Abstract
Physiological aging is a complex process, influenced by a plethora of genetic and environmental factors. While being far from fully understood, a number of common aging hallmarks have been elucidated in recent years. Among these, transcriptomic alterations are hypothesized to represent a crucial early manifestation of aging. Accordingly, several transcription factors (TFs) have previously been identified as important modulators of lifespan in evolutionarily distant model organisms. Based on a set of TFs conserved between nematodes, zebrafish, mice, and humans, we here perform a RNA interference (RNAi) screen in C. elegans to discover evolutionarily conserved TFs impacting aging. We identify a basic helix-loop-helix TF, named HLH-2 in nematodes (Tcf3/E2A in mammals), to exert a pronounced lifespan-extending effect in C. elegans upon impairment. We further show that its impairment impacts cellular energy metabolism, increases parameters of healthy aging, and extends nematodal lifespan in a ROS-dependent manner. We then identify arginine kinases, orthologues of mammalian creatine kinases, as a target of HLH-2 transcriptional regulation, serving to mediate the healthspan-promoting effects observed upon impairment of hlh-2 expression. Consistently, HLH-2 is shown to epistatically interact with core components of known lifespan-regulating pathways, i.e. AAK-2/AMPK and LET-363/mTOR, as well as the aging-related TFs SKN-1/Nrf2 and HSF-1. Lastly, single-nucelotide polymorphisms (SNPs) in Tcf3/E2A are associated with exceptional longevity in humans. Together, these findings demonstrate that HLH-2 regulates energy metabolism via arginine kinases and thereby affects the aging phenotype dependent on ROS-signaling and established canonical effectors.
Keywords: Aging, Transcription, ROS, Redox, Arginine kinase, Creatine kinase
Highlights
-
•
A C. elegans RNAi screen identifies conserved aging-related transcription factors.
-
•
Impairment of transcription factor hlh-2 has the most pronounced effect on lifespan.
-
•
C. elegans HLH-2 affects cellular energy homeostasis and redox signaling.
-
•
HLH-2 modulates arginine kinase to interact with downstream longevity pathways.
-
•
Polymorphisms (SNPs) in the hlh-2 orthologue Tcf3/E2A are linked to human longevity.
1. Introduction
Aging is defined as decrease in physiological function over time. Age-related transcriptomic changes, e.g. due to the aberrant action of transcription factors or chromatin modifiers, are proposed to represent a crucial early manifestation of biological aging [[1], [2], [3]]. Therefore, in addition to environmental influences, initial changes in gene expression networks can be considered as one of the main drivers of the aging phenotype and its associated diseases, leading to persistent and detrimental changes in cellular physiology and, ultimately, death. Of note, one of the genes consistently linked to human longevity in genetic association studies is the evolutionarily conserved transcription factor (TF) Foxo3 [[4], [5], [6], [7]], which was also found to promote lifespan-extension in several animal aging models [8]. Likewise, the evolutionarily conserved TF Nrf2, a master regulator of cellular detoxification and stress response pathways, has been shown to positively influence lifespan in different model organisms [9].
As exemplified by Foxo3 and Nrf2, TFs are particularly interesting candidates to consider as trans-species master regulators of aging, since they frequently display considerable evolutionary conservation [10], and can influence complex transcriptional networks of functionally related genes [11,12]. Thus, changes in the regulation and activity of a single TF and/or its accessory proteins have the potential to influence the age-dependent transcriptomic alterations that are occurring in virtually all animal species displaying an aging phenotype [3]. Conceptually, TFs can be classified as pro-aging if their absence or decreased activity benefits health and survival of the organism, while their increased or unregulated activity might lead to harmful consequences. Inversely, anti-aging TFs can be defined by being able to protect organisms from age-dependent physiological decline when increasingly active, while their absence or decreased activity is potentially detrimental.
Whether pro-aging genes in particular exert their detrimental effects due to aberrant developmental processes, stochastically acquired mutations that alter their function, or hypothesized phenomena such as antagonistic pleiotropy, is still open to debate [13]. Pragmatically, the identification of pro- and anti-aging TFs should aid in designing appropriate experiments to better understand the relevance of transcriptomic alterations for organismal aging and is an important step in establishing interventions targeted at aging-associated diseases and aging in general, e.g. by establishing specific TFs as targets for pharmacological manipulation [14].
The nematodal model organism C. elegans has been used in aging research for several decades [15] and thus far served to elucidate a number of evolutionarily conserved molecular pathways, e.g. impaired insulin/IGF1 signalling, that influence life- and healthspan in eukaryotic organisms, including mammals [16]. Most transcription factors acting as regulators of aging known to date were either identified in C. elegans or have subsequently been shown to impact nematodal aging [17]. Therefore, C. elegans is a promising model to identify as yet unrecognized pro- and anti-aging TFs with possible roles conserved across species.
Given the fact that more than 750 individual TFs exist in C. elegans, many of which have potential homologues in other eukaryotic species [18], and that each of these TFs could impact aging upon either increased or impaired activity, it is possible to combine hands-on experimental approaches with sensible pre-selection criteria and appropriate bioinformatic analyses to hone in on candidates of interest. In a previous study and as a basis for the current publication, a total of 3,608 differentially expressed genes during nematodal aging (aging DEGs) were identified [19]. Analysing the promoter regions of such aging DEGs provides one option to use in silico analyses for an initial assesment of potential candidate TFs.
We here focus on a set of 16 TFs evolutionarily conserved across eukaryotic species with known and experimentally confirmed DNA-binding properties at the time of analysis. After verifying that they indeed are predicted to bind to nematodal aging DEGs, as identified before [19], we systematically tested their impact on the aging phenotype of C. elegans upon impairment by RNA interference. Thereby, several novel pro- and anti-aging TF candidates were identified. Focusing on HLH-2, showing the strongest impact on lifespan of all 16 TFs tested, we demonstrate that this particular TF is a negative regulator of aging and healthspan in C. elegans, i.e. a pro-aging TF, and elucidate its ROS-dependent mechanism of action.
2. Materials and methods
2.1. C. elegans strains and maintenance
The following C. elegans strains used for this publication were provided by the Caenorhabditis Genetics Center (CGC at the University of Minnesota, USA): Wild-type N2 (Bristol), EU31 skn-1(zu135) IV/nT1 [unc-?(n754) let-?] (IV; V), EU1 skn-1(zu67) IV/nT1 [unc-?(n754) let-?] (IV; V), PS3551 hsf-1(sy441) I, RB754 aak-2(ok524) X, MQ887 isp-1(qm150) IV, TU282 lin-32(u282) X, GA800 wuIs151 [ctl-1(+) + ctl-2(+) + ctl-3(+) + myo-2p:GFP], CL802 smg-1(cc546) I; rol-6(su1006) II, CL4176 smg-1(cc546) I; dvIs27 [myo-3p:A-Beta (1–42):let-851 3′UTR) + rol-6(su1006)] X, and CL2006 dvIs2 [pCL12(unc-54/human Abeta peptide 1–42 minigene) + pRF4]. For maintenance, nematodes were grown on Nematode Growth Medium (NGM) agar plates in 90 mm petri dishes at 20 °C using E. coli OP50 bacteria as a food source [20]. NGM agar plates, after pouring, were dried at room temperature for 1–2 days and then stored at 4 °C until further use.
2.2. E. coli strains and culturing
E. coli OP50 bacteria (CGC) were streaked out on DYT agar plates and single colonies picked from such plates were cultured overnight at 37 °C and constant shaking in Erlenmeyer flasks containing liquid DYT medium. Bacterial overnight cultures were concentrated by centrifugation for 30 min at 3,200 g and 4 °C. The prepared bacteria were spotted on NGM agar plates and allowed to grow for 16–24 h prior to use.
