Abstract
Coral reefs are one of the most diverse and productive ecosystems on the planet, yet they have suffered tremendous losses due to anthropogenic disturbances and are predicted to be one of the most adversely affected habitats under future climate change conditions. Coral reefs can be viewed as microbially driven ecosystems that rely on the efficient capture, retention, and recycling of nutrients in order to thrive in oligotrophic waters. Microorganisms play vital roles in maintaining holobiont health and ecosystem resilience under environmental stress; however, they are also key players in positive feedback loops that intensify coral reef decline, with cascading effects on biogeochemical cycles and marine food webs. There is an urgent need to develop a fundamental understanding of the complex microbial interactions within coral reefs and their role in ecosystem acclimatization, and it is important to include microorganisms in reef conservation in order to secure a future for these unique environments.
Subject Areas: Microbiology, Microbiome, Biogeoscience, Global Nutrient Cycle
Graphical Abstract
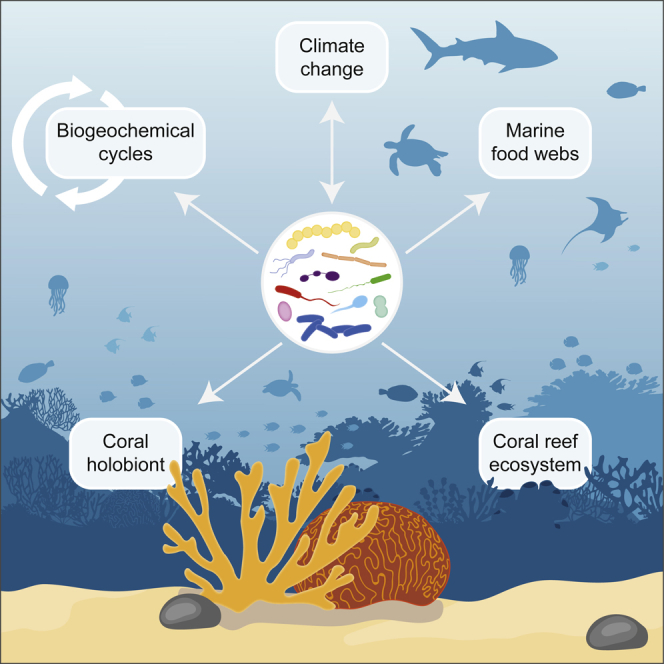
Microbiology; Microbiome; Biogeoscience; Global Nutrient Cycle
Introduction
Coral reefs are facing significant challenges from anthropogenic and natural stressors, including declining water quality, overfishing, physical destruction, disease, and climate change (Figure 1) (Hoegh-Guldberg et al., 2017, Hughes et al., 2017a, Pawlik et al., 2016). These ecosystems are one of the most diverse and productive environments on the planet, offering shelter, food, and habitat for at least 25% of known marine species and providing important economic services, including coastal protection, tourism, and fisheries (Figure 1) (Bourne et al., 2016, Moberg and Folke, 1999, Silveira et al., 2017, Torda et al., 2017). Despite their global importance, coral reefs have suffered unprecedented losses in the recent past and are critically threatened by the continued increase in sea surface water temperature and ocean acidification resulting from climate change (Hughes et al., 2017b, Pawlik et al., 2016). The first mass coral bleaching event was recorded in 1988 (Cziesielski et al., 2019), and over the past 4 years alone, prominent coral reef ecosystems like the Great Barrier Reef have lost as much as 50% of shallow-water corals (Hoegh-Guldberg et al., 2019). A thorough understanding of the mechanisms involved in coral bleaching and resilience is needed, in combination with immediate global action to reduce carbon emissions in order to avoid further coral reef decline (Sully et al., 2019).
Figure 1.
Impacts of Human Activities on the Important Services Provided by Coral Reefs
Overview of ecosystems services provided by a healthy coral reef ecosystem (left) and anthropogenic disturbances that lead to coral reef decline (right). Top left, counter clockwise: Habitat and biodiversity, biogeochemical cycling, tourism, shoreline protection, and fisheries. Top right, clockwise: Climate change, nutrient and sediment run-off from agricultural practices and deforestation, reduced water quality due to waste(water) pollution, physical destruction (e.g., due to shipping and dredging), and overfishing. The photos represent the Great Barrier Reef (left, 2008) and the Scott Reef lagoon (right, 2016).
Coral reefs rely on a diverse consortium of free-living and host-associated microorganisms for the capture, retention, and recycling of nutrients and trace elements that allow these ecosystems to thrive in the marine equivalent of a desert (Cardini et al., 2014, de Goeij et al., 2013). Benthic organisms, such as scleractinian (stony) corals, i.e. the foundation species of coral reefs (see Box 1 for full definition of this term and others used in this review article), and sponges, form close associations with a wide diversity of eukaryotes, prokaryotes, and viruses, collectively termed the holobiont (Bourne and Webster, 2013). The importance of endosymbiotic dinoflagellates in providing nutrients and energy to animal hosts such as corals has been well established (Stat et al., 2006). However, there is increasing support for the fundamental role of prokaryotic members of the holobiont in maintaining host fitness, with functions ranging from nutrient supply and recycling to protection against invading pathogens (Bourne et al., 2016). Climate change conditions can trigger a destabilization of the natural microbiome, leading to a state of dysbiosis with the emergence of opportunistic and potentially pathogenic taxa and resulting in a rising incidence of disease, bleaching, and ultimately host mortality (Littman et al., 2011, van Oppen and Blackall, 2019). It has therefore been postulated that the microbiome could play a central role in coral reef restoration (van Oppen and Blackall, 2019) and that microbial community changes in response to disturbances could act as early warning signals since these shifts may precede visual signs of bleaching and tissue necrosis (Bourne et al., 2008, Glasl et al., 2017, Lee et al., 2016).
Box 1. Glossary.
Assisted evolution: a range of approaches that involve human intervention to accelerate the rate of naturally occurring evolutionary processes.
Benthic-pelagic coupling: the exchange of energy, mass, and nutrients between benthic and pelagic habitats; critical to nutrient cycling in aquatic environments and energy transfer in food webs.
Bioindicator: biological processes, species, or communities that are used to screen the quality and health of an ecosystem in the environment and how this changes over time.
Diazotrophs: bacteria and archaea capable of fixing elemental nitrogen to produce ammonia.
Disease: a shift away from a healthy state that may be caused by exogenous (external, environmental) or endogenous (internal, from the organism itself) factors.
Dysbiosis: an imbalance of the natural microbiome that results in a disruption of the symbiotic relationship between the host and associated microorganisms, which in turn could result in disease.
Endosymbionts: organisms that form a symbiotic (often mutualistic) relationship with another cell or organism. They can be found either within cells (intracellular) or attached to the surface of cells (extracellular).
Foundation species: species that structure the habitat, determine the biodiversity, control the dynamics, and are fundamental to the ecosystem resilience; often referred to as ecosystem engineers.
Functional redundancy: multiple species from a variety of taxonomic groups that fulfill similar functions within an ecosystem.
Functionome: a comprehensive set of functions in individual organisms or a community of organisms in a specific environment.
Holobiont: an ecological unit comprising the host and all associated organisms.
Nested ecosystems: an ecological concept that considers each ecosystem as a whole and, at the same time, as nested within a larger system. Changes within each of these systems will have cascading effects on consecutively larger ecosystems.
Probiotics: microorganisms that provide health benefits when administered to a host.
Resilience: the rate of recovery following a disturbance.
Resistance: the level of insensitivity to a disturbance.
Selfish symbiont: symbionts that favor using their products of photosynthesis for selfish reproduction over translocating photosynthates to their host; ultimately this could lead to a form of parasitism.
Viral shunt: a pathway that releases carbon and nutrients back into the environment and away from secondary consumers through viral lysis of microbial cells.
Coral reef microorganisms can also contribute to ecosystem phase shifts that occur due to changes in competitiveness of the dominant benthic organisms following anthropogenic disturbances, with cascading effects on nutrient cycles, energy use, and higher trophic levels (Kelly et al., 2018). An example is the shift from coral- to algae-dominated states, which has been observed on reefs worldwide following mass coral bleaching and/or disease (Meirelles et al., 2018). This transition is accelerated and intensified through a microbially driven positive feedback loop that leads to increased coral mortality and microbialization of the ecosystem (Haas et al., 2016, Silveira et al., 2017). A holistic microbial ecosystem approach is therefore needed to accurately evaluate and predict the impacts of future climate conditions on coral reefs, integrating across scales from individual microbes and holobiont functioning to ecosystem-wide food webs and biogeochemical processes (Garren and Azam, 2012, Kelly et al., 2018).
In this review, we discuss the central role that microorganisms play in coral reef ecosystems, focusing on bacteria and archaea. We link microbial community dynamics to animal host physiology (corals and sponges) and biogeochemical transformations in order to unravel the impacts of climate change on the fate of coral reefs. By discussing microbially driven effects on holobiont health in relation to the broader ecosystem and food webs through benthic-pelagic coupling and ecosystem phase shifts, we strive to provide a holistic view of microbial interactions in coral reef environments. Finally, we assess the use of microorganisms as biomarkers of ecosystem health and their potential application in marine conservation and reef restoration practices. Given the critical role of microorganisms in driving global climate change, we advocate that microbes become an integral component of climate change research and decision-making in conservation and natural resource management.
Nutrient Cycling within the Coral Holobiont
Functional Roles of the Coral Microbiome
Corals obtain a fraction of their nutrients via heterotrophic feeding, i.e., predation of zooplankton, but largely depend on their microbiome for proficient nutrient acquisition and recycling in nutrient-poor seawaters (Figure 2) (Bourne et al., 2016). Although the endosymbiotic Symbiodiniaceae translocates photosynthates to the coral host, there is increasing support for an active role of prokaryotes in meeting the host's nutritional needs (Peixoto et al., 2017). For example, genes encoding enzymes involved in carbon fixation via the Calvin cycle, reverse Krebs cycle, and modified hydroxypropionate/hydroxybutyrate pathway have been identified in coral microbiomes (Kimes et al., 2010, Robbins et al., 2019). In addition, coral-associated microorganisms are capable of utilizing simple and complex carbohydrates (Wegley et al., 2007), which would allow them to recycle a portion of the carbon and energy invested in mucus production (Bourne et al., 2016, Kimes et al., 2010).
