Abstract
Background
Nucleotide metabolism is a critical pathway that generates purine and pyrimidine molecules for DNA replication, RNA synthesis, and cellular bioenergetics. Increased nucleotide metabolism supports uncontrolled growth of tumors and is a hallmark of cancer. Agents inhibiting synthesis and incorporation of nucleotides in DNA are widely used as chemotherapeutics to reduce tumor growth, cause DNA damage, and induce cell death. Thus, the research on nucleotide metabolism in cancer is primarily focused on its role in cell proliferation. However, in addition to proliferation, the role of purine molecules is established as ligands for purinergic signals. However, so far, the role of the pyrimidines has not been discussed beyond cell growth.
Scope of the review
In this review we present the key evidence from recent pivotal studies supporting the notion of a non-proliferative role for pyrimidine metabolism (PyM) in cancer, with a special focus on its effect on differentiation in cancers from different origins.
Major conclusion
In leukemic cells, the pyrimidine catabolism induces terminal differentiation toward monocytic lineage to check the aberrant cell proliferation, whereas in some solid tumors (e.g., triple negative breast cancer and hepatocellular carcinoma), catalytic degradation of pyrimidines maintains the mesenchymal-like state driven by epithelial-to-mesenchymal transition (EMT). This review further broadens this concept to understand the effect of PyM on metastasis and, ultimately, delivers a rationale to investigate the involvement of the pyrimidine molecules as oncometabolites. Overall, understanding the non-proliferative role of PyM in cancer will lead to improvement of the existing antimetabolites and to development of new therapeutic options.
Keywords: Cancer, Pyrimidine metabolism, Epithelial-to-mesenchymal transition, Chemoresistance
Abbreviations: PyM, pyrimidine metabolism; EMT, epithelial-to-mesenchymal transition; CSC, cancer stem cells; dNTP, deoxyribonucleoside triphosphate; NSCLC, non-small-cell lung cancer; TNBC, triple-negative breast cancer; HCC, hepatocellular carcinoma
1. Synthesizing pyrimidines for epithelial-to-mesenchymal transition?
Pyrimidine metabolism (PyM) is a complex enzymatic network that integrates nucleoside salvage, de novo nucleotide synthesis, and catalytic degradation of pyrimidines. Cancer cells, unlike resting cells, rely on the de novo pathway to ensure a continuous supply of deoxyribonucleoside triphosphates (dNTPs) and, thereby, support the uncontrolled tumor growth [1,2]. PyM has been implicated in differentiation of leukemic cells, but not much has been conceptualized in context of the differentiation status of solid tumors [3]. Emerging literature has linked PyM genes to epithelial-to-mesenchymal transition (EMT), a genetic and molecular program that is associated with loss of morphological and/or functional epithelial-like phenotype of solid carcinomas, increased resistance to therapy, and enhanced stem-like features of the cancer cells [4].
EMT observed during cancer progression is an aberrantly reactivated embryonic process that drives loss of differentiation that defines the epithelial lineage, and renders cells in a mesenchymal-like state [5,6]. It is a type-3 EMT, which is functionally distinct from type-1 and type-2 EMT programs observed during embryogenesis and the wound-healing process respectively [7]. EMT is experimentally characterized by change in morphology and assayed by gain of functions such motility and invasiveness [8,9], aggressiveness [[10], [11], [12]], stemness [[13], [14], [15], [16], [17], [18], [19]], and chemoresistance [[20], [21], [22], [23], [24], [25], [26]]. However, in some cases EMT could be restricted to acquisition of a combination of these functional properties in absence of distinct morphological change.
EMT associated with cancer is orchestrated by extracellular factors [e.g., tumor growth factor-beta (TGF-β) [[27], [28], [29]], bone morphogenetic protein (BMP) [30,31], and hypoxia [32]] that activate EMT master transcription factors (TFs) like TWIST [33], SNAI1 [34,35], and ZEB1/2 [6,36]. Several micro-RNAs (miRNAs) also regulate EMT, some of which do so by forming a feedback-inhibition loop with EMT-TFs [37]. One of the most studied feedback loop is mediated by members of the miR-200 family and ZEB1/2 [[38], [39], [40]], but other similar mechanisms have been described to determine EMT-associated alterations in cancer [[41], [42], [43]]. The transcriptional activation of EMT is relayed to an array of diverse downstream effector pathways that are essential for the resultant functional and morphological mesenchymal-like phenotypes. Metabolic adjustments and role of metabolites, especially from the tricarboxylic acid cycle, have been established as one of the important downstream effectors of EMT [[44], [45], [46], [47], [48]]. EMT induction has also been associated with alteration in PyM [49]. However, the exact role of PyM in EMT is not comprehensively understood.