E. coli HT115(DE3) bacteria (CGC), containing either the L4440 control vector or a L4440-derived RNAi vector, were streaked out on LB agar plates with 100 μg/ml Ampicillin and 12.5 μg/ml Tetracycline and single colonies picked from such plates were cultured overnight at 37 °C and constant shaking in Erlenmeyer flasks containing liquid LB medium with 100 μg/ml Ampicillin. Bacterial overnight cultures were concentrated by centrifugation for 30 min at 3,200 g and 4 °C. The prepared bacteria were spotted on NGM agar plates additionally containing 100 μg/ml Ampicillin and 1 mM Isopropyl-beta-d-thiogalactopyranoside (IPTG) and allowed to grow for 16–24 h prior to use (reagents from AppliChem, Darmstadt, Germany).
2.3. RNA-mediated gene inhibition (RNAi)
For RNAi-mediated gene knockdown experiments, E. coli HT115 bacteria were applied to nematodes by feeding as previously described [21]. The clones for hlh-2/M05B5.5, hlh-15/C43H6.8, mxl-1/T19B10.11, mdl-1/R03E9.1, mxl-3/F46G10.6, lin-32/T14F9.5, ceh-22/F29F11.5, ces-2/ZK909.4, hlh-11/F58A4.7, pha-4/F38A6.1, daf-16/R13H8.1, daf-12/F11A1.3, blmp-1/F25D7.3, skn-1/T19E7.2, argk-1/F44G3.2, let-363/B0261.2, trx-1/B0228.5, and trx-2/B0024.9 were obtained from the Vidal RNAi ORFeome library v1.1 (Thermo Fisher Scientific, Waltham, MA, USA). The clone for efl-1/Y102A5C.18 was constructed by inserting a sequence matching 248 bp of the efl-1/Y102A5C.18 mRNA, starting at position 161, into the L4440 empty vector. The clone for grh-1/Y48G8AR.1 was constructed by inserting a sequence covering most of exon 1 of grh-1/Y48G8AR.1, matching the C. elegans DNA on chromosome I at positions 1,267,785–1,268,085, into the L4440 empty vector. All clones, including the commercially obtained ones, were verified by sequencing prior to use.
Incubation with RNAi clones started 64 h after synchronization of the population, by washing the synchronized, young adult nematodes and then transferring them to the respective treatment plates using S-Buffer [22]. To maintain synchronized populations during long-term experiments, nematodes were washed off the plates into 15 ml tubes every day of the reproductive period, allowed to settle and then washed again repeatedly until the supernatant was free of progeny. The clean nematodes were then transferred to the freshly prepared treatment plates.
2.4. Lifespan assays and compound treatment
All C. elegans lifespan assays were performed at 20 °C according to standard protocols as previously described, explicitly omitting FUdR [21]. Briefly, adult nematodes were allowed to lay eggs for 4 h and the resulting eggs incubated for 64 h at 20 °C on NGM agar plates inoculated with OP50 to obtain a synchronized population of young adult nematodes. For a typical lifespan assay, 250 young adult nematodes per condition were manually transferred to NGM agar plates (50 nematodes per 55 mm petri dish) that were inoculated with the respective E. coli bacteria as indicated. For the first 10–12 days, nematodes were transferred daily and afterwards every 2–3 days. Nematodes showing no reaction to gentle stimulation were scored as dead. Nematodes that crawled off the plates or that displayed internal hatching or a protruding vulva were censored.
N-acetyl cysteine (NAC) and butylated hydroxy-anisole (BHA) were dissolved in water (for NAC, 500-fold stock solution, 500 mM) and DMSO (for BHA, 1,000-fold stock solution, 25 mM), respectively. Nematodes were propagated on agar plates containing the respective antioxidants or a vehicle control for two generations before start of the respective interventions.
2.5. Aging pigment analysis
Nematodes were synchronized and treated for 10 days with either control vector or hlh-15/C43H6.8 or lin-32/T14F9.5 RNAi starting at L4 larvae stage. On day 10, nematodes were washed off the plates and distributed on 8 wells of a 96-well plate (Bioswisstec 96-well CG black with glass bottom, #5241). Age pigment auto-fluorescence was measured using a fluorescence plate reader (FLUOstar Omega, BMG Labtech, Offenburg, Germany) (excitation/bandpass: 340/10 nm, emission: 440/80 nm). We normalized the age pigment fluorescence to the stable auto-fluorescence signal of the nematodes (excitation/bandpass: 290/10 nm, emission: 330/10 nm) as described [19].
2.6. Fertility assay
To determine fertility, nematodes were synchronized as described above. Single L4 larvae were transferred onto individual plates inoculated with bacteria carrying the control vector or hlh-2/M05B5.5 or lin-32/T14F9.5 RNAi bacteria (10 plates per condition) and subsequently onto fresh plates every day of the reproductive period. Progeny were allowed to hatch and counted manually.
2.7. Locomotion assay
Nematodes were synchronized and treated for 5 days with either control vector L4440 or hlh-2/M05B5.5, lin-32/T14F9.5, or argk-1/F44G3.2 RNAi bacteria. After 5 days, 10–15 nematodes were transferred to fresh plates and 30 s video clips were recorded (Leica M165FC microscope with Leica camera DFC 3000G). Subsequently, 20 independent videos per condition were analyzed regarding the number of body bends per worm.
2.8. Amplex Red-based quantification of hydrogen peroxide
Synchronized nematodes from 5 plates were washed with S-buffer, pooled, and incubated in an Eppendorf tube in 100 μl of 50 mM sodium-phosphate buffer, pH 7.4, with 100 μM Amplex Red (Invitrogen, Carlsbad, USA) and 0.2 U/ml of horseradish peroxidase. Following incubation in the dark for 3 h with constant gentle shaking, fluorescence intensity in the supernatant was measured (FLUOstar Omega, BMG Labtech, Offenburg, Germany; excitation/bandpass: 544/10 nm, emission: 590/10 nm). Afterwards, nematodes were washed with S-buffer, sonicated on ice, and centrifuged at 12000 g for 15 min at 4 °C. The protein content was determined for normalization using a standard BCA assay.
2.9. ATP determination
Nematodes were harvested and immediately shock frozen in liquid nitrogen. The frozen pellet was ground in a liquid nitrogen-chilled mortar to a fine powder. A solution of 4 M Guanidinium-HCl was freshly prepared, heated to 100 °C, and then mixed with the frozen powder to abolish ATPase activity and to further lyse the samples. The mixture was boiled for 15 min at 100 °C and then immediately centrifuged for 30 min at 13,200 g at 4 °C. The supernatant was diluted with ddH2O at a 1:200 ratio and analyzed using a commercially available kit (CellTiter Glo; Promega, Fitchburg, WI, USA) according to the manufacturer's instructions. For normalization of the luminescence signal, protein was determined using a standard BCA assay.
2.10. RNA extraction and RT-qPCR
Total RNA was isolated using QIAzol (Qiagen, Hilden, Germany) based on the phenol-chloroform extraction method. Concentration of the resulting RNA was measured using an LVis Plate on a CLARIOstar microplate reader (BMG LABTECH, Ortenberg, Germany) and stored at −80 °C until further use. Reverse transcription of RNA to cDNA was performed using the High-Capacity cDNA Reverse Transcription Kit together with Oligo(dT) primers following the manufacturer's protocol. Quantitative real-time PCR was carried out on a ViiA 7 Real-Time PCR System using the SYBR Select Master Mix in 384 well plates according to the manufacturer's instructions, and with the recommended cycling conditions (all reagents from Thermo Fisher Scientific, Waltham, MA, USA). Samples were analyzed by the ddCT method, with normalization to the reference gene Y45F10D.4 [23].
The following primers were used for this study:
hlh-2/M05B5.5 primers.
FWD TGCCTGATTATTGGAGTGGA;
REV CTACGGAAGCAGGGGTTTC;
lin-32/T14F9.5 primers.
FWD TCAGTCAGACCAACCATGAGC;
REV GGCTTTGCAGTGGAGTTGTC;
argk-1/F44G3.2 primers.
FWD CGGCTCCAAACAGCATGAAG;
REV TTTTGCACCATCAGGACCGT;
argk-2/W10C8.5 primers.
