Graphical abstract
Keywords: Machine-learning, Essential genes, Lethal, Drosophila, Essentiality prediction, Homo sapiens
Abstract
Genes are termed to be essential if their loss of function compromises viability or results in profound loss of fitness. On the genome scale, these genes can be determined experimentally employing RNAi or knockout screens, but this is very resource intensive. Computational methods for essential gene prediction can overcome this drawback, particularly when intrinsic (e.g. from the protein sequence) as well as extrinsic features (e.g. from transcription profiles) are considered. In this work, we employed machine learning to predict essential genes in Drosophila melanogaster. A total of 27,340 features were generated based on a large variety of different aspects comprising nucleotide and protein sequences, gene networks, protein-protein interactions, evolutionary conservation and functional annotations. Employing cross-validation, we obtained an excellent prediction performance. The best model achieved in D. melanogaster a ROC-AUC of 0.90, a PR-AUC of 0.30 and a F1 score of 0.34. Our approach considerably outperformed a benchmark method in which only features derived from the protein sequences were used (P < 0.001). Investigating which features contributed to this success, we found all categories of features, most prominently network topological, functional and sequence-based features. To evaluate our approach we performed the same workflow for essential gene prediction in human and achieved an ROC-AUC = 0.97, PR-AUC = 0.73, and F1 = 0.64.
In summary, this study shows that using our well-elaborated assembly of features covering a broad range of intrinsic and extrinsic gene and protein features enabled intelligent systems to predict well the essentiality of genes in an organism.
1. Introduction
Essential genes are necessary for viability and replication, and thus knowledge about essentiality is central for a broad range of life science research, most prominently for drug target identification [1], but also synthetic biology [2], evolutionary studies [3] and cancer research [4]. Specifically, knowledge about essential genes in insects is highly relevant for health care and the agriculture, since this group of organisms comprises the most important vectors for infectious diseases like malaria, dengue, sleeping sickness as well as crop pests. Specific insect genera are responsible for this large medical and economic damage, like Anopheles [5], Aedes [6], [7], the tsetse fly [8] or Sitophilus [9]. This burden is typically approached by insecticides used e.g. for indoor spraying or coating of mosquito nets. However, the vectors develop resistances fast, making it mandatory to develop new insecticides [10], [11]. This is an intriguing scientific field of research and can be effectively approached by identifying so far unexplored essential genes providing targets for novel insecticides.
However, to identify essential genes experimentally on a large scale is resource intensive, and may not be feasible for all organisms and genes, as typically, for each gene, a knock-out or knockdown strain needs to be generated. With the advent of genomics there has been an increasing interest in the identification of essential genes, which was vigorously stimulated computationally using machine learning (ML) algorithms. Moreover, ML algorithms facilitated comparative analyses of features revealing characteristics of essential genes [12]. In general, essential gene prediction can base on intrinsic features from nucleotide [13] or protein sequences [14] (e.g. GC content, codon usage, protein length) and combinations of both [15]. Intrinsic in this context denotes features, which can be directly derived from DNA and protein sequences. In addition, characteristics extrinsic to a gene sequence like network topology (e.g. degree centrality and clustering coefficient), homology (e.g. number of homologs), gene expression (e.g. co-expression networks, fluctuations in gene-expression), cellular localization of the expressed protein and functional domains have been used as predictors for essentiality [15], [16], [17], [18]. We elaborated on a comprehensive integration of all these aspects. Notably, also Campos et al. hypothesized recently [14] that combining intrinsic and extrinsic features should improve prediction performance.
As a case study, we predicted essential genes in Drosophila (D.) melanogaster based on the integration of sequence derived, topology, homology and functional features. We used D. melanogaster because several knock-out and knock-down experiments have been performed with this model insect providing a plethora of excellent data and its capacity to serve as a model for further applications targeting insecticide discovery of the above described disease transmitting vectors. In order to test if our comprehensive feature selection approach performs in other eukaryotes as good as in D. melanogaster, we performed the same workflow on human data and achieved even better results. This indicates the generalizability of our approach.
2. Materials and methods
2.1. Defining the gold standard
We assembled a list of essential genes selected from the databases Online GEne Essentiality (OGEE) [19] and the Database of Essential Genes (DEG) [20]. OGEE contains essential gene annotations based on two RNAi screens from cultured cells and whole organisms of D. melanogaster [21], [22]. In total, OGEE collected essentiality information of 13,852 genes. 13,781 of which were provided by Boutros et al. basing on experiments using cell lines [21] and 437 by Chen et al. observing whole organisms [22]. 46 genes had different essentiality status between these two studies and were excluded from our analyses (they were neither in the list of essential nor non-essential genes of our gold standard). In total 249 genes were obtained from OGEE to be essential.
DEG contains 339 essential genes derived from a p-element insertion screen on whole organisms [23]. However, DEG does not provide information about non-essential genes in D. melanogaster. To work with a comprehensive set of essential genes we combined the lists of essential genes from OGEE and DEG (union) and used the list of non-essential genes from OGEE. This resulted in 441 essential and 11,788 non-essential genes. For these genes, features were derived as described in the following.
2.2. Feature generation
A main hypothesis of this work was, that a broad collection of intrinsic and extrinsic gene features from a large variety of different data sources should outperform the usage of a narrower spectrum, such as only protein sequence features in the prediction of essential genes in eukaryotes.
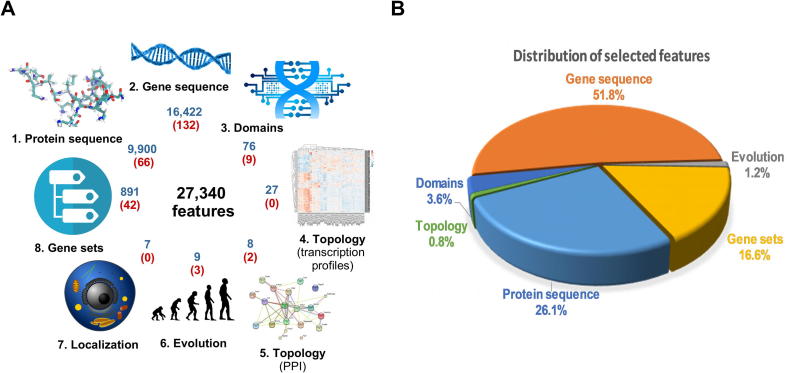
A large set of initial features was generated based on eight different sources including (1) protein sequence, (2) gene sequence, (3) functional domains of the proteins, (4) topology features derived from transcription profiles, (5) topological features derived from protein interactions, (6) evolution/conservation, (7) protein subcellular localization, and (8) gene sets from Gene Ontology and KEGG, depicted in Fig. 1A.
