Abstract
Research studies of infectious disease outbreaks in wild species of the cat family Felidae have revealed unusual details regarding forces that shape population survival and genetic resistance in these species. A highly virulent feline coronavirus epidemic in African cheetahs, a disease model for human SARS, illustrates the critical role of ancestral population genetic variation. Widespread prevalence of species specific feline immunodeficiency virus (FIV), a relative of HIV–AIDS, occurs with little pathogenesis in felid species, except in domestic cats, suggesting immunological adaptation in species where FIV is endemic. Resolving the interaction of host and pathogen genomes can shed new light on the process of disease outbreak in wildlife and in humankind. The role of disease in endangered populations and species is difficult to access as opportunities to monitor outbreaks in natural populations are limited. Conservation management may benefit greatly from advances in molecular genetic tools developed for human biomedical research to assay the biodiversity of both host species and emerging pathogen. As these examples illustrate, strong parallels exist between disease in human and endangered wildlife and argue for an integration of the research fields of comparative genomics, infectious disease, epidemiology, molecular genetics and population biology for an effective proactive conservation approach.
Keywords: FIV, SARS, AIDS, CDV
1. Introduction
It is becoming increasingly clear that understanding the components of disease processes, particularly infectious disease, will be critical for understanding the survival or demise of free ranging mammalian species. Genomic variation of both host species and infectious agent is of critical importance in the outcome of a disease outbreak, and even more so in endangered species. Species with reduced genetic variation, particularly in genes involved with disease resistance, may be less able to mount an effective immune response against an emerging pathogen. Representing carnivores, the cat family Felidae offers numerous examples of reduced genetic variation in natural populations common to endangered species including Asian lion (Panthera leo persica) (Gilbert et al., 1991), cheetah (Acinonyx jubatus) (Menotti-Raymond and O’Brien, 1993), tiger (P. tigris) (Luo et al., 2004), leopard (P. pardus) (Uphyrkina et al., 2001, Uphyrkina et al., 2002) and the North American populations of puma (Puma concolor) (Culver et al., 2000, Roelke et al., 1993) that may signify increased susceptibility to opportunistic infectious disease. In addition, the pathogens themselves evolve, constantly developing new genetic based strategies to overcome or abrogate the immune defenses of the host species. The consequence of the co-evolution of pathogen and host are a dynamic ratchet of constantly improving virulence of the agent and resistance of the host. Both the pathogens and the host are survivors of ancient deadly struggles and the exquisite strategies that have been retained are only now being deciphered.
Conservation management of endangered carnivores has limited opportunities to detect, monitor and contain emergent pathogens due to logistic concerns involved with the continued monitoring of the health status of natural populations. As a result, we know most about disease processes in humans, Homo sapiens (Garrett, 1994), so understanding human–pathogen interactions can help us correctly interpret examples from wildlife. There are far more people and more human medical researchers than there are for most large charismatic mammals, so more human disease outbreaks have been tracked. Tools of molecular biology, genetics, immunology and epidemiology were developed with human disease in mind, but they work very well in other mammals. Thus, efforts in characterizing the genetics, evolution, and pattern of transmission of pathogens, and the resultant conservation implications for the host species, benefit directly from advances in human genomic research.
One of the most significant advances in the genomics era integral to conservation genetics is the whole genome sequence of a score of mammals. Starting with human, mouse, rat, and chimp, these projects now capture the phylogenetic divergence across all the orders of placental mammals by inclusion of dog, cat, elephant, cow and others (NHGRI website: www.genome.gov). Further, a subset of orthologous genes sequenced in representatives of 5400 species determined conclusively the evolutionary pattern of divergence of placental mammals during the last 105 MY (Murphy et al., 2001a, Murphy et al., 2004, Murphy et al., 2001b, O’Brien et al., 1999a). The utility of the mammalian phylogeny in conservation ranges from defining taxonomic units to using the evolutionary tree as a reference guide to detect patterns of mutation, adaptation, and selection in endangered species. For example, whole genome sequences of human, chimpanzee (Pan troglodytes), mouse (Mus musculus) and dog (Canis familiaris) indicate that not only do genomes evolve at different rates (Cooper et al., 2004), but also categories of genes experience different regimes of selection to either diversify or maintain function among primates, rodents and carnivores. In addition, human and chimpanzee, separated by only 5–7 MY of evolution (Enard and Paabo, 2004) exhibit profound differences in chromosome recombination (Winckler et al., 2005), gene expression (Hill and Walsh, 2005) and differential selection for biological function (Clark et al., 2003). Further, adaptive evolution of genes involved with the immune response to infectious disease can be estimated only by comparison across species. For example, the major histocompatibility complex (MHC) is a large multi-gene complex responsible for adaptive immune response in mammals and is integral to host resistance to emerging pathogens (Kumanovics et al., 2003). Whole genome sequence of MHC reveals differences in gene composition, gene order and putative gene function between primates, rodents and carnivores (Belov et al., 2004, Kumanovics et al., 2003, Yuhki et al., 2003). Thus, conservation genetic strategies for targeting informative genes benefit from a wealth of sequence data defining unique differences for these gene families among taxonomic groups.
Our ongoing research into host–pathogen interactions in the cat family Felidae offers additional insights on how the application of molecular genomic technologies to non-human animal species not traditionally studied in research laboratories holds real promise in conservation. There are 38 species of felids nearly all of which are listed as threatened or near threatened with extinction (www.iucnredlist.org). Recently, we have defined the pattern of divergence of the eight major lineages of cats from a common origin in Asia and a series of global migration events over the past 10 MY (Johnson et al., 2006) (Fig. 1 ) which provides an important evolutionary context for the analysis of host–pathogen adaptation. In the following, we illustrate how our investigations into the genomic, evolutionary and population studies of endangered cats species reviewed in O’Brien and Johnson (2005) uncovered feline disease that resembled those in human and vice versa. The first involves the SARS epidemic that devastated human populations in east Asia in 2003 and a parallel outbreak in the cheetah (A. jubatus) years before (Pearks Wilkerson et al., 2004). The second focuses on the lentivirus genus, which a generation ago (Korber et al., 2000) leapt from chimpanzee to humans-likely through the bush meat trade in western Africa (Hahn et al., 2000, Sharp et al., 1999, Sharp et al., 2005), and precipitated perhaps the most deadly scourge in recorded history, HIV–AIDS . The cat family Felidae is afflicted with a close relative of HIV, feline immunodeficiency virus (FIV). In domestic cat Felis catus, FIV infection results in disease progression and outcome similarly to that of HIV in humans, and offers a natural model to AIDS (Bendinelli et al., 1995). Here, we compare and contrast FIV genetics among additional cat species to yield new perspectives on host–pathogen adaptation.
Fig. 1.
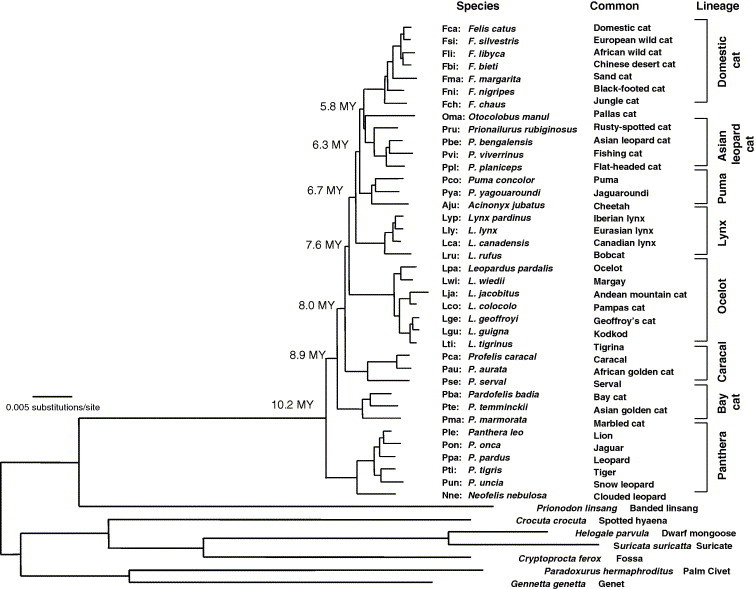
Cat family phylogeny from Johnson et al., 2006. Shown is the maximum likelihood tree using the GTR +I model of sequence evolution from 18,853 bp of data from autosome, X and Y linked genes. Terminal nodes are labelled with three-letter codes, scientific name, common name and grouped in to eight major lineages within Felidae.
