Abstract
An epidemiological survey on Blastocystis was carried out enrolling a total of 2524 subjects referred to the Umberto I Academic Hospital in Rome, for the routine parasitological exams, during 2017–2018. The studied population included a sample of immunocompromised individuals (N = 130) followed at the same hospital.
DNA sequencing of the small subunit rRNA gene (SSU rDNA) locus was performed on samples positive to the coproparasitological analysis to molecular characterize the Blastocystis-subtypes.
Microscopical analysis detected Blastocystis in 192/2524 (7.6%) of the enrolled subjects. It was the organism most frequently identified in the analysed faecal samples diagnosed in single infection (5.6%) or in co-infection with other enteric protozoa (2%). Furthermore, it was found mainly in immunocompromised patients (22.3%) compared to immunocompetent ones (6.8%).
As expected, ST3 was the most occurring subtype identified in 40% of the subjects, followed by ST1 (29%), ST2 (16%), ST4 (12%), and ST7 (3%).
Next-generation sequencing (NGS) of the 16S rDNA was performed on a sub-sample of Blastocystis-ST3-carriers, homogenous by age and gender, as well as on Blastocystis-free subjects, to profile and compare their gut bacterial composition. A higher bacterial diversity was found in ST3-Blastocystis-carriers, which exhibited a high abundance of Prevotella, Methanobrevibacter and Ruminococcus while, a high percentage of Bacteroides was found in Blastocystis-free subjects.
This study evidenced the presence of Blastocystis in 7.6% of faecal samples in Italy and a high circulation of the protist among immunocompromised patients (22.3%). Molecular characterization of positive samples evidenced the occurrence of five different subtypes, including zoonotic ST such as the ST7, highlighting the risk of transmission from animals. Study of the gut microbiota composition confirms previous evidences according to which, the colonisation by Blastocystis would be linked with an eubiotic gut characterized by potentially beneficial species such as Prevotella and Ruminococcus, rather than with a dysbiotic state, with a high abundance of Enterobacteriaceae, and corroborated the role of the protist as “an old friend” of the human gut.
Keywords: Blastocystis, Subtypes, ST3, Human gut microbiota, Italy
Highlights
-
•
Microscopical analysis detected Blastocystis in 7.6% subjects in Italy.
-
•
Molecular methods allowed the identification of 5 STs (ST1, ST2, ST3, ST4, ST7).
-
•
ST3 resulted the most frequent subtype.
-
•
NGS of the 16S rDNA gene was performed in Blastocystis-ST3-carriers.
-
•
Blastocystis-ST3 resulted correlated to an eubiotic gut characterized by potentially beneficial species.
1. Introduction
Blastocystis is currently known as belonging to the Stramenopiles, a complex and heterogeneous evolutionary assemblage of heterotrophic and photosynthetic protozoa (Silberman et al., 1996; Arisue et al., 2002; Riisberg et al., 2009). It colonises intestine of humans and wide range of animals. In humans, it exhibits a worldwide distribution, with a prevalence from 0.5%–30% and 30–76% in industrialized and developing countries, respectively (Alfellani et al., 2013). Molecular study based on the small subunit ribosomal RNA (SSU-rDNA) evidenced an extensive genetic heterogeneity allowing the identification in mammalian and avian hosts of at least 26 divergent lineages, termed subtypes (STs), which could be considered arguably separate species (Alfellani et al., 2013; Zhao et al., 2017; Maloney et al., 2019). Ten of the 26 subtypes, ST-1 to ST-9 and ST-12, have been identified in human samples, and all, except for ST-9, have also been reported in animals (Ramírez et al., 2016; Stensvold and Clark, 2016). Despite the pathogenetic role of Blastocystis has been the subject of many investigations (Sheehan et al., 1986; Dogruman-Al et al., 2009; Poirier et al., 2012; Wawrzyniak et al., 2013; Azizian et al., 2016; Mattiucci et al., 2016; Stensvold et al., 2009), it's so far remaining unclear. Several authors suggested that it could be linked to the impact of the protist on the gut microbiota. Analysis of the microbial communities in healthy individuals or affected by intestinal disorders evidenced three main microbiota patterns, named enterotypes, characterized by three dominant bacteria clusters: Bacteroides (enterotype I), Prevotella (enterotype II), or Ruminococcus (enterotype III) (Arumugam et al., 2011). Each enterotype harbours different bacteria genera, defines a distinctive way of generating energy and seem to be linked to dietary habits (Rinninella et al., 2019). Recent NGS-based studies associated Blastocystis with higher diversity of gut microbiota, low Body Mass Index (BMI), and the enterotype II and III (Andersen et al., 2015; Andersen and Stensvold, 2016). Furthermore Blastocystis was also linked to an eubiotic state, characterized by a preponderance of potentially beneficial species, belonging mainly to the phyla Firmicutes and Bacteoides, instead of those of the phylum Protebacteria, since significantly higher Faecalibacterium prausnitzii-Escherichia coli ratio was found in Blastocystis-positive than in Giardia-positive individuals (Iebba et al., 2016a).