Fig. 1.
Features for gene essentiality prediction in D. melanogaster were assembled from various resources. (A) The generated features included intrinsic (e.g. protein and DNA sequence), as well as extrinsic features (e.g. topology of co-expression and protein-protein interaction networks). The number of features derived from individual categories are shown in blue and the selected ones for machine learning are shown in red. (B) Distribution of the selected features across all categories. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Protein and gene sequence features (feature categories 1 and 2): Protein and DNA sequences were obtained from FlyBase [24] (version 2019_02). For deriving the protein and gene sequence features, various numerical representations characterizing the nucleotide and amino acid sequences and compositions of the query gene were calculated using seqinR [25], protr [26], CodonW [27] and rDNAse [28].
With seqinR [25] the number and fraction of individual amino acids and other simple protein sequence information including the number of residues, the percentage of physico-chemical classes and the theoretical isoelectric point were calculated. Most protein sequence features were obtained using protr [26] including autocorrelation, CTD, conjoint triad, quasi-sequence order and pseudo amino acid composition. CodonW [27] was used to calculate simple gene characteristics like length and GC content but also frequency of optimal codons and effective number of codons. With rDNAse [28] gene descriptors like auto covariance or pseudo nucleotide composition, and kmer frequencies (n = 2–7) were calculated.
Domain features (feature category 3): For deriving domain features BioMart [29] was used to obtain pfam domains, number of coiled coils, trans membrane helices and signal peptides. In addition, the number and length of UTRs were obtained from BioMart.
Topology features (feature categories 4 and 5): Topology features were computed based on two types of interaction data, i.e., from protein-protein interaction (PPI) and transcription profiles. The PPI network was assembled using the PPI information for D. melanogaster listed in BioGrid [30], IntAct [31], HitPredict [32] and DroID [33]. We selected a PPI only if we found it in at least two of these databases. By this, we got 18,265 PPIs with which an undirected graph was generated and topology features (including degree, degree distribution, betweenness, closeness and clustering coefficient) were calculated using ProNet [34]. For the co-expression network, RPKM values from 124 RNA-Seq experiments of modENCODE [35], [36] were obtained from FlyBase [24] (version 2019_02). The data comprises transcription profiles from developmental stages, different tissues, and a variety of different treatments and cell lines. RPKM values were used to perform a weighted Pearson correlation network analysis and to generate topology features using WGCNA [37].
Evolution/conservation features (feature category 6): The number of homologous proteins was derived blasting the protein sequence of the query against the complete RefSeq database [38] using PSI-BLAST [39]. The number of proteins found with e-value cutoffs from 1e−5 to 1e−100 were used as features.
Localization features (feature category 7): To predict the subcellular localization of the query protein, we used the Bologna Unified Subcellular Component Annotator (BUSCA) [40], which assigns one of the nine subcellular compartments described for eukaryotic cells (nucleus, cytoplasm, mitochondrion, extracellular space, endomembrane system, plasma membrane, organelle membrane, mitochondrial membrane and outer membrane) to the protein.
Gene set features (feature category 8): We collected 8388 Gene Ontology (GO) terms including biological process, cellular localization and molecular function from FlyBase [24]. Gene sets from Gene Ontology were discarded if they showed high redundancy according to the following method.
The gene overlap of each pair of gene sets A and B was quantified calculating Jaccard similarity coefficients,
| (1) |
Pairs with J(A, B) above a threshold (threshold = 0.3) were included in the model and represented as an undirected graph, G = (X, E), with the gene sets as vertices X and the pairs above the threshold as edges E. With this, we formulated the optimization problem to select at most one gene set from each pair in such a way that the overall number of non-redundant gene sets was maximized. This optimization problem was formulated as an mixed integer linear programming problem and solved using Gurobi (version 7.5.1, https://www.gurobi.com), leading to 2627 gene sets. Furthermore, too specific gene sets with less than 16 genes were discarded yielding 770 GO terms in final. In addition, 121 gene sets from the KEGG map definitions [41] (corresponding to the investigated genes) were obtained from g:Profiler [42], leading to a total 770 + 121 = 891 gene sets. Recently, Chen et al. predicted essential genes using the information about enrichments of gene sets defined by Gene Ontology and KEGG combing this information with a gene network [43]. By this, not only the characterization of the query gene is taken into account, but also of its neighbors in the protein association network making the features more robust against false gene set annotations. We followed a similar approach and assembled the nearest neighbors of the query gene employing the gene network definitions for D. melanogaster of STRING [44]. For this gene set and each of the 891 above described gene sets, an enrichment test was performed employing Fisher’s exact test. P-values were binarised, p-values < 0.05 were set to one indicating a significant enrichment, and zero otherwise.