The impact of emerging pathogens in felids in these cases provides a cautionary tale of the importance of disease outbreaks in free-ranging species. The unpredictable nature of these outbreaks argues for conservation management strategies to guard against the introduction of disease by domestic animals or introduction of infected individuals into naïve populations. However, these cases show how in humans, rapid genetic and genomic characterization of both pathogen and host has been effective in development of treatment, intervention, and therapy, and offers a paradigm for disease research in conservation of endangered species.
2. SARS
SARS (severe acute respiratory syndrome) first appeared as a flu-like disease caused by a new human coronavirus in Guangdong Province in Southern China (Drazen, 2003, Drosten et al., 2003, Holmes, 2003). In the space of nine months the virus traveled to 29 countries, infected over 8000 people, and caused nearly 800 deaths (CDC, 2004). The virus spread with alarming speed among health care workers, through casual contacts, and across the globe, causing human suffering and huge economic costs. Published reports indicate a virus phylogenetically close to the SARS virus was discovered in samples collected in Chinese food markets from Himalayan palm civets Paguma lavarta (Guan et al., 2003). Further screening of wildlife has now identified one or more species of bat as the presumptive host reservoir for SARS-like coronaviruses (SL-CoV); (Lau et al., 2005, Li et al., 2005, Poon et al., 2005). The epidemic subsided by May 2003, presumably consequent of Draconian quarantine measures. There is still little clear understanding of the precise mode of transmission, and while there are promising advances in research of SARS protease inhibitors (Savarino, 2005, Wei et al., 2006) there is still no vaccine or efficacious treatment for infected patients.
The SARS outbreak caught many by surprise, since human coronaviruses are well known as the cause of one third of common colds and are rarely deadly. Further, veterinary virologists have commonly studied coronaviruses in livestock, dogs, cats and poultry and find these viruses seldom cause fatal diseases (Holmes, 2003). However, exceptions have occurred, for example in pigs, where a single nucleotide variant of porcine coronavirus leads to virulent pathogenic enteric coronavirus (Ballesteros et al., 1997, Sanchez et al., 1999). The second exception involved a devastating feline coronavirus outbreak in cheetahs A. jubatus documented in a wild animal park, Wildlife Safari, in rural Winston, Oregon (Heeney et al., 1990, O’Brien et al., 1985).
Wildlife Safari was then the most prolific A. jubatus breeding facility in the world, holding some 60 cheetahs. In May 1982, two young cheetahs arrived from the Sacramento Zoo in California and rapidly developed symptoms of fever, severe diarrhea, jaundice, and neurological spasms. Both died and were diagnosed at necropsy with the wet form of feline infectious peritonitis (FIP) a disease caused by feline coronavirus in domestic cats (Heeney et al., 1990). Within six months, every cheetah in the park developed antibodies to the FIP virus (FIPV), and exhibited diarrhea, jaundice, weight loss, gingivitis, and renal and hepatic pathology (Fig. 2 ). Within two years 60% of the cheetahs had died of FIP (Evermann et al., 1988, Heeney et al., 1990). To our knowledge, this was the worst outbreak of FIP in any cat species. In reported domestic cat outbreaks, mortality seldom exceeds 5% (Foley et al., 1997).
Fig. 2.
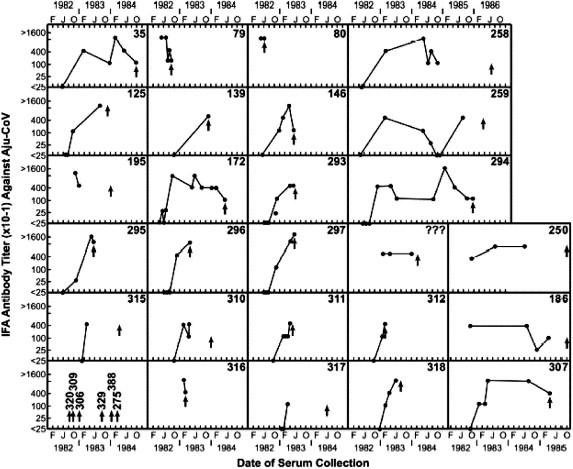
Time course of the antibody titers against FCoV in 35 cheetahs living in a wildlife park in Winston, Oregon in 1982–1986 (modified Fig. 1 from Heeney et al., 1990). Titers are based on immunofluorescent assay of cultured cells. Numbers are from the North American cheetah studbook numbers for individual cheetahs (Marker and O’Brien, 1989). Arrows represent date of death by Aju-CoV for each animal.
The Winston FIP outbreak preceded PCR and advanced molecular phylogenetic methods, but when SARS appeared in 2003, archival specimens were revisited to characterize the nature of the cheetah coronavirus (Pearks Wilkerson et al., 2004). Sequences of three viral genes from five cheetahs were PCR amplified. Phylogenetic analyses of the cheetah strains and other known coronaviruses placed the cheetah FIPV within the monophyletic Group I lineage as close relatives of domestic cat FIPV, porcine transmissible gastroenteritis virus (TGEV) and canine coronavirus (CcoV) (Fig. 3 a). Furthermore, the polyphyletic arrangement of the cheetah strains interspersed with domestic cat FIPV suggested few differences between the deadly cheetah virus and the more innocuous domestic cat virus (Fig. 3b). Given the high genetic similarity between domestic cat FIPV and cheetah FIPV, and the fact that several lions (P. leo) at Winston Park became infected with the virus simultaneously but did not succumb to FIP, we suggested the reason for the extremely high morbidity and mortality was due to host genetics.
Fig. 3.
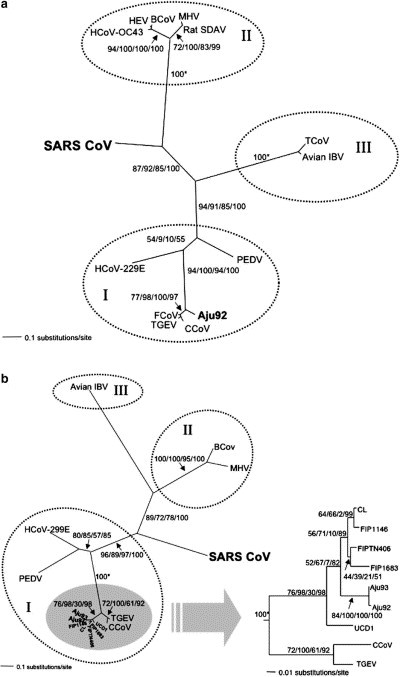
(a) Phylogenetic analysis of cheetah coronavirus Aju-CoV pol 1b (439 bp) sequences from archived liver and kidney tissues from cheetahs living at Wildlife Safari during the coronavirus outbreak unrooted maximum likelihood tree under the GTR+I model of sequence evolution (−ln likelihood = 3347.49). Numbers plotted along the branches indicate bootstrap values and Bayesian posterior probabilities shown as percentages depicted in the following order, maximum likehood/maximum parsimony/minimum evolution/Bayesian. The three major coronavirus antigenic groups are supported by the genomic data and are indicated by hatched circles and roman numerals. Abbreviations are as follows, human coronavirus 229E (HCoV-229E), canine coronavirus (CCoV), feline coronavirus (FCoV), porcine transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), human coronavirus OC43 (HCoV-OC43), bovine coronavirus (BCoV), porcine hemagglutinating encephalomyelitis virus (HEV), rat sialodacryoadenitis (SDAV), mouse hepatitis virus (MHV), turkey coronavirus (TCoV), avian infectious bronchitis virus (avian IBV), SARS coronavirus from human (SARSCoV) and from palm civet (SZ16). (b) Phylogenetic analyses based on pol 1a (405 bp). Inset depicts portions of the unrooted maximum likelihood tree for pol 1a (405 bp) under the GTR + I model of sequence evolution (−ln likelihood = 3368.65). CL, FIP1146, FIPTN406, FIP1683 and UCD1 are FCoV strains from domestic cats. Cheetah isolates from Winston, Oregon outbreak are indicated as Aju. Details of the sequences and experimental procedures are provided in Pearks Wilkerson et al. (2004).