FWD CACTCTCACTCCACAGCAACTT;
REV TCCGAGCTTTGTACGCTTAGT;
argk-5/ZC434.8 primers.
FWD GAACGAACCGCCTGTTACAC;
REV TCCTTGGAGTTTCGTGTAGACTT;
2.11. Next-generation sequencing (RNAseq)
Total RNA was inspected for degradation using Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). For library preparation an amount of 2 μg of total RNA per sample was processed using Illumina's TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer's instruction. Sequencing was done on a HiSeq2500 in SR/50 bp/high output mode. Libraries were multiplexed in five per lane. Sequencing yielded ~35 Mio reads per sample.
Sequence data were extracted in FastQ format and used for mapping. The FastQ files were mapped using Tophat versus the reference genome obtained from Ensembl [24]. Uniquely mapped reads were counted for all genes using featureCounts [25]. RPKM values were computed using exon lengths provided by featureCounts and the sum of all mapped reads per sample. DEGs were identified using edgeR [26] and DESeq2 [27]. Both packages provide statistical routines for determining differential expression in gene expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted using the Benjamini and Hochberg approach to control for the false discovery rate (FDR). If both FDR values (by edgeR and DESeq2) were below P = 0.05, genes were counted as differentially expressed.
The datasets generated for this study can be found in the NCBI's Gene Expression Omnibus, GEO Series accession number GSE144118.
2.12. Promoter analysis
A FASTA file containing 1 kb upstream of the ATG of each differentially expressed nematodal gene [19], as a set of putative promoter regions, was obtained using WormMart. The sequence files were scanned for one or more matches to the position weight matrices using the matrix-scan function of the pattern-matching program RSAT [28]. The position-specific scoring matrices contain a nucleotide frequency at each position within the binding sites and were obtained from either the Jaspar database [29,30] and/or individual publications (see Table S1). The threshold P-value, which indicates the risk of false positive predictions, was set to 0.0001.
2.13. Tcf3 polymorphism analysis
A detailed description of the cohorts used and the analysis itself can be found in the 2017 study of Ben-Avraham and colleagues [31]. Briefly, participants were recruited through publicity, and stated age was verified by checking birth certificates or U.S. passports in all participants. Medical history, demographic characteristics, and clinical data were obtained uniformly using a structured questionnaire. All participants underwent a physical examination and provided a blood sample. Informed written consent was obtained in accordance with the policy of the Committee on Clinical Investigation of the Albert Einstein College of Medicine.
2.14. Statistical analyses
Statistical analyses for all data except for lifespan assays was carried out using Student's t-test (unpaired, two-tailed) or ANOVA after testing for equal distribution of the data and equal variances within the data set. If the samples had unequal variance, the data was log-transformed before carrying out Student's t-test.
Experiments were performed in triplicate except where stated otherwise. For comparing significant distributions between different groups in the lifespan assays the statistical calculations were performed using JMP software version 9.0 (SAS Institute Inc., Cary, North Carolina, USA), applying the log-rank test. All of the other calculations were performed using Excel 2010 (Microsoft, Albuquerque, NM, USA). A P-value <0.05 was considered statistically significant. Data are expressed as mean ± standard deviation (SD) unless otherwise indicated.
3. Results
3.1. Identification of evolutionarily conserved transcription factors impacting C. elegans aging
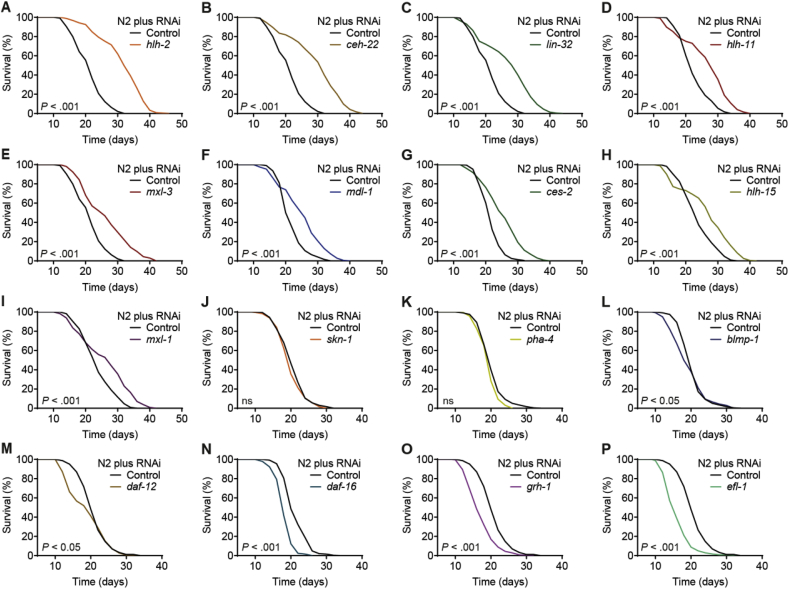
We focused on transcription factors (TFs) that are evolutionarily conserved between C. elegans, zebrafish, mice, and humans, and only considered those with experimentally verified nematodal position weight matrices (PWMs) available at the time of our analysis, which resulted in a shortlist of 16 candidate TFs. Scanning the promoter regions of 3,608 differentially expressed genes during nematodal aging (aging DEGs) [19] with the respective C. elegans PWMs predicted the different TFs to regulate between 16%–34% of these genes (Table S1). Each individual TF was then impaired by RNA interference (RNAi) beginning post-developmentally in young adult worms and effects on lifespan were assayed. Fig. 1 depicts representative outcomes of individual experiments. Detailed lifespan data and statistical analyses of all lifespan experiments performed in this study are given in Table S2.
Fig. 1.
Identification of evolutionarily conserved transcription factors impacting C. elegans aging. (A–P) Representative outcomes of lifespan assays following post-developmental impairment of the indicated transcription factors by RNAi in nematodes. All assays used wild-type N2 nematodes and control in each case refers to treatment with the L4440 control vector. P-values are as indicated in the graphs. See Table S2 for corresponding detailed data and statistical analyses of lifespan assays.
Individual impairment of nine TFs, namely ceh-22, ces-2, hlh-2, hlh-11, hlh-15, lin-32, mdl-1, mxl-1, and mxl-3, significantly extended mean lifespan compared to control (Fig. 1A–I). To date, four of these TFs, i.e. HLH-15 19, MDL-1 and MXL-1 [32], as well as MXL-3 [33], have been previously linked to the regulation of aging in nematodes. Impairment of two C. elegans TFs previously described to act as regulators of longevity, namely skn-1 [34] and pha-4 [35], were found to have no effect on lifespan in our experiments, i.e. when being impaired in the post-developmental state (Fig. 1J and K). RNAi impairment of the remaining five TFs (blmp-1, daf-12, daf-16, efl-1, and grh-1) significantly reduced the lifespan of C. elegans (Fig. 1L–P). Out of these, BLMP-1 [36], DAF-12 [37,38], and DAF-16 [39] have previously been shown to affect lifespan in a similar way as observed here.
Overall, our initial experiments revealed that 14 of the tested TFs impact aging in C. elegans when impaired by RNAi. The strongest lifespan-extending effects were observed upon impairment of hlh-2 and ceh-22, while interference with efl-1 and grh-1 expression led to the most pronounced lifespan-shortening effects. Notably, at the time of our analysis, none of these TFs had so far been linked to the regulation of lifespan. Thus, HLH-2 (mammalian Tcf3/E2A) and CEH-22 (mammalian Nkx2-2) are promising novel candidates to act as potent pro-aging TFs in C. elegans, whereas EFL-1 (mammalian E2f4) and GRH-1 (mammalian Grhl1) may oppose aging in this organism.
In the present study, we focused in detail on the transcription factor HLH-2, which showed the strongest relative effect on lifespan upon RNAi impairment. Interestingly, HLH-2 is known to form heterodimers with at least 14 other bHLH TFs in C. elegans [40], two of which, LIN-32 (mammalian Atoh1) and HLH-15 (mammalian Nhlh1), were also part of our analysis and likewise extended lifespan when impaired (Fig. 1C and H, and for HLH-15 reference [19]), with lin-32 RNAi having a more pronounced effect.