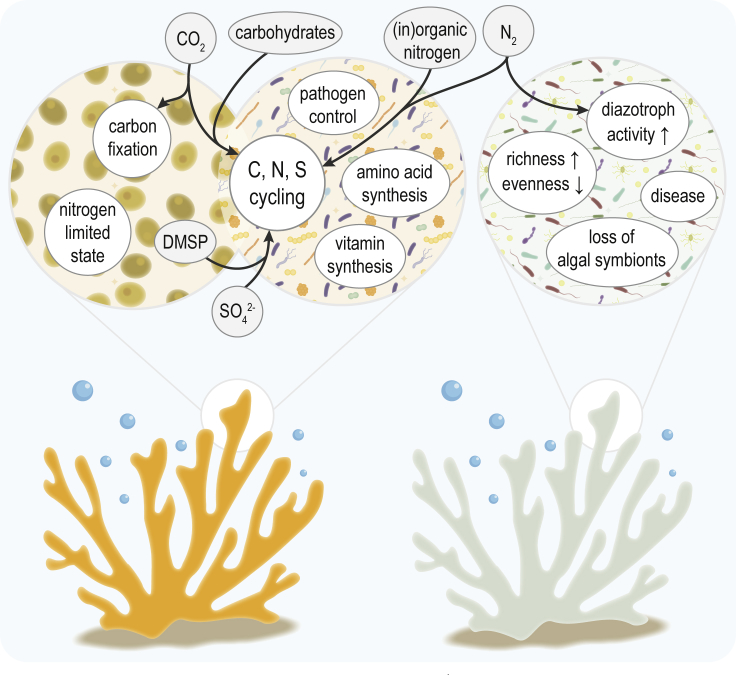
Figure 2.
Microbial Processes and Climate Change Impacts within the Coral Holobiont
Microbially mediated processes within the coral holobiont in a healthy state (left) compared with a state of dysbiosis (right) resulting from increased seawater temperature and/or ocean acidification. Microorganisms in healthy corals are involved in nutrient cycling, production of essential amino acids and vitamins, and maintaining pathogen control. Stress events can lead to a shift in the microbiome, with an increase in opportunistic and potentially pathogenic species that could result in coral disease. Furthermore, elevated temperatures may result in increased nitrogen fixation, which disrupts internal nutrient balances and may enhance coral bleaching.
Metabolic potential for complete nitrogen cycling has been detected in microbiomes of multiple coral species, suggesting a key role for nitrogen in coral holobiont functioning (Radecker et al., 2015). Inorganic and organic nitrogen sources may be actively taken up from the surrounding seawater via ammonia and urea transporters (Robbins et al., 2019), or alternatively, nitrogen fixation could serve as a mechanism to counteract environmental nitrogen deficits (Pogoreutz et al., 2017). Potential N2-fixing bacteria (diazotrophs) have been detected across all coral life stages and are persistent over time and space (Benavides et al., 2017). Their ubiquity indicates that diazotrophs likely represent symbiotic partners interacting in a close biological relationship with the holobiont (Hernandez-Agreda et al., 2017). However, this needs to be further validated since there is currently only one genome-centric study of a coral holobiont, which showed that nitrogen fixation genes were either absent or present at very low abundance within the Porites lutea microbiome (Robbins et al., 2019). Various nitrifying, denitrifying, and ammonifying bacteria and archaea have also been found within coral microbiomes (Robbins et al., 2019, Siboni et al., 2008, Yang et al., 2013). Although their specific roles remain understudied, potential involvement in the removal of ammonia waste produced by the host (Siboni et al., 2008) and in maintaining stable coral-Symbiodianiceae symbiosis by controlling nitrogen availability to the algae have been proposed (Morris et al., 2019, Peixoto et al., 2017).
Corals are the largest producers of dimethylsulfoniopropionate (DMSP) in the marine environment (Raina et al., 2013). Bacteria capable of using DMSP and its breakdown product dimethylsulfide (DMS) as sole carbon sources have been observed in coral microbiomes, indicating substantial involvement in dissolved sulfur cycling on the reef (Frade et al., 2016, Hernandez-Agreda et al., 2017, Raina et al., 2010). Moreover, DMSP can function as an osmolyte or cryoprotectant, whereas DMS can act as a scavenger of reactive oxygen species (ROS) via conversion to dimethylsulfoxide (DMSO) (Ainsworth et al., 2015). A recent study showed the presence of DMSO reductase genes associated with Endozoicomonas, a likely constituent of the coral core microbiome (Box 2), highlighting a potential role in the regeneration of this antioxidant (Robbins et al., 2019). As various microorganisms can show chemotaxis toward DMSP, it may also be involved in the acquisition of symbionts, such as Rhizobiales and Roseobacter, or even potential pathogens like Vibrio spp (Bourne et al., 2016, Garren et al., 2014). The presence of micro-niches within corals (Box 2) may facilitate anaerobic processes to occur, such as sulfate reduction potentially coupled to methane oxidation and methanogenesis (Bourne and Webster, 2013, Kimes et al., 2010); however, these assumed functions remain to be validated.
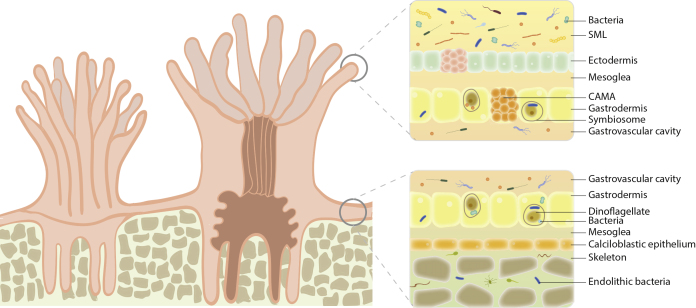
Box 2. The Coral Holobiont.
Scleractinian corals are marine invertebrates that form close associations with a wide diversity of microorganisms, including photoautotrophic dinoflagellates (Symbiodiniaceae), protozoa, fungi, bacteria, archaea, and viruses. This symbiotic consortium is termed the coral holobiont and acts as an integrated unit to maintain organismal function under constant environmental pressure (Morrow et al., 2018). Coral-associated microorganisms frequently form intimate species-specific relationships with their host (Forest et al., 2002), but they can also vary across different temporal and spatial scales, the latter ranging from biogeographical regions to the many microbial niches of a coral colony (Hernandez-Agreda et al., 2018). Although most contemporary coral microbiome research has analyzed composite coral samples, it is important to distinguish between the different microhabitats, including the coral mucus, tissues, skeleton, and gastric cavity. Within each of these habitats, microorganisms can reside as transient communities or established symbionts with positive, neutral, or negative interactions (Wada et al., 2019). The surface mucous layer (SML) is the interface between the surrounding seawater and the coral epithelium and plays an essential role in coral reef nutrient cycling, heterotrophic feeding, sediment removal, pathogen defense, and protection against stress (Bourne et al., 2016). The microbial community within this layer is expected to be dynamic because of the highly variable environment and more loosely associated with the host owing to continuous shedding and replenishing of the SML (Hernandez-Agreda et al., 2017). Coral-associated microbial aggregates (CAMAs) on the other hand are prevalent and abundant within coral tissues (epidermal and gastrodermal epithelia). They are often co-localized with Symbiodiniaceae and are likely integrated into shared metabolic pathways central to maintaining coral fitness (Wada et al., 2019). Finally, the coral skeleton is highly environmentally stable yet harbors a vastly diverse endolithic community, comprising bacteria, fungi, and filamentous algae, and is structured down to millimeter scales (Pernice et al., 2019, van Oppen and Blackall, 2019). Several attempts have been made to identify a core coral microbiome in order to establish a baseline from which to decipher key determinants of holobiont well-being and functioning under future environmental change (Ainsworth et al., 2015, Hernandez-Agreda et al., 2018, Trevathan-Tackett et al., 2019). This has, for example, led to an alleged classification of the microbiome into three components: an environmentally responsive community (mostly transient), a resident microbiome (species specific), and a small conserved core microbiome (possibly symbiotic) (Hernandez-Agreda et al., 2018). Putative beneficial microorganisms for corals have also been established that may be used to improve coral health (Peixoto et al., 2017). As a level of functional redundancy has been suggested among members of the coral holobiont (Hernandez-Agreda et al., 2018, Robbins et al., 2019), it may, however, be more insightful to define a core functionome consisting of metabolic traits that are essential to establishing a symbiotic relationship and maintaining holobiont fitness.
Box 2 Figure. The microhabitats of the coral holobiont (adapted from Hernandez-Agreda et al. [2017]; van Oppen and Blackall [2019]).
Other putative roles for coral-associated microorganisms include provision of essential amino acids and cofactors, such as B vitamins, to the coral host and algal endosymbionts as they cannot produce these compounds themselves (Figure 2) (Matthews et al., 2020, Robbins et al., 2019). The native microbiome can also exert pathogen control through colonization of the coral surface, competition for nutrients and/or space, and production of antimicrobial compounds (Peixoto et al., 2017).