Conversly, Nourrisson and colleagues evidenced a decrease of faecal microbiota protective bacteria in Blastocystis-colonised individuals (Nourrisson et al., 2014). Moreover, it has been suggested that microbiota composition in Blastocystis-carriers may be dependent on the organism's ST and that certain isolates of Blastocystis, as the ST7, potentially lead to an imbalance of the gut microbiota (Yason et al., 2019; Tito et al., 2019). Finally, it has been proposed that some populations are more susceptible to Blastocystis colonisation (Wawrzyniak et al., 2013). Thus, this protozoon is frequently found in immunocompromised individuals, such as those with human immunodeficiency virus/acquired immunodeficiency syndrome or cancer (Piranshahi et al., 2018; Kurniawan et al., 2009; Tan et al., 2009). Studies of gut microbiota in HIV-infected patients evidenced a change in the Bacteroides:Prevotella ratio (Williams et al., 2016), which could also be linked to sexual practice and lifestyle rather than HIV infection and Highly Active AntiRetroviral Therapy (HAART)-treatment (Noguera-Julian et al., 2016).
However, no specific investigations have been so far conducted to investigate on the relationship between the presence of Blastocystis and the faecal microbiota composition in such group of patients.
Aim of this study was to: i) assess the circulation of Blastocystis among patients refereed to the Academic Hospital "Policlinico Umberto I" for the routine parasitological exams; ii) molecular characterize the Blastocystis-STs from positive subjects; iii) analyse the faecal microbial composition from a cohort of patients, including immunocompromised subjects, found positive to Blastocystis-ST3.
2. Materials and methods
2.1. Faecal samples collection
Faecal samples were collected from subjects refereed to the Diagnostic Parasitology laboratory and to the Department of Translation and Precision Medicine of the Academic Hospital “Policlinico Umberto I”, during the years 2017–2018. This epidemiological survey was carried out after the previously study on Blastocystis STs from 189 isolates, collected during the years 2012–2014 among mildly symptomatic patients, or those affected by inflammatory bowel disease (IBD), irritable bowel syndrome (IBS) or chronic diarrhoea, or otherwise immunosuppressed referred at the same hospital (Mattiucci et al., 2016). In such survey six subtypes (ST1, 2, 3, 4, 6, 8) were detected and significant occurrence of Blastocystis ST4 in patients suffering from IBS, IBD or chronic diarrhoea was observed (Mattiucci et al., 2016).
Demographic data were obtained from subjects included in the study, as well as their written informed consent. The study was approved with respect to the Helsinki Declaration by the Ethical Committee of the Academic Hospital “Policlinico Umberto I” (license number: n. 4836).
2.2. Laboratory procedures
From one to three faecal samples were collected from each subject and submitted to the microscopic observation of the wet smears stained with Lugol, directly and after Ridley concentration (Ridley and Hawgood, 1956). Genomic DNA was then extracted from samples positive to the microscopic observation and submitted to PCR amplification using primers previously described (Scicluna et al., 2006), which target a fragment of about 600 bp from the Blastocystis-SSU rDNA gene, following PCR protocol and conditions as previously described (Mattiucci et al., 2016). The resulting chromatograms were analysed and edited by using the software Chromas version 2.33 (Technelysium Pty Ltd., Australia). The sequences obtained were compared to those of Blastocystis STs previously deposited in GenBank using the BLAST application (www.ncbi.nlm.nih.gov/BLAST). The STs were identified by determining the exact match or closest identity (99%), according to the classification given by Stensvold et al. 2007 (https://pubmlst.org/blastocystis/). NGS of the bacterial 16S rRNA was thus carried out on a subsample of subjects Blastocystis-free and Blastocystis-carriers homogeneous by age (<40 years-old), gender (male) and found positive for the same subtype (ST3). NGS and data analysis was supported by BMR Genomics (Padua, Italy). Briefly, V3-V4 regions of 16S rRNA gene were amplified using the primers Pro341F (5′-CCTACGGGNBGCASCAG-3′) and Pro805R (5′-GACTACNVGGGTATCTAATCC-3′) (Takahashi et al., 2014). Primers were modified with forward overhang: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-[locus-specific sequence]-3′ and with reverse overhang: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACA-[locus-specific sequence]-3′ necessary for dual index library preparation, following Illumina protocol (https://web.uri.edu/gsc/files/16s-metagenomic-library-prep-guide-15044223-b.pdf). PCR mix was composed as follows: 5 μl DNA, 6.2 μl of mix (Taq Platinum-Thermo Fisher, Primer and MgSO4) and 13.8 μl DNA-se free water. Amplification conditions were: 1 cycle at 94 °C for 1 min, 25 cycles at 94 °C for 1 min, 55° for 30 s, 68° for 45 s and 1 final cycle at 68 °C for 7 min. Amplicons of expected size (about 476 bp) were purified by means Beads Amplure XP 0,8×, normalized, pooled and run on Illumina MiSeq with 2 × 300 bp approach in order to produce about 50000 reads (±20%).
The sequencing reads were filtered for average quality (Q > 30), and R1 and R2 were merged using FLASH with default parameters. QIIME software version 1.9.1 was used to perform the full analysis of OTUs selected for the statistical analysis using the pick_closed_reference_otus.py wrapper from the Greengenes reference database (13.8 version) (Caporaso et al., 2010).
2.3. Data analysis
To test the distribution of different Blastocystis subtypes among different age groups and gender, the Wilcoxon signed-rank non-parametric test was used. Data were analysed using IBM SPSS, version 21.0 (SPSS Inc., Chicago, USA). Alpha (within a community) and beta (between communities) diversity metrics, as well as taxonomic community assessments, were produced using QIIME 1.9.1 scripts on the normalized read counts. The following indexes were calculated for each sample: Chao1 bias-corrected and observed OTUs. Alpha-diversity rarefaction curves were produced by plotting these several diversity metrics against the number of sequences considered from a sample. Differences in diversity between Blastocystis-carriers (both immunocompetent and immunocompromised) and Blastocystis-free subjects were tested by Kruskal-Wallis test using the Stats Direct Statistical software v. 3.1.20 (http://www.statsdirect.com. England: StatsDirect Ltd. 2013). Multivariate analysis was done through the compare_category.py script of QIIME. Differences in beta diversity (weighted UniFrac distances) were identified using Analysis of Similarity (ANOSIM) and the effect of size indicated by an R-value (between −1 and +l), with a value of 0 representing the null hypothesis1. Adonis (ANOVA tests group) was applied to verify variations within a category and between categories. Analyses were performed among the required groups and on different metrics: weighted and unweighted. MatagenomeSeq was used to calculate species differential abundance between the groups. The Mann-Whitney test was then applied to evaluate the significance of the comparison. P-values <0.05 were considered statistically significant.