3.2. The transcription factor HLH-2 is a negative regulator of C. elegans healthspan
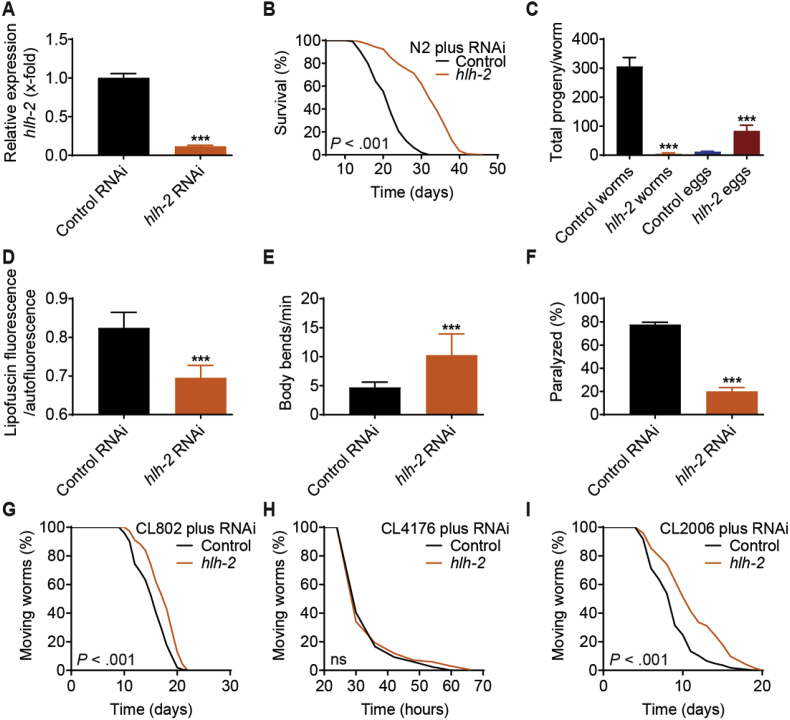
RNAi impairment of hlh-2 in wild-type nematodes reduced hlh-2 expression on the mRNA level by almost 90% when compared to feeding with the L4440 control vector (Fig. 2A). This reduced hlh-2 expression was accompanied by a significantly increased lifespan in several independent experiments (Figs. 1A and 2B and Table S2). Impairment of hlh-2 was further found to affect fertility of the nematodes, increasing the number of unhatched eggs laid per worm in a 24 h time period and greatly decreasing the number of live progeny (Fig. 2C). This demonstrates a reduced fertility of nematodes treated with hlh-2 RNAi and reflects the relevance of this TF for early development. To exclude the possibility that nematodes treated with hlh-2 RNAi, while clearly being longer lived than their control-treated counterparts, are generally less fit, i.e. live longer simply due to developmental defects causing a prolonged sickspan, we next assayed several parameters associated with healthy aging.
Fig. 2.
The transcription factor HLH-2 is a negative regulator of C. elegans healthspan. (A–E) Experimental outcomes of wild-type N2 nematodes treated with hlh-2 RNAi, analyzed regarding (A) hlh-2 mRNA levels as determined by RT-qPCR (n = 3), (B) effect on lifespan in a representative assay (also see Fig. 1A), (C) fertility as determined by counting the number of live progeny and unhatched eggs per worm (n = 10), (D) quantification of “aging pigments” by measuring lipofuscin fluorescence normalized to autoflourescence (n = 8), and (E) locomotion as determined by the number of body bends per min (n = 30). (F) Percentage of paralyzed AM44 mutant worms after treatment with hlh-2 RNAi (n = 30). (G–I) Effect of hlh-2 RNAi on lifespan of the indicated C. elegans strains used as a model for Alzheimer's disease (control strain CL802, rapid paralysis strain CL4176, slow paralysis strain CL2006). Control in all cases refers to treatment with the L4440 vector. P-values for lifespan assays are as indicated in the graphs. See Table S2 for corresponding detailed data and statistical analyses of lifespan assays. Data in histograms are represented as mean ± SD.
ns P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Comparing age-matched nematodes treated with either hlh-2 RNAi or the L4440 control, we found that impairment of hlh-2 reduced the degree of intestinal autofluorescence (Fig. 2D), used as a marker for health and rate of aging in C. elegans [41,42]. Furthermore, hlh-2 RNAi-treated animals were more active than control treated ones, displaying an increased number of body bends over time (Fig. 2E). In a C. elegans mutant model of polyglutamine neurodegenerative diseases, in which polyQ proteins are expressed throughout the nematodal nervous system of the strain AM44, treatment with hlh-2 RNAi was found to protect from paralysis-inducing effects (Fig. 2F). Additionally, in a set of C. elegans strains used as a model for Alzheimer's disease [43,44], we found that impairment of hlh-2, while only mildly affecting the control strain CL802 (Fig. 2G) and having no impact on the rapid paralysis strain CL4176 (Fig. 2H), had beneficial effects on the lifespan of the slow paralysis strain CL2006, which expresses human Aβ1-42 under control of a muscle-specific promoter (Fig. 2I). Overall, these findings indicate that impairment of hlh-2 in C. elegans not only prolongs lifespan, but also has pronounced health-beneficial effects for the organism while negatively affecting its fertility, implicating a trade-off between individual healthspan and overall reproductive fitness. In designing mid-to late-life interventions aimed at staving of the effects of aging and its associated diseases, this is clearly a point that warrants careful consideration when attempting to translate such findings to, ultimately, humans.
Notably, RNAi impairment of the C. elegans bHLH TF lin-32, a known interaction partner of HLH-2 [40], also led to a pronounced lifespan extension in our initial experiments (Fig. 1C). This prompted us to investigate whether lin-32 RNAi had similar effects on healthspan as observed for hlh-2 RNAi and whether these TFs possibly act in concert as pro-aging TFs. Impairment of lin-32 expression in C. elegans had, overall, very similar health-beneficial effects to those observed for treatment of nematodes with hlh-2 RNAi (Fig. S1). However, and clearly distinct from the findings for hlh-2 (Fig. 2C), we did not observe any reduction in fertility of nematodes treated with lin-32 RNAi when compared to control (Fig. S1C). Furthermore, hlh-2 RNAi applied to a lin-32 deletion strain could still extend nematodal lifespan (Fig. S1J) and co-feeding of hlh-2 and lin-32 RNAi to the C. elegans wild-type strain revealed additive effects on lifespan (Fig. S1K). In other words, simultaneous knockdown of both TF together extended lifespan in our co-feeding experiment to a greater extent than knockdown of each TF alone. This indicates that, while both TFs have comparable effects on nematodal life- and healthspan, they act in a distinct fashion. We thus decided to focus on HLH-2 as our main candidate, for the reasons stated above.
The choice of HLH-2 is further supported by the fact that within the genome of a previously described cohort of Ashkenazi-Jewish centenarians [6,31,45,46], several single-nucleotide polymorphisms (SNPs) within the mammalian HLH-2 homologue Tcf3/E2A display significantly different frequencies when compared to control individuals (Table S3), indicating a potential link of this TF to the human aging phenotype. It will be interesting to characterize the putative functional relevance of these SNPs on the expression and/or function of Tcf3/E2A in more detail. While two of the SNPs are located within coding exons of Tcf3/E2A, two others are located within an intronic region. Interestingly, intronic SNPs of the TF Foxo3 that are associated with human longevity have previously been shown to impact Foxo3 gene expression [47].
3.3. Impairment of hlh-2 reduces cellular ATP and generates a health-beneficial ROS signal
Having confirmed that knockdown of hlh-2 leads to positive effects on C. elegans life- and healthspan, we sought to better understand the cellular and molecular basis for this phenomenon. Low degrees of metabolic stress, e.g. in the form of mild energy depletion or ROS generation, have repeatedly been shown to be associated with benefical outcomes through activation of protective pathways, while more severe degrees of metabolic stress usually have harmful consequences [48,49].