Coral Holobiont Response to Climate Change
Microorganisms can respond rapidly to changing conditions, and environmental stress linked to climate change can alter the coral microbiome, generally leading to an increased richness, lower evenness (higher variability), and reduced stability (McDevitt-Irwin et al., 2017, van Oppen and Blackall, 2019) (Figure 2). Most studies report that, upon exposure to higher ocean temperature and/or acidification, there is a shift in dominance from putatively beneficial bacterial taxa, e.g., Endozoicomonas, to opportunistic and potentially pathogenic groups, such as Alteromonadaceae and Vibrionaceae (Bourne et al., 2016, Littman et al., 2011, McDevitt-Irwin et al., 2017, O'Brien et al., 2016, Thurber et al., 2009). This microbial shift typically coincides with an increased prevalence of stress resistance and virulence genes (Littman et al., 2011, Nguyen et al., 2015) and may result in elevated incidence of disease, including black band disease, yellow band disease, and dark spot syndrome, particularly when corals are exposed to multiple stressors (Zaneveld et al., 2016). Furthermore, several studies have reported shifts in microbiome metabolism, such as a transition from predominantly autotrophic to heterotrophic metabolism, along with changes in secondary metabolite profiles, sulfur and nitrogen metabolism, motility and chemotaxis, and fatty acid and lipid utilization (Littman et al., 2011, Thurber et al., 2009). There is also increasing support for the role of nitrogen-fixing bacteria in maintaining the stability of the coral-Symbiodiniaceae symbiosis, and thus in regulating bleaching (Morrow et al., 2018). Elevated temperatures could increase diazotroph activity and abundance, with a resultant rise in nitrogen availability (Cardini et al., 2016, Pogoreutz et al., 2017, Santos et al., 2014). This can be beneficial for corals since moderate levels of ammonia have been shown to increase thermal tolerance (Morris et al., 2019). However, as nitrogen fixation continues to increase under heat-stressed conditions, the finely tuned internal nutrient balance within the coral holobiont may shift, thereby releasing Symbiodiniaceae from its nutrient limited state and inducing phosphate starvation (Morris et al., 2019, Pogoreutz et al., 2017). This in turn could lead to reduced translocation of photosynthates to the coral host - also known as the selfish symbiont (Baker et al., 2018) - and increased susceptibility to photodamage and subsequent bleaching (Morrow et al., 2018, Pogoreutz et al., 2017, Radecker et al., 2015, Wiedenmann et al., 2013).
It is well recognized that climate change conditions can destabilize the microbiome, initially leading to a state of dysbiosis, which can develop into an alternate stable state characterized by disease, bleaching, and mortality (Egan and Gardiner, 2016, van Oppen and Blackall, 2019). Nonetheless, the underlying cause-effect pathway is still underexplored. Some coral species show resistance to environmental changes and are capable of maintaining a stable microbiome at lower pH (Meron et al., 2012) or elevated temperatures (Epstein et al., 2019, Grottoli et al., 2018, Tracy et al., 2015), whereas other species maintain a stable microbiome until a critical temperature threshold is exceeded (Salerno et al., 2011). Compositional and functional changes in the microbiome can also be decoupled as a result of functional redundancy within the holobiont (Glasl et al., 2017). Adding to the complexity is the observation that some coral microbiomes can return to their original state once the stressor has been removed (Bourne et al., 2008, Ziegler et al., 2019), whereas others change irreversibly into a new stable state that can be either beneficial or detrimental to the holobiont (Tracy et al., 2015).
Microbial Contributions to Coral Acclimatization
It has been suggested that the microbiome may help increase the resilience of reef-building corals to future climate conditions upon pre-exposure to sub-lethal elevated temperatures, e.g., via seasonal variations or natural anomalies (Ziegler et al., 2017). Furthermore, microorganisms could positively or negatively impact the long-term resilience of coral holobionts and reef ecosystems to environmental change through their rapid response to disturbances (McDevitt-Irwin et al., 2017, Webster and Reusch, 2017). The extensive taxonomic and metabolic diversity of the microbiome, and notably shorter generation times compared to the coral host, provide considerable potential for microorganisms to contribute to the adaptive response of the holobiont (Torda et al., 2017). Variation in microbial symbionts can occur via acquisition of novel microbial strains (symbiont switching), microbial community shifts (symbiont shuffling), lifestyle changes (host-associated versus free-living, mutualistic versus parasitic), horizontal gene transfers (McDaniel et al., 2010), or genetic mutations (Torda et al., 2017; Webster and Reusch, 2017). Advantageous modifications to the microbiome could potentially be vertically transferred to subsequent coral generations, thereby increasing the overall population fitness (Quigley et al., 2019, Webster and Reusch, 2017). Importantly, however, although there is theoretical potential for the microbiome to contribute to coral acclimatization, to date there is little empirical evidence to support these ideas, and further research into the role of the microbiome in coral reef adaptation is therefore urgently needed.
Impacts of Climate Change at the Coral Reef Ecosystem Level
Microbially Driven Nested Ecosystems
Diverse holobionts like reef-building corals are themselves considered as complex ecosystems in which the (inter)actions of the individual members shape the overall functioning and fitness of the holobiont (Pita et al., 2018). However, these meta-organisms do not occur in isolation but are nested within communities of neighboring holobionts that, in turn, interact with and influence successively larger and more complex communities and marine environments (McFall-Ngai et al., 2013). This concept is referred to as nested ecosystems and implies that the actions of individual constituents of the coral microbiome can have effects far beyond that of the holobiont (Costello et al., 2012, Pita et al., 2018). The impacts of climate change on the coral microbiome can therefore have ramifications at the coral reef ecosystem level, including potentially transformative effects on primary production, nutrient cycling, disease ecology, and benthic community structure (Hoegh-Guldberg et al., 2017).
Tight benthic-pelagic coupling occurs in shallow, well-mixed coral reefs with strong interlinkages between microbial communities associated with different benthic organisms, the sediment, and the overlaying water column (Bourne and Webster, 2013). Considering these connections, predictions of the resilience of coral reef ecosystems to future ocean conditions could benefit from linking microbial community dynamics to host physiology and biogeochemical transformations (Garren and Azam, 2012, Kelly et al., 2018). An example of an important energetic and ecological link between the coral holobiont and its surrounding environment is coral mucus (Wild et al., 2004). It provides a nutritious medium on which a diverse assemblage of coral-associated microbes thrive and interact with the environment via quorum sensing (Garren et al., 2014) and the production of allelochemicals (Ritchie, 2006). The mucus simultaneously operates as a defensive barrier against invasive pathogenic microbes from the surrounding seawater through the production of antimicrobial compounds and the colonization of this bordering niche by resident microorganisms (Glasl et al., 2016, Peixoto et al., 2017, Ritchie, 2006). Coral mucus is also continuously or periodically - depending on the coral species and mucus age - sloughed off from the coral surface into the reef environment. This represents one of the largest sources of organic matter in the coral reef ecosystem (Rix et al., 2017, Wild et al., 2004). Mucus can dissolve in seawater (dissolved organic matter, DOM) where it fuels the microbial loop (Box 3) (Fonvielle et al., 2015, Wild et al., 2004), get consumed by detritivores (see sponge loop) (de Goeij et al., 2013), or sink to be degraded by sediment microbial communities, all of which result in the recycling of essential nutrients and energy back into the reef system (Bourne and Webster, 2013, Garren and Azam, 2012, Wild et al., 2004). Coral bleaching has been shown to change the production, structure, and composition of coral mucus (Wright et al., 2019), which will have flow-on consequences for microbially driven nutrient cycles and food web dynamics within coral reefs (Hoegh-Guldberg et al., 2017, Morrow et al., 2018).
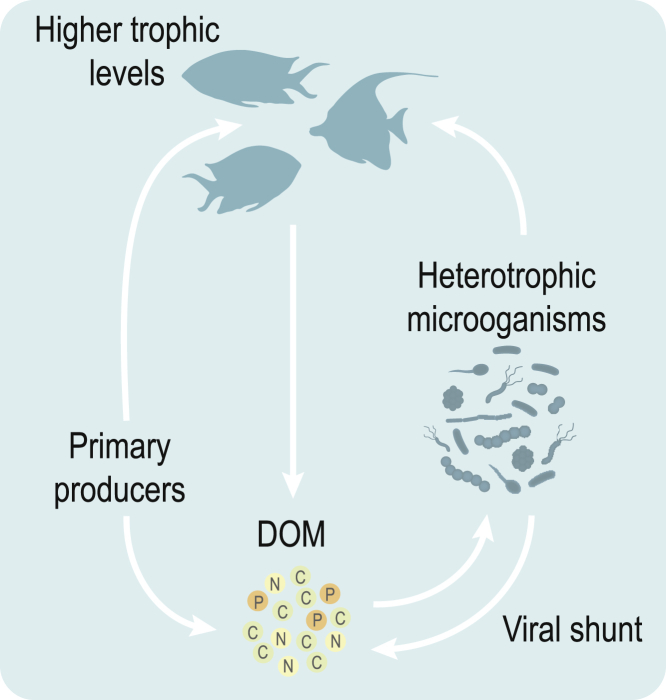
Box 3. The Microbial Loop and Viral Shunt.
The microbial loop is recognized as an important mechanism for the retention and recycling of valuable nutrients and energy within all marine environments, including coral reefs (Silveira et al., 2017). It was first described in 1983 to represent the missing link between DOC resulting from phytoplankton primary production and its transfer to higher trophic levels within the open ocean (Azam et al., 1983). Heterotrophic marine microorganisms utilize DOC as an energy source and transfer carbon up the food web to flagellates and microzooplankton, which in turn are fed upon by progressively larger macroorganisms. The microbial loop has since expanded to incorporate other groups, including photosynthetic prokaryotes, mixotrophic protists, and viruses (Fenchel, 2008). Viruses are the most abundant organisms in global ecosystems, and evidence suggests that they represent important agents of mortality for marine microorganisms (Payet et al., 2014, Payet and Suttle, 2013, Thurber et al., 2017). Viral infection and lysis of microbial cells releases DOM and nutrients back into the seawater DOM pool, retaining them within the microbial food web (Wilhelm and Suttle, 1999). This process is referred to as the viral shunt and diverts microbial biomass away from higher trophic levels while strongly impacting marine nutrient cycles (Danovaro et al., 2011, Weitz and Wilhelm, 2012). Viruses also exert control on microbial community structure and diversity through “kill-the-winner” or “piggy-back-the-winner” mechanisms (Coutinho et al., 2017, Gregory et al., 2019, Silveira and Rohwer, 2016, Thingstad, 2000, Weitz and Wilhelm, 2012). Climate change conditions are expected to influence the interactions between viruses and their hosts, with profound impacts on biogeochemical cycles and ecosystem functioning (Danovaro et al., 2011 ). For example, viral lysis of microbial blooms in algal-dominated reefs results in amplified carbon and nutrient levels within the ecosystem, with concomitant impacts on microbial populations and viral production, leading to a positive feedback on coral disease and mortality (Thurber et al., 2017).