3. Results
3.1. Study subjects and coproparasitological results
During the years 2017–2018 a total of 2524 faecal samples were screened for intestinal protozoa among the patients admitted at the Diagnostic Parasitology laboratory, “Policlinico Umberto I”, Rome, Italy. The study population included 2394 subjects referred at the hospital for the routinely copro-parasitological analyses, without any indication about their clinical status, and 130 HIV-positive patients regularly followed at the Department of Translation and Precision Medicine of the same hospital.
Microscopical analysis detected Blastocystis in 192/2524 (7.6%) of the enrolled subjects. In detail, Blastocystis was frequently found in the group of immunocompromised patients (22.3%) compared to immunocompetent ones (6.8%).
As expected, it was the most frequent organism identified in the faecal samples diagnosed in single infection (5.6%) or in co-infection with other enteric protozoa (2%), followed by Endolimax nana (2.5%), Entamoeba coli (1.7%), Giardia intestinalis (1.2%), Entamoeba complex (0.5%), Iodamoeba butschlii (0.3%).
Blastocystis-carriers were mainly males (65%), mean age 47.35 (95% CI, 40.94–54.74), 62% were from Italy.
3.2. Molecular characterization of Blastocystis subtypes
A total of 142/192 stool samples positive to the microscopical analysis were successfully DNA extracted and subtyped by sequences analysis of the SSU-rDNA (barcode region). BLAST searches allowed the identification of 5 STs, with different percentage values.
The most common subtype was the ST3 identified in 40% of the subjects, followed by ST1 (29%), ST2 (16%), ST4 (12%), and ST7 (3%). No significant correlation was found between STs distribution and patients' gender. Regarding age, some significant difference was observed in the distribution of ST1 and ST4 between different age groups, being more prevalent in subjects <40 years than in the older ones (p = 0.0048 and p = 0.0068, respectively).
3.3. Bacterial microbiota composition of Blastocystis-ST3 faecal samples
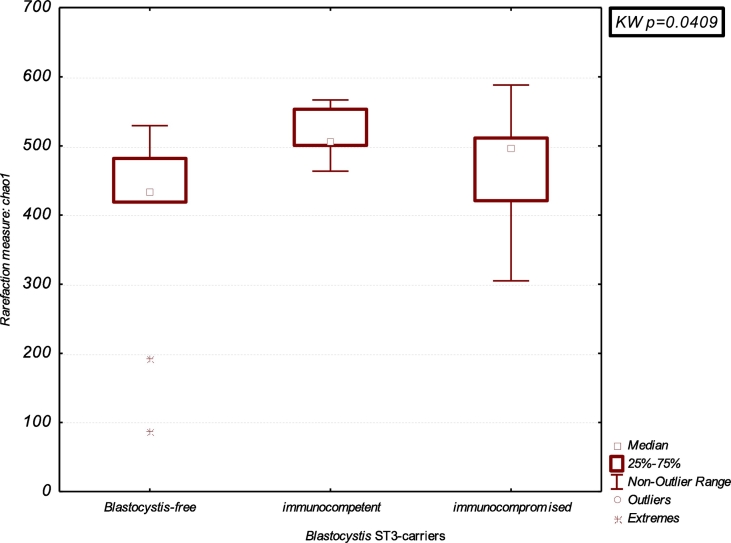
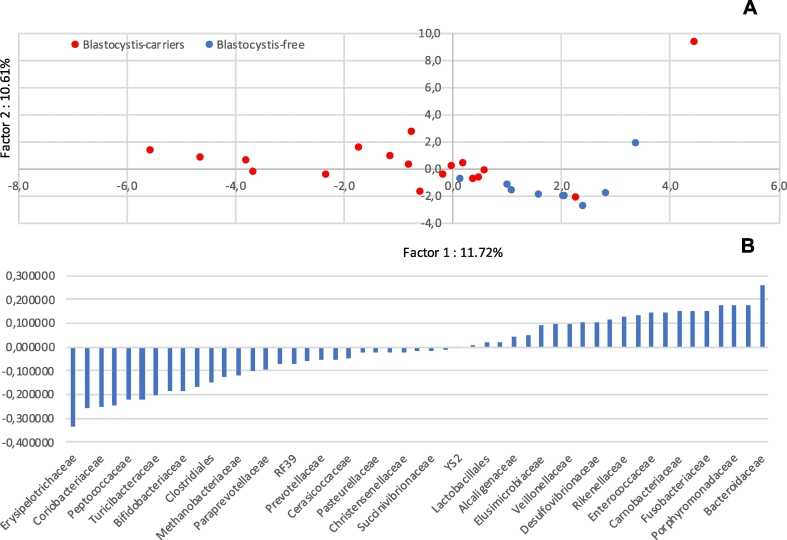
The gut microbiota composition of Blastocystis-colonised patients was investigated by a cross-sectional study on subsample of patients, as above described, homogeneous by age (<40 years-old) and gender (male) and found positive for the same subtype (ST3). Thus, 16S rDNA sequencing was performed on a total of 27 male subjects, <40 years old, divided in Blastocystis-ST3-carriers immunocompetent and immunocompromised (N = 18) and Blastocystis-free subjects (N = 9, control group). A higher bacterial diversity in faecal microbiota was evidenced in the Blastocystis-ST3 carriers. Indeed, the Chao1 richness index and the observed OTUs in colonised patients resulted significantly higher than in Blastocystis-free subjects (p = 0.040 and p = 0.038, respectively) (Table 1 and Fig. 1). The Principal Component Analysis (PCA) plot obtained from the beta-diversity calculation in QIIME demonstrated a relative clustering of samples, whereby the scores for Factor1 and Factor2 account for 22.33% of the variance in the data. This difference between the bacterial communities resulted significant, as determined using the ANOSIM nonparametric statistical test analysis of similarity, where R = 0.18 (p = 0.025) (Fig. 2A).