The cheetah is one of the most genetically homogeneous species within the cat family. The cheetah has as its closest relatives two North and South American species of puma (P. concolor) and jaguarondi (P. yagouaroundi) and together form the puma lineage (Fig. 1). Fossil records indicate that cheetah once had a nearly global distribution, compared with its current distribution of sub-Saharan Africa and a relict population in Iran (Nowell and Jackson, 1996). A series of genetic studies of cheetah populations describe a species depauperate in genomic variation and this loss in genetic heterogeneity was most likely due to a severe population bottleneck that felled scores of large mammal species in North America, Europe, Asia and Australia 10,000–12,000 years ago (Driscoll et al., 2002, Menotti-Raymond and O’Brien, 1993, O’Brien et al., 1987). The near extirpation led to generations of close inbreeding during the cheetah’s ancestry reducing overall genomic diversity 10–100-fold (Menotti-Raymond and O’Brien, 1993). This reduction included variation in the immune response genes within the major histocompatibility complex (O’Brien et al., 1985, O’Brien and Yuhki, 1999b, Yuhki and O’Brien, 1990). The genetic basis for disease resistance is that inherent population genetic diversity provides a broad moving target for evolving pathogens. Thus, when a microbe evolves a strategy to abrogate immune defenses of an individual, the genetically diverse population may still be protected. However, once virulence was achieved in the first cheetah, the conditions were set for transmission, pathogenesis and morbidity in the other individuals within the population made vulnerable by shared genetic and immunologic homogeneity.
The parallels and lessons for SARS have relevance to conservation of biodiversity in wildlife. First, the deadly coronaviruses in both human and cheetah represent profound consequences that may accompany viral emergence into a new host. Phylogenetic analyses of genetic data clearly define the domestic cat origin of cheetah FIPV (Fig. 3a, b) and that the virus appeared to be highly pathogenic upon entering the new host. The similarity of the pattern of high pathogenicity with SARS infection of humans suggests that the virus is potentially more benign in the reservoir species of bats, albeit this remains unconfirmed (Lau et al., 2005, Li et al., 2005). Even so, the pattern of genetic changes within the pathogen among the intermediate host of palm civet P. lavarta was very similar to those observed in human strains, and is consistent with the expectation of rapid evolution in the pathogen to adapt to the new host immune response (Song et al., 2005). Second, while mortality in humans with SARS symptoms (CDC, 2004) and in house cats with FIPV (Foley et al., 1997) was low (10% in humans, 5% in cats), infected cheetahs exhibited the opposite extreme with 90% morbidity and over 60% mortality (Fig. 2). As for cheetahs, the combined evidence suggests that genetic uniformity makes epidemics much worse. It is likely that variation in immune defense genes of humans and domestic cats protected them from a deadly disease, to which most exposed and genetically impoverished cheetahs succumbed (Fig. 2). Thus, genomic characterization of both pathogen and host are key for conservation of biodiversity and prevention of disease outbreak. Lastly, FIPV in cheetahs and SARS in humans are highly contagious, spreading rapidly in close quarters in weeks, if not days (Fig. 2; (Shaw, 2006)). This inherent ability of emergent pathogens to rapidly infect endangered populations should be an integral consideration for management strategies of re-introduction.
3. AIDS and FIV
AIDS had first appeared in humans in the early 1980s as a clustering of patients from homosexual communities in Los Angeles and New York City with a rare cancer, Kaposi’s sarcoma, and pneumonia. AIDS is caused by a lentivirus, a genus within the family Retroviridae, termed human immunodeficiency virus or HIV that infects and destroys CD4-bearing T-cells. There are two forms of HIV (types 1 and 2) and since its recognition in the early 1980s, HIV-1 has spread across the globe infecting 65 million people and killing over 22 million (www.unaids.org). AIDS is potentially the most deadly disease in recorded history because it is a disease with no vaccine and a treatment that slows the virus but does not clear it from sequestered cellular reservoirs, and has infected 65 million and continues to infect new cohorts within the world populations (Rathbun et al., 2006) (www.unaids.org).
The origin of the ongoing HIV-1 pandemic is recent and the virus entered humans early in the 20th century, originating from a subspecies of chimpanzee P. troglodytes troglodytes in central West Africa (Gao et al., 1999). Wild chimpanzees are infected with SIVcpz (simian immunodeficiency virus) and genetic study affirms this strain is the closest genetic relative to HIV (Gao et al., 1999). Additional evolutionary studies indicated these cross-species transmission events occurred more than once between chimpanzee and human to form HIV-1 subtypes M, N and O (Gao et al., 1999) and at least once between sooty mangabey (Cercocebus atys) and humans to form HIV-2 (Gao et al., 1992). In total, primate lentiviruses infect at least 23 species and like SIVcpz are host-specific (Hahn et al., 2000, Sharp et al., 1999, Sharp et al., 2005). All naturally occurring SIV are in African primates and these species seem resistant to AIDS-like pathology. By contrast, Asian species are naïve, and do develop AIDS when infected with the SIV from African primates (Hirsch, 2004).
The sole naturally occurring model for AIDS disease in humans is the feline homologue of HIV, feline immunodeficiency virus (FIV). Although related lentiviruses are found in other species such as sheep and goats (caprine arthritis encephalitis virus – CAEV), horse (equine infectious anemia virus – EIAV), and cattle (bovine immunodeficiency virus – BIV), only FIV in domestic cats causes AIDS-like disease. FIV was first discovered in 1986 in a house cat with a wasting-like disease (Pedersen et al., 1987). Currently, FIV is epidemic among feral domestic cats throughout the world and is associated with an Aids-like syndrome of immune depletion, increased susceptibility to rare cancers and opportunistic disease, and death (Bendinelli et al., 1995, Willett et al., 1997). FIV has a small viral RNA genome of about 9000 nucleotides that is similar in sequence, gene content, and gene arrangement to that of HIV-1 (Miyazawa et al., 1994).