Box 3 Figure. A simplified overview of DOM—consisting of dissolved organic carbon (DOC), nitrogen (DON), phosphorous (DOP)—cycling in marine environments via the microbial loop.
Sponges as Potential Winners under Climate Change Conditions
Marine sponges are ecologically important constituents of global benthic environments, often exceeding the diversity of corals and algae (Bell, 2008, Maldonado et al., 2017). Sponges provide habitat for a wide range of reef fauna and are known to host dense and highly specific microbial communities, which can comprise up to 40% of the sponge biomass (Webster and Taylor, 2012). As such, sponge-microbial symbioses represent one of the most diverse and complex holobionts in the marine environment (Moitinho-Silva et al., 2017, Thomas et al., 2016). Depending on the abundance of their associated microorganisms, sponge species are typically classified as having high (HMA) or low (LMA) microbial abundance (Gloeckner et al., 2014). The sponge microbiome is involved in autotrophic and/or heterotrophic carbon metabolism, nitrogen metabolism (e.g., the oxidation of ammonia waste from the host), sulfur cycling, storage of polyphosphate granules, and the biosynthesis of essential vitamins (Engelberts et al., 2020, Zhang et al., 2019). A remarkable property of the sponge holobiont is the capacity to produce a diverse range of secondary metabolites that may provide protection against predators and epibionts (Pawlik, 2011, Slaby et al., 2017).
Sponges represent a textbook example of functionally important nested ecosystems within coral reefs owing to their impressive filtering capacity (Pita et al., 2018), which provides a vital link between the benthos and ambient seawater (i.e., benthic-pelagic coupling) (Glasl et al., 2017, Maldonado et al., 2017). Although sponges have been thought to primarily feed on picoplankton, a number of encrusting, excavating, and massive reef sponges are also able to assimilate DOM, and this can comprise up to 90% of the holobiont's total heterotrophic carbon uptake (Achlatis et al., 2019, Pita et al., 2018, Rix et al., 2017). The DOM pool predominantly consists of coral and algal exudates and represents the largest resource of organic matter produced on reefs (de Goeij et al., 2013). A substantial fraction of the assimilated dissolved organic carbon (DOC), nitrogen (DON), and phosphorous (DOP) is subsequently released by the sponge as particulate organic matter (POM or detritus) via the rapid turnover and shedding of sponge cells (de Goeij et al., 2013) or alternatively incorporated into sponge biomass (McMurray et al., 2018). POM and biomass are in turn readily consumed by small detritivores and spongivores, respectively, who themselves are fed upon by larger animals higher in the food web (de Goeij et al., 2013, McMurray et al., 2018, Webster and Thomas, 2016). Sponges thus retain organic matter within reef ecosystems that would otherwise be lost to the open ocean and transfer the energy and nutrients stored in the DOM pool to higher trophic levels, a phenomenon referred to as the sponge loop (de Goeij et al., 2013, Rix et al., 2018). Both sponge cells and sponge-associated microorganisms can assimilate DOM (de Goeij et al., 2013, Pita et al., 2018), and it has been shown that they may utilize different fractions of the reef DOM pool depending on DOM availability, quality, and origin (Rix et al., 2017).
The sponge microbiome is reportedly highly stable across different geographic locations and natural environmental gradients and over time (Astudillo-García et al., 2017). There is also an increasing body of evidence suggesting that sponge holobionts are generally resistant to local and global stressors, including ocean warming and acidification (Botte et al., 2019, Simister et al., 2012), with some studies suggesting sponges will be potential winners under future climate scenarios (Bell et al., 2018a, Glasl et al., 2018). However, other studies have demonstrated shifts in the microbiome following disturbances, leading to a loss of metabolic potential and dysbiosis and eventually resulting in sponge disease and/or mortality (Botte et al., 2019, Fan et al., 2013, Lesser et al., 2016, Pita et al., 2018). Changes in the sponge microbiome have been detected prior to visual signs of bleaching, whereas other studies have observed stable microbial communities until the very late stages of thermal stress when the host itself becomes necrotic (Ramsby et al., 2018). Considering this variability in sensitivity, it is apparent that the impact of climate change on sponge holobionts will be species specific, as well as dependent on the type, intensity, and duration of the disturbance. At the ecosystem level, this could result in an alternate stable state with higher abundance yet lower diversity of resistant/resilient sponges (Bell et al., 2015, Glasl et al., 2017, Pawlik et al., 2016, Pita et al., 2018). A relative increase in sponge dominance at the expense of coral abundance would cause the benthic community to shift from being primarily dominated by photoautotrophic organisms to heterotrophic species (Bell et al., 2018a, Pawlik et al., 2016). As this may also result in an overall loss of complexity, it is unclear if higher trophic levels can remain supported under these conditions (Bell et al., 2018b). Considering the vital functional roles that sponge holobionts fulfill in reef ecosystems, changes in the benthic communities in response to climate change will have significant consequences for food webs and biogeochemical cycles on coral reefs (Rix et al., 2017).
Ecosystem Phase Shifts from Coral- to Algae-Dominated Reefs
Anthropogenic disturbances have led to worldwide shifts from coral-dominated reefs to reef systems characterized by turf and fleshy macroalgae (Barott and Rohwer, 2012, Pawlik et al., 2016). Coral demise due to bleaching and disease triggers a positive feedback loop of coral degradation and increasing algal dominance that is further accelerated and intensified by coral reef microorganisms (Radecker et al., 2015). It has been hypothesized that algae can transfer harmful pathogens to corals upon contact and that labile algal-derived DOC stimulates the growth of copiotrophic and potentially pathogenic microorganisms (Barott and Rohwer, 2012, Meirelles et al., 2018, Nelson et al., 2013). It has also been hypothesized that elevated levels of DOC combined with temperature stress can enhance bleaching through proliferation of diazotrophs (Pogoreutz et al., 2017, Radecker et al., 2015). Furthermore, increased microbial metabolism resulting from higher amounts of bioavailable DOC can create hypoxic zones at the coral-algal interface leading to additional coral mortality (Bourne et al., 2016, Silveira et al., 2017). The mechanisms by which both disease and microbial respiration favor algal overgrowth are collectively described in the DDAM model (DOC, disease, algae, and microbes) (Barott and Rohwer, 2012, Haas et al., 2016).
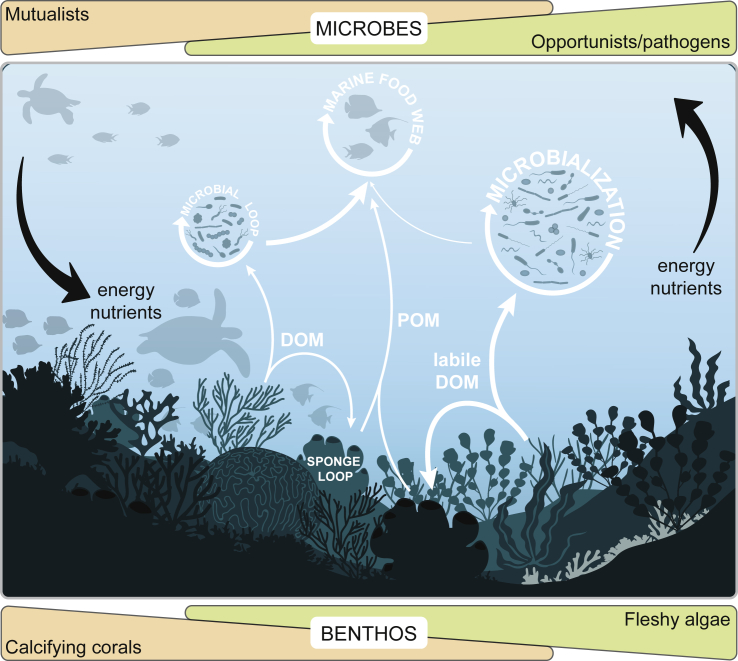
Greater quantities of labile algal-derived DOC can stimulate the microbial loop and cause a shift in ecosystem trophic structure toward significantly higher microbial biomass (Figure 3) (Kelly et al., 2018). This is referred to as microbialization and involves a depletion of the DOC standing stock, as well as a transition in central carbohydrate metabolism from the energy-efficient Embden-Meyerhof-Parnas pathway to the less efficient but faster Enter-Doudoroff and pentose phosphate pathways (Haas et al., 2013, Haas et al., 2016). This yield-to-power switch is a common ecological strategy employed by microorganisms that allows them to sacrifice energetic efficiency in order to outgrow competitors and deprive them of food. However, it also means resources are locked within the microbial compartments of the reef at the expense of energy partitioning toward higher trophic levels (Haas et al., 2016, Silveira et al., 2017). Elevated temperatures further stimulate microbial metabolism, and ocean acidification and hypoxia are additionally predicted to enhance nitrogen fixation while limiting nitrification, resulting in a net increase in nitrogen availability in the reef (Hutchins and Fu, 2017). Together with local impacts from overfishing and eutrophication (Hughes et al., 2007), these effects can further exacerbate algal overgrowth and the positive feedback loop toward microbialization.
Figure 3.
Cycling of Dissolved and Particulate Organic Matter through the Microbial and Sponge Loops
As coral reef ecosystems shift toward an algae-dominated state after coral bleaching and/or disease, significant amounts of readily available DOM are released into the environment, which stimulates growth of copiotrophic and potentially pathogenic microorganisms. Although microbes in healthy reefs dominated by calcifying corals excel at capturing and recycling energy and nutrients within the reef—thereby supporting high primary production and a diverse marine food web—microbialization limits the flow of energy to higher trophic levels and can lead to an overall loss of energy from the ecosystem under future climate change conditions.