Table 1.
Comparison of the bacterial richness and diversity in the group of ST3-carriers both immunocompromised and immunocompetent ones and Blastocystis-free subjects. Statistical analysis was performed using the Kruskal-Wallis test. Significant results were marked with*.
| Group of patients (N) | Richness estimator |
|||
|---|---|---|---|---|
| Observed OTUs | p-value K-W | Chao 1 | p-value K-W | |
| Blastocystis-free (9) | 340.37 | 0.038⁎ | 365.95 | 0.040⁎ |
| Blastocystis-ST3 carriers immunocompromised (9) | 402.54 | 432.33 | ||
| Blastocystis-ST3 carriers immunocompetent (9) | 454.38 | 479.82 | ||
Fig. 1.
Boxplot of Chao1 diversity index between Blastocystis-free and Blastocystis-ST3-carriers divided in immunocompromised and immunocompetent subjects. Statistical analysis was performed using the Kruskal-Wallis (K-W) test.
Fig. 2.
Analyses of the variance in bacterial microbiota composition at the family taxonomic rank observed in Blastocystis-ST3 carriers and Blastocystis-free subjects A) PCA plot of microbial communities in Blastocystis-ST3 carriers (red dots) and Blastocystis-free subjects (blue dots); B) eigenvectors of correlation matrix of microbial communities in the 2 groups of subjects. RF39: unclassified Enterococcaceae; YS2: Cyanobacteria.
The composition of gut microbial of Blastocystis-ST3-carriers and Blastocystis-free subjects was further examined evidencing a relative clustering of the group of subjects (Blastocystis-ST3-carriers and Blastocystis-free) linked to the bacterial communities (Fig. 2B).
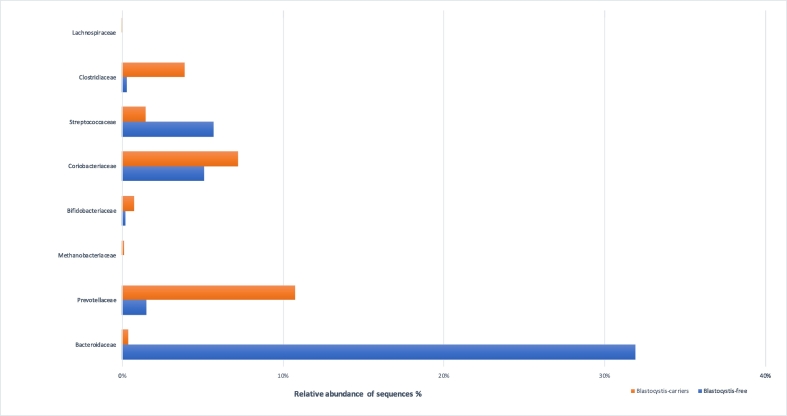
In detail, at the phylum level, Firmicutes, Bacteroidetes and Proteobacteria were the most predominant phyla in all patients' faecal samples with different prevalence values in colonised and not-colonised patients with Blastocystis (53.1% vs 38.2%; 28.3% vs 45.4%; 8.1% vs 14.3%, respectively). The differences in the relative abundances of OTUs were further analysed using the Mann-Whitney test at different taxa levels. When comparing Blastocystis-ST3-carriers with free ones, at the class level, a higher abundance of Bacteroidia was found in Blastocystis-free subjects in comparison with the colonised ones (p = 0.013); while, at the family level, the presence of Blastocystis is significantly associated with an increased relative abundance of Prevotellaceae, Methanobacteriaceae, Clostridiaceae Lachnospiraceae, Erysipelotrichaceae and Pasteurellaceae (p = 0.044; p = 0.022; p = 0.005; p = 0.04; p = 0.04; p < 0.001; p = 0.03, respectively), and low level of Bacteroidaceae and Veillonecellaceae (p < 0.001; p = 0.04, respectively) (Fig. 3).
Fig. 3.
Comparisons of the median values of the relative abundance at the level of bacterial family (level 5) for Bacteroidaceae, Prevotellaceae, Methanobacteriaceae, Clostridiaceae, Lachnospiraceae, Veillonellaceae, Erysipelotrichaceae, Pasteurellaceae in Blastocystis-carriers and Blastocystis-free subjects (orange and blue bars, respectively). The Mann-Whitney test was used to evaluate the 2 groups.