As with primate lentiviruses, FIV naturally infects multiple species of cat in the wild. Western blot screening of antibodies against FIV in free-ranging non-domestic cat species (Fig. 4 ) revealed that several were exposed to the virus (Carpenter and O’Brien, 1995, Olmsted et al., 1989, Olmsted et al., 1992, Troyer et al., 2005). A recent comprehensive survey of serum and lymphocyte specimens from 3055 individuals within 35 Felidae and 3 Hyenidae species used three FIV antigens isolated from domestic cat, lion, and puma as targets of western blot plus PCR-based FIV genome sequence amplification as a validation of FIV infection (Troyer et al., 2005). Those results revealed that at least 11 free ranging Felidae species harbor FIV antibodies and FIV viral genomes (Fig. 4). These species are distributed across Africa including lion, leopard (P. pardus), and cheetah along with two Hyenidae species of spotted hyena (Crocuta crocuta) and striped hyena (Hyaena hyaena). FIV positive species occur in North and South America including puma, jaguaroundi (P. onca), jaguarondi, ocelot (Leopardus pardalis), margay (L. weidii), Geoffroy’s cat (L. geoffroyi), tigrina (L. tigrina) and in Asia with pallas cat (Otocolobus manul). Eight other species had western blot signals of antibodies, but did not yield FIV sequences using multiple PCR primers which is likely due to divergence within the primer binding sites, exceedingly low proviral load, or both (Fig. 4). Overall, inclusion of FIV western blot results from captive individuals indicates that as many as 31 species of cat are susceptible to FIV infection and may harbor species specific viruses or be infected with non-native virus (Troyer et al., 2005) (Fig. 4). Further, the distribution of FIV is not uniform among cat species, being endemic in Africa and North and South America, but rare in species from Europe and Asia (Fig. 4).
Fig. 4.
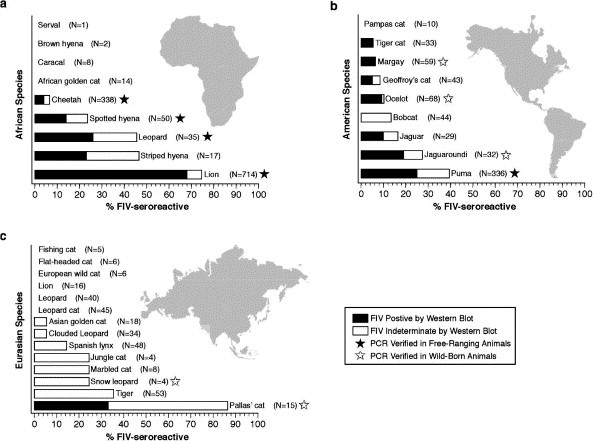
Prevalence and distribution of FIV seroreactivity in free-ranging, and captive but wild-born, animals (Troyer et al., 2005) The dark portion of the bar represents seropositives and the white represents seroindeterminates, giving conservative and liberal estimates of total seroprevalence of FIV. The number of animals tested for each species is given in parentheses. A black star indicates that PCR verification has been obtained from at least one free-ranging individual for that species. A white star indicates that PCR verification has been obtained from at least one captive, but wild-born individual for that species. (a) Percent seroreactivity in African carnivores (excluding African golden cat, sand cat, and black footed cat, for which there were no wild-born samples). (b) Percent seroreactivity in American felids (excluding kodkod and Canadian lynx, for which there were no wild-born samples). (c) Percent seroreactivity in Eurasian felids (excluding lynx, bay cat, rusty spotted cat and Chinese dessert cat, for which there were no wild-born samples).
Phylogenetic analyses of the pol-RT region (476 bp) isolated in Felidae species reveal the sequences form species-specific monophyletic clades (Fig. 5 ). This monophyletic pattern suggests that FIV does not spread among species easily. Rather, FIV infected an ancestor of each host and then evolved within that species. There have been exceptions to this finding, but only in captive animals and these occur rarely. For example, the captive snow leopard (P. uncia) FIV sequence is well within the African lion FIV clade A (Fig. 5). Another example is a captive puma infected with a domestic cat strain of FIV (Carpenter et al., 1996). However, in the wild, opportunities for inter-species transmission are limited and thus the virus becomes adapted to its particular host. This evolutionary pattern resembles that of naturally occurring SIV in African primates, where SIV forms monophyletic lineages that are specific to each species (Allan et al., 1991, Sharp et al., 2005) and are host-dependent. Yet, FIV transmission may have a geographic component whereby species in close proximity share more similar FIV strains. This is suggested in the common node shared by leopard FIV and cheetah FIV (Fig. 5).
Fig. 5.
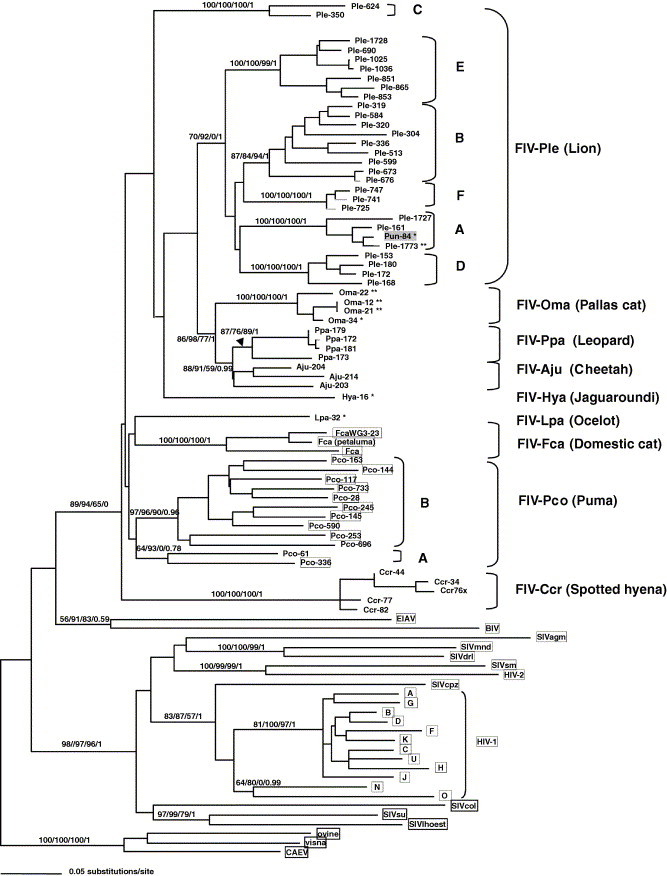
Phylogenetic tree for FIV pol-RT nucleotide sequences (476 bp included in analysis) isolates from indicated Felidae species. Shown here is the single maximum likelihood (ML) tree. Maximum parsimony (MP), minimum evolution (ME; Neighbor joining based on the Tajima Nei algorithm for distance measure) and Bayesian (BA) trees gave similar topologies. Bootstrap values and posterior probabilities are included at all nodes with bootstrap support (ML/ME/MP/Bayesian). Taxa are designated with the species code followed by the source animal ID#. Sequence downloaded from GenBank for comparison are boxed, all other sequences are novel. A single asterix (∗) indicates the sequence was obtained from a captive, but wild-born animal, a double asterix (∗∗) indicates sequence from captive-born individuals. All other novel sequences are from free-ranging animals. Analysis used empirical base frequencies, an estimated shape parameter of 0.6618, an estimated substitution matrix as follows: A/C = 2.0642, A/G = 7.2581, A/T = 1.4798, C/G = 4.0169, C/T = 14.0129 and an estimated proportion of invariant sites of 0.1525. Shown here is the single maximum likelihood tree with maximum likelihood bootstraps from 1000 replicates.
Clinical consequences for FIV infection in most non-domestic Felidae species is not known, and there has been little convincing acute disease or patterned disease symptoms associated with FIV infection as is observed consistently for domestic cats. Only one example of FIV-related mortality has been documented in a species other than domestic cat, a captive lion (Poli et al., 1995). The lack of documentation of clinical signs of FIV infection in wild populations may reflect logistic obstacles and expenses in routine physicals for these species. Alternatively, these species-specific FIV strains may have undergone a prolonged period of co-adaptation with their respective host and may be less pathogenic as a consequence (Carpenter et al., 1996, Carpenter and O’Brien, 1995). In the following study with lions, we describe how current conservation efforts with endangered cat species are integrating genetic studies of FIV in natural populations.
