretin of Aeromonas hydrophila . Mol Microbiol. 2006;59:1062–1072. [DOI] [PubMed] [Google Scholar]
- 125. Michel GP, Aguzzi A, Ball G, Soscia C, Bleves S, Voulhoux R. Role of fimV in type II secretion system‐dependent protein secretion of Pseudomonas aeruginosa on solid medium. Microbiology. 2011;157:1945–1954. [DOI] [PubMed] [Google Scholar]
- 126. Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol. 2010;76:889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Buensuceso RN, Nguyen Y, Zhang K, et al. The conserved tetratricopeptide repeat‐containing C‐terminal domain of Pseudomonas aeruginosa FimV is required for its cyclic AMP‐dependent and ‐independent functions. J Bacteriol. 2016;198:2263–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Balasingham SV, Collins RF, Assalkhou R, et al. Interactions between the lipoprotein PilP and the secretin PilQ in Neisseria meningitidis . J Bacteriol. 2007;189:5716–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Berry JL, Phelan MM, Collins RF, et al. Structure and assembly of a trans‐periplasmic channel for type IV pili in Neisseria meningitidis . PLoS Pathog. 2012;8:e1002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kachlany SC, Planet PJ, Desalle R, Fine DH, Figurski DH, Kaplan JB. flp‐1, the first representative of a new pilin gene subfamily, is required for non‐specific adherence of Actinobacillus actinomycetemcomitans . Mol Microbiol. 2001;40:542–554. [DOI] [PubMed] [Google Scholar]
- 131. Ryan KR, Taylor JA, Bowers LM. The BAM complex subunit BamE (SmpA) is required for membrane integrity, stalk growth and normal levels of outer membrane beta‐barrel proteins in Caulobacter crescentus . Microbiology. 2010;156:742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ross JA and Plano GV. A C‐terminal region of Yersinia pestis YscD binds the outer membrane secretin YscC. J. Bacteriol. 2011; 193:2276–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]