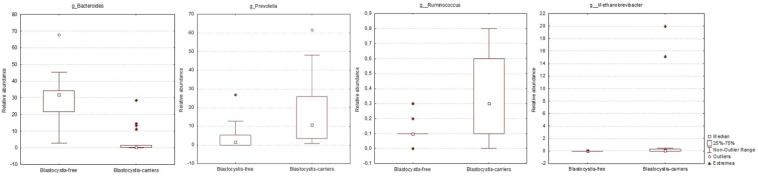
At the genus level, Prevotella, Methanobrevibacter, Ruminococcus were significantly more abundant in Blastocystis-carriers (p = 0.04; p = 0.02; p = 0.01, respectively), while Bacteroides showed higher abundance in those Blastocystis-free subjects (p < 0.001) (Fig. 4). As expected, all Blastocystis-ST3-carriers, both immunocompetent and immunocompromised subjects, showed similar faecal microbiota composition, despite a significantly higher relative abundance of Bifidobacteriaceae (p = 0.03) was detected in immunocompetent individuals compared to those immunocompromised patients, here analysed.
Fig. 4.
Relative abundances of bacterial genera that significantly differ between Blastocystis-carriers and Blastocystis-free subjects. The Mann-Whitney test was used to evaluate the 2 groups.
4. Discussion
This study aimed to describe the occurrence of Blastocystis in patients, mainly Italians, admitted at the Academic Hospital “Policlinico Umberto I”, Rome, Italy and submitted to microscopical analysis of faecal samples. The protist was detected in about 8% of the subjects and, as expected, resulted the organism more frequently diagnosed in faecal samples. Despite the microscopy is the classical procedure for diagnosing parasitic infections and is still considered the primary and cheapest test offered by most routine diagnostic services, it exhibits well-known limits regarding the skills and accuracy of the microscopist and the sensitivity (85.9% vs PCR = 100%) (Formenti et al., 2017). In this study the stool microscopy was performed on one to three sample/patient and a double blinded analysis of each smear was done to assure the diagnostic reliability. Despite a higher prevalence should be expected using molecular diagnostic tools, the epidemiological scenario described in this study resulted in line with that from a previously survey (prevalence = 7.1%) on Italian population (Masucci et al., 2011). Molecular methods, herein applied to 142 out of 192 (74%) microscopy-positive samples, confirmed the copro-parasitological analysis and allowed the subtype identification. In this study, in addition to the four most common STs identified in humans such as ST1, ST2, ST3, ST4, already identified in our previous survey (Mattiucci et al., 2016), the ST7 was also detected. This subtype, as the ST5, ST6, ST8 is rarely identified in humans and therefore, it has been suggested that these rare subtypes in humans are of zoonotic origin (Stensvold and Clark, 2016).
The occurrence of such STs in Italy was expected, as ST3 was already identified in symptomatic patients (Mattiucci et al., 2016; Meloni et al., 2011) as well as the ST7 has been preliminarily identified in farmed and edible animals (Gabrielli et al., 2018), highlighting the risk of transmission of Blastocystis, through animal handling.
Furthermore, in the recent years, several studies in order to address the issue of Blastocystis pathogenicity, suggested that it could be related to genetic differences on the subtype- or strain-level (Stensvold et al., 2009; Wu et al., 2014), correlating the ST-1, 4, and ST-7 with pathological alterations in humans, while ST-2 and ST-3 has been identified as non-pathogenic (Stensvold et al., 2009; Domínguez-Márquez et al., 2009; Eroglu et al., 2009). Also, the presence of both pathogenic and apathogenic strains within one subtype has been reported (Hussein et al., 2008). A recent multi-locus sequence typing analysis of Blastocystis ST-3 and ST-4 has provided valuable insight into genetic variation within and between the two subtypes, evidencing high or low level of genetic diversity for ST-3 and ST4, respectively (Stensvold et al., 2012). Similar results have been obtained in our previous survey where 3 haplotypes (H1, H3, H7) have been identified in ST3 isolates, while a single haplotype (H2) was observed in ST4 symptomatic patients (Mattiucci et al., 2016). Thus, we suggested that the intra-subtype diversity showed by ST3 and ST4 could be linked to the evolutionary history of Blastocystis subtypes and ST3 may have co-evolved with human hosts over a longer period than ST4, which may has extended its range to humans more recently. Furthermore, it has been demonstrated that Blastocystis ST7 caused a decrease in beneficial bacteria, such as Bifidobacterium and Lactobacillus (Yason et al., 2019).
In this study, focused on the ST3, Blastocystis was associated to an eubiotic state characterized by a preponderance of potentially beneficial species, belonging to the phyla Firmicutes and Bacteroidetes, such as those of the genera Ruminococcus and Prevotella, respectively (Iebba et al., 2016b). Furthermore, our results are in line with that reported in other studies (Andersen et al., 2015; Nieves-Ramírez et al., 2018), which showed the protist significantly more abundant in subjects with Ruminococcus or Prevotella-driven enterotype and less common in individuals with Bacteroides-driven enterotype (Fig. 4). They also evidenced that Blastocystis colonisation was linked with higher bacterial richness and lower BMI (Andersen et al., 2015).
Our study confirms and supports this hypothesis evidencing a higher bacterial diversity in Blastocystis-ST3 carriers compared to that identified in Blastocystis-free individuals (Fig. 1). Moreover, at the family level, Prevotellaceae, Methanobacteriaceae, Clostridiaceae were also more abundant, whereas Bacteroidaceae were enriched in Blastocystis-free subjects (Fig. 2, Fig. 3). As expected, several taxa detected in Blastocystis-carriers included bacteria strictly anaerobes and known to produce butyrate, which is considered one of the most important metabolite for maintaining health of colon tract in humans, as it serves as the major energy source of colonocytes, possesses anti-inflammatory properties, and it regulates gene expression, differentiation and apoptosis in host cells. Butyrate is also required to activate oxidative metabolism and, therefore, to provide low level of oxygen in the gut lumen. Such eubiotic environment supports continued colonisation by Blastocystis, as was recently elucidated (Stensvold and van der Giezen, 2018).