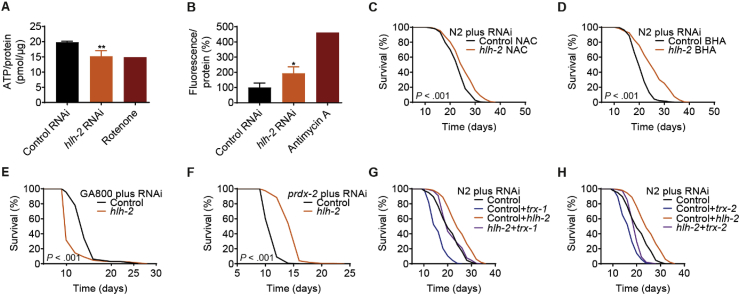
We observed that treatment with hlh-2 RNAi caused a significant drop in ATP levels (Fig. 3A), notably to a similar degree as treatment with the complex I inhibitor rotenone, which has been shown to extend lifespan of C. elegans [50] and N. furzeri [51] at low doses. In addition, hlh-2 RNAi caused an increase in H2O2 formation, as quantified by the Amplex Red assay (Fig. 3B). As cellular ROS and in particular hydrogen peroxide, aside from acting as damaging agents above certain thresholds, often serve an important signaling function [48,52], we next analyzed whether H2O2 generated upon impairment of hlh-2 played a role in mediating the observed lifespan extension.
Fig. 3.
Impairment of hlh-2 reduces cellular ATP and generates a health-beneficial ROS signal. (A–D) Experimental outcomes of wild-type N2 nematodes treated with hlh-2 RNAi, analyzed regarding (A) ATP levels normalized to protein levels (n = 3), with Rotenone used as a positive control, (B) percentage Amplex Red-based fluorescence normalized to protein levels (n = 3) as a measure of hydrogen peroxide quantity, with Antimycin A used as a positive control, and (C) effect on lifespan in the presence of the antioxidant NAC or (D) BHA. (E) Effect of hlh-2 RNAi on the lifespan of the catalase-deficient strain GA800. (F) Effect of hlh-2 RNAi on the lifespan of a strain deficient for the nematodal peroxiredoxin prdx-2. (G) Lifespan assay of wild-type N2 nematodes upon co-feeding of RNAi against hlh-2 and the nematodal thioredoxin trx-1 or (H) trx-2. Control in all cases refers to treatment with the L4440 vector. P-values for lifespan assays are as indicated in the graphs or described in detail in Table S2. Data in histograms are represented as mean ± SD.
ns P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Co-treatment of nematodes with hlh-2 RNAi and two different compounds with well described antioxidative properties, namely N-acetyl cysteine (NAC) and butylated hydroxyl-anisole (BHA), showed that NAC strongly reduced the lifespan-extending effect upon hlh-2 knockdown (Fig. 3C), whereas BHA had a smaller but still noticeable impact in reducing the extended lifespan of hlh-2 RNAi treated animals (Fig. 3D). Interestingly, in the already short-lived mutant strain GA800, deficient for all three C. elegans catalase homologues, hlh-2 RNAi further reduced lifespan (Fig. 3E). These observations support the notion that H2O2 generated following hlh-2 knockdown is, at least partially, necessary to induce the observed phenotype. In the absence of catalases, the main cellular H2O2 detoxifying enzymes, damage caused by H2O2-derived hydroxyl radicals likely outweighs the beneficial effects of ROS-induced stress response pathways, parts of which are the catalases themselves. Interestingly, in a short-lived peroxiredoxin 2 C. elegans deletion strain, hlh-2 RNAi had almost the same relative lifespan-extending effect as in wild-type nematodes (Fig. 3F). Simultaneous knockdown of hlh-2 and thioredoxin 1 or 2, which are mainly involved in the reduction of cysteine disulfide bridges in oxidatively damaged proteins, either not at all (Fig. 3G) or only partially (Fig. 3H) blocked the relative lifespan-extending effect of hlh-2 RNAi. This indicates, that in the presence of catalases and possibly other primary ROS-detoxifying enzymes, ROS-derived protein damage upon hlh-2 knockdown only occurs to a minor extent.
Taken together, these results demonstrate that treatment with hlh-2 RNAi affects cellular energy and ROS homeostasis in C. elegans, and indicate that H2O2 likely serves an important function as a signaling molecule mediating the lifespan-extension resulting from impaired hlh-2 expression.
3.4. Arginine kinases are a target of HLH-2 and their impairment has similar effects on phenotype
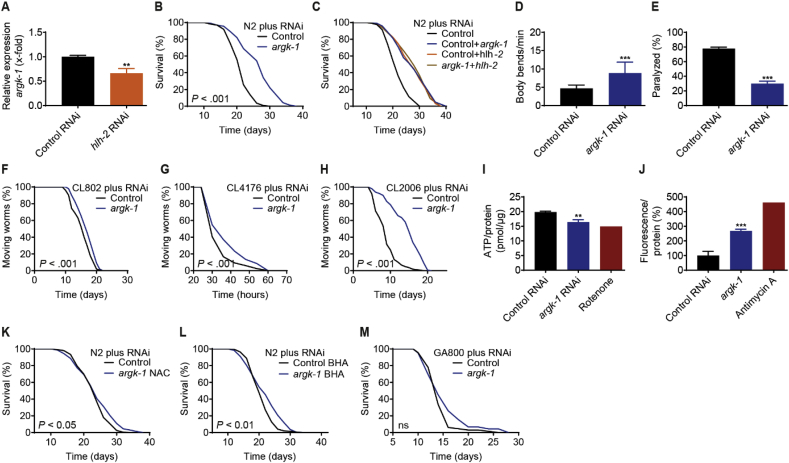
While HLH-2 has been analyzed in terms of its interacting transcription factors [40], only very limited information is available on its target genes. For an unbiased identification of HLH-2 target genes, we performed RNAseq of nematodes treated with hlh-2 RNAi compared to L4440 control. Also included were nematodes treated with RNAi against lin-32, a known interacting transcription factor of HLH-2 40, as we here have demonstrated that lin-32 knockdown leads to similiar outcomes on life- and healthspan as knockdown of hlh-2 (Fig. S1). The top downregulated transcript upon both hlh-2 or lin-32 impairment was that of the arginine kinase argk-1 (Table S4), suggesting that argk-1 is a target gene of HLH-2 transcriptional regulation. Possibly confounding this result to some extent, it should be noted that five different arginine kinase homologues with very similar sequence properties are known to exist in C. elegans [53]. Indeed, the transcript levels of the arginine kinases argk-2, argk-3, argk-4, and argk-5 were all found to be slightly downregulated upon impairment of hlh-2 or lin-32, but only that of argk-2 significantly so under both conditions and that of argk-3 only upon lin-32 impairment (Table S4). To further address this issue experimentally, we assayed the effect of the commercially available argk-1 RNAi clone on the expression of three different C. elegans arginine kinase homologues, i.e. argk-1, argk-2, and argk-5 (Fig. S2). We observed that argk-1 RNAi, while most strongly reducing mRNA levels of argk-1, also significantly impaired expression of argk-2 and argk-5. Therefore, while all following experiments were performed with a clone designated as argk-1 RNAi in the Vidal RNAi ORFeome library v1.1, it should be taken into account that the effects described below may not be specific to the impairment of argk-1 in C. elegans and rather might reflect impairment of nematodal arginine kinases in general.