2.3. Data normalization and feature selection
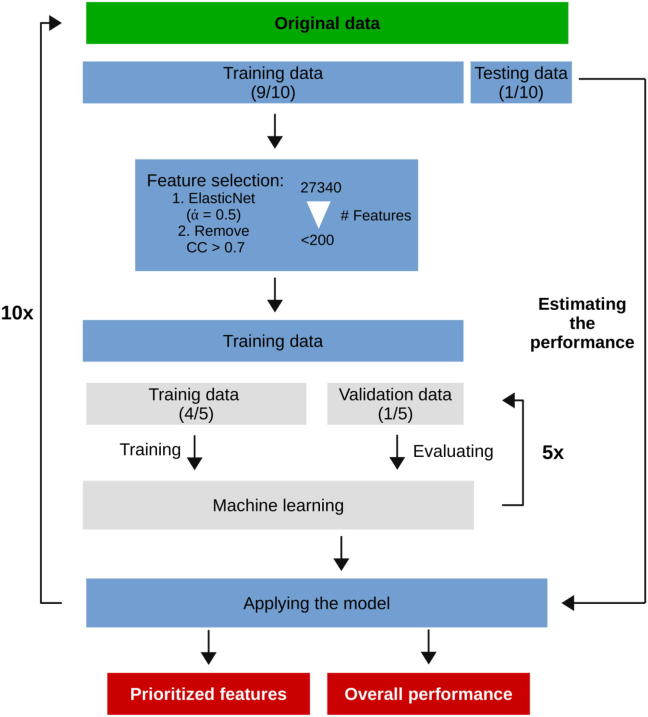
In total, we generated 27,340 features, that were assembled from the eight categories (Fig. 1A). Each feature was z-score transformed for normalization. Next, we performed two steps for feature selection prior to ML training. After splitting of the training (9/10) and testing data (1/10) as a first step we applied ElasticNet (Fig. 2). ElasticNet uses a modification of Least Absolute Shrinkage and Selection Operator (LASSO) by adding Ridge regression into the optimization criterion. ElasticNet was used from the “glmnet” package in R [45] (cv.glmnet function with parameters alpha = 0.5, type.measure = “auc”). To avoid over-fitting, feature selection was performed only on the training data, leaving the testing set unseen (Fig. 2). Interestingly the selected features came from six out of the eight categories (Fig. 1A). In step two, highly correlating features with Pearson correlation coefficients > 0.70 were removed avoiding collinearity [46], [47].
Fig. 2.
Schematic overview of the computational workflow (see text).
2.4. Sub-sampling, ML training and performance evaluation
To overcome class imbalances when training the classifiers, we used the Synthetic Minority Over-sampling Technique (SMOTE). SMOTE is a frequently used sampling method that creates synthetic, non-duplicated samples of the minority class balancing the total number of samples [48]. For each sample of the minority class, SMOTE calculates the k nearest neighbors of the same class and randomly creates multiple synthetic samples between the observation and the nearest neighbors depending on the number of additional samples needed. As classification methods, we used Generalised Linear Model (GLM), Support-Vector Machines (SVM), Random Forests (RF), Artificial Neural Networks (NNET) and Extreme Gradient Boosting (XGB) from the caret package in R [49]. For GLM tuning, alpha was held at 1 and lambda was sequentially increased from 0.001 to 0.1, in 0.001 steps. The SVM tune-grid consisted of sigma = 0.005, 0.01, 0.02, 0.05 and 0.1, and C = 0.1, 0.75 and 0.9. For RF and NNET tuning, the tuneLength parameter in the train function was set to 10 resulting in 10 mtry values for RF (number of variables randomly sampled as candidates at each split) and 100 combinations of size and decay values for NNET. Thereby size is the number of units in the hidden layer (NNET fit a single hidden layer neural network) and decay is the regularization parameter to avoid over-fitting. For XGB eta, nrounds, max_depth, min_child_weight and colsample_bytree were optimized in a tune-grid whereas gamma and subsample parameters were held constant at 0 and 1, respectively. This resulted in 216 different parameter combinations for XGB tuning.
To improve generalizability, we performed a stratified randomized 10-fold cross validation (CV). 90% of the data was used for feature selection and training of the classifiers, and 10% for testing. Within the training step, features were selected, the model was learned and evaluated in a 5-fold CV (inner loop). Fig. 2 sketches an overview of the process.
3. Results
3.1. Combination of a variety of features outperforms protein sequence features alone in D. melanogaster
Our comprehensive assembly of features resulted in 27,340 features for 12,229 genes of D. melanogaster. These comprised eight categories including protein sequence, DNA sequence, protein domains, topology features from gene expression data and a PPI network, homology, subcellular localization, and gene set features (Fig. 1). Essential gene information was obtained from DEG and OGEE databases. To improve generalizability, feature selection was performed leading to less than 200 features. For ML, a nested cross-validation scheme was applied, first to train and optimize the model in which the imbalances in the class labels were corrected based on training data (Fig. 2). Finally, the overall performance was estimated using the testing dataset.
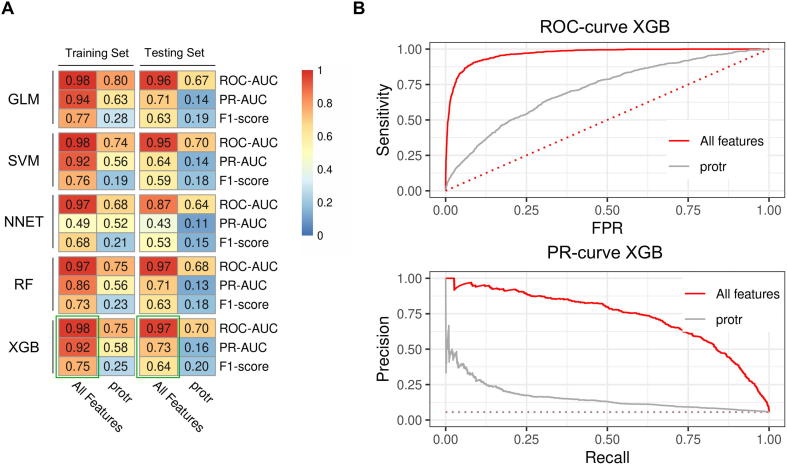
Five ML algorithms were applied for the classification of essential genes i.e. GLM. SVM, NNET, RF and XGB. In general, all five approaches yielded very good performance results, but XGB performed slightly better than the others in both, the training and testing sets (Fig. 3). For benchmarking our approach, we used a study recently published by Campos et al. [14], which predicted essential genes of D. melanogaster and other model organisms using (only) intrinsic protein features. Their gold standard based on essential gene information of OGEE alone. For D. melanogaster, they achieved a ROC-AUC of appr. 0.81 and a PR-AUC of appr. 0.15 in the testing set using a gradient boosting method for ML. Using also only these protein-based features (protr features), essential gene information from OGEE and gradient boosting for ML, we observed a similar ROC-AUC of 0.836 and PR-AUC of 0.186 (Fig. 3A).
Fig. 3.