Similarly, to that reported for Blastocystis and confirmed also in our survey, several studies evidenced an increase of bacterial diversity in patients infected by helminths as hookworms and a shift in the composition of the human gut microbiota towards a ‘healthy’ phenotype, as well as increased levels of metabolites with anti-inflammatory properties. According to the Hygiene or Old Friends hypothesis' (Rook, 2010), certain STs of Blastocystis, such as ST3, would be considered as ‘old friends’ (Mattiucci et al., 2016), as in the case hookworms. They may be required for maintaining the intestinal homeostasis and/or play a role as a marker of intestinal homeostasis (Stensvold and van der Giezen, 2018).
Although Blastocystis was never considered as an opportunistic organism, it has also been frequently found in immunocompromised individuals presenting diarrhoea, with prevalence value ranging from 15% to 72.4% (Tan et al., 2009). In this study, Blastocystis was more frequently identified in faecal samples from HIV-positive patients (22%) than in immunocompetent subjects (7%) reporting values in line with previously surveys (Piranshahi et al., 2018; Tan et al., 2009). However, no significant differences in faecal microbiota composition was found between HIV-positive and immunocompetent subjects, despite immunocompromised individuals showed low level of bacterial richness (Fig. 1). Several Authors have evaluated the intestinal microbiome in HIV-positive subjects, with somewhat inconsistent or controversial results (Williams et al., 2016). This may be because of small sample sizes, lack of appropriate controls, and/or regional differences in dietary and environmental factors. Regardless, many authors evidenced a change in the ratio Bacteroides:Prevotella; this finding has been suggested to be linked to the antiretroviral therapy (Vesterbacka et al., 2017; Ling et al., 2016), and/or to the sexual practice and lifestyle (Noguera-Julian et al., 2016).
Faecal samples from our HIV-positive patients exhibited the so-called Prevotella enterotype (or enterotype II), which characterizes the HIV-positive treated patients (Lozupone et al., 2013; Vujkovic-Cvijin et al., 2013) and, as above described, favours the Blastocystis colonisation. Taking these observations into account, it appears that the high prevalence of Blastocystis observed in these patients is a consequence of the microbiota composition which highly supports the colonisation by the protist.
In conclusion, despite large-scale surveys using molecular approaches would be needed to assess the Blastocystis epidemiology in Italy, the present study characterized Blastocystis-subtypes from several faecal samples collected from subjects with different immunological status, evidencing a high circulation of the protist in immunocompromised individuals. Zoonotic STs as ST7 were identified in humans highlighting the risk of Blastocystis transmission from animals. Despite the study of the microbiota composition was performed on few subjects mainly Italians colonised by the ST3, the results so far obtained confirm the association between the presence of the protist, and a greater bacterial diversity with the high occurrence of Prevotella and Ruminococcus-enterotype and with a eubiotic gut. Surely, more investigations on the relationship between the intra-subtype variability or haplotypes and the gut microbiota are needed to confirm this positive association.
As the subtype identification and intra-subtype variability seems to be essential for assessing the relationship between Blastocystis, microbiota profile and human disease, further studies on the gut microbiota in patients colonised by different STs at the inter- and intra-subtype level should be enhanced, as well as affected by other immunological disorders in order to add knowledge on the potential role played by this protist in the human gut microbiota composition.
Acknowledgments
Acknowledgements
The authors are grateful to Eleonora Sattin from BMR-Genomics for the technical support in the bioinformatic analysis of the NGS. We thank Referee and Editor for their valuable comments that have helped to improve the MS.
Funding
This study was supported by Sapienza University-Progetti di Ricerca Ateneo- years 2016, 2017 and 2018, Grants number: RM11615506189BE0, RM11715C81DDD2F3, RM11816432E08126.
Declaration of competing interest
The authors declare that they have no competing interests.