Thus, we revisit the PyM-mediated differentiation model in leukemia, systemically scrutinize the evidence to establish a role of PyM in EMT-driven changes in differentiation in carcinoma and weigh a possibility of a similar mechanism in non-epithelial cancers. We further assess pathological and clinical consequences of differentiation, such as metastasis and chemoresistance, in context of PyM. As a corollary to the established role of PyM enzymes in EMT, we finally aim to propose pyrimidine nucleotides as oncometabolites. Overall, we aim to initiate the follow-up studies that will explore the mechanism behind the PyM in the context of metastasis and chemoresistance and will result in improving therapies that target PyM pathways.
2. Oncogenes upregulate de novo pyrimidine synthesis and the catabolic pathway
2.1. Pathways of pyrimidine metabolism
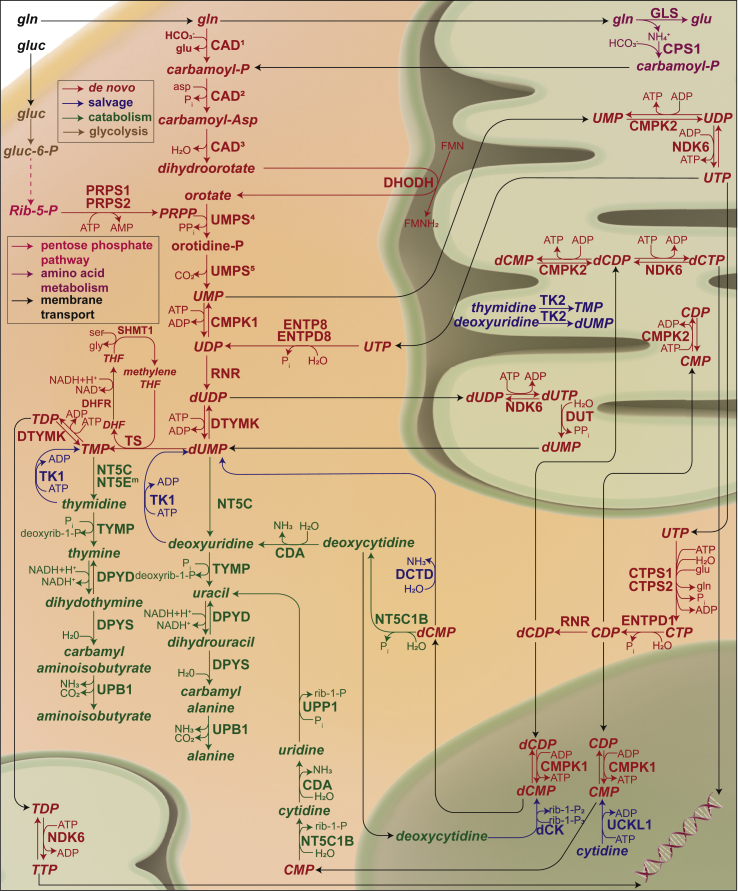
PyM is a branch of nucleotide metabolism that produces nucleosides and deoxy/ribonucleotides of the pyrimidine bases (cytosine, thymine, and uracil). Together with purine metabolism, it generates the deoxyribonucleotide pool required for cell proliferation. PyM consists of three pathways, as follows: 1) salvaging of free nucleosides and bases, 2) the de novo synthesis from amino acids and ribose precursors, and 3) the catabolism of excess nucleoside and nucleotides (illustrated in Figure 1) [50]. Salvaging of bases and nucleosides is a key feature of the terminally differentiated or resting cells [51]. Cells assimilate free nucleosides and pyrimidine bases from the extracellular space and convert them into their respective ribonucleotides and deoxyribonucleotides. The salvage enzymes, such as thymidine kinase, can also convert thymidine from nucleotide catabolism to thymidylate [50].
Figure 1.
Pathway map of pyrimidine metabolism depicting de novo, salvage, and catalytic pathways. CAD: carbamoyl-phosphate synthetase 21, aspartate transcarbamylase2, and dihydroorotase³, UMPS: uridine monophosphate synthetase (orotate phosphorybosyl transferase4, and orotidylate decarboxylase5), CMPK1/2: cytidine/uridine monophosphate kinase 1/2, RNR: ribonucleotide reductase, DTYMK: deoxythymidylate kinase, ENTPD8: ectonucleoside triphosphate diphosphohydrolase 8, NDK6: nucleoside diphosphate kinase 6, DUT: deoxyuridine triphosphatase, CTPS1/2: CTP synthase 1/2, ENTPD1: ectonucleoside triphosphate diphosphohydrolase 1, PRPS1/2: phosphoribosyl pyrophosphate synthetase 1/2, TS: thymidylate synthase, SHMT1: serine hydroxymethyltransferase 1, DHFR: dihdyrofolate reductase, NT5C: 5′,3′-nucleotidase, cytosolic, TYMP: thymidine phosphorylase, DPYD: dihydropyrimidine dehydrogenase, DPYS: dihydropyrimidinase, UPB1: beta-ureidopropionase 1, NT51B: 5′-nucleotidase, cytosolic IB, CDA: cytidine deaminase, UPP1: uridine phosphorylase 1, CDA: cytidine deaminase, TK1/2: thymdine kinase 1/2 dCK: deoxycytidine kinase, UCKL1: uridine-cytidine kinase 1 like 1, DCTD: dCMP deaminase, GLS: glutaminase, CPS1: carbamoyl-phosphate synthase.