A less studied phase shift is the potential transition to sponge-dominated reefs and the competitive interactions between coral, algae, and sponges (Bell et al., 2018a). On the one hand, sponges could benefit from an increased food supply in the form of algal exudates as it has been shown that algal-derived DOM is more rapidly assimilated by sponges compared with coral-derived DOM (Rix et al., 2017). Although stimulated nutrient retention via the sponge loop would support coral reef primary production, elevated nitrogen release to the benthos may lead to eutrophication, which would further stimulate algal overgrowth and have negative consequences for coral health (Pawlik et al., 2016, Rix et al., 2017). Similar to the microbial loop, labile algal-derived DOM may preferentially fuel microbial metabolism within the sponge holobiont, and this could potentially occur at the expense of the animal host (Rix et al., 2017). Given the complexity of microbial interactions within coral reefs, and the ongoing degradation from local and global disturbances, it is imperative that we investigate these nested microbial ecosystems in order to predict how future reefs will function (Bell et al., 2018b).
Microbial Ecology in Coral Reef Management
Microbes as Early Warning Indicators to Avert Coral Reef Decline
Coral reef monitoring is currently based on visual signs of health deterioration in corals (e.g., bleaching, disease, and mortality) and shifts in benthic community structure in response to climate change (Hill and Wilkinson, 2004). Since these signs often only become evident once the reef is in an advanced stage of stress and decline, sensitive and rapid markers for ecosystem stress would be highly advantageous (Glasl et al., 2017). Microorganisms have previously been raised as potential early warning bioindicators given their rapid and often specific responses to environmental disturbances (Faust et al., 2015, Glasl et al., 2019, Semenza et al., 2017). For example, total coliform counts are typically used as a measure of fecal contamination in surface waters and drinking water supplies (Tallon et al., 2005). To assess human impacts on coral reefs, a microbialization score has been proposed as a measure of metabolic rates of microbial communities and reef-associated fishes (McDole et al., 2012). Presence/absence and shifts in relative abundance of free-living or host-associated microorganisms in coral reefs could also be of high diagnostic value (Glasl et al., 2017, McDevitt-Irwin et al., 2017). Recent research has revealed that seawater microbial communities rather than host-associated microbiomes have a strong potential to reflect environmental conditions owing to their high sensitivity and predictability (Glasl et al., 2019). However, further research is needed to establish robust baselines for microbial communities in all compartments of the reef (Egan and Gardiner, 2016) and to elucidate their structural and functional responses to local and global stressors by incorporating microbes in long-term reef monitoring initiatives (Faust et al., 2015, Glasl et al., 2019).
The Central Role of the Microbiome in Coral Reef Restoration
Reef conservation strategies have predominantly focused on the establishment and management of marine protected areas and preservation of water quality, but massive global coral decline in response to climate change (Hughes et al., 2017b) has prompted a rise in efforts toward reef restoration (Silveira et al., 2017). Corals have been the focus of restoration practices such as coral aquaculture (“gardening”) or fragmentation followed by transplantation onto the reef (Rinkevich, 2008, Silveira et al., 2017). However, a growing appreciation for the role that microorganisms play in maintaining animal health and ecosystem functioning, together with a realization of possible implications for conservation and management of threatened species (West et al., 2019), has led some researchers to explore whether manipulation of the microbiome could be used to assist coral reef restoration (Silveira et al., 2017). For example, natural microbiomes of healthy/stress-resistant corals, synthetic communities consisting of beneficial microorganisms for corals (BMC as probiotics) or even genetically modified bacterial strains could be used to inoculate new or diseased coral colonies in an attempt to transfer health benefits and stress resistance (Peixoto et al., 2019, Rosado et al., 2019, van Oppen and Blackall, 2019). Another possibility could be the development of coral stocks with enhanced stress tolerance through acceleration of coral microbiome evolutionary changes, an approach referred to as assisted evolution (van Oppen et al., 2017). It remains to be established, however, whether long-term modifications of the microbiome can be achieved, if upscaling is at all feasible and what the implications will be for the ecosystem as a whole (van Oppen and Blackall, 2019). An alternative approach could be the identification and application of microbial cues either to increase coral larvae settlement (Hadfield, 2011) or to avoid the growth of fouling microorganisms that would otherwise compete for space (Wood et al., 2016). Finally, phage therapy has been proposed to control disease progression in corals (Cohen et al., 2013). Nonetheless, the majority of these methods are still in their infancy and require significant research and development to determine their suitability for active reef restoration efforts.
Microorganisms in Climate Change Research and Conservation Management
The distribution, abundance, diversity, interactions, and functions of microbial communities are directly and indirectly impacted by climate change (Hutchins et al., 2019), with altered nutrient cycles and resource allocations in coral reefs being just one of the many examples. Given that the microbial world constitutes the life support system of the biosphere, the potential consequences of diversity and biomass loss, local extinction, and community shifts are substantial and cannot be ignored (Hutchins and Fu, 2017, McFall-Ngai et al., 2013). Although microorganisms directly contribute to climate change through the production of greenhouse gases, they also offer important opportunities for remediating human-induced change (Cavicchioli et al., 2019). We need to understand how marine microbiomes respond and potentially adapt to climate change in order to define tolerance thresholds and identify climate-ecosystem feedbacks in coral reefs (Hutchins et al., 2019, Webster et al., 2018). Despite the overwhelming evidence of the fundamental role of microorganisms in climate change biology, they have rarely been the focus of climate change research thus far (Cavicchioli et al., 2019). It is time for microbial ecologists, conservationists, and policy makers to work together to construct an integrated understanding of coral reef functioning that includes microorganisms, which is vital for the long-term survival and conservation of these diverse environments (Kelly et al., 2018, West et al., 2019).
Conclusions and Future Perspectives
Coral reefs support tremendous biodiversity and provide essential ecosystem services, yet they are suffering from rapid degradation in response to climate change and local anthropogenic disturbances (Hughes et al., 2017a). There is substantial research supporting the vital role of the microbiome in maintaining coral health and in regulating ecosystem resilience. Microorganisms are capable of buffering or exacerbating cumulative anthropogenic impacts via their role in holobiont fitness as well as by modifying nutrient and energy flows within coral reefs (Bourne et al., 2016). Recent advances in DNA-based molecular approaches have provided unprecedented insights into the vast taxonomic and metabolic diversity of the microbial world on coral reefs, although the majority of microbial functions have merely been inferred from 16S rRNA gene analyses. In-depth functional analyses using metatranscriptomics, metaproteomics, and metabolomics combined with isotopic labeling experiments and advanced microscopy methods are thus needed to validate predicted holobiont functioning and responses to environmental stress, and to determine how these relate to modified biogeochemical fluxes within the ecosystem (Trevathan-Tackett et al., 2019). A holistic ecosystem microbiology approach needs to be adopted that views coral reefs as integrated systems, from genes and microbes to ecosystem-scale processes (Garren and Azam, 2012). Microbial responses to global and local stressors also need to be evaluated over different time scales, ranging from immediate physiological acclimatization to prolonged adaptive evolution (Hutchins et al., 2019). Furthermore, the incorporation of microbial monitoring in long-term coral reef monitoring projects would provide invaluable insights into microbial baselines and community dynamics in relation to biogeochemical parameters, natural anomalies, and anthropogenic disturbances. Robust predictive models should include microbial processes to interpolate and extrapolate observed microbial interactions within the coral reef environment in order to predict future reef outcomes under different disturbance regimes as well as the capacity for reef acclimatization (Hutchins et al., 2019). Finally, interdisciplinary and collaborative research is the key to successful coral reef conservation and is essential in order to guarantee a future for these unique and critical ecosystems.
Acknowledgments
The authors would like to acknowledge Eric Matson and James Gilmour from the Australian Institute of Marine Science for providing the coral reef photos in Figure 1.
Author Contributions
I.V. and N.S.W. conceived the review topic and scoped the sections. I.V. wrote the manuscript, and N.S.W. edited the manuscript.