4. FIV in African lions
Natural populations of African lions represent one of the longest running studies of FIV infected felids in the wild (Brown et al., 1994, Troyer et al., 2004). A western blot survey of lions throughout Africa (Fig. 6 ) indicates FIV antibody seroprevalence can range from 0% (in Namibia and Kalahari) to >94% in the Serengeti in Tanzania and Kruger Park in South Africa. This east–west discordance in FIV seroprevalence may be a consequence of geographic barriers such as the Kalahari Desert. However, it is not clear whether these geographic differences in FIV prevalence (most were sampled only once) reflect an ongoing spread or a stable restriction of FIV spread in the low incidence regions.
Fig. 6.
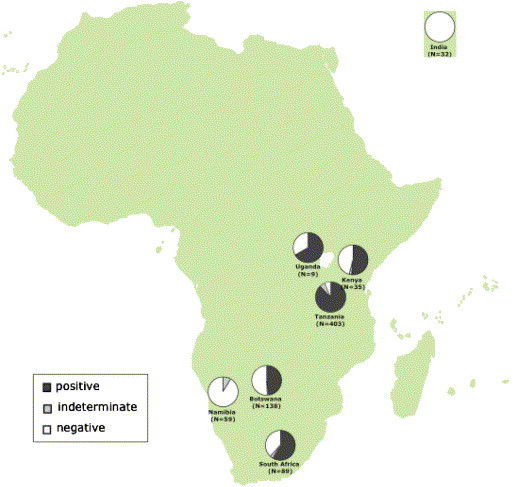
Prevalence of FIV antibodies among lions living in wild populations in Africa. The dark gray portion of the circle represents seropositives and the light grey represents seroindeterminates, giving conservative and liberal estimates of total seroprevalence of FIV. The white portion represents FIV negative lions. The number of animals tested for each population is given in parentheses.
Phylogenetic analysis of FIV RT-pol sequences in lions reveal the persistence of at least six principal viral clades or subtypes (A–F) represented in different populations of lions in Africa. Some are unique to certain populations (e.g. Subtype E in Botswana, Subtype C in the Serengeti and subtype F in Kenya) while others are present in multiple populations (Fig. 7 ). The genetic distances among these lion FIV subtypes are as great as the distance observed between FIV isolates from different cat species (Fig. 5, Fig. 7). This level of separation would suggest that the clades evolved in isolation. The common monophyletic node uniting all lion FIV strains suggests that FIV infected a common ancestor early in lion population history. The virus subtypes then evolved in isolation with specific lion populations as observed with subtypes C, D, and E. In the case of subtype A and B, it is likely that these evolved in formerly geographically isolated lion populations that recently came together allowing the transmission of the virus subtypes currently observed. Because there is little known of disease associations with FIV infection in lions it is difficult to know whether there are now (or ever were) virulence distinctions among the clades.
Fig. 7.
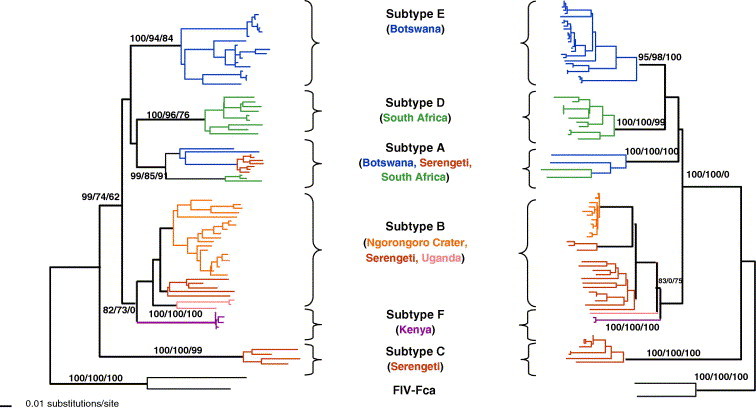
Phylogenetic tree for FIV pol-RT (520 bp included in analysis) and gag (400 bp) nucleotide sequences isolated from lion populations sampled in Fig. 6. Shown here is the single maximum likelihood (ML) tree. Maximum parsimony (MP), and minimum evolution (ME; Neighbor joining based on the Tajima Nei algorithm for distance measure) trees gave similar topologies. Bootstrap values are included at all nodes with bootstrap support (ML/ME/MP). Taxa are colored by the country of origin of the lion sampled. FIV–FCA is the outgroup strain of domestic cat FIV.
One of the most detailed lion population studies is that of the Serengeti in Tanzania. This large panmictic population over 3000 individuals has provided many useful insights into lion pride structure, population dynamics, and transmission of infectious diseases (Gilbert et al., 1991, Hofmann-Lehmann et al., 1996, Muller-Graf et al., 1999, Munson et al., 1996, Packer et al., 2005). Long-term surveillance of lions in the Serengeti has not revealed any evidence of AIDS like illnesses, decreased viability, or decreased fecundity in this population where seroprevalence among adult lions is 100%. These observations of (relatively) benign infection are similar to those for FIV in North American puma (Biek et al., 2006, Biek et al., 2003) and consistent with the notion that FIV has persisted in these host species longer than in domestic cat and that significant host adaptation has already occurred (Carpenter et al., 1996, Carpenter and O’Brien, 1995). However, recently we have shown that lions infected with FIV have significantly lower CD4 counts than uninfected animals suggesting that there may be sub-clinical immunological effects associated with FIV-infection in this population (Roelke et al., in press). At one stage we also suspected that infection with one clade of FIV might be a natural vaccine against infection with another or the mutational occurrence of a virulent deadly FIV variant (Carpenter and O’Brien, 1995). However, Troyer et al. (2004) inspected the pattern of FIV subtype distribution among 13 Serengeti lion prides and discovered multiple clade infections (i.e., two or three subtypes of virus present in the same lion) in 43% of the infected lions, an observation that would argue against acquired immunity by infection with a single strain.
In 1994, the Serengeti lions became the target of a deadly infectious disease outbreak that caused whisker twitches, neurological seizures, and death to over 1000 lions, a third of the population. FIV was excluded as the primary cause since several afflicted lions were FIV negative (Roelke-Parker et al., 1996). The etiological agent was a hyper-virulent form of a canine distemper virus (CDV) a virus previously known to infect but seldom cause morbidity in Felidae species (Roelke-Parker et al., 1996). This time, in the most deadly reported outbreak of the CDV among African carnivore species, a virulent transmissible CDV variant spread from domestic dogs living around the Serengeti park to hyena, bat eared foxes Otocyon megalotis, and to lions (Carpenter et al., 1998).
Although FIV was not the primary explanation for the 1994 Serengeti lion mortality, it may have played a supporting role. HIV infection in humans causes increased susceptibility to opportunistic infections of normally benign infectious agents (like HHV8/Kaposi’s sarcoma, Pneumocystis carnii pneumonia or cytomegalovirus) as a consequence of CD4 T-lymphocyte depletion and immune collapse (Levy, 1993). Nearly all the lions in the Serengeti carried FIV, and FIV has been shown to be associated with depleted CD4-lymphocyte counts of 200 cells/μl compared with counts of 800–1000 cells/μl in uninfected individuals (Roelke et al., in press). One could speculate that adult lions may have done better in defending against CDV had they not been afflicted with lentivirus that depletes their principal defense against viruses, the CD4 lymphocyte population. While there were FIV-negative animals that died, these were all juveniles (Roelke-Parker et al., 1996) and it is possible that their immature immune system may have accounted for their susceptibility to the new virus.