The de novo synthesis of pyrimidines is a sequential synthesis of pyrimidine bases from amino acids and priming the resulting base to a ribose moiety from phosphoribosyl pyrophosphate (PRPP). In the first step of the de novo pyrimidine synthesis, the trifunctional protein carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase (CAD) fixes cytosolic glutamine to dihydroorotate. Dihydroorotate is converted to orotate by the mitochondrial enzyme dihydroorotate dehydrogenase (DHODH). The cytosolic enzyme uridine monophosphate synthase (UMPS) uses dihydroorotate as a precursor for the synthesis of UMP. The ribose moiety for UMP is donated by PRPP. UMP is then sequentially converted to CTP. Deoxyribonucleotides are synthesized from ribonucleotides by action of the enzyme ribonucleotide reductase (RNR) that converts diphosphates of the ribonucleotides to respective deoxyribonucleotides. As there is no ribonucleotide precursor for thymidylate, it is synthesized by thymidylate synthase (TS)-mediated methylation of deoxyuridine monophosphate [50].
Cells also have an endogenous mechanism to eliminate excess nucleotides and nucleosides that are generated by degradation of the nucleic acids. Pyrimidine catabolism uses the same set of enzymes for all pyrimidine molecules, as TMP and dUMP are sequentially dephosphorylated to their respective bases and then cleaved into open chain amino acids. Cytidine enters the catabolic pathway via deamination to uridine. The key enzymes of this pathway are thymidine phosphorylase (TYMP), 5′,3′-nucleotidase, cytosolic (NT5C), and dihydropyrimidine dehydrogenase (DPYD) [50].
2.2. Oncogenic activation of pyrimidine metabolism in cancer
A continuous supply of dNTPs is essential for survival of the cancer cells and, therefore, the perpetual activation of de novo PyM genes becomes incumbent for a growing tumor. Not surprisingly, the expression of de novo pathway genes (also nucleoside salvage pathway genes) is regulated by key oncogenes. Mutant p53 (p53MUT) regulates the expression of an array of PyM genes and impacts the flux of the ribonucleotides phosphates to the dNTP pool by increasing the expression of the regulatory subunit of RNR [52]. p53MUT-regulated expression of PyM has also been implicated in resistance against the drugs that target TS [53]. MYC (protein: c-myc), another key oncogene, binds to the regulatory region of several PyM genes and increases their expression [54,55] to maintain an elevated dNTP pool [56]. mTORC1 is a protein complex that is activated by loss of the tumor suppressor PTEN or activation of PI3K-AKT signaling. It integrates the oncogenic signals to mediate the major cancer-associated metabolic shifts. It phosphorylates S6K and the transcription factor E2F1 [57] to increase PyM. S6K post-translationally activates carbamoyl-phosphate synthase 2 (a component of CAD) [58,59] by phosphorylating Ser1859 and subsequent oligomerization [59], whereas E2F1 upregulates the expression of pyrimidine anabolic genes such as TYMS (gene coding TS) and TK and catabolic genes such as DPYD [60] at the transcriptional level. mTOR also controls the efflux of mitochondrial UTP to cytosol to support proliferation [61]. RAS [62] and EGFR [63] mediate the activation of MAPK phosphorylate CAD at Tyr456, resulting in desensitization to the UTP inhibition and increased sensitivity to the activation by PRPP . KRAS has been shown to drive tumor growth in the pancreatic cancer by activating PyM [64], and DHODH inhibition has been shown to be selectively responsive to the cells with KRAS mutation [65].
Thus, cancer cells can employ different mechanisms not only to activate the PyM genes but also to desensitize feedback regulatory pathways to circumvent allosteric inhibition, ensuring a continuous flow of the cellular nitrogen toward pathways that generate ribonucleotide phosphates and dNTPs. Since fueling of proliferation is the prime role of PyM in cancer cells, other functions have been underappreciated. In the following sections, corroborations from literature will be discussed to emphasize the role of PyM in differentiation in leukemia and solid tumors.