Machine learning using a broad range of features leads to high classification performance in D. melanogaster. (A) Heatmaps showing accuracy metrics for the performance evaluation of essential gene classification. Five ML approaches were used (Generalised linear model [GLM], Support-Vector Machines [SVM], Neural Network [NNET], Random Forests [RF], and Extreme Gradient Boosting [XGB]). In addition, essentiality information was derived from two databases (DEG and OGEE). The performance was measured for the training and testing sets. Features were generated following our new approach including features basing on a broad range of aspects (All Features) and compared to the benchmarking data based on protein sequence features only (protr). The algorithm with the best performance (highest harmonic mean) was XGB, indicated by a green frame. (B) Receiver operating characteristic curve and (C) Precision-Recall curve from XGB classifier measured on the testing sets, both showing the performance difference between all features (All) and protein sequence features only (protr). Random classification is indicated by dotted lines. Using all features and essentially information from two databases (red) yielded distinctively better results compared to the benchmarking approaches (black and grey). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Strikingly, we achieved a considerably increased performance (p < 0.001) when using all features, including intrinsic and extrinsic features (Fig. 3A). XGB performed best yielding an ROC-AUC = 0.922, PR-AUC = 0.278 and F1 = 0.265, when considering essentially information from OGEE only. Furthermore, we observed even better performance when using essential gene information from OGEE and DEG (ROC-AUC = 0.902, PR-AUC = 0.296, F1 = 0.335). PR-AUC and F1 measure the performance of the positive prediction against total positive observation, where the higher the score the better the model, especially when predicting the positive class is the focus of the analysis as we have in this study. Fig. 3B and C shows the results in more detail. Especially, when compared to the benchmarking approach the increase in ROC-AUC and PR-AUC can be seen. In the following, we investigated the results based on this setup, i.e. when using all features, both databases for the gold standard and XGB for essential gene prediction.
3.2. Investigating the features with high discriminative power in D. melanogaster
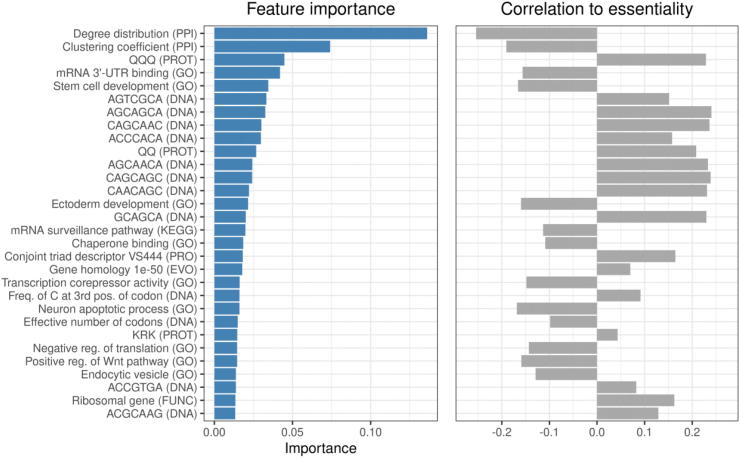
We were interested in which features contributed most to the good classification performance. For this, we estimated the “importance” of a feature by a bootstrapping approach obtaining the accuracy of each tree in the forest using the out-of-bag samples as validation. The labels of the feature are permuted and the average decrease in accuracy is used to obtain the importance score (varImp function of the caret package [49]). The 30 most important features and their correlation to essentiality in D. melanogaster are shown in Fig. 4. Among these most important features, we found features covering most (six out of eight) categories supporting our broad-spectrum approach. The most important feature was degree distribution, which describes the fraction of nodes with the same degree of the query gene. It correlates negatively with degree centrality. Biological networks such as metabolic or PPI networks show a scale-free degree distribution in which the majority of nodes (enzymes, proteins or genes) are only sparsely connected to other nodes (orphans), whereas a few nodes (hubs) are connected to many other nodes in the network. Specifically, the degree distribution follows a negative linear function in a double logarithmically scaled projection of the data [50], [51]. It has been shown that nodes with higher degree are more likely to be essential [52] confirming our observation of a negative correlation of essentiality for the degree distribution (Fig. 4). The second most important feature was clustering coefficient (CC). It was negatively correlated with essentiality, i.e. the lower the CC the higher the possibility of being essential. CC describes the connectedness of the neighbors of the query gene. We speculate that a higher CC make it more likely for the signaling stream via the affected gene/protein in the network to be replaced by a signaling cascade among its neighbors. Interestingly, we observed the same phenomenon in metabolic networks previously [16].
Fig. 4.
Both intrinsic and extrinsic features contributed substantially to the predictions in D. melanogaster. Features were ranked based on their discriminative power. The categories of the features are stated in brackets. The direction and correlation (positive or negative) to essentiality is shown in the right panel.
Other features were positively correlated with essentiality such as certain peptide triplets (QQQ and KRK), DNA heptamers (e.g. AGTCGCA), a homolog feature (number of homologs with an e-value cutoff of 1e-50) or the biological process feature of query genes coding for ribosomal proteins. In general, the high-ranking gene set terms, like mRNA 3′UTR binding or Stem cell development, were negatively correlated with essentiality, i.e. genes in these sets are more likely to be non-essential. Still, some genes in these sets were also essential, which is illustrated in the density distributions of the 30 most important features (Fig. S1).