The African lion study provides some useful insights on the impact of infectious disease in endangered populations. Development of detection methods and effective monitoring of the health of animals in the wild is problematic, and as in the case of the CDV outbreak, was documented only due to the presence of researchers already in place to collect samples and observe the final outcome. Although this CDV outbreak was marked by relatively high mortality estimates (Roelke-Parker et al., 1996), subsequent outbreaks were less virulent (Packer et al., 1999) within the same population. Until we fully understand the immunological effects of infection with prevalent FIV strains, the interaction of pathogens such as CDV and FIV, and the role of the host immune system in modulating these viral infections, the question of their importance and collaborations in pathogenesis will continue.
One approach is to continue to adapt and apply new technologies developed from human biomedical research to conservation of wildlife. For example, the massive research effort into understanding HIV in humans offers new technologies to apply towards related viruses such as FIV in endangered cat species. Recognition that FIV causes AIDS-like disease in domestic cat and that this is a natural model for HIV in humans, has led to extensive surveillance of FIV in other species (Biek et al., 2006, Biek et al., 2003, Carpenter et al., 1996, Carpenter and O’Brien, 1995, Olmsted et al., 1992, Troyer et al., 2004, Troyer et al., 2005), an essential step in determining the genetic diversity of the pathogen. Knowledge of key amino acid motifs in the env gene of HIV linked with evading host immune surveillance (Wyatt and Sodroski, 1998, Yusim et al., 2002) is in turn targets for comparative genomic research of virulence in FIV (de Parseval et al., 2006). Understanding how genetic diversity within MHC in humans is linked with either resistance to HIV or increased susceptibility (Carrington and O’Brien, 2003) provides an incentive to sequence MHC in multiple mammalian taxa, including cats (Yuhki et al., 2003, Yuhki and O’Brien, 1990). The links between HIV in humans and FIV in endangered cat species illustrate the utility of biomedical approaches and discoveries that is directly applicable to elucidating the role of disease in endangered species.
5. Conclusions
We are just now beginning to understand the complex interplay of host genetic background, immune competence, and pathogen evolution that contribute to the course of an epidemic when a disease agent enters a new species or population. The advances in molecular biology, immunology and genetics provide a diverse collection of effective tools for unraveling the secrets to survival. As the tools that are used in human medicine become available to wildlife research, new approaches and understanding of disease outbreaks and effective host control of infection are increasingly possible.
Unfortunately, in wildlife populations, few disease outbreaks are followed from start to finish and the vast majority remain completely unobserved and undocumented [but see Randall et al. on Ethiopian wolves and other papers in this special edition]. For this reason, our understanding of the effects of population demographics, genetics, mating and migration patterns on the spread of (and ultimately clearance of and/or adaptation to) disease is limited.
These examples of FIPV and FIV illustrate a critical need for consistent programs designed to monitor and survey endangered populations. Conservation strategies that incorporate biological sampling yield invaluable genetic profiles of disease and disease resistance within endangered species. Genomic analyses provide insight into the history of a population and its pathogen(s) that can assist in documenting patterns of co-evolution and adaptation. Yet, these examples underscore the unpredictable aspects of epidemics within natural populations of threatened or endangered species and there is need for much more comprehensive and long-term data. Finally, conservation management would clearly benefit from a better knowledge of viral evolution, adaptation, and cross-species infections to plan protected areas, inform strategies of re-introduction and relocation, and provide effective interventions in cases of potential disease epidemics. By resolving the root causes of human and animal outbreaks and their outcomes, we shall continue to learn lessons such as the cats have shared on SARS and AIDS.
References
- Allan J.S., Short M., Taylor M.E., Su S., Hirsch V.M., Johnson P.R., Shaw G.M., Hahn B.H. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J. Virol. 1991;65:2816–2828. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros M.L., Sanchez C.M., Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227:378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov K., Lam M.K., Colgan D.J. Marsupial MHC class II beta genes are not orthologous to the eutherian beta gene families. J. Hered. 2004;95:338–345. doi: 10.1093/jhered/esh049. [DOI] [PubMed] [Google Scholar]
- Bendinelli M., Pistello M., Lombardi S., Poli A., Garzelli C., Matteucci D., Ceccherini-Nelli L., Malvaldi G., Tozzini F. Feline immunodeficiency virus: an interesting model for AIDS studies and an important cat pathogen. Clin. Microbiol. Rev. 1995;8:87–112. doi: 10.1128/cmr.8.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R., Rodrigo A.G., Holley D., Drummond A., Anderson C.R., Jr., Ross H.A., Poss M. Epidemiology, genetic diversity, and evolution of endemic feline immunodeficiency virus in a population of wild cougars. J. Virol. 2003;77:9578–9589. doi: 10.1128/JVI.77.17.9578-9589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R., Drummond A.J., Poss M. A virus reveals population structure and recent demographic history of its carnivore host. Science. 2006;311:538–541. doi: 10.1126/science.1121360. [DOI] [PubMed] [Google Scholar]
- Brown E.W., Yuhki N., Packer C., O’Brien S.J. A lion lentivirus related to feline immunodeficiency virus: epidemiologic and phylogenetic aspects. J. Virol. 1994;68:5953–5968. doi: 10.1128/jvi.68.9.5953-5968.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter M.A., O’Brien S.J. Coadaptation and immunodeficiency virus: lessons from the Felidae. Curr. Opin. Genet. Dev. 1995;5:739–745. doi: 10.1016/0959-437x(95)80006-q. [DOI] [PubMed] [Google Scholar]
- Carpenter M.A., Brown E.W., Culver M., Johnson W.E., Pecon-Slattery J., Brousset D., O’Brien S.J. Genetic and phylogenetic divergence of feline immunodeficiency virus in the puma (Puma concolor) J. Virol. 1996;70:6682–6693. doi: 10.1128/jvi.70.10.6682-6693.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter M.A., Appel M.J., Roelke-Parker M.E., Munson L., Hofer H., East M., O’Brien S.J. Genetic characterization of canine distemper virus in Serengeti carnivores. Vet. Immunol. Immunopathol. 1998;65:259–266. doi: 10.1016/s0165-2427(98)00159-7. [DOI] [PubMed] [Google Scholar]
- Carrington M., O’Brien S.J. The influence of HLA genotype on AIDS. Annu. Rev. Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- CDC, 2004. Update: Severe Acute Respiratory Syndrome – Worldwide and United States. Page 664 in C. f. D. C. a. Prevention, editor. MMWR. [PubMed]