To confirm that argk-1 is indeed regulated by HLH-2, we assayed argk-1 mRNA levels in nematodes treated with hlh-2 RNAi by qPCR and thereby confirmed a significant reduction in relative argk-1 expression compared to control (Fig. 4A). Next, we assayed the effect of post-developmental argk-1 RNAi treatment on nematodal lifespan and found that it significantly increased lifespan compared to control (Fig. 4B). Notably, co-treatment of wild-type nematodes with hlh-2 and argk-1 RNAi did not lead to a further increase of lifespan compared to knockdown with each RNAi alone (Fig. 4C), indicating that arginine kinases are downstream effectors of the phenotype observed upon hlh-2 impairment. Similar to what was observed for hlh-2 RNAi (Fig. 2), argk-1 RNAi treated animals were more active and protected from polyQ protein induced paralysis (Fig. 4D and E). In the aforementioned model strains for Alzheimer's disease, argk-1 RNAi, again similar to hlh-2 RNAi, markedly prolonged lifespan of the slow paralysis strain CL2006, while not or only slightly affecting CL802 and CL4176 (Fig. 4F–H). Therefore, impaired expression of argk-1 and other arginine kinase homologues has lifespan-extending and overall health beneficial effects in C. elegans, comparable to that following the impaired expression of hlh-2.
Fig. 4.
Arginine kinases are a target of HLH-2 and their impairment has similar effects on phenotype. (A) argk-1 mRNA levels in wild-type N2 nematodes treated with hlh-2 RNAi as determined by RT-qPCR (n = 3). (B) Effect of argk-1 RNAi on the lifespan of wild-type N2 nematodes. (C) Lifespan assay of wild-type N2 nematodes upon co-feeding of RNAi against argk-1 and hlh-2. (D) Effect of argk-1 RNAi on locomotion of wild-type N2 nematodes, as determined by the number of body bends per min (n = 30). (E) Percentage of paralyzed AM44 mutant worms after treatment with argk-1 RNAi (n = 30). (F–H) Effect of argk-1 RNAi on lifespan of the indicated C. elegans strains used as a model for Alzheimer's disease (control strain CL802, rapid paralysis strain CL4176, slow paralysis strain CL2006). (I) ATP levels normalized to protein levels in wild-type N2 nematodes treated with argk-1 RNAi (n = 3), with Rotenone used as a positive control. (J) Percentage Amplex Red-based fluorescence normalized to protein levels, in wild-type N2 nematodes treated with argk-1 RNAi (n = 3), as a measure of hydrogen peroxide quantity, with Antimycin A used as a positive control. (K) Effect of argk-1 RNAi on the lifespan of wild-type N2 nematodes in the presence of the antioxidant NAC or (L) BHA. (M) Effect of argk-1 RNAi on the lifespan of the catalase-deficient strain GA800. Control in all cases refers to treatment with the L4440 vector. P-values for lifespan assays are as indicated in the graphs or described in detail in Table S2. Data in histograms are represented as mean ± SD.
ns P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Invertebrate arginine kinases are the functional orthologues of vertebrate creatine kinases, which all fulfill an essential function in rapid and reversible ATP formation [53,54]. Accordingly, and as for hlh-2 impaiment (Fig. 3A), we observed a decrease in nematodal ATP levels upon impairment of arginine kinases with argk-1 RNAi (Fig. 4I). Furthermore, argk-1 RNAi also increased H2O2 (Fig. 4J), again similar to what was observed for hlh-2 impairment (Fig. 3B). Co-treatment of nematodes with argk-1 RNAi together with NAC or BHA revealed that both compounds almost completely blocked the lifespan-extension following knockdown of arginine kinases (Fig. 4K, L). Lastly, lifespan extension upon argk-1 RNAi was also blocked in absence of catalases in the strain GA800 (Fig. 4 M), whereas hlh-2 RNAi in this strain was found to even reduce lifespan (Fig. 3E).
In summary, these results indicate that arginine kinases are targets of HLH-2 transcriptional regulation, and that impairment of arginine kinases by the argk-1 RNAi resembles the phenotype of hlh-2 RNAi. This places arginine kinases downstream of HLH-2, strongly indicating that the phenotype observed upon hlh-2 RNAi is, at least in part, caused by impaired cellular energy metabolism as a consequence of their reduced expression.
3.5. Lifespan extension following hlh-2 and arginine kinase impairment requires known longevity pathways
Having established that hlh-2 impairment likely induces healthspan extension by a decrease in arginine kinase expression, which in turn results in alterations of energy metabolism and ROS homeostasis, we aimed to further validate these experimental conclusions and to identify downstream effectors necessary for the mediation of the observed health-beneficial effects.
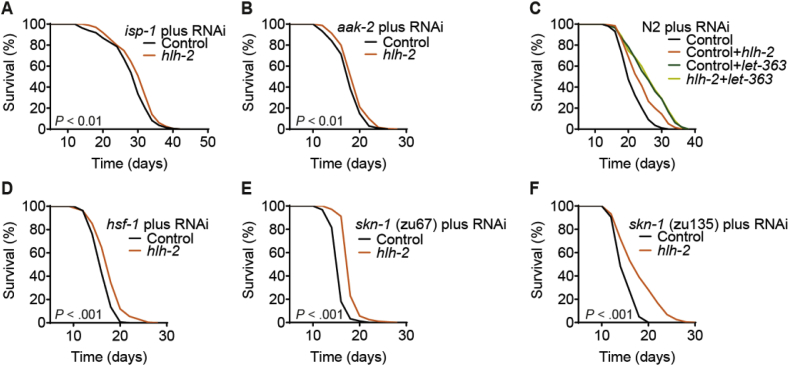
Supporting the notion that a limited energy deficit is causative in lifespan-extension upon hlh-2 impairment, we found that the already extended lifespan of a C. elegans knockout strain of isp-1 [55], an essential component of mitochondrial complex III, was only marginally extended by either hlh-2 (Fig. 5A) or argk-1 RNAi (Fig. S3A). Moreover, in a loss-of-function mutant of aak-2, the C. elegans homologue of the mammalian energy sensor AMP-dependent kinase [56], hlh-2 RNAi only slightly extended lifespan (Fig. 5B) and argk-1 RNAi had no effect (Fig. S3B). Notably, co-feeding experiments of nematodes with RNAi against hlh-2 and let-363, the C. elegans homologue of mTOR, revealed that hlh-2 RNAi was not able to further extend nematodal lifespan upon simultaneous knockdown of let-363, which by itself already has a pronounced lifespan-extending effect [57] (Fig. 5C). Since let-363 is located downstream of aak-2, particularly under conditions of energy deprivation [58], this further supports the notion that the longevity phenotype upon hlh-2 impairment is caused, at least in part, by an impairment in cellular energy metabolism. We further observed that lifespan extension following both hlh-2 and argk-1 RNAi was almost fully dependent on the presence of the known lifespan-regulating transcription factor HSF-1 [59] (Fig. 5D and Fig. S3C) and at least in part on SKN-1 (Fig. 5E, F and Figs. S3D and E), the C. elegans homologue of mammalian Nrf2, another well-established regulator of nematodal lifespan [60], known to be activated by i.a. disturbances in ROS homeostasis [61].
Fig. 5.
Lifespan extension following hlh-2 impairment requires known longevity pathways. (A and B) Effect of hlh-2 RNAi on the lifespan of mutant strains deficient for isp-1 (A) or aak-2 (B). (C) Lifespan assay of wild-type N2 nematodes upon co-feeding of RNAi against hlh-2 and the nematodal mTOR homologue let-363. (D–F) Effect of hlh-2 RNAi on the lifespan of mutant strains deficient for hsf-1 (D) or skn-1 (E and F). Control in all cases refers to treatment with the L4440 vector. P-values for lifespan assays are as indicated in the graphs or described in detail in Table S2.
4. Discussion
Transcriptional regulation of gene expression not only controls cell fate during development [62] and adaptation to changing environmental conditions [63], but also is associated with organismal aging [1,3]. To which extent alterations in the transcriptomic landscape occurring with increasing age represent cause or effect of the aging process seems dauntingly complex to disentangle. Nevertheless, age-related changes in the transcriptome are proposed to represent a crucial early manifestation of biological aging [3] and evidence is accumulating that individual transcription factors can act as evolutionarily conserved master regulators of aging [8,9]. Expanding the repertoire of TFs able to influence aging across species should serve to aid current efforts to define transcriptomic signatures associated with accelerated or delayed aging, to identify additional pathways regulating aging, and to establish pharmacological targets that allow translation of findings in model organisms to human therapy, i.e. to stave of aging-associated diseases and, ultimately, the aging process itself.