References
- Alfellani M.A., Stensvold C.R., Vidal-Lapiedra A., Onuoha E.S., Fagbenro-Beyioku A.F., Clark C.G. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013;126:11–18. doi: 10.1016/j.actatropica.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Andersen L.O., Stensvold C.R. Blastocystis in health and disease: are we moving from a clinical to a public health perspective? J. Clin. Microbiol. 2016;54:524–528. doi: 10.1128/JCM.02520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen L.O., Bonde I., Nielsen H.B., Stensvold C.R. A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiol. Ecol. 2015;91:1–9. doi: 10.1093/femsec/fiv072. [DOI] [PubMed] [Google Scholar]
- Arisue N., Hashimoto T., Yoshikawa H., Nakamura Y., Nakamura G., Nakamura F. Phylogenetic position of Blastocystis hominis and of Stramenopiles inferred from multiple molecular sequence data. J. Eukaryot. Microbiol. 2002;49:42–53. doi: 10.1111/j.1550-7408.2002.tb00339.x. [DOI] [PubMed] [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizian M., Basati G., Abangah G., Mahmoudi M.R., Mirzaei A. Contribution of Blastocystis hominis subtypes and associated inflammatory factors in development of irritable bowel syndrome. Parasitol. Res. 2016;115:2003–2009. doi: 10.1007/s00436-016-4942-4. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogruman-Al F., Kustimur S., Yoshikawa H., Tuncer C., Simsek Z., Tanyuksel M. Blastocystis subtypes in irritable bowel syndrome and inflammatory bowel disease in Ankara, Turkey. Mem. Inst. Oswaldo Cruz. 2009;104:724–727. doi: 10.1590/s0074-02762009000500011. [DOI] [PubMed] [Google Scholar]
- Domínguez-Márquez M.V., Guna R., Muñoz C., Gómez-Muñoz M.T., Borrás R. High prevalence of subtype 4 among isolates of Blastocystis hominis from symptomatic patients of a health district of Valencia (Spain) Parasitol. Res. 2009;105:949–955. doi: 10.1007/s00436-009-1485-y. [DOI] [PubMed] [Google Scholar]
- Eroglu F., Genc A., Elgun G., Koltas I.S. Identification of Blastocystis hominis isolates from asymptomatic and symptomatic patients by PCR. Parasitol. Res. 2009;105:1589–1592. doi: 10.1007/s00436-009-1595-6. [DOI] [PubMed] [Google Scholar]
- Formenti F., Valerio M., Guerriero M., Perandin F., Pajola B., Mistretta M. Molecular biology can change the classic laboratory approach for intestinal protozoan infections. Front. Microbiol. 2017;8:2191. doi: 10.3389/fmicb.2017.02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielli S., Brianti E., Furzi F., Gaglio G., Napoli E., Mattiucci S. 14th International Congress of Parasitology (ICOPA) 19–24 august. 2018. Molecular epidemiology of Blastocystis in domestic and farmed animals in Italy: preliminary results. Daegu, Korea. [Google Scholar]
- Hussein E.M., Hussein A.M., Eida M.M., Atwa M.M. Pathophysiological variability of different genotypes of human Blastocystis hominis Egyptian isolates in experimentally infected rats. Parasitol. Res. 2008;102:853–860. doi: 10.1007/s00436-007-0833-z. [DOI] [PubMed] [Google Scholar]
- Iebba V., Santangelo F., Totino V., Pantanella F., Monsia A., Di Cristanziano V. Gut microbiota related to Giardia duodenalis, Entamoeba spp. and Blastocystis hominis infections in humans from Côte d'Ivoire. J. Infect. De Ctries. 2016;10:1035–1041. doi: 10.3855/jidc.8179. [DOI] [PubMed] [Google Scholar]
- Iebba V., Totino V., Gagliardi A., Santangelo F., Cacciotti F., Trancassini M. Eubiosis and dysbiosis: the two sides of the microbiota. New Microbiol. 2016;39:1–12. [PubMed] [Google Scholar]
- Kurniawan A., Karyadi T., Dwintasari S.W., Sari I.P., Yunihastuti E., Djauzi S., Smith H.V. Intestinal parasitic infections in HIV/AIDS patients presenting with diarrhoea in Jakarta, Indonesia. Trans. R. Soc. Trop. Med. Hyg. 2009;103:892–898. doi: 10.1016/j.trstmh.2009.02.017. [DOI] [PubMed] [Google Scholar]
- Ling Z., Jin C., Xie T., Cheng Y., Li L., Wu N. Alterations in the fecal microbiota of patients with HIV-1 infection: an observational study in a Chinese population. Sci. Rep. 2016;6:30673. doi: 10.1038/srep30673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C.A., Li M., Campbell T.B., Flores S.C., Linderman D., Gebert M.J. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney J.G., Lombard J.E., Urie N.J., Shivley C.B., Santín M. Zoonotic and genetically diverse subtypes of Blastocystis in US pre-weaned dairy heifer calves. Parasitol. Res. 2019;118:575–582. doi: 10.1007/s00436-018-6149-3. [DOI] [PubMed] [Google Scholar]
- Masucci L., Graffeo R., Bani S., Bugli F., Boccia S., Nicolotti N. Intestinal parasites isolated in a large teaching hospital, Italy, 1 May 2006 to 31 December 2008. Euro Surveill. 2011;16:19891. doi: 10.2807/ese.16.24.19891-en. [DOI] [PubMed] [Google Scholar]
- Mattiucci S., Crisafi B., Gabrielli S., Paoletti M., Cancrini G. Molecular epidemiology and genetic diversity of Blastocystis infection in humans in Italy. Epidemiol. Infect. 2016;144:635–646. doi: 10.1017/S0950268815001697. [DOI] [PubMed] [Google Scholar]
- Meloni D., Sanciu G., Poirier P., El Alaoui H., Chabé M., Delhaes L. Molecular subtyping of Blastocystis sp. isolates from symptomatic patients in Italy. Parasitol. Res. 2011;109:613–619. doi: 10.1007/s00436-011-2294-7. [DOI] [PubMed] [Google Scholar]