- Clark A.G., Glanowski S., Nielsen R., Thomas P.D., Kejariwal A., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Ferriera S., Wang G., Zheng X., White T.J., Sninsky J.J., Adams M.D., Cargill M. Inferring nonneutral evolution from human–chimp–mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Cooper G.M., Brudno M., Stone E.A., Dubchak I., Batzoglou S., Sidow A. Characterization of evolutionary rates and constraints in three Mammalian genomes. Genome Res. 2004;14:539–548. doi: 10.1101/gr.2034704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver M., Johnson W.E., Pecon-Slattery J., O’Brien S.J. Genomic ancestry of the American puma (Puma concolor) J. Hered. 2000;91:186–197. doi: 10.1093/jhered/91.3.186. [DOI] [PubMed] [Google Scholar]
- de Parseval A., Grant C.K., Sastry K.J., Elder J.H. Sequential CD134-CXCR4 interactions in feline immunodeficiency virus (FIV): soluble CD134 activates FIV Env for CXCR4-dependent entry and reveals a cryptic neutralization epitope. J. Virol. 2006;80:3088–3091. doi: 10.1128/JVI.80.6.3088-3091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazen J.M. SARS – looking back over the first 100 days. New Engl. J. Med. 2003;349:319–320. doi: 10.1056/NEJMp038118. [DOI] [PubMed] [Google Scholar]
- Driscoll C.A., Menotti-Raymond M., Nelson G., Goldstein D., O’Brien S.J. Genomic microsatellites as evolutionary chronometers: a test in wild cats. Genome Res. 2002;12:414–423. doi: 10.1101/gr.185702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Preiser W., Gunther S., Schmitz H., Doerr H.W. Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol. Med. 2003;9:325–327. doi: 10.1016/S1471-4914(03)00133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W., Paabo S. Comparative primate genomics. Annu. Rev. Genomics Hum. Genet. 2004;5:351–378. doi: 10.1146/annurev.genom.5.061903.180040. [DOI] [PubMed] [Google Scholar]
- Evermann J.F., Heeney J.L., Roelke M.E., McKeirnan A.J., O’Brien S.J. Biological and pathological consequences of feline infectious peritonitis virus infection in the cheetah. Arch. Virol. 1988;102:155–171. doi: 10.1007/BF01310822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J.E., Poland A., Carlson J., Pedersen N.C. Risk factors for feline infectious peritonitis among cats in multiple-cat environments with endemic feline enteric coronavirus. J. Am. Vet. Med. Assoc. 1997;210:1313–1318. [PubMed] [Google Scholar]
- Gao F., Yue L., White A.T., Pappas P.G., Barchue J., Hanson A.P., Greene B.M., Sharp P.M., Shaw G.M., Hahn B.H. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature. 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- Gao F., Bailes E., Robertson D.L., Chen Y., Rodenburg C.M., Michael S.F., Cummins L.B., Arthur L.O., Peeters M., Shaw G.M., Sharp P.M., Hahn B.H. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- Garrett L. Penguin; New York: 1994. The Coming Plague. [Google Scholar]
- Gilbert D.A., Packer C., Pusey A.E., Stephens J.C., O’Brien S.J. Analytical DNA fingerprinting in lions: parentage, genetic diversity, and kinship. J. Hered. 1991;82:378–386. doi: 10.1093/oxfordjournals.jhered.a111107. [DOI] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Hahn B.H., Shaw G.M., De Cock K.M., Sharp P.M. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- Heeney J.L., Evermann J.F., McKeirnan A.J., Marker-Kraus L., Roelke M.E., Bush M., Wildt D.E., Meltzer D.G., Colly L., Lukas J. Prevalence and implications of feline coronavirus infections of captive and free-ranging cheetahs (Acinonyx jubatus) J. Virol. 1990;64:1964–1972. doi: 10.1128/jvi.64.5.1964-1972.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R.S., Walsh C.A. Molecular insights into human brain evolution. Nature. 2005;437:64–67. doi: 10.1038/nature04103. [DOI] [PubMed] [Google Scholar]
- Hirsch V. What can natural infection of African monkeys with simian immunodeficiency virus tell us about the pathogenesis of AIDS? AIDS Rev. 2004;6:40–53. [PubMed] [Google Scholar]
- Hofmann-Lehmann R., Fehr D., Grob M., Elgizoli M., Packer C., Martenson J.S., O’Brien S.J., Lutz H. Prevalence of antibodies to feline parvovirus, calicivirus, herpesvirus, coronavirus, and immunodeficiency virus and of feline leukemia virus antigen and the interrelationship of these viral infections in free-ranging lions in east Africa. Clin. Diagn. Lab. Immunol. 1996;3:554–562. doi: 10.1128/cdli.3.5.554-562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. SARS – associated coronavirus. New Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- Johnson W.E., Eizirik E., Pecon-Slattery J., Murphy W.J., Antunes A., Teeling E., O’Brien S.J. The late Miocene radiation of modern Felidae: a genetic assessment. Science. 2006;311:73–77. doi: 10.1126/science.1122277. [DOI] [PubMed] [Google Scholar]
- Korber B., Muldoon M., Theiler J., Gao F., Gupta R., Lapedes A., Hahn B.H., Wolinsky S., Bhattacharya T. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- Kumanovics A., Takada T., Lindahl K.F. Genomic organization of the mammalian MHC. Annu. Rev. Immunol. 2003;21:629–657. doi: 10.1146/annurev.immunol.21.090501.080116. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J.A. HIV and host immune responses in AIDS pathogenesis. J. Clin. Apher. 1993;8:19–28. doi: 10.1002/jca.2920080106. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Luo S.J., Kim J.H., Johnson W.E., van der Walt J., Martenson J., Yuhki N., Miquelle D.G., Uphyrkina O., Goodrich J.M., Quigley H.B., Tilson R., Brady G., Martelli P., Subramaniam V., McDougal C., Hean S., Huang S.Q., Pan W., Karanth U.K., Sunquist M., Smith J.L., O’Brien S.J. Phylogeography and genetic ancestry of tigers (Panthera tigris) PLoS Biol. 2004;2:e442. doi: 10.1371/journal.pbio.0020442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marker L., O’Brien J.S. Captive breeding of the cheetah (Acinonyx jubatus) in North American zoos (1971–1986) Zoo Biol. 1989;8:3–16. [Google Scholar]
- Menotti-Raymond M., O’Brien S.J. Dating the genetic bottleneck of the African cheetah. Proc Natl Acad Sci USA. 1993;90:3172–3176. doi: 10.1073/pnas.90.8.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa T., Tomonaga K., Kawaguchi Y., Mikami T. The genome of feline immunodeficiency virus. Arch. Virol. 1994;134:221–234. doi: 10.1007/BF01310563. [DOI] [PubMed] [Google Scholar]
- Muller-Graf C.D., Woolhouse M.E., Packer C. Epidemiology of an intestinal parasite (Spirometra spp.) in two populations of African lions (Panthera leo) Parasitology. 1999;118(Pt 4):407–415. doi: 10.1017/s0031182098003813. [DOI] [PubMed] [Google Scholar]
- Munson L., Brown J.L., Bush M., Packer C., Janssen D., Reiziss S.M., Wildt D.E. Genetic diversity affects testicular morphology in free-ranging lions (Panthera leo) of the Serengeti Plains and Ngorongoro Crater. J. Reprod. Fertil. 1996;108:11–15. doi: 10.1530/jrf.0.1080011. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Eizirik E., Johnson W.E., Zhang Y.P., Ryder O.A., O’Brien S.J. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Stanyon, R., O’Brien, S.J., 2001b. Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol. 2, REVIEWS0005. [DOI] [PMC free article] [PubMed]
- Murphy W.J., Pevzner P.A., O’Brien S.J. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Nowell, K., Jackson, P., 1996. Status Survey and Conservation Action Plan, Wild Cats. International Union for Conservation of Nature and Natural Resources, Gland, Switzerland.