References
- Achlatis M., Pernice M., Green K., de Goeij J.M., Guagliardo P., Kilburn M.R., Hoegh-Guldberg O., Dove S. Single-cell visualization indicates direct role of sponge host in uptake of dissolved organic matter. J. Exp. Mar. Biol. Ecol. 2019;516:140–149. doi: 10.1098/rspb.2019.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth T.D., Krause L., Bridge T., Torda G., Raina J.B., Zakrzewski M., Gates R.D., Padilla-Gamino J.L., Spalding H.L., Smith C. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015;9:2261–2274. doi: 10.1038/ismej.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astudillo-García C., Bell J.J., Webster N.S., Glasl B., Jompa J., Montoya J.M., Taylor M.W. Evaluating the core microbiota in complex communities: a systematic investigation. Environ. Microbiol. 2017;19:1450–1462. doi: 10.1111/1462-2920.13647. [DOI] [PubMed] [Google Scholar]
- Azam F., Fenchel T., Field J.G., Gray J.S., Meyer-Reil L.A., Thingstad F. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 1983;10:257–263. [Google Scholar]
- Baker D.M., Freeman C.J., Wong J.C.Y., Fogel M.L., Knowlton N. Climate change promotes parasitism in a coral symbiosis. ISME J. 2018;12:921–930. doi: 10.1038/s41396-018-0046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barott K.L., Rohwer F.L. Unseen players shape benthic competition on coral reefs. Trends Microbiol. 2012;20:621–628. doi: 10.1016/j.tim.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Bell J.J. The functional roles of marine sponges. Estuarine, Coastal Shelf Sci. 2008;79:341–353. [Google Scholar]
- Bell J.J., Bennett H.M., Rovellini A., Webster N.S. Sponges to be winners under near-future climate scenarios. BioScience. 2018;68:955–968. [Google Scholar]
- Bell J.J., McGrath E., Biggerstaff A., Bates T., Cárdenas C.A., Bennett H. Global conservation status of sponges. Conserv. Biol. 2015;29:42–53. doi: 10.1111/cobi.12447. [DOI] [PubMed] [Google Scholar]
- Bell J.J., Rovellini A., Davy S.K., Taylor M.W., Fulton E.A., Dunn M.R., Bennett H.M., Kandler N.M., Luter H.M., Webster N.S. Climate change alterations to ecosystem dominance: how might sponge-dominated reefs function? Ecology. 2018;99:1920–1931. doi: 10.1002/ecy.2446. [DOI] [PubMed] [Google Scholar]
- Benavides M., Bednarz V.N., Ferrier-Pages C. Diazotrophs: overlooked key players within the coral symbiosis and tropical reef ecosystems? Front. Mar. Sci. 2017;4:10. [Google Scholar]
- Botte E.S., Nielsen S., Wahab M.A.A., Webster J., Robbins S., Thomas T., Webster N.S. Changes in the metabolic potential of the sponge microbiome under ocean acidification. Nat. Commun. 2019;10:4134. doi: 10.1038/s41467-019-12156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D.G., Iida Y., Uthicke S., Smith-Keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008;2:350–363. doi: 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- Bourne D.G., Morrow K.M., Webster N.S. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 2016;70:317–340. doi: 10.1146/annurev-micro-102215-095440. [DOI] [PubMed] [Google Scholar]
- Bourne D.G., Webster N.S. Coral reef bacterial communities. In: Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F.L., editors. The Prokaryotes: Prokaryotic Communities and Ecophysiology. Springer Berlin Heidelberg; 2013. pp. 163–187. [Google Scholar]
- Cardini U., Bednarz V.N., Foster R.A., Wild C. Benthic N2 fixation in coral reefs and the potential effects of human-induced environmental change. Ecol. Evol. 2014;4:1706–1727. doi: 10.1002/ece3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardini U., van Hoytema N., Bednarz V.N., Rix L., Foster R.A., Al-Rshaidat M.M.D., Wild C. Microbial dinitrogen fixation in coral holobionts exposed to thermal stress and bleaching. Environ. Microbiol. 2016;18:2620–2633. doi: 10.1111/1462-2920.13385. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R., Ripple W.J., Timmis K.N., Azam F., Bakken L.R., Baylis M., Behrenfeld M.J., Boetius A., Boyd P.W., Classen A.T. Scientists' warning to humanity: microorganisms and climate change. Nat. Rev. Microbiol. 2019;17:569–586. doi: 10.1038/s41579-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y., Joseph Pollock F., Rosenberg E., Bourne D.G. Phage therapy treatment of the coral pathogen Vibrio coralliilyticus. Microbiol. Open. 2013;2:64–74. doi: 10.1002/mbo3.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E.K., Stagaman K., Dethlefsen L., Bohannan B.J.M., Relman D.A. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho F.H., Silveira C.B., Gregoracci G.B., Thompson C.C., Edwards R.A., Brussaard C.P.D., Dutilh B.E., Thompson F.L. Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans. Nat. Commun. 2017;8:15955. doi: 10.1038/ncomms15955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cziesielski M.J., Schmidt-Roach S., Aranda M. The past, present, and future of coral heat stress studies. Ecol. Evol. 2019;9:10055–10066. doi: 10.1002/ece3.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danovaro R., Corinaldesi C., Dell'Anno A., Fuhrman J.A., Middelburg J.J., Noble R.T., Suttle C.A. Marine viruses and global climate change. FEMS Microbiol. Rev. 2011;35:993–1034. doi: 10.1111/j.1574-6976.2010.00258.x. [DOI] [PubMed] [Google Scholar]
- de Goeij J.M., van Oevelen D., Vermeij M.J.A., Osinga R., Middelburg J.J., de Goeij A.F.P., Admiraal W. Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science. 2013;342:108–110. doi: 10.1126/science.1241981. [DOI] [PubMed] [Google Scholar]
- Egan S., Gardiner M. Microbial dysbiosis: rethinking disease in marine ecosystems. Front. Microbiol. 2016;7:991. doi: 10.3389/fmicb.2016.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberts J.P., Robbins S.J., de Goeij J.M., Aranda M., Bell S.C., Webster N.S. Characterization of a sponge microbiome using an integrative genome-centric approach. ISME J. 2020:1–10. doi: 10.1038/s41396-020-0591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein H.E., Torda G., van Oppen M.J.H. Relative stability of the Pocillopora acuta microbiome throughout a thermal stress event. Coral Reefs. 2019;38:373–386. [Google Scholar]
- Fan L., Liu M., Simister R., Webster N.S., Thomas T. Marine microbial symbiosis heats up: the phylogenetic and functional response of a sponge holobiont to thermal stress. ISME J. 2013;7:991–1002. doi: 10.1038/ismej.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K., Lahti L., Gonze D., de Vos W.M., Raes J. Metagenomics meets time series analysis: unraveling microbial community dynamics. Curr. Opin. Microbiol. 2015;25:56–66. doi: 10.1016/j.mib.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Fenchel T. The microbial loop – 25 years later. J. Exp. Mar. Biol. Ecol. 2008;366:99–103. [Google Scholar]
- Fonvielle J.A., Reynaud S., Jacquet S., LeBerre B., Ferrier-Pages C. First evidence of an important organic matter trophic pathway between temperate corals and pelagic microbial communities. PLoS One. 2015;10:e0139175. doi: 10.1371/journal.pone.0139175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forest R., Victor S., Farooq A., Nancy K. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 2002;243:1–10. [Google Scholar]
- Frade P.R., Schwaninger V., Glasl B., Sintes E., Hill R.W., Simó R., Herndl G.J. Dimethylsulfoniopropionate in corals and its interrelations with bacterial assemblages in coral surface mucus. Environ. Chem. 2016;13:252–265. [Google Scholar]
- Garren M., Azam F. New directions in coral reef microbial ecology. Environ. Microbiol. 2012;14:833–844. doi: 10.1111/j.1462-2920.2011.02597.x. [DOI] [PubMed] [Google Scholar]
- Garren M., Son K., Raina J.-B., Rusconi R., Menolascina F., Shapiro O.H., Tout J., Bourne D.G., Seymour J.R., Stocker R. A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 2014;8:999–1007. doi: 10.1038/ismej.2013.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasl B., Bourne D.G., Frade P.R., Thomas T., Schaffelke B., Webster N.S. Microbial indicators of environmental perturbations in coral reef ecosystems. Microbiome. 2019;7:94. doi: 10.1186/s40168-019-0705-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasl B., Herndl G.J., Frade P.R. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 2016;10:2280–2292. doi: 10.1038/ismej.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasl B., Smith C.E., Bourne D.G., Webster N.S. Exploring the diversity-stability paradigm using sponge microbial communities. Sci. Rep. 2018;8:8425. doi: 10.1038/s41598-018-26641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasl B., Webster N.S., Bourne D.G. Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems. Mar. Biol. 2017;164:91. [Google Scholar]
- Gloeckner V., Wehrl M., Moitinho-Silva L., Gernert C., Schupp P., Pawlik J.R., Lindquist N.L., Erpenbeck D., Wörheide G., Hentschel U. The HMA-LMA dichotomy revisited: an electron microscopical survey of 56 sponge species. Biol. Bull. 2014;227:78–88. doi: 10.1086/BBLv227n1p78. [DOI] [PubMed] [Google Scholar]
- Gregory A.C., Zayed A.A., Conceição-Neto N., Temperton B., Bolduc B., Alberti A., Ardyna M., Arkhipova K., Carmichael M., Cruaud C. Marine DNA viral macro- and microdiversity from pole to pole. Cell. 2019;177:1109–1123. doi: 10.1016/j.cell.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grottoli A.G., Dalcin Martins P., Wilkins M.J., Johnston M.D., Warner M.E., Cai W.-J., Melman T.F., Hoadley K.D., Pettay D.T., Levas S. Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS One. 2018;13:e0191156. doi: 10.1371/journal.pone.0191156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A.F., Fairoz M.F.M., Kelly L.W., Nelson C.E., Dinsdale E.A., Edwards R.A., Giles S., Hatay M., Hisakawa N., Knowles B. Global microbialization of coral reefs. Nat. Microbiol. 2016;1:16042. doi: 10.1038/nmicrobiol.2016.42. [DOI] [PubMed] [Google Scholar]
- Haas A.F., Nelson C.E., Rohwer F., Wegley-Kelly L., Quistad S.D., Carlson C.A., Leichter J.J., Hatay M., Smith J.E. Influence of coral and algal exudates on microbially mediated reef metabolism. PeerJ. 2013;1:1–28. doi: 10.7717/peerj.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield M.G. Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Annu. Rev. Mar. Sci. 2011;3:453–470. doi: 10.1146/annurev-marine-120709-142753. [DOI] [PubMed] [Google Scholar]