3.3. Putative essential genes in D. melanogaster are associated with lethality and other drastic phenotype alterations
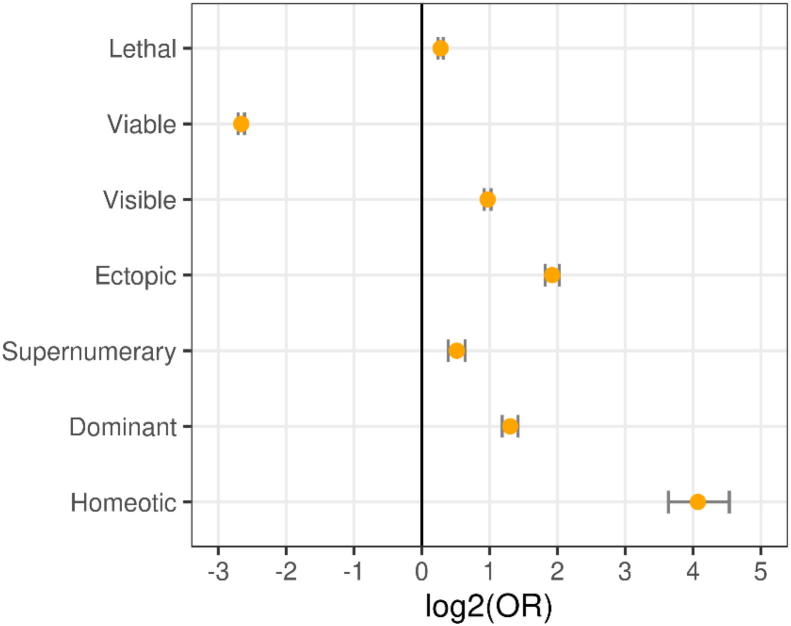
The gold standard (OGEE + DEG) consisted of n = 441 essential genes, from which we predicted 424 correctly to be essential and 11,017 genes correctly to be non-essential (sensitivity = 0.93, specificity = 0.88). However, quite a considerable number of genes (n = 771) were predicted to be essential which were, according to the databases, non-essential (therefore false positives). Table 1 lists the top ten predictions of non-essential genes according to OGEE + DEG and Table S1 shows all genes predicted to be essential in D. melanogaster. We hypothesized that these genes may also considerably contribute to viability even though not identified to be essential in the experimental screens our gold standard based on. We call these putative essential genes (PEGs) in the following. Table 1 lists the top ten and Table S1 all PEGs. Assuming that most mutations cause rather a loss than a gain of function [53], we compared the phenotypes of animals with mutations in PEGs to the phenotypes of animals with mutations in non-essential genes. For this, we interrogated FlyBase [24], which contains a comprehensive collection of 316,967 allele mutations in 11,352 genes from 15,473 studies, provided with an excellent controlled vocabulary describing the traits. Fig. 5 shows the odds ratios of the most prominent phenotypic descriptions highly over- or under-represented in PEGs (P < 1e−15 for each of these phenotypes, Fisher’s exact test). The phenotypic descriptions of these mutations in FlyBase did not contain the term “essential”. Instead, we found “lethal”, defined in FlyBase as a phenotype of a population in which all animals die at some stage or stages prior to becoming a mature adult. Therefore, the FlyBase term lethal for a mutation can be regarded as essential. Indeed, we found PEGs to be highly enriched in genes described by FlyBase to be lethal when mutated (n = 636, P < 1e−21). In line, the phenotype viable (survival until mature adulthood) was under-represented in PEGs compared to the non-essential genes (Fig. 5). Moreover, mutations in PEGs were more often found to be visible meaning that phenotypes are macroscopic and therefore show an anatomic aberration. In concordance, animals with mutations in PEGs show more often extra copies of an anatomical structure (i.e. wing, leg) at the normal (supernumerary) or abnormal (ectopic) location. Interestingly, PEGs were found to be more often genetically dominant, i.e. the phenotypes were manifested also in heterozygotes. The largest enrichment was observed for homeotic phenotypes in which one or more segments were transformed to another segment, like the transformation (also partially) of the antennas to legs, pointing to severe phenotypic aberrations.
Table 1.
Top ten predicted essential genes not found in OGEE or DEG database.
| Ranking | Gene | Function | Number of references |
P-value* | |
|---|---|---|---|---|---|
| Viable | Lethal | ||||
| 1 | RpL13 | Structural constituent of ribosome | 0 | 0 | – |
| 2 | pnr | Transcription factor; activator of proneural achaete-scute complex genes | 11 | 37 | <0.001 |
| 3 | Prosα4 | Proteasome α4 subunit | 7 | 0 | <0.001 |
| 4 | RpS2 | Ribosomal protein S2 | 2 | 16 | <0.001 |
| 5 | Hsc70-3 | Heat shock 70-kDa protein cognate 3 | 4 | 32 | <0.001 |
| 6 | RpS11 | Ribosomal protein S11 | 2 | 2 | NS |
| 7 | ct | Homeoprotein that functions as a transcriptional factor | 24 | 230 | <0.001 |
| 8 | CG4374 | Transcription factor | 0 | 1 | NS |
| 9 | lilli | Transcription factor | 7 | 32 | <0.001 |
| 10 | N | Notch signaling pathway core component | 30 | 314 | <0.001 |
Significance of enrichment (Fisher’s exact test) of references describing the gene as lethal compared to the number of references describing the gene as viable.
Fig. 5.
Mutations in 771 putatively essential genes (PEGs) are associated with severe phenotype alterations. The odds ratios of significantly (P < 1e−15) over- and under-represented prominent phenotypic descriptions obtained from FlyBase are shown. The error bars show the 95% confidence intervals.
Furthermore, we investigated the ten PEGs with the highest essentiality score according to our machines (listed in Table 1). To estimate their essentiality/importance for viability, we again went into FlyBase and counted how many references labeled a mutation in the according gene as lethal or viable. Of the top 10 PEGs, seven (pnr, Prosα4, RpS2, Hsc70-3, ct, lilli and N) were significantly more often found to be lethal than viable. Two genes did not yield significant results. The gene with our highest essentiality score was RpL13, which is a central structural component of the ribosome. For this gene no statistics could be obtained as a phenotype due to its mutation has not been described yet.
In summary, PEGs are associated with increased lethality, reduced viability and severe phenotypic representations such as a severely altered anatomy. This highlights the indispensable functions of most of the identified PEGs in D. melanogaster for viability, development and fitness. Most of the predictions of false positives rather hint to essentiality than non-essentiality supporting the strategy of our approach and suggesting further, more detailed experimental investigations on the essentiality of these genes.