3. Pyrimidine metabolism, leukemia, and proliferation-dependent terminal differentiation
The role of PyM in differentiation was first reported in late 1980 in leukemic cells, where the interplay between thymidine catabolic and anabolic pathways was shown to induce terminal differentiation [3,66]. These studies focused on acute myeloid lymphoma (AML) cells that originate from the hematopoietic progenitor cells (blast cells) that fail to differentiate and, therefore, rapidly proliferate [67]. A higher enzymatic activity of PyM anabolic enzymes (thymidine kinase and thymidylate synthase) was maintained in the leukemic cells compared with the normal leukocytes. In contrast, inducing differentiation in the leukemic cell line U-937 using 12-O-tetradecanoylphorbol-13-acetate (TPA) led to higher DPYD activity, indicating that the cells tend to eliminate thymidine via catabolic degradation [3]. In fact, treatment of AML cells with thymidine and inhibition of uridylate conversion to cytidine (thereby limiting the dNTP pool) promoted differentiation and maturation of leukemic cells [68]. Similarly, inhibition of DHODH [[69], [70], [71]] and UMPS [70] in AML released the blast cells from differentiation blockade (inability to differentiate) and resulted in their maturation.
Considered together, these data present a proliferation-dependent model of PyM-mediated leukemic differentiation. In summary, as normal lymphoblasts undergo oncogenic transformation, they direct thymidine toward thymidylate synthesis and maintain a balanced dNTP pool for proliferation. Whereas, following differentiation, thymidine is fluxed to catabolism, thus limiting the availability of thymidylate for cellular proliferation. However, as it will be discussed in the following section, the mode of PyM-mediated differentiation in solid tumors considerably varies from this mechanism.
4. Pyrimidine metabolism, solid tumors, and proliferation-independent differentiation
4.1. PyM maintains the EMT phenotypes in epithelial tumors
The notion that EMT-driven aggressive epithelial tumors rely on PyM to maintain a functional phenotype is now well established in the literature. For instance, in aggressive lung cancers that are driven by simultaneous KRAS activation and loss of tumor suppressor STK11 (serine/threonine kinase 11; protein: liver kinase B1/LKB1), the mitochondrial carbamoyl phosphate from the urea cycle is diverted toward pyrimidine synthesis rather than the excretory pathway [72]. LKB1 has been previously shown to block EMT by inhibiting ZEB1 in lung [73] and hepatic cancer [74] and to co-localize with E-cadherin to promote the epithelial phenotype in breast cancer [75]. This might be the first indication that diversion of the mitochondrial carbamoyl phosphate to the production of pyrimidine could be a consequence of aggressive mesenchymal phenotype associated with the loss of LKB1. Indeed, several genes from the PyM correlate with the aggressive phenotypes in many tumors [[76], [77], [78]], but their impact on cancer malignancy has been exclusively attributed to their role in proliferation.
In an attempt to establish a fundamental link between the functional phenotypes of EMT and PyM in carcinoma, our laboratory discovered a strong correlation between TYMS mRNA (a gene that is central to the de novo pathway) and key markers of epithelial and mesenchymal-like phenotypes from a transcriptomic dataset from the NCI (NCI-60 panel, spanning nine different types of cancers). Among the EMT markers investigated, we could validate a functional association between TS and ZEB1 and observed that shRNA-mediated ZEB1 knockdown could not only suppress EMT, but also decrease TS protein level. Further, we identified a role for miR-375 in TYMS regulation, and the existence of a ZEB1/miR-375/TS axis in regulating EMT in non-small-cell lung cancer (NSCLC) cells. However, more surprisingly, in the same work, a similar effect of EMT reversal was observed in cancer cells from different tumor types by direct shRNA-mediated targeting of TYMS mRNA, which was shown to suppress migration in a proliferation-independent manner [79].