Starting with a RNAi screen in C. elegans, we here newly identify several evolutionarily conserved TFs acting in a pro-aging (CEH-22/Nkx2-2, CES-2/Hlf, HLH-2/Tcf3/E2A, HLH-11/Tfap4, LIN-32/Atoh1) or anti-aging (EFL-1/E2f4, GRH-1/Grhl1) fashion in nematodes. It should be noted that in parallel to our initial analysis of these TFs, EFL-1/E2f4 was identified to be involved into the aging process by others [64]. We decided to focus in detail on HLH-2/Tcf3/E2A since it had the most pronounced impact on nematodal lifespan in our screen.
Murine Tcf3/E2A has been described as a transcriptional regulator of anterior-posterior axis development in the early embryo [65] and as an important factor of embryonic stem cell self-renewal and skin cell fate determination [[66], [67], [68], [69]]. Indeed, several transcription factors, such as those regulating Wnt-signalling [70] or of the Fox TF family [71], are recognized to have dual roles in development and in influencing later-life phenotypes, though it is still unclear to what extent this is an evolved feature or an unintended consequence of what has been termed selection shadow. In addition, Tcf3/E2A is an important regulator of B cell development, and altered Tcf3/E2A expression has been hypothesized to contribute to the defects in humoral immunity in senescent mice [72].
In C. elegans, we here show HLH-2 to impact cellular energy and ROS homeostasis, at least in part as a consequence of HLH-2 regulating the expression of arginine kinases, the nematodal equivalents of mammalian creatine kinases. In this regard it should be noted that a previous publication on the effects of argk-1 RNAi on C. elegans lifespan showed different outcomes (i.e., no effect on the lifespan of wild-type animals, whereas the lifespan of rsks-1/S6K mutants was shortened, consistent with phenotypes observed in argk-1 genetic mutants) [54] when compared to our findings as reported herein. One possible explanation for the discrepancy between these results could be the use of different N2 strains, i.e., CGC-derived N2 hermaphrodites in the current publication, versus the use of the N2 Tuck strain in the Hansen lab study (unpublished results). Whether e.g. the recently identified fln-2 mutation contributes to this remains to be identified [73].
Of note, defects in creatine kinase function and altered creatine levels have been, at least tentatively, linked to the aging process [74]. In general, mild levels of oxidative stress and energy depletion, as observed upon impaired hlh-2 expression, have repeatedly been demonstrated as associated with favorable outcomes on lifespan and parameters of healthy aging [21,49,75,76], which is congruent with our finding that RNAi of hlh-2 is health-beneficial. This seemingly paradoxical phenomenon is, according to the current state of research, mainly explained by the activation of different metabolic, stress response, and detoxification pathways in an attempt re-establish cellular homeostasis, which together overcompensate the negative effects of the initial insult and thus produce a net positive outcome for the organism, a process named mitohormesis [77,78]. In line with this interpretation, we observe that the extended lifespans upon both hlh-2 and argk-1 RNAi are epistatically dependent on different established nematodal longevity pathways and aging-related transcription factors. Furthermore, hydrogen peroxide generated upon hlh-2 RNAi is shown to serve an important signalling function mediating lifespan extension.
In summary, we here identify HLH-2/Tcf3/E2A as a novel evolutionarily conserved TF that, at least in the nematodal aging model C. elegans, impacts life- and healthspan. We further establish that the positive effects upon hlh-2 impairment are linked to mild disturbances of cellular energy metabolism by virtue of reduced expression of arginine kinases, necessitate a ROS signal, and are dependent on known longevity pathways. It will be interesting to probe to what extent these findings also hold true for the mammalian HLH-2 homologue Tcf3/E2A, specific variants of which are at least tentatively linked to the aging phenotype of human centenarians (Table S3), and whether this transcription factor can serve as a target for pharmacological manipulation to achieve health-beneficial outcomes.
Author contributions
Mi.R. conceived and supervised the project.
L.R. performed the majority of experiments with additional experiments and data analyses performed by Me.R., G.G., J.M. and K.Z.
N.B. and G.A. analyzed and provided data on Tcf3/E2A SNPs in a centenarian cohort, which was further analyzed by M.C.Z.
L.R. and F.F. analyzed and visualized the data.
F.F. and Mi.R. wrote the manuscript with editorial input from most authors.
Declaration of competing interest
None declared.
Acknowledgements
We thank Dr. Malene Hansen for exchange of materials and performing experiments on argk-1 RNAi (see Discussion section), and for important comments on the manuscript. We thank Dr. Marco Groth for the primary analysis of the raw RNASeq data. C. elegans strains used in this work were provided by the Caenorhabditis Genetics Centre (Univ. of Minnesota, USA), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This study was funded by the following grants: The Swiss National Science Foundation (31003A_176127) (Mi.R.), the Nathan Shock Center of Excellence for the Basic Biology of Aging (P30AG038072) (N.B.), the Einstein-Paul Glenn Foundation for Medical Research Center for the Biology of Human Aging (N.B.), NIH/NIA-1 R01 AG044829 (N.B.), NIH/NIA-1 R01 AG042188-01 (N.B., G.A.), NIH-1 R01 AG 046949–01 (N.B.). The Ristow laboratory is supported by the Horizon 2020 program of the European Union (Ageing with Elegans, grant 633589).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101448.
Contributor Information
Fabian Fischer, Email: fabian-fischer@ethz.ch.
Michael Ristow, Email: michael-ristow@ethz.ch.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Overview of Evolutionarily Conserved Aging-Related Transcription Factors Studied in RNAi Lifespan Assays.
Detailed Information and Statistical Analyses of all Lifespan Assays.
Single-Nucleotide Polymorphisms of Tcf3/E2A in a Centenarian Aging Cohort.
RNAseq Raw Data and Analyses for Differentially Expressed Genes upon hlh-2 and lin-32 RNAi.