3.4. Prediction of essential genes in human
In order to test if our comprehensive feature selection approach performs in other eukaryotes as good as in D. melanogaster, we performed the same workflow on human data. OGEE [19] contains essentiality information for 21,556 human genes assembled from 18 studies. Of these only 183 were labeled to be “essential”, 14,388 “non-essential”, and 6985 were labeled “conditional”. “Conditional” stands for contradicting information from different studies. In order to include more genes in the analysis a majority voting scheme for essentially labeling was performed. For this, all genes analyzed by at least 5 studies and 3 times more often reported to be “essential” than “non-essential” were considered to be essential. This led to 833 essential genes and 13,743 non-essential genes for which all features could be generated as for D. melanogaster (described in Section 2)
Fig. 6A shows the performance of the five classifiers on the training and testing sets in human. Incorporation of a variety of features surpass usage of protein sequence features alone. Overall, the performance was high for all algorithms and the classifier with the best performance was, as in D. melanogaster, XGB. XGB achieved a ROC-AUC of 0.969 and a PR-AUC of 0.729. The benchmarking approach by Campos et al. [14] based on protein sequence features led to a much lower ROC-AUC of 0.699 and a PR-AUC of 0.155. In Fig. 6B the ROC- and the PR-curves of the best performing classifier (XGB) are shown. As for D. melanogaster, features from all categories (except co-expression network features) were selected by ElasticNet (Fig. S2). A difference between D. melanogaster and human was the proportion of selected features in the categories. In human the most often selected category was “Protein sequence”, whereas in D. melanogaster the most often selected category was “Gene sequence”. A collection of all essential gene predictions in human is shown in Table S2.
Fig. 6.
Machine learning using a broad range of features leads to high classification performance in human. (A) Heatmaps showing accuracy metrics for the performance evaluation of essential gene classification in human. Five ML approaches were used (Generalised linear model [GLM], Support-Vector Machines [SVM)], Neural Network [NNET], Random Forests [RF], and Extreme Gradient Boosting [XGB]). The performance was measured for the training and testing sets. Features were generated following our new approach including a broad range of features (All Features) and the performance was benchmarked to protein sequence features only (protr). The algorithm with the best performance (highest harmonic mean) was XGB, indicated by a green frame. (B) Receiver operating characteristic and Precision-Recall curves from XGB classifier measured on the testing sets. Using all features (red) yielded distinctively better results compared to the results from the protein sequence features only (grey). Results from a random classification is indicated by dotted lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
When predicting human essential genes with the classifier trained on the D. melanogaster data a ROC-AUC of 0.75 was achieved (compared to 0.58 when using protein sequence features only). The prediction of D. melanogaster essential genes with the human classifier yielded a ROC-AUC of 0.61 (compared to 0.54 when using protein sequence features only) indicating that our approach outperformed protein features only. Furthermore, these results show that essential gene prediction worked well for predicting essential genes in human and there might the possibility to use a classifier across organisms.
4. Discussion
Recently, Campos et al. [14] made methodologically an intriguing contribution towards essentiality predictions using features from protein sequences. Their model achieved a good prediction performance for D. melanogaster. We used this work for benchmarking our results (Figs. 3 and 6). In the present study, we demonstrated that a well-defined and elaborated assembly of intrinsic and extrinsic features from a large range of sources covering a broad range of intrinsic and extrinsic aspects of a gene considerably outperforms the approach based solely on protein sequence features. Additionally, a better performance for D. melanogaster was achieved when combining the complementary databases OGEE and DEG reflecting the more comprehensiveness of the gold standard.
Our classifier performed very well in terms of ROC-AUC in D. melanogaster. Notably, the class distribution in our study was very skewed (ratio of 1:27, 441 essential versus 11,788 non-essential genes when using the gold standard based on OGEE + DEG). Hence, regarding the ROC-AUC alone may not be sufficient to estimate the performance of a model on such an imbalanced dataset [54]. Accordingly, we also estimated the performance by regarding the PR-AUC basing on precision and recall, hence not balancing for the classes. The best performance in terms of PR-AUC was 0.296 (Fig. 3A). To get a correct perspective for this value, one may compare it to results based on random guessing. Random guessing would yield a PR-AUC of 0.036 for D. melanogaster, an indeed considerably lower value (Fig. 3C).
Furthermore, it could be shown that the same approach for feature generation and selection as used for D. melanogaster led to very good performances for human i.e. ROC-AUC = 0.969 and PR-AUC = 0.729 (Fig. 6A and B). Interestingly, features from most categories (except co-expression network features) were selected to be most discriminative in D. melanogaster and human (Figs. 1B and S2). Overall, the good results for two distantly related species indicate the generalizability of our approach.
In total 1195 genes were predicted to be essential in D. melanogaster containing also 771 false positives. These “false positives” led to the relatively low PR-AUC. Assuming them to be enriched in new essential genes, we denoted them as putative essential genes (PEGs) and surveyed the properties of the phenotypes caused by mutations in these genes. One reason for the PEGs to be denoted as non-essential is that this information came from a cell line screen [21] that cannot completely capture essentiality on an organism level. Indeed, mutations in these PEGs were significantly more often associated with lethal and significantly less often with viable phenotypes (Fig. 5), compared to non-essential genes. Strikingly, mutations in PEGs led to drastic phenotype alterations (extra or missing anatomical structures) most likely accompanied by considerably reduced fitness, supporting our ML approach. Interestingly, the PEG with the highest score was RpL13 (Table 1). RpL13 is a central structural component of the ribosome and the phenotype of a loss of function of this gene has not been described yet. We suggest further experimental investigations testing the essentiality of this gene. Altogether, these results demonstrate that these PEGs are highly associated with lethality. Moreover, genes might be conditional essential (i.e. essential under experimental, developmental or environmental conditions) meaning that the genes not identified by the studies [21], [22], [23] are not necessarily non-essential in other, maybe even more natural conditions.
When interrogating the literature, besides the work by Campos et al., we found several other very recent studies about essential gene prediction. These studies were not used for benchmarking but to relate the performance of the presented approach (D. melanogaster: ROC-AUC = 0.90 and PR-AUC = 0.30; human: ROC-AUC = 0.97 and PR-AUC = 0.73) and give an overview of existing approaches. In 2017, Guo et al. [13] predicted essential genes in human cells based on nucleotide composition and essentiality information for the genes from DEG [20]. They achieved an ROC-AUC of 0.85. Currently, the same eukaryotic essentiality-related data are present in both OGEE and DEG [14]. The results for human presented in this work are based on OGEE, but 95% of genes had the same essentiality status in both databases. Accordingly, we achieved an ROC-AUC of 0.96 when basing the human gold standard on DEG and including all features and an ROC-AUC of 0.84, when considering DNA sequence based features only. In another study, Azhagesan et al. achieved an ROC-AUC of 0.86 predicting essential genes for various prokaryotes [55]. They used network and sequence-based features. In the same year, Tian et al. predicted genes being essential during the development of mice [56]. They used several intrinsic and extrinsic features resulting in an ROC-AUC of 0.80. Campos et al. achieved an ROC-AUC of 0.85 for human and 0.81 for D. melanogaster. Our approach basing on a comprehensive set of features performs better, but future investigations are necessary covering also other organisms.