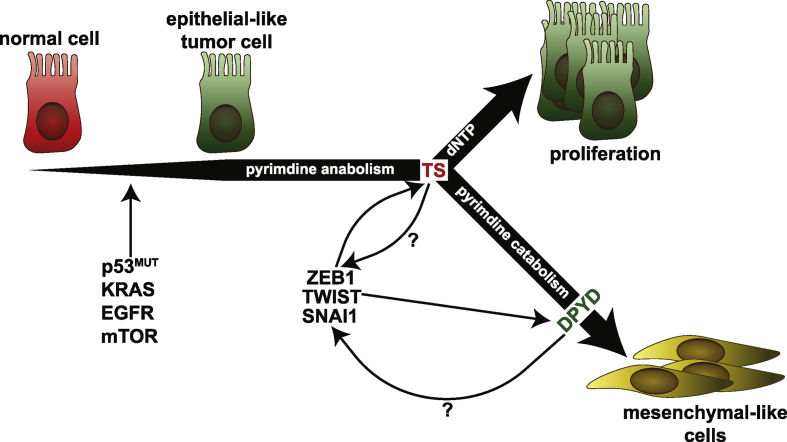
When the model of poorly differentiated and mesenchymal-like triple-negative breast cancer (TNBC, breast cancer subtype with aggressive clinical behavior and predominantly basal-like phenotype) was tested, knockdown of TS led to an increase in the CD24+ population (markers of differentiated breast cancer cells) and reduced the migration across the wound in a monolayer culture [80]. The PyM-mediated EMT in breast epithelial cells further involves the enzymes for the catabolic degradation of thymidine, and a prominent role of DPYD, the only reversible enzyme in the catabolic pathway, has been recognized [80,81]. EMT induction in human breast epithelial cells by three independent overexpressions of EMT-TFs (TWIST, SNAI1, and GCS-goosecoid) led to an increased accumulation of the pyrimidine catabolic products [49]. Shaul et al. identified DPYD from a metabolic signature associated with EMT in a panel of 978 human cancer cell lines from different tumors, and demonstrated the importance of its catabolic products, dihydrothymine and dihydrouracil, in EMT in breast cancer [81]. The significance of DPYD in EMT has been further shown in the hepatocellular carcinomas (HCCs), where DPYD was shown to regulate the expression of EMT-TF SNAI1 through nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) signaling [82]. Hence, these data tender a model with an anabolism/catabolism axis for EMT in aggressive cancers. Data from our laboratory further reinforce this notion, as we have shown that TS enzymatic activity is essential for the maintenance of the EMT phenotype of the TNBC cells and linked it to the catalytic degradation of thymidylate operated by DPYD [80]. We could further identify NFκB signaling as one of the most differentially regulated pathways in breast cancer cell lines with TS knockdown. Along with the involvement of the DPYD/NFκB axis in HCC [82], these observations suggest a TS/DPYD axis in the maintenance of an aggressive phenotype via regulation of EMT-TFs (Figure 2).
Figure 2.
Interplay between EMT-TFs and pyrimidine metabolism genes. As the normal cells undergo oncogenic transformation, oncogenes activate and upregulate PyM genes. PyM in cancer is essential for proliferation, but the catabolic activity of PyM enzymes is further required to maintain aggressive cells in a mesenchymal-like state. This review aims to open a discussion that could lead to identification of possible feedback loops that connect the PyM back to EMT. The question marks in the figure indicate possibility of an existing feedback activation.
4.2. PyM affects aggressive behavior of non-epithelial tumors
Having professed a role for PyM in EMT in epithelial tumors, we wanted to understand its impact on non-epithelial tumors such as glioblastoma and melanoma, which have also been found to be driven by EMT-related processes [[83], [84], [85]]. In fact, in glioblastoma, the anabolic rate-limiting enzymes CAD and DHODH have been reported to be essential for self-renewal and maintenance of stem cells. A pyrimidine metabolic signature has been identified in glioblastoma, which stratified patients with higher grade from lower grades, and higher levels of pyrimidine metabolites were found in glioblastoma stem cells compared with the differentiated cells [86]. Further, ENPP1, a peripheral PyM enzyme, has been shown to regulate E2F1 to maintain stem cells in glioblastoma [87]. White et al. showed the importance of DHODH in self-renewal of neural-crest stem cells in development of zebrafish embryos. Interestingly, melanoma cells, which originate from the neural crest [88] and are characterized by stem cell phenotypes [89], showed a marked decrease in growth in vivo and in vitro when treated with DHODH inhibitors [90].
Further, even in multiple myeloma stem cells, identified by low CD138 expression and high aldehyde dehydrogenase 1 family member A1 (ALDH1, a marker for CSCs) activity, inhibition of TS stimulated the proliferation and simultaneously broke the dormant phenotype associated with stemness [91]. Briefly, treatment of the CD27+CD138- stem cells with a sub-lethal dose of 5-fluoro-2′-deoxyuridine (FdUrd, a competitive inhibitor of TS) resulted in a reduction in ALDH1 activity, marked the increase in the population of the CD138+ differentiated cells and increased the sensitivity to 125I-ITdU (an Auger electron emitting thymidine analogue [92]) [91]. Loss of the stem cells phenotype with TS inhibition, concurrent with an unexpected increase in proliferation, strongly indicated a proliferation-independent role of TS in stem cell phenotype.