References
- 1.Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy B.K. Geroscience: linking aging to chronic disease. Cell. 2014;159:709–713. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booth L.N., Brunet A. The aging epigenome. Mol. Cell. 2016;62:728–744. doi: 10.1016/j.molcel.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawlikowska L. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8:460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budovsky A. LongevityMap: a database of human genetic variants associated with longevity. Trends Genet. 2013;29:559–560. doi: 10.1016/j.tig.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Bae H. Effects of FOXO3 polymorphisms on survival to extreme longevity in four centenarian studies. J. Gerontol. Biol. Med. Sci. 2018;73:1439–1447. doi: 10.1093/gerona/glx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tacutu R. Human ageing genomic resources: new and updated databases. Nucleic Acids Res. 2018;46:D1083–D1090. doi: 10.1093/nar/gkx1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martins R., Lithgow G.J., Link W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 2016;15:196–207. doi: 10.1111/acel.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis K.N., Mele J., Hayes J.D., Buffenstein R. Nrf2, a guardian of healthspan and gatekeeper of species longevity. Integr. Comp. Biol. 2010;50:829–843. doi: 10.1093/icb/icq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nitta K.R. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. eLife. 2015;4 doi: 10.7554/eLife.04837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tonelli C., Chio I.I.C., Tuveson D.A. Transcriptional regulation by Nrf2. Antioxidants Redox Signal. 2018;29:1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milan G. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat. Commun. 2015;6:6670. doi: 10.1038/ncomms7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin K. Modern biological theories of aging. Aging Dis. 2010;1:72–74. [PMC free article] [PubMed] [Google Scholar]
- 14.Tarkhov A.E. A universal transcriptomic signature of age reveals the temporal scaling of Caenorhabditis elegans aging trajectories. Sci. Rep. 2019;9:7368. doi: 10.1038/s41598-019-43075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tissenbaum H.A. Using C. elegans for aging research. Invertebr. Reprod. Dev. 2015;59:59–63. doi: 10.1080/07924259.2014.940470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bitto A., Wang A.M., Bennett C.F., Kaeberlein M. Biochemical genetic pathways that modulate aging in multiple species. Cold Spring Harb. Perspect. Med. 2015;5 doi: 10.1101/cshperspect.a025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uno M., Nishida E. Lifespan-regulating genes in C. elegans. Nature. 2016;8 doi: 10.1038/npjamd.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narasimhan K. Mapping and analysis of Caenorhabditis elegans transcription factor sequence specificities. eLife. 2015;4 doi: 10.7554/eLife.06967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansfeld J. Branched-chain amino acid catabolism is a conserved regulator of physiological ageing. Nat. Commun. 2015;6:e10043. doi: 10.1038/ncomms10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulz T.J. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metabol. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. (07)00256-2 [pii], S1550-4131. [DOI] [PubMed] [Google Scholar]
- 22.Zarse K. Impaired insulin/IGF1-signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metabol. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoogewijs D., Houthoofd K., Matthijssens F., Vandesompele J., Vanfleteren J.R. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 2008;9(9):1471–2199. doi: 10.1186/1471-2199-9-9. 9-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 26.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. btp616 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turatsinze J.V., Thomas-Chollier M., Defrance M., van Helden J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat. Protoc. 2008;3:1578–1588. doi: 10.1038/nprot.2008.97. nprot.2008.97 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Mathelier A. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Avraham D. The GH receptor exon 3 deletion is a marker of male-specific exceptional longevity associated with increased GH sensitivity and taller stature. Sci. Adv. 2017;3:e1602025. doi: 10.1126/sciadv.1602025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson D.W. The Caenorhabditis elegans Myc-Mondo/Mad complexes integrate diverse longevity signals. PLoS Genet. 2014;10:e1004278. doi: 10.1371/journal.pgen.1004278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Rourke E.J., Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 2013;15:668–676. doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackwell T.K., Steinbaugh M.J., Hourihan J.M., Ewald C.Y., Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 2015 doi: 10.1016/j.freeradbiomed.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panowski S.H., Wolff S., Aguilaniu H., Durieux J., Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 36.Greer E.L. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. nature09195 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsen P.L., Albert P.S., Riddle D.L. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerisch B., Weitzel C., Kober-Eisermann C., Rottiers V., Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- 39.Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R.A.C. Elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 40.Grove C.A. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerstbrein B., Stamatas G., Kollias N., Driscoll M. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans. Aging Cell. 2005;4:127–137. doi: 10.1111/j.1474-9726.2005.00153.x. ACE153 [pii] [DOI] [PubMed] [Google Scholar]
- 42.Pincus Z., Mazer T.C., Slack F.J. Autofluorescence as a measure of senescence in C. elegans: look to red, not blue or green. Aging. 2016;8:889–898. doi: 10.18632/aging.100936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Link C.D. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Link C.D. Transgenic invertebrate models of age-associated neurodegenerative diseases. Mech. Ageing Dev. 2001;122:1639–1649. doi: 10.1016/s0047-6374(01)00291-3. [DOI] [PubMed] [Google Scholar]
- 45.Sebastiani P. Limitations and risks of meta-analyses of longevity studies. Mech. Ageing Dev. 2017 doi: 10.1016/j.mad.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deelen J. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat. Commun. 2019;10:3669. doi: 10.1038/s41467-019-11558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flachsbart F. Identification and characterization of two functional variants in the human longevity gene FOXO3. Nat. Commun. 2017;8:2063. doi: 10.1038/s41467-017-02183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ristow M., Schmeisser K. Dose-Response; 2014. Mitohormesis: Promoting Health and Lifespan by Increased Levels of Reactive Oxygen Species (ROS) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gems D., Partridge L. Stress-response hormesis and aging: "that which does not kill us makes us stronger. Cell Metabol. 2008;7:200–203. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Schmeisser S. Neuronal ROS signaling rather than AMPK/sirtuin-mediated energy sensing links dietary restriction to lifespan extension. Mol. Metabol. 2013;2:92–102. doi: 10.1016/j.molmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baumgart M. Longitudinal RNA-seq analysis of vertebrate aging identifies mitochondrial complex I as a small-molecule-sensitive modifier of lifespan. Cell Syst. 2016;2:122–132. doi: 10.1016/j.cels.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 52.Gough D.R., Cotter T.G. Hydrogen peroxide: a Jekyll and Hyde signalling molecule. Cell Death Dis. 2011;2:e213. doi: 10.1038/cddis.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fraga D., Aryal M., Hall J.E., Rae E., Snider M. Characterization of the arginine kinase isoforms in Caenorhabditis elegans. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2015;187:85–101. doi: 10.1016/j.cbpb.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 54.McQuary P.R. C. elegans S6K mutants require a creatine-kinase-like effector for lifespan extension. Cell Rep. 2016;14:2059–2067. doi: 10.1016/j.celrep.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng J., Bussiere F., Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 56.Curtis R., O'Connor G., DiStefano P.S. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. 2006;5:119–126. doi: 10.1111/j.1474-9726.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- 57.Vellai T. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 58.Canto C., Auwerx J. Calorie restriction: is AMPK a key sensor and effector? Physiology. 2011;26:214–224. doi: 10.1152/physiol.00010.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsu A.L., Murphy C.T., Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 60.Tullet J.M. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaspar J.W., Niture S.K., Jaiswal A.K. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwafuchi-Doi M., Zaret K.S. Cell fate control by pioneer transcription factors. Development. 2016;143:1833–1837. doi: 10.1242/dev.133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lopez-Maury L., Marguerat S., Bahler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat. Rev. Genet. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 64.Xie Q. E2F transcription factor 1 regulates cellular and organismal senescence by inhibiting forkhead box O transcription factors. J. Biol. Chem. 2014;289:34205–34213. doi: 10.1074/jbc.M114.587170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merrill B.J. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- 66.Merrill B.J., Gat U., DasGupta R., Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001;15:1688–1705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yi F., Pereira L., Merrill B.J. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cell. 2008;26:1951–1960. doi: 10.1634/stemcells.2008-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen H., Rendl M., Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 69.Pereira L., Yi F., Merrill B.J. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lezzerini M., Budovskaya Y. A dual role of the Wnt signaling pathway during aging in Caenorhabditis elegans. Aging Cell. 2014;13:8–18. doi: 10.1111/acel.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Golson M.L., Kaestner K.H. Fox transcription factors: from development to disease. Development. 2016;143:4558–4570. doi: 10.1242/dev.112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Riley R.L., Blomberg B.B., Frasca D. B cells, E2A, and aging. Immunol. Rev. 2005;205:30–47. doi: 10.1111/j.0105-2896.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- 73.Zhao Y., Wang H., Poole R.J., Gems D. A fln-2 mutation affects lethal pathology and lifespan in C. elegans. Nat. Commun. 2019;10:5087. doi: 10.1038/s41467-019-13062-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sumien N., Shetty R.A., Gonzales E.B. Creatine, creatine kinase, and aging. Subcell. Biochem. 2018;90:145–168. doi: 10.1007/978-981-13-2835-0_6. [DOI] [PubMed] [Google Scholar]
- 75.Bonawitz N.D., Chatenay-Lapointe M., Pan Y., Shadel G.S. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metabol. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Urban N. Non-linear impact of glutathione depletion on C. elegans life span and stress resistance. Redox Biol. 2017;11:502–515. doi: 10.1016/j.redox.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ristow M. Mitohormesis explains ROS-induced health benefits. Nat. Med. 2014;20:709–711. doi: 10.1038/nm.3624. [DOI] [PubMed] [Google Scholar]
- 78.Shadel G.S., Horvath T.L. Mitochondrial ROS signaling in organismal homeostasis. Cell. 2015;163:560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of Evolutionarily Conserved Aging-Related Transcription Factors Studied in RNAi Lifespan Assays.
Detailed Information and Statistical Analyses of all Lifespan Assays.
Single-Nucleotide Polymorphisms of Tcf3/E2A in a Centenarian Aging Cohort.
RNAseq Raw Data and Analyses for Differentially Expressed Genes upon hlh-2 and lin-32 RNAi.