When predicting human essential genes with the classifier trained on the D. melanogaster and vice versa we achieved ROC-AUCs of 0.75 and 0.61 respectively. This is significantly higher than using a classifier based on protein sequence features only but it also indicates that essential gene classification works very well within a selected species but the trained machines cannot simply be applied to distant relatives. In order to predict essential genes in a new organism we would recommend training of the classifier directly on the organism at hand.
Another interesting application of the presented approach for human may be the association of predicted essential genes to human diseases. Recently, Zhao et al. analyzed topological features of the top ten human disease genes [57]. They found that, on average, these show a higher betweenness centrality, a smaller average shortest path length, and a smaller clustering coefficient than non-disease genes. In line, we also found topological features related to centrality, such as degree distribution and clustering coefficient to have a high impact on the essentiality predictions. Still, there seems to be a generic, intrinsic difference between essential and disease-causing genes. None of the ten genes investigated by Zhao et al. were annotated to be essential in OGEE. This may indicate that the function of these top ten disease genes can be partly compensated by other genes making them non-essential, but the functional alterations still cause diseases. In the past years, besides OMIM [58], databases like DisGeNET [59] emerged assembling associations of human genes to diseases. As a study for the future, it can be very intriguing to associate sequence, topology and functional features to genetic diseases, allowing the identification of so far non-studied novel mechanisms causing disease manifestations.
5. Conclusion
Using only intrinsic features limit the success for essential gene prediction. The presented approach provides a means to better predict essential genes utilizing a broad collection of intrinsic and extrinsic gene features from a large variety of different data sources. As a case study, the method was applied to predict essential genes in D. melanogaster and in human. The method considerably outperformed a comparable approach reported very recently, which based only on intrinsic features, and has potential to be applied to other organisms.
CRediT authorship contribution statement
Olufemi Aromolaran: Conceptualization, Software, Data curation, Visualization, Writing - original draft, Writing - review & editing. Thomas Beder: Conceptualization, Methodology, Software, Data curation, Visualization, Writing - original draft, Writing - review & editing. Marcus Oswald: Conceptualization. Jelili Oyelade: Supervision. Ezekiel Adebiyi: Conceptualization, Supervision, Writing - review & editing. Rainer Koenig: Conceptualization, Methodology, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (https://www.dfg.de/) within the project KO 3678/5-1, and the German Federal Ministry of Education and Research (BMBF) within the project Center for Sepsis Control and Care (CSCC, 01EO1002 and 01EO1502).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.02.022.
Contributor Information
Olufemi Aromolaran, Email: olufemi.aromolaran@stu.cu.edu.ng.
Thomas Beder, Email: Thomas.Beder@med.uni-jena.de.
Marcus Oswald, Email: Marcus.Oswald@med.uni-jena.de.
Jelili Oyelade, Email: ola.oyelade@covenantuniversity.edu.ng.
Ezekiel Adebiyi, Email: ezekiel.adebiyi@covenantuniversity.edu.ng.
Rainer Koenig, Email: Rainer.Koenig@uni-jena.de.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Nature Putting gene essentiality into context. Nat Rev Genet. 2018;19:1. doi: 10.1038/nrg.2017.141. [DOI] [PubMed] [Google Scholar]
- 2.Lartigue C., Glass J.I., Alperovich N., Pieper R., Parmar P.P., Hutchison C.A. Genome transplantation in bacteria: changing one species to another. Science (80-) 2007;317:632–638. doi: 10.1126/science.1144622. [DOI] [PubMed] [Google Scholar]
- 3.Albalat R., Cañestro C. Evolution by gene loss. Nat Rev Genet (Nature Publishing Group) 2016:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 4.Sharma A.K., Eils R., König R. Copy number alterations in enzyme-coding and cancer-causing genes reprogram tumor metabolism. Cancer Res. 2016;76:4058–4067. doi: 10.1158/0008-5472.CAN-15-2350. [DOI] [PubMed] [Google Scholar]
- 5.Caraballo H., King K. Emergency department management of mosquito-borne illness: malaria, dengue, and West Nile virus. Emerg Med Pract. 2014;16:1–23. [PubMed] [Google Scholar]
- 6.Lanciotti R.S., Kosoy O.L., Laven J.J., Velez J.O., Lambert A.J., Johnson A.J. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyer O. British Medical Journal Publishing Group; 2015. Zika virus spreads across Americas as concerns mount over birth defects. [DOI] [PubMed] [Google Scholar]
- 8.Meyer A., Holt H.R., Oumarou F., Chilongo K., Gilbert W., Fauron A. Integrated cost-benefit analysis of tsetse control and herd productivity to inform control programs for animal African trypanosomiasis. Parasit Vect. 2018;11:154. doi: 10.1186/s13071-018-2679-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sallam M. 2013. INSECT DAMAGE: damage on post-harvest. [Google Scholar]
- 10.Ranson H., N’guessan R., Lines J., Moiroux N., Nkuni Z., Corbel V. Pyrethroid resistance in African anopheline mosquitoes: what are the implications for malaria control? Trends Parasitol. 2011;27:91–98. doi: 10.1016/j.pt.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt M., Hrabcova V., Jun D., Kuca K., Musilek K. Vector control and insecticidal resistance in the African malaria mosquito Anopheles gambiae. Chem Res Toxicol. 2018;31:534–547. doi: 10.1021/acs.chemrestox.7b00285. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X., Acencio M.L., Lemke N. Predicting essential genes and proteins based on machine learning and network topological features: a comprehensive review. Front Physiol. 2016;7:75. doi: 10.3389/fphys.2016.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo F.-B., Dong C., Hua H.-L., Liu S., Luo H., Zhang H.-W. Accurate prediction of human essential genes using only nucleotide composition and association information. Bioinformatics. 2017;33:1758–1764. doi: 10.1093/bioinformatics/btx055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campos T.L., Korhonen P.K., Gasser R.B., Young N.D. An evaluation of machine learning approaches for the prediction of essential genes in eukaryotes using protein sequence-derived features. Comput Struct Biotechnol J. 2019 doi: 10.1016/j.csbj.