Therefore, the above discussion supports that PyM can impact the aggressive nature of tumors that do not originate from the epithelial lineage. Although the exact mechanism has not yet been elucidated, a mechanistic overlap between carcinoma and non-epithelial aggressive tumors could be speculated in the context of PyM and validated experimentally.
4.3. PyM-mediated EMT and proliferation are independent modalities
Taking the discussed arguments into account, a strong case could be advocated for the role of PyM in cancer cell differentiation, which is a function of PyM anabolism as well as catabolism. It is of further interest to speculate about the connection between proliferation and EMT in the TS/DPYD model, and our observations strongly suggest that proliferation and differentiation are two independent modalities in epithelial tumors [70]. In our TNBC models, the effect on proliferation correlated with level of TS knockdown. Complete shRNA-induced ablation of TS resulted in growth arrest and cell death; strong TS reduction caused a proliferation defect; whereas, mild reduction in TS did not affect cell growth at all. Nevertheless, TS depletion always attenuated the mesenchymal-like differentiation, irrespective of loss in proliferation. When dNTPs were quantified in the TS knocked-down MDA-MB-231 cells with loss of CD24-negativity and suppression of EMT signature genes but without a detectable defect in proliferation, no imbalance in the dNTP pool was observed. These observations clearly decouple the TS-mediated EMT from proliferation, despite the necessity of TS enzymatic activity for maintenance of the mesenchymal-like state. These observations substantiate a model in which some highly aggressive cancer cells maintain thymidylate in excess of the threshold level required for proliferation. Excess thymidylate is then degraded by DPYD to promote and keep the EMT state intact [80,81].
The concept of the pyrimidine-mediated EMT appends another dimension to the existing studies that have shown that some metabolic pathways can act upstream of the EMT-TFs. For instance, fumarate acts as an epigenetic regulator of EMT [93], and excess glucose can enter the polyol pathway to regulate EMT by activating the autocrine TGF-β signaling [94]. As increase in glucose uptake is a salient feature of EMT [95,96], the existence of a polyol pathway could serve as a feedback channel to maintain it. Based on the discussed evidence, a reciprocal loop could be hypothesized between the EMT-induced PyM metabolism and sustained activation of EMT by activated PyM at transcriptional level. However, more studies are required to understand the downstream mechanism in detail, to uncover the whole network that transduces the PyM-mediated differentiation program, and to explore its possible connection with EMT-TFs such as ZEB1, TWIST, and SNAI1 (Figure 2). Moreover, the role of the peripheral PyM genes, such as ENPP1, a transmembrane protein that catalyzes the conversion of extracellular NTPs to NMPs, could be investigated in the context of its enzymatic activity, as it has been shown to promote the stem cell phenotype of breast cancer cells by promoting the cell surface localization of ABCG2 (ATP-binding cassette subfamily G member 2) transporter [97].
5. PyM can tune differentiation status to impact metastasis
Differentiation is an important determinant of metastasis; thus, the effect of PyM on the metastatic outcome is a rational supposition. Indeed, in vivo mouse models have shown that suppression of PyM genes influences metastasis. In a colorectal cancer model, the reduction of DHODH did not affect the primary tumor growth from two different cell lines, but dramatically reduced liver metastasis (indicating that loss of DHODH affected the metastatic colonization although there was no proliferative defect in the primary tumors) [98]. In pancreatic cancer, TS knocked-down cells showed a marked reduction in metastatic colonization, whereas the supplementation of thymidylate rescued the effect of TS knockdown [99]. Similarly, DPYD knockdown in immortalized breast cells showed reduced metastatic behavior in vivo [81]. However, in our model from TNBC breast cancer cell lines, TS knockdown resulted in increased metastasis [80]. The variable outcome of PyM ablation on metastasis can be explained by the model of partial EMT and its association with differentiation status of the cells.
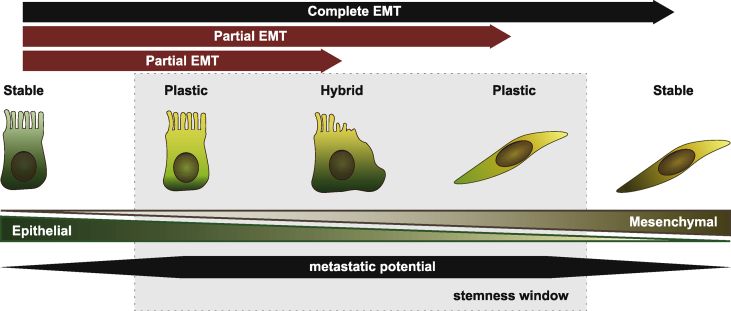
EMT is a non-binary process that includes multiple genetic and phenotypic alterations [100]. As epithelial cells lose their differentiation via EMT, they may acquire a complete mesenchymal phenotype, or they may acquire some of the several changes that define EMT [100]. The latter case is termed a partial/hybrid EMT state, and has now been reported in different cancer types [[101], [102], [103], [104]]. Partial EMT does not only allow the transit to mesenchymal phenotype through selected functional changes, but the degree to which these changes are attained is also variable. This associates it with epithelial plasticity, and allows the tumor cells to exist in different states of differentiation [104]. The cells that undergo complete EMT are believed to lose their ability to interchange phenotypes and seed new tumors [101,103]. Thus, there exists a window of stemness, a zone marked by cells in partial EMT, that determines the metastatic potential of carcinomas [101] (Figure 3). A correlation between partial EMT and enhanced metastasis is in fact now substantially documented in several in vivo models [105]. In addition, in certain cancers, such as breast cancer, cells with partial EMT have been shown as the main driver of metastasis [103,105,106].