- Hernandez-Agreda A., Gates R.D., Ainsworth T.D. Defining the core microbiome in corals' microbial soup. Trends Microbiol. 2017;25:125–140. doi: 10.1016/j.tim.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Hernandez-Agreda A., Leggat W., Bongaerts P., Herrera C., Ainsworth T.D. Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. mBio. 2018;9:e00812–e00818. doi: 10.1128/mBio.00812-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J., Wilkinson C. Global Coral Reef Monitoring Network; 2004. Methods for Ecological Monitoring of Coral Reefs; pp. 1–123. [Google Scholar]
- Hoegh-Guldberg O., Jacob D., Taylor M.W., Guillén Bolaños T., Bindi M., Brown S., Camilloni I.A., Diedhiou A., Djalante R., Ebi K. The human imperative of stabilizing global climate change at 1.5°C. Science. 2019;365:1–13. doi: 10.1126/science.aaw6974. [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg O., Poloczanska E.S., Skirving W., Dove S. Coral reef ecosystems under climate change and ocean acidification. Front. Mar. Sci. 2017;4:158. [Google Scholar]
- Hughes T.P., Barnes M.L., Bellwood D.R., Cinner J.E., Cumming G.S., Jackson J.B.C., Kleypas J., van de Leemput I.A., Lough J.M., Morrison T.H. Coral reefs in the anthropocene. Nature. 2017;546:82–90. doi: 10.1038/nature22901. [DOI] [PubMed] [Google Scholar]
- Hughes T.P., Kerry J.T., Álvarez-Noriega M., Álvarez-Romero J.G., Anderson K.D., Baird A.H., Babcock R.C., Beger M., Bellwood D.R., Berkelmans R. Global warming and recurrent mass bleaching of corals. Nature. 2017;543:373–377. doi: 10.1038/nature21707. [DOI] [PubMed] [Google Scholar]
- Hughes T.P., Rodrigues M.J., Bellwood D.R., Ceccarelli D., Hoegh-Guldberg O., McCook L., Moltschaniwskyj N., Pratchett M.S., Steneck R.S., Willis B. Phase shifts, herbivory, and the resilience of coral reefs to climate change. Curr. Biol. 2007;17:360–365. doi: 10.1016/j.cub.2006.12.049. [DOI] [PubMed] [Google Scholar]
- Hutchins D.A., Fu F.X. Microorganisms and ocean global change. Nat. Microbiol. 2017;2:17058. doi: 10.1038/nmicrobiol.2017.58. [DOI] [PubMed] [Google Scholar]
- Hutchins D.A., Jansson J.K., Remais J.V., Rich V.I., Singh B.K., Trivedi P. Climate change microbiology - problems and perspectives. Nat. Rev. Microbiol. 2019;17:391–396. doi: 10.1038/s41579-019-0178-5. [DOI] [PubMed] [Google Scholar]
- Kelly L.W., Haas A.F., Nelson C.E. Ecosystem microbiology of coral reefs: linking genomic, metabolomic, and biogeochemical dynamics from animal symbioses to reefscape processes. Msystems. 2018;3 doi: 10.1128/mSystems.00162-17. e00162–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimes N.E., Van Nostrand J.D., Weil E., Zhou J., Morris P.J. Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ. Microbiol. 2010;12:541–556. doi: 10.1111/j.1462-2920.2009.02113.x. [DOI] [PubMed] [Google Scholar]
- Lee S.T.M., Davy S.K., Tang S.-L., Kench P.S. Mucus sugar content shapes the bacterial community structure in thermally stressed Acropora muricata. Front. Microbiol. 2016;7:371. doi: 10.3389/fmicb.2016.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser M.P., Fiore C., Slattery M., Zaneveld J. Climate change stressors destabilize the microbiome of the Caribbean barrel sponge, Xestospongia muta. J. Exp. Mar. Biol. Ecol. 2016;475:11–18. [Google Scholar]
- Littman R., Willis B.L., Bourne D.G. Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ. Microbiol. Rep. 2011;3:651–660. doi: 10.1111/j.1758-2229.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- Maldonado M., Aguilar R., Bannister R., Bell J., Conway J., Dayton P., Diaz C., Gutt J., Kelly M., Kenchinton E. Sponge grounds as key marine habitats: a synthetic review of types, structure, functional roles, and conservation concerns. In: Rossi S., Bramanti L., Gori A., Saco del Valle C.O., editors. Marine Animal Forests: The Ecology of Benthic Biodiversity Hotspots. Springer International Publishing; 2017. pp. 145–183. [Google Scholar]
- Matthews J.L., Raina J.-B., Kahlke T., Seymour J.R., van Oppen M.J.H., Suggett D.J. Symbiodiniaceae-bacteria interactions: rethinking metabolite exchange in reef-building corals as multi-partner metabolic networks. Environ. Microbiol. 2020:1–13. doi: 10.1111/1462-2920.14918. [DOI] [PubMed] [Google Scholar]
- McDaniel L.D., Young E., Delaney J., Ruhnau F., Ritchie K.B., Paul J.H. High frequency of horizontal gene transfer in the oceans. Science. 2010;330:50. doi: 10.1126/science.1192243. [DOI] [PubMed] [Google Scholar]
- McDevitt-Irwin J.M., Baum J.K., Garren M., Vega Thurber R.L. Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 2017;4:262. [Google Scholar]
- McDole T., Nulton J., Barott K.L., Felts B., Hand C., Hatay M., Lee H., Nadon M.O., Nosrat B., Salamon P. Assessing coral reefs on a pacific-wide scale using the microbialization score. PLoS One. 2012;7:e43233. doi: 10.1371/journal.pone.0043233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M., Hadfield M.G., Bosch T.C.G., Carey H.V., Domazet-Lošo T., Douglas A.E., Dubilier N., Eberl G., Fukami T., Gilbert S.F. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U S A. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray S.E., Stubler A.D., Erwin P.M., Finelli C.M., Pawlik J.R. A test of the sponge-loop hypothesis for emergent Caribbean reef sponges. Mar. Ecol. Prog. Ser. 2018;588:1–14. [Google Scholar]
- Meirelles P.M., Soares A.C., Oliveira L., Leomil L., Appolinario L.R., Francini R.B., de Moura R.L., Almeida R.T.D., Salomon P.S., Amado G.M. Metagenomics of coral reefs under phase shift and high hydrodynamics. Front. Microbiol. 2018;9:2203. doi: 10.3389/fmicb.2018.02203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meron D., Rodolfo-Metalpa R., Cunning R., Baker A.C., Fine M., Banin E. Changes in coral microbial communities in response to a natural pH gradient. ISME J. 2012;6:1775–1785. doi: 10.1038/ismej.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg F., Folke C. Ecological goods and services of coral reef ecosystems. Ecol. Econ. 1999;29:215–233. [Google Scholar]
- Moitinho-Silva L., Nielsen S., Amir A., Gonzalez A., Ackermann G.L., Cerrano C., Astudillo-Garcia C., Easson C., Sipkema D., Liu F. The sponge microbiome project. Gigascience. 2017;6:1–7. doi: 10.1093/gigascience/gix077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris L.A., Voolstra C.R., Quigley K.M., Bourne D.G., Bay L.K. Nutrient availability and metabolism affect the stability of coral-symbiodiniaceae symbioses. Trends Microbiol. 2019;27:678–689. doi: 10.1016/j.tim.2019.03.004. [DOI] [PubMed] [Google Scholar]
- Morrow K.M., Muller E., Lesser M.P. How does the coral microbiome cause, respond to, or modulate the bleaching process? In: VanOppen M.J.H., Lough J.M., editors. Coral Bleaching Ecological Studies (Analysis and Synthesis) Springer; 2018. pp. 153–+. [Google Scholar]
- Nelson C.E., Goldberg S.J., Wegley Kelly L., Haas A.F., Smith J.E., Rohwer F., Carlson C.A. Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J. 2013;7:962–979. doi: 10.1038/ismej.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H., Bouvier T., Bouvier C., Bui V.N., Le-Lan H., Bettarel Y. Viral and bacterial epibionts in thermally-stressed corals. J. Mar. Sci. Eng. 2015;3:1272–1286. [Google Scholar]
- O'Brien P.A., Morrow K.M., Willis B.L., Bourne D.G. Implications of ocean acidification for marine microorganisms from the free-living to the host-associated. Front. Mar. Sci. 2016;3:1–14. [Google Scholar]
- Pawlik J.R. The chemical ecology of sponges on caribbean reefs: natural products shape natural systems. BioScience. 2011;61:888–898. [Google Scholar]
- Pawlik J.R., Burkepile D.E., Thurber R.V. A vicious circle? altered carbon and nutrient cycling may explain the low resilience of caribbean coral reefs. Bioscience. 2016;66:470–476. [Google Scholar]
- Payet J.P., McMinds R., Burkepile D.E., Vega Thurber R.L. Unprecedented evidence for high viral abundance and lytic activity in coral reef waters of the South Pacific Ocean. Front. Microbiol. 2014;5:493. doi: 10.3389/fmicb.2014.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payet J.P., Suttle C.A. To kill or not to kill: the balance between lytic and lysogenic viral infection is driven by trophic status. Limnol. Oceanogr. 2013;58:465–474. [Google Scholar]
- Peixoto R.S., Rosado P.M., Leite D.C.D., Rosado A.S., Bourne D.G. Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Front. Microbiol. 2017;8:341. doi: 10.3389/fmicb.2017.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto R.S., Sweet M., Bourne D.G. Customized medicine for corals. Front. Mar. Sci. 2019;6:686. [Google Scholar]
- Pernice M., Raina J.-B., Rädecker N., Cárdenas A., Pogoreutz C., Voolstra C.R. Down to the bone: the role of overlooked endolithic microbiomes in reef coral health. ISME J. 2019;14:325–334. doi: 10.1038/s41396-019-0548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pita L., Rix L., Slaby B.M., Franke A., Hentschel U. The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome. 2018;6:46. doi: 10.1186/s40168-018-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoreutz C., Radecker N., Cardenas A., Gardes A., Voolstra C.R., Wild C. Sugar enrichment provides evidence for a role of nitrogen fixation in coral bleaching. Glob. Change Biol. 2017;23:3838–3848. doi: 10.1111/gcb.13695. [DOI] [PubMed] [Google Scholar]
- Quigley K.M., Willis B.L., Kenkel C.D. Transgenerational inheritance of shuffled symbiont communities in the coral Montipora digitata. Sci. Rep. 2019;9:13328. doi: 10.1038/s41598-019-50045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radecker N., Pogoreutz C., Voolstra C.R., Wiedenmann J., Wild C. Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol. 2015;23:490–497. doi: 10.1016/j.tim.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Raina J.-B., Dinsdale E.A., Willis B.L., Bourne D.G. Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol. 2010;18:101–108. doi: 10.1016/j.tim.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Raina J.-B., Tapiolas D.M., Forêt S., Lutz A., Abrego D., Ceh J., Seneca F.O., Clode P.L., Bourne D.G., Willis B.L. DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature. 2013;502:677–680. doi: 10.1038/nature12677. [DOI] [PubMed] [Google Scholar]