- O’Brien S.J., Johnson W.E. Big cat genomics. Annu. Rev. Genomics Hum. Genet. 2005;6:407–429. doi: 10.1146/annurev.genom.6.080604.162151. [DOI] [PubMed] [Google Scholar]
- O’Brien S.J., Yuhki N. Comparative genome organization of the major histocompatibility complex: lessons from the Felidae. Immunol. Rev. 1999;167:133–144. doi: 10.1111/j.1600-065X.1999.tb01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien S.J., Roelke M.E., Marker L., Newman A., Winkler C.A., Meltzer D., Colly L., Evermann J.F., Bush M., Wildt D.E. Genetic basis for species vulnerability in the cheetah. Science. 1985;227:1428–1434. doi: 10.1126/science.2983425. [DOI] [PubMed] [Google Scholar]
- O’Brien S.J., Wildt D.E., Bush M., Caro T.M., FitzGibbon C., Aggundey I., Leakey R.E. East African cheetahs: evidence for two population bottlenecks? Proc. Natl. Acad. Sci. USA. 1987;84:508–511. doi: 10.1073/pnas.84.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien S.J., Menotti-Raymond M., Murphy W.J., Nash W.G., Wienberg J., Stanyon R., Copeland N.G., Jenkins N.A., Womack J.E., Marshall Graves J.A. The promise of comparative genomics in mammals. Science. 1999;286:458–462. doi: 10.1126/science.286.5439.458. 479–481. [DOI] [PubMed] [Google Scholar]
- Olmsted R.A., Hirsch V.M., Purcell R.H., Johnson P.R. Nucleotide sequence analysis of feline immunodeficiency virus: genome organization and relationship to other lentiviruses. Proc. Natl. Acad. Sci. USA. 1989;86:8088–8092. doi: 10.1073/pnas.86.20.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted R.A., Langley R., Roelke M.E., Goeken R.M., Adger-Johnson D., Goff J.P., Albert J.P., Packer C., Laurenson M.K., Caro T.M. Worldwide prevalence of lentivirus infection in wild feline species: epidemiologic and phylogenetic aspects. J. Virol. 1992;66:6008–6018. doi: 10.1128/jvi.66.10.6008-6018.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer C., Altizer S., Appel M.J., Brown E.W., Martenson J., Roelke-Parker M.E., Hofmann-Lehmann R., Lutz H. Viruses of the Serengeti: patterns of infection and mortality in African lions. J. Anim. Ecol. 1999;68:1161–1178. [Google Scholar]
- Packer C., Hilborn R., Mosser A., Kissui B., Borner M., Hopcraft G., Wilmshurst J., Mduma S., Sinclair A.R. Ecological change, group territoriality, and population dynamics in Serengeti lions. Science. 2005;307:390–393. doi: 10.1126/science.1105122. [DOI] [PubMed] [Google Scholar]
- Pearks Wilkerson A.J., Teeling E.C., Troyer J.L., Bar-Gal G.K., Roelke M., Marker L., Pecon-Slattery J., O’Brien S.J. Coronavirus outbreak in cheetahs: lessons for SARS. Curr. Biol. 2004;14:R227–R228. doi: 10.1016/j.cub.2004.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Ho E.W., Brown M.L., Yamamoto J.K. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- Poli A., Abramo F., Cavicchio P., Bandecchi P., Ghelardi E., Pistello M. Lentivirus infection in an African lion: a clinical, pathological, and virologic study. J. Wildlife Dis. 1995;31:70–74. doi: 10.7589/0090-3558-31.1.70. [DOI] [PubMed] [Google Scholar]
- Poon L.L., Chu D.K., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H., Lau S.K., Woo P.C., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S. Identification of a novel coronavirus in bats. J. Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathbun R.C., Lockhart S.M., Stephens J.R. Current HIV treatment guidelines – an overview. Curr. Pharm. Des. 2006;12:1045–1063. doi: 10.2174/138161206776055840. [DOI] [PubMed] [Google Scholar]
- Roelke M.E., Martenson J.S., O’Brien S.J. The consequences of demographic reduction and genetic depletion in the endangered Florida panther. Curr. Biol. 1993;3:340–350. doi: 10.1016/0960-9822(93)90197-v. [DOI] [PubMed] [Google Scholar]
- Roelke, M.E., Pecon-Slattery, J., Taylor, S., Citino, S., Brown, E.W., Packer, C., VandeWoude, S., O’Brien S, J., in press. T-Lymphocyte profiles in FIV infected wild lions and pumas reveal CD4 depletion. J. Wildlife Dis. [DOI] [PubMed]
- Roelke-Parker M.E., Munson L., Packer C., Kock R., Cleaveland S., Carpenter M., O’Brien S.J., Pospischil A., Hofmann-Lehmann R., Lutz H. A canine distemper virus epidemic in Serengeti lions (Panthera leo) Nature. 1996;379:441–445. doi: 10.1038/379441a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C.M., Izeta A., Sanchez-Morgado J.M., Alonso S., Sola I., Balasch M., Plana-Duran J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarino A. Expanding the frontiers of existing antiviral drugs: possible effects of HIV-1 protease inhibitors against SARS and avian influenza. J. Clin. Virol. 2005;34:170–178. doi: 10.1016/j.jcv.2005.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Bailes E., Robertson D.L., Gao F., Hahn B.H. Origins and evolution of AIDS viruses. Biol. Bull. 1999;196:338–342. doi: 10.2307/1542965. [DOI] [PubMed] [Google Scholar]
- Sharp P.M., Shaw G.M., Hahn B.H. Simian immunodeficiency virus infection of chimpanzees. J. Virol. 2005;79:3891–3902. doi: 10.1128/JVI.79.7.3891-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw K. The 2003 SARS outbreak and its impact on infection control practices. Public Health. 2006;120:8–14. doi: 10.1016/j.puhe.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer J.L., Pecon-Slattery J., Roelke M.E., Black L., Packer C., O’Brien S.J. Patterns of feline immunodeficiency virus multiple infection and genome divergence in a free-ranging population of African lions. J. Virol. 2004;78:3777–3791. doi: 10.1128/JVI.78.7.3777-3791.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer J.L., Pecon-Slattery J., Roelke M.E., Johnson W., VandeWoude S., Vazquez-Salat N., Brown M., Frank L., Woodroffe R., Winterbach C., Winterbach H., Hemson G., Bush M., Alexander K.A., Revilla E., O’Brien S.J. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J. Virol. 2005;79:8282–8294. doi: 10.1128/JVI.79.13.8282-8294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uphyrkina O., Johnson W.E., Quigley H., Miquelle D., Marker L., Bush M., O’Brien S.J. Phylogenetics, genome diversity and origin of modern leopard, Panthera pardus. Mol. Ecol. 2001;10:2617–2633. doi: 10.1046/j.0962-1083.2001.01350.x. [DOI] [PubMed] [Google Scholar]
- Uphyrkina O., Miquelle D., Quigley H., Driscoll C., O’Brien S.J. Conservation genetics of the Far Eastern leopard (Panthera pardus orientalis) J. Hered. 2002;93:303–311. doi: 10.1093/jhered/93.5.303. [DOI] [PubMed] [Google Scholar]
- Wei P., Fan K., Chen H., Ma L., Huang C., Tan L., Xi D., Li C., Liu Y., Cao A., Lai L. The N-terminal octapeptide acts as a dimerization inhibitor of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 2006;339:865–872. doi: 10.1016/j.bbrc.2005.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett B.J., Flynn J.N., Hosie M.J. FIV infection of the domestic cat: an animal model for AIDS. Immunol. Today. 1997;18:182–189. doi: 10.1016/s0167-5699(97)84665-8. [DOI] [PubMed] [Google Scholar]
- Winckler W., Myers S.R., Richter D.J., Onofrio R.C., McDonald G.J., Bontrop R.E., McVean G.A., Gabriel S.B., Reich D., Donnelly P., Altshuler D. Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- Wyatt R., Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Yuhki N., O’Brien S.J. DNA variation of the mammalian major histocompatibility complex reflects genomic diversity and population history. Proc. Natl. Acad. Sci. USA. 1990;87:836–840. doi: 10.1073/pnas.87.2.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuhki N., Beck T., Stephens R.M., Nishigaki Y., Newmann K., O’Brien S.J. Comparative genome organization of human, murine, and feline MHC class II region. Genome Res. 2003;13:1169–1179. doi: 10.1101/gr.976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusim K., Kesmir C., Gaschen B., Addo M.M., Altfeld M., Brunak S., Chigaev A., Detours V., Korber B.T. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 2002;76:8757–8768. doi: 10.1128/JVI.76.17.8757-8768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