- Nieves-Ramírez M.E., Partida-Rodríguez O., Laforest-Lapointe I., Reynolds L.A., Brown E.M., Valdez-Salazar A. Asymptomatic intestinal colonization with protist Blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems. 2018;3:e00007–e00018. doi: 10.1128/mSystems.00007-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera-Julian M., Rocafort M., Guillén Y., Rivera J., Casadellà M., Nowak P. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine. 2016;5:135–146. doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourrisson C., Scanzi J., Pereira B., NkoudMongo C., Wawrzyniak I., Cian A. Blastocystis is associated with decrease of faecal microbiota protective bacteria: comparative analysis between patients with irritable bowel syndrome and control subjects. PLoS One. 2014;9:e111868. doi: 10.1371/journal.pone.0111868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piranshahi A.R., Tavalla M., Khademvatan S. Genomic analysis of Blastocystis hominis isolates in patients with HIV-positive using locus SSU-rDNA. J. Parasit. Dis. 2018;42:28–33. doi: 10.1007/s12639-017-0957-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier P., Wawrzyniak I., Vivarès C.P., Delbac F., Alaoui H.E. New insights into Blastocystis spp.: a potential link with irritable bowel syndrome. PLoS Pathog. 2012;8:e1002545. doi: 10.1371/journal.ppat.1002545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez J.D., Sánchez A., Hernández C., Flórez C., Bernal M.C., Giraldo J.C. Geographic distribution of human Blastocystis subtypes in South America. Infect. Genet. Evol. 2016;41:32–35. doi: 10.1016/j.meegid.2016.03.017. [DOI] [PubMed] [Google Scholar]
- Ridley D.S., Hawgood B.C. The value of formolether concentration of faecal cysts and ova. J. Clin. Pathol. 1956;9:74–76. doi: 10.1136/jcp.9.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riisberg I., Orr R.J., Kluge R., Shalchian-Tabrizi K., Bowers H.A., Patil V. Seven gene phylogeny of heterokonts. Protist. 2009;160:191–204. doi: 10.1016/j.protis.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Rinninella E., Raoul P., Cintoni M., Franceschi F., Miggiano G.A.D., Gasbarrini A., Mele M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7:E14. doi: 10.3390/microorganisms7010014. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook G.A. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Darwinian medicine and the ‘hygiene’ or ‘old friends’ hypothesis. Clin. Exp. Immunol. 2010;160:70–79. doi: 10.1111/j.1365-2249.2010.04133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scicluna S.M., Tawari B., Clark C.G. DNA barcoding of Blastocystis. Protist. 2006;157:77–85. doi: 10.1016/j.protis.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Sheehan D.J., Raucher B.G., McKitrick J.C. Association of Blastocystis hominis with signs and symptoms of human disease. J. Clin. Microbiol. 1986;24:548–550. doi: 10.1128/jcm.24.4.548-550.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberman J.D., Sogin M.L., Leipe D.D., Clark C.G. Human parasite finds taxonomic home. Nature. 1996;380:398. doi: 10.1038/380398a0. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Clark C.G. Current status of Blastocystis: a personal view. Parasitol. Int. 2016;65:763–771. doi: 10.1016/j.parint.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., van der Giezen M. Associations between gut microbiota and common luminal intestinal parasites. Trends Parasitol. 2018;34:369–377. doi: 10.1016/j.pt.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Suresh G.K., Tan K.S., Thompson R.C., Traub R.J., Viscogliosi E. Terminology for Blastocystis subtypes – a consensus. Trends Parasitol. 2007;23:93–96. doi: 10.1016/j.pt.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Lewis H.C., Hammerum A.M., Porsbo L.J., Nielsen S.S., Olsen K.E. Blastocystis: unravelling potential risk factors and clinical significance of a common but neglected parasite. Epidemiol. Infect. 2009;137:1655–1663. doi: 10.1017/S0950268809002672. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Alfellani M., Clark C.G. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect. Genet. Evol. 2012;12:263–273. doi: 10.1016/j.meegid.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Tomita J., Nishioka K., Hisada T., Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One. 2014;9 doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T.C., Ong S.C., Suresh K.G. Genetic variability of Blastocystis sp. isolates obtained from cancer and HIV/AIDS patients. Parasitol. Res. 2009;105 doi: 10.1007/s00436-009-1551-5. 1283-1236. [DOI] [PubMed] [Google Scholar]
- Tito R.Y., Chaffron S., Caenepeel C., Lima-Mendez G., Wang J., Vieira-Silva S. Population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut. 2019;68:1180–1189. doi: 10.1136/gutjnl-2018-316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesterbacka J., Rivera J., Noyan K., Parera M., Neogi U., Calle M. Richer gut microbiota with distinct metabolic profile in HIV infected elite controllers. Sci. Rep. 2017;7:6269. doi: 10.1038/s41598-017-06675-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I., Dunham R.M., Iwai S., Maher M.C., Albright R.G., Broadhurst M.J. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 2013 doi: 10.1126/scitranslmed.3006438. 193-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzyniak I., Poirier P., Viscogliosi E., Dionigia M., Texier C., Delbac F., Alaoui H.E. Blastocystis, an unrecognized parasite: an overview of pathogenesis and diagnosis. Ther. Adv. Infect. Dis. 2013;1:167–178. doi: 10.1177/2049936113504754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B., Landay A., Presti R.M. Microbiome alterations in HIV infection a review. Cell. Microbiol. 2016;18:645–651. doi: 10.1111/cmi.12588. [DOI] [PubMed] [Google Scholar]
- Wu Z., Mirza H., Tan K.S. Intra-subtype variation in enteroadhesion accounts for differences in epithelial barrier disruption and is associated with metronidazole resistance in Blastocystis Subtype-7. PLoS Negl. Trop. Dis. 2014;8:e2885. doi: 10.1371/journal.pntd.0002885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yason J.A., Liang Y.R., Png C.W., Zhang Y., Tan K.S. Interactions between a pathogenic Blastocystis subtype and gut microbiota: in vitro and in vivo studies. Microbiome. 2019;7:30. doi: 10.1186/s40168-019-0644-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G.H., Hu X.F., Liu T.L., Hu R.S., Yu Z.Q., Yang W.B. Molecular characterization of Blastocystis sp. in captive wild animals in Qinling Mountains. Parasitol. Res. 2017;116:2327–2333. doi: 10.1007/s00436-017-5506-y. [DOI] [PubMed] [Google Scholar]