- Ramsby B.D., Hoogenboom M.O., Whalan S., Webster N.S. Elevated seawater temperature disrupts the microbiome of an ecologically important bioeroding sponge. Mol. Ecol. 2018;27:2124–2137. doi: 10.1111/mec.14544. [DOI] [PubMed] [Google Scholar]
- Rinkevich B. Management of coral reefs: we have gone wrong when neglecting active reef restoration. Mar. Pollut. Bull. 2008;56:1821–1824. doi: 10.1016/j.marpolbul.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Ritchie K.B. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 2006;322:1–14. [Google Scholar]
- Rix L., de Goeij J.M., van Oevelen D., Struck U., Al-Horani F.A., Wild C., Naumann M.S. Differential recycling of coral and algal dissolved organic matter via the sponge loop. Funct. Ecol. 2017;31:778–789. [Google Scholar]
- Rix L., Goeij J., Oevelen D., Struck U., Al-Horani F.A., Wild C., Naumann M.S. Reef sponges facilitate the transfer of coral-derived organic matter to their associated fauna via the sponge loop. Mar. Ecol. Prog. Ser. 2018;589:85–96. [Google Scholar]
- Robbins S.J., Singleton C.M., Chan C.X., Messer L.F., Geers A.U., Ying H., Baker A., Bell S.C., Morrow K.M., Ragan M.A. A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nat. Microbiol. 2019;4:2090–2100. doi: 10.1038/s41564-019-0532-4. [DOI] [PubMed] [Google Scholar]
- Rosado P.M., Leite D.C.A., Duarte G.A.S., Chaloub R.M., Jospin G., Nunes da Rocha U.P., Saraiva J., Dini-Andreote F., Eisen J.A., Bourne D.G. Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J. 2019;13:921–936. doi: 10.1038/s41396-018-0323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno J.L., Reineman D.R., Gates R.D., Rappé M.S. The effect of a sublethal temperature elevation on the structure of bacterial communities associated with the coral porites compressa. J. Mar. Biol. 2011;2011:1–9. [Google Scholar]
- Santos H.F., Carmo F.L., Duarte G., Dini-Andreote F., Castro C.B., Rosado A.S., van Elsas J.D., Peixoto R.S. Climate change affects key nitrogen-fixing bacterial populations on coral reefs. ISME J. 2014;8:2272–2279. doi: 10.1038/ismej.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza J.C., Trinanes J., Lohr W., Sudre B., Löfdahl M., Martinez-Urtaza J., Nichols G.L., Rocklöv J. Environmental suitability of vibrio infections in a warming climate: an early warning system. Environ. Health Perspect. 2017;125:107004. doi: 10.1289/EHP2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siboni N., Ben-Dov E., Sivan A., Kushmaro A. Global distribution and diversity of coral-associated Archaea and their possible role in the coral holobiont nitrogen cycle. Environ. Microbiol. 2008;10:2979–2990. doi: 10.1111/j.1462-2920.2008.01718.x. [DOI] [PubMed] [Google Scholar]
- Silveira C.B., Cavalcanti G.S., Walter J.M., Silva-Lima A.W., Dinsdale E.A., Bourne D.G., Thompson C.C., Thompson F.L. Microbial processes driving coral reef organic carbon flow. FEMS Microbiol. Rev. 2017;41:575–595. doi: 10.1093/femsre/fux018. [DOI] [PubMed] [Google Scholar]
- Silveira C.B., Rohwer F.L. Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Microbiomes. 2016;2:16010. doi: 10.1038/npjbiofilms.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister R., Taylor M.W., Tsai P., Webster N. Sponge-microbe associations survive high nutrients and temperatures. PLoS One. 2012;7:e52220. doi: 10.1371/journal.pone.0052220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaby B.M., Hackl T., Horn H., Bayer K., Hentschel U. Metagenomic binning of a marine sponge microbiome reveals unity in defense but metabolic specialization. ISME J. 2017;11:2465–2478. doi: 10.1038/ismej.2017.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stat M., Carter D., Hoegh-Guldberg O. The evolutionary history of Symbiodinium and scleractinian hosts - symbiosis, diversity, and the effect of climate change. Perspect. Plant Ecol. Evol. Syst. 2006;8:23–43. [Google Scholar]
- Sully S., Burkepile D.E., Donovan M.K., Hodgson G., van Woesik R. A global analysis of coral bleaching over the past two decades. Nat. Commun. 2019;10:1264. doi: 10.1038/s41467-019-09238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallon P., Magajna B., Lofranco C., Leung K.T.J. Microbial indicators of faecal contamination in water: a current perspective. Int. J. Environ. Res. Public Health. 2005;166:139–166. [Google Scholar]
- Thingstad T.F. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 2000;45:1320–1328. [Google Scholar]
- Thomas T., Moitinho-Silva L., Lurgi M., Björk J.R., Easson C., Astudillo-García C., Olson J.B., Erwin P.M., López-Legentil S., Luter H. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 2016;7:11870. doi: 10.1038/ncomms11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber R.V., Payet J.P., Thurber A.R., Correa A.M.S. Virus–host interactions and their roles in coral reef health and disease. Nat. Rev. Microbiol. 2017;15:205. doi: 10.1038/nrmicro.2016.176. [DOI] [PubMed] [Google Scholar]
- Thurber R.V., Willner-Hall D., Rodriguez-Mueller B., Desnues C., Edwards R.A., Angly F., Dinsdale E., Kelly L., Rohwer F. Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 2009;11:2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Torda G., Donelson J.M., Aranda M., Barshis D.J., Bay L., Berumen M.L., Bourne D.G., Cantin N., Foret S., Matz M. Rapid adaptive responses to climate change in corals. Nat. Clim. Change. 2017;7:627–636. [Google Scholar]
- Tracy A.M., Koren O., Douglas N., Weil E., Harvell C.D. Persistent shifts in Caribbean coral microbiota are linked to the 2010 warm thermal anomaly. Environ. Microbiol. Rep. 2015;7:471–479. doi: 10.1111/1758-2229.12274. [DOI] [PubMed] [Google Scholar]
- Trevathan-Tackett S.M., Sherman C.D.H., Huggett M.J., Campbell A.H., Laverock B., Hurtado-McCormick V., Seymour J.R., Firl A., Messer L.F., Ainsworth T.D. A horizon scan of priorities for coastal marine microbiome research. Nat. Ecol. Evol. 2019;3:1509–1520. doi: 10.1038/s41559-019-0999-7. [DOI] [PubMed] [Google Scholar]
- van Oppen M.J.H., Blackall L.L. Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 2019;17:557–567. doi: 10.1038/s41579-019-0223-4. [DOI] [PubMed] [Google Scholar]
- van Oppen M.J.H., Gates R.D., Blackall L.L., Cantin N., Chakravarti L.J., Chan W.Y., Cormick C., Crean A., Damjanovic K., Epstein H. Shifting paradigms in restoration of the world's coral reefs. Glob. Change Biol. 2017;23:3437–3448. doi: 10.1111/gcb.13647. [DOI] [PubMed] [Google Scholar]
- Wada N., Ishimochi M., Matsui T., Pollock F.J., Tang S.-L., Ainsworth T.D., Willis B.L., Mano N., Bourne D.G. Characterization of coral-associated microbial aggregates (CAMAs) within tissues of the coral Acropora hyacinthus. Sci. Rep. 2019;9:14662. doi: 10.1038/s41598-019-49651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N.S., Reusch T.B.H. Microbial contributions to the persistence of coral reefs. ISME J. 2017;11:2167–2174. doi: 10.1038/ismej.2017.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N.S., Taylor M.W. Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 2012;14:335–346. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- Webster N.S., Thomas T. The sponge hologenome. mBio. 2016;7 doi: 10.1128/mBio.00135-16. e00135-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N.S., Wagner M., Negri A.P. Microbial conservation in the anthropocene. Environ. Microbiol. 2018;20:1925–1928. doi: 10.1111/1462-2920.14124. [DOI] [PubMed] [Google Scholar]
- Wegley L., Edwards R., Rodriguez-Brito B., Liu H., Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 2007;9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- Weitz J.S., Wilhelm S.W. Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol. Rep. 2012;4:17. doi: 10.3410/B4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.G., Waite D.W., Deines P., Bourne D.G., Digby A., McKenzie V.J., Taylor M.W. The microbiome in threatened species conservation. Biol. Conserv. 2019;229:85–98. [Google Scholar]
- Wiedenmann J., D’Angelo C., Smith E.G., Hunt A.N., Legiret F.-E., Postle A.D., Achterberg E.P. Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nat. Clim. Change. 2013;3:160–164. [Google Scholar]
- Wild C., Huettel M., Klueter A., Kremb S.G., Rasheed M.Y.M., Jørgensen B.B. Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature. 2004;428:66–70. doi: 10.1038/nature02344. [DOI] [PubMed] [Google Scholar]
- Wilhelm S.W., Suttle C.A. Viruses and Nutrient Cycles in the Sea: viruses play critical roles in the structure and function of aquatic food webs. BioScience. 1999;49:781–788. [Google Scholar]
- Wood T.L., Guha R., Tang L., Geitner M., Kumar M., Wood T.K. Living biofouling-resistant membranes as a model for the beneficial use of engineered biofilms. Proc. Natl. Acad. Sci. U S A. 2016;113:2802–2811. doi: 10.1073/pnas.1521731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R.M., Strader M.E., Genuise H.M., Matz M. Effects of thermal stress on amount, composition and antibacterial properties of coral mucus. PeerJ. 2019;7:1–16. doi: 10.7717/peerj.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Sun W., Zhang F., Li Z. Phylogenetically diverse denitrifying and ammonia-oxidizing bacteria in corals Alcyonium gracillimum and Tubastraea coccinea. Mar. Biotechnol. 2013;15:540–551. doi: 10.1007/s10126-013-9503-6. [DOI] [PubMed] [Google Scholar]
- Zaneveld J.R., Burkepile D.E., Shantz A.A., Pritchard C.E., McMinds R., Payet J.P., Welsh R., Correa A.M.S., Lemoine N.P., Rosales S. Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat. Commun. 2016;7:11833. doi: 10.1038/ncomms11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Jonas L., Lin H., Hill R.T. Microbially mediated nutrient cycles in marine sponges. FEMS Microbiol. Ecol. 2019;95:1–14. doi: 10.1093/femsec/fiz155. [DOI] [PubMed] [Google Scholar]
- Ziegler M., Grupstra C.G.B., Barreto M.M., Eaton M., BaOmar J., Zubier K., Al-Sofyani A., Turki A.J., Ormond R., Voolstra C.R. Coral bacterial community structure responds to environmental change in a host-specific manner. Nat. Commun. 2019;10:3092. doi: 10.1038/s41467-019-10969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler M., Seneca F.O., Yum L.K., Palumbi S.R., Voolstra C.R. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat. Commun. 2017;8:14213. doi: 10.1038/ncomms14213. [DOI] [PMC free article] [PubMed] [Google Scholar]