Figure 3.
Correlation between different EMT states and stemness. Cancer cells that are locked in either epithelial or mesenchymal phenotype lose their plasticity to switch between EMT states and seed new tumors. Whereas, cells that undergo partial EMT exist within the frame of stemness window and could readily switch between differentiation phenotypes. The stemness window defines the zone that supports metastasis in cancers such as breast.
Therefore, for a successful metastasis, reduction of PyM must direct the cells into the stemness window that is defined by the partial EMT state. Thus, in an experimental model with a knockdown approach, metastatic colonization depends on the initial EMT state of the cells and on the extent of change in EMT status after knockdown. For instance, in highly mesenchymal MDA-MB-231 cells, TS knockdown led to the loss of mesenchymal differentiation with a dramatic increase in the CD24+ cells and reduction of migration [80]. However, these cells intravasated and extravasated more readily compared with the control cells. Also, they colonized more in the lungs, indicating that the changes induced by the TS knockdown rendered the cells in partial EMT state with high metastatic potential compared with highly mesenchymal parental controls. This model is supported by the fact that overexpression of miR-200c, a potent EMT inhibitor, promotes metastasis [15,107]; and, other studies have shown simultaneous loss of EMT in vitro and increased metastasis in vivo (reviewed in [108]).
These results also have important clinical implications, because anti-PyM drugs are frequently administered to patients with aggressive cancers, and expression level of PyM genes, such as TS, could be important determinants of metastasis-free survival. Hence, a spontaneous metastatic model is clearly needed to understand how PyM directs the metastasis in vivo, and detailed clinical investigations need to be undertaken to comprehend how anti-PyM drugs correlate with the metastasis-free survival in patients with aggressive cancers. As a result, the EMT status of cancers could be considered in the therapeutic decision making and used to stratify the patients to be treated with PyM-directed agents.
6. Pyrimidines as potential oncometabolites
The arguments presented in the preceding section establish a role of PyM genes in maintaining the EMT status of the cells. However, it is of great interest to understand if the pyrimidine metabolites play a direct and essential role in the process. Intracellular and extracellular accumulations of the pyrimidine metabolites are reported but not functionally described in detail. An oncogenic role of the pyrimidine metabolites could be anticipated, as purine molecules have been shown to act as receptor ligands in the microenvironment of many tumors [109]. Extracellular purines generate signaling, classified as purinergic signaling, through a dedicated set of cell surface receptors [110] and can affect proliferation and metastasis [[110], [111], [112]]. A similar function could also be speculated and explored for pyrimdines.
The endogenous accumulation of pyrimidine metabolites has been reported in the glycine decarboxylase-driven CD166+ CSCs from NSCLC [113]. In the hTERT-transformed mammary epithelial cells, elevated levels of dihydrothymine were reported in TWIST-induced EMT, and accumulation of dihydrothymine was shown to be essential for EMT [81]. However, the exact contribution of these metabolites has never been estimated.
Notably, in vitro the exogenous supplementation of pyrimidine metabolites has been used to rescue the EMT reversal induced by the knockdown of PyM genes [80,81], suggesting that the extracellular pyrimidine metabolites can be easily uptaken and work as oncometabolites. Along this line, recent reports in pancreatic cancer [114,115] have shown that tumor microenvironment can be enriched with pyrimidines secreted by non-tumor entities such as tumor-associated macrophages (TAMs) and stromal stellar cells. The macrophages secrete a broader spectrum of pyrimidines (thymidine, thymine, dCMP, cytosine, cytidine, dUMP, uridine, and uracil) with the highest secretion of thymidine [115], whereas stellar cells mainly secrete deoxycytidine (dC) with a detectable level of thymidine and deoxyuridine [114]. Although in both cases the assimilation of the deoxycytidine by the cancer cells was shown as a strategy to increase resistance against gemcitabine (a chemical analogue of dC), these observations open several possibilities in relation to metabolic control of EMT, such as cell intrinsic regulation, extracellular signaling, and assimilation of extracellular nucleotides. Although there is a lack of empirical data, based on the purinergic signals, a concept of pyrimidine-mediated cellular crosstalk can be hypothesized and validated experimentally. Therefore, it could be interesting to investigate the non-cancer components that might be involved in the enriching tumor microenvironment with pyrimidine metabolites and understand the mode of action of extracellular pyrimidines in differentiation.