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng J., Deng L., Su S., Zhang M., Lin X., Wei L. Investigating the predictability of essential genes across distantly related organisms using an integrative approach. Nucleic Acids Res. 2011;39:795–807. doi: 10.1093/nar/gkq784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plaimas K., Eils R., König R. Identifying essential genes in bacterial metabolic networks with machine learning methods. BMC Syst Biol. 2010;4:56. doi: 10.1186/1752-0509-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H., Zhang Z., Jiang S., Li R., Li W., Zhao C. New insights on human essential genes based on integrated analysis and the construction of the HEGIAP web-based platform. Brief Bioinform. 2019 doi: 10.1093/bib/bbz072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plaimas K., Mallm J.-P., Oswald M., Svara F., Sourjik V., Eils R. Machine learning based analyses on metabolic networks supports high-throughput knockout screens. BMC Syst Biol. 2008;2:67. doi: 10.1186/1752-0509-2-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W.-H., Lu G., Chen X., Zhao X.-M., Bork P. OGEE v2: an update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 2016:gkw1013. doi: 10.1093/nar/gkw1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo H., Lin Y., Gao F., Zhang C.T., Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014;42:D574–D580. doi: 10.1093/nar/gkt1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boutros M., Kiger A.A., Armknecht S., Kerr K., Hild M., Koch B. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science (80-) 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 22.Chen S., Zhang Y.E., Long M. New genes in Drosophila quickly become essential. Science (80-) 2010;330:1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spradling A.C., Stern D., Beaton A., Rhem E.J., Laverty T., Mozden N. The Berkeley Drosophila genome project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thurmond J., Goodman J.L., Strelets V.B., Attrill H., Gramates L.S., Marygold S.J. FlyBase 2.0: the next generation. Nucleic Acids Res. 2018;47:D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charif D., Lobry J.R. Springer; 2007. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. Structural approaches to sequence evolution; pp. 207–232. [Google Scholar]
- 26.Xiao N., Cao D.-S., Zhu M.-F., Xu Q.-S. protr/ProtrWeb: R package and web server for generating various numerical representation schemes of protein sequences. Bioinformatics. 2015;31:1857–1859. doi: 10.1093/bioinformatics/btv042. [DOI] [PubMed] [Google Scholar]
- 27.Peden J. Univ Nottingham; 1997. CodonW. [Google Scholar]
- 28.Zhu M., Dong J., Cao D.-S. 2016. rDNAse: R package for generating various numerical representation schemes of DNA sequences. [DOI] [PubMed] [Google Scholar]
- 29.Smedley D., Haider S., Ballester B., Holland R., London D., Thorisson G. BioMart–biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oughtred R., Stark C., Breitkreutz B.J., Rust J., Boucher L., Chang C. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47:D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López Y., Nakai K. Patil A (2015) HitPredict version 4: comprehensive reliability scoring of physical protein–protein interactions from more than 100 species. Database. 2015 doi: 10.1093/database/bav117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murali T., Pacifico S., Yu J., Guest S., Roberts G.G., Finley R.L. DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 2010;39:D736–D743. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X.Y., Xia X.Y. ProNet: biological network construction, visualization and analyses. R Packag version. 2015. p. 1. [Google Scholar]
- 35.Graveley B.R., Brooks A.N., Carlson J.W., Duff M.O., Landolin J.M., Yang L. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown J.B., Boley N., Eisman R., May G.E., Stoiber M.H., Duff M.O. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savojardo C., Martelli P.L., Fariselli P., Profiti G., Casadio R. BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M. The KEGG database. Silico Simul Biol Process. 2002;247:91–103. [PubMed] [Google Scholar]
- 42.Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H. g: Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L., Zhang Y.-H., Wang S., Zhang Y., Huang T., Cai Y.-D. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon N., Friedman J., Hastie T., Tibshirani R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dormann C.F., Elith J., Bacher S., Buchmann C., Carl G., Carré G. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography (Cop) 2013;36:27–46. [Google Scholar]
- 47.Meloun M., Militký J., Hill M., Brereton R.G. Crucial problems in regression modelling and their solutions. Analyst. 2002:433–450. doi: 10.1039/b110779h. [DOI] [PubMed] [Google Scholar]
- 48.Chawla N.V., Bowyer K.W., Hall L.O., Kegelmeyer W.P. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321–357. [Google Scholar]
- 49.Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28:1–26. [Google Scholar]
- 50.Jeong H., Mason S.P., Barabási A.-L., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 51.Guardiola X., Guimera R., Arenas A., Diaz-Guilera A., Streib D., Amaral L.A.N. 2002. [cited 30 Nov 2019] Macro- and micro-structure of trust networks.http://arxiv.org/abs/cond-mat/0206240 Available: [Google Scholar]
- 52.Acencio M.L., Lemke N. Towards the prediction of essential genes by integration of network topology, cellular localization and biological process information. BMC Bioinf. 2009;10:290. doi: 10.1186/1471-2105-10-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lodish H., Berk A., Zipursky S.L., Matsudaira P., Baltimore D., Darnell J. 2000. Mutations: types and causes. [Google Scholar]
- 54.Saito T., Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. Brock G, editor. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azhagesan K., Ravindran B., Raman K. Network-based features enable prediction of essential genes across diverse organisms. Mande SC, editor. PLoS One. 2018;13 doi: 10.1371/journal.pone.0208722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian D., Wenlock S., Kabir M., Tzotzos G., Doig A.J., Hentges K.E. Identifying mouse developmental essential genes using machine learning. DMM Dis Model Mech. 2018;11 doi: 10.1242/dmm.034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao X., Liu Z.-P. Analysis of topological parameters of complex disease genes reveals the importance of location in a biomolecular network. Genes (Basel) 2019;10:143. doi: 10.3390/genes10020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piñero J., Bravo À., Queralt-Rosinach N., Gutiérrez-Sacristán A., Deu-Pons J., Centeno E. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016:gkw943. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.