7. The link between PyM, chemoresistance, stemness, and EMT
As the cancer cells are addicted to the de novo pathway, competitive inhibitors of the de novo enzymes, such as 5-flurouracil (5-FU), capecitabine, and pemetrexed, have been used clinically for inducing DNA damage and cell death [[116], [117], [118], [119], [120]]. Although these drugs are widely used, their clinical efficacy is limited by chemoresistance attributed to the increased expression of the targeted PyM genes [[121], [122], [123]] and the reprogramming of the cells to the stem-like phenotype that is regulated by EMT [124].
Induction of the PyM genes is a common feature of the cells with the resistant phenotype. Several studies have reported an increase in the cellular levels of TS after cells are subjected to chemotherapeutic drugs [[125], [126], [127], [128]]. Exposure of a TNBC cell line to doxorubicin has shown phosphorylation of CAD and induction of the de novo pyrimidine synthesis, leading to an increase in cellular triphosphate of cytidine and thymidine [129]. Colon cancer cells increased the expression of DPYD by suppressing miR-494 after exposure to 5-FU gradient at a sub-lethal dose [130].
However, chemoresistance has been shown to be promoted by EMT, which reprograms epithelial cells to mesenchymal-like cells with stem cell properties [[131], [132], [133]]. Many of the EMT TFs, TWIST [[134], [135], [136], [137]], ZEB1/2 [[138], [139], [140], [141]], SNAI1 [[142], [143], [144], [145]], and SNAI2 [[146], [147], [148]], are upregulated in resistant cells and play a functional role in desensitizing cells to chemotherapy. Many of them do so by increasing the expression of the PyM genes. An EMT-promoting gene, astrocyte-elevated gene-1 (AEG-1) [149,150], confers resistance in HCC by inducing the expression of DPYD [151]. miRNAs that are associated with EMT have also been shown to regulate expression of the PyM genes and simultaneously modulate the drug response. For instance, miR-203, an important EMT inhibitor [152], binds directly to TS and sensitizes cells to 5-FU [153]. On the other hand, chemotherapeutic drugs induce the expression of EMT-TF; for example, gemcitabine has been shown to induce EMT through ZEB1 [154].
Since PyM plays a role in EMT, it would be interesting to evaluate the PyM-mediated EMT as a mechanism of acquired chemoresistance. In this direction, our laboratory has demonstrated that exposing NSCLC cell lines to increasing doses of 5-FU up-regulated the expression of its target, TS. The cells developed resistance against the drug and underwent EMT with an increase in ALDH1 activity. However, TS knockdown blocked these effects, indicating that TS is critical for the drug-induced cancer stem cell phenotype in these cells [79]. Cells with the stem-like phenotype have been found to be more resistant to antimetabolites that target the PyM pathway compared with the non-stem cells [[155], [156], [157], [158]], and these antimetabolites, in turn, may enhance the stem-cell phenotype [159,160], similarly to other chemotherapeutic agents [161]. Therefore, there exists a clear network between EMT, stemness, PyM, and chemoresistance that could be investigated, with the possibility of feedback loops that regulate chemoresistance via PyM-mediated EMT.
8. Therapeutic impact
Oncogenic activation and elevation of the PyM genes are key features of the tumor cell, and PyM anabolic and catabolic pathways are associated with differentiation apart from maintaining a continuous proliferative state. The studies presented in this review strongly support a model in which some aggressive tumor types maintain their mesenchymal-like state by channeling the pyrimidine metabolites to the catabolic pathway, which is mediated via DPYD. Interestingly, diminution of the PyM can have different effects on metastasis in vivo, depending on the cellular context and level of suppression.
Therefore, there is a need to extensively probe the role of PyM in differentiation and its subsequent effect on metastasis. This is essential, for instance, in the case of TS, which is an important clinical drug target, and TS-inhibiting drugs are often given to patients with highly aggressive cancers [120]. The effects of TS inhibition on the aggressive tumors also need to be re-evaluated in light of the new evidence. Simultaneously, chemotherapeutic inhibition of DHODH and other PyM enzymes could be tested for repressing EMT and metastatic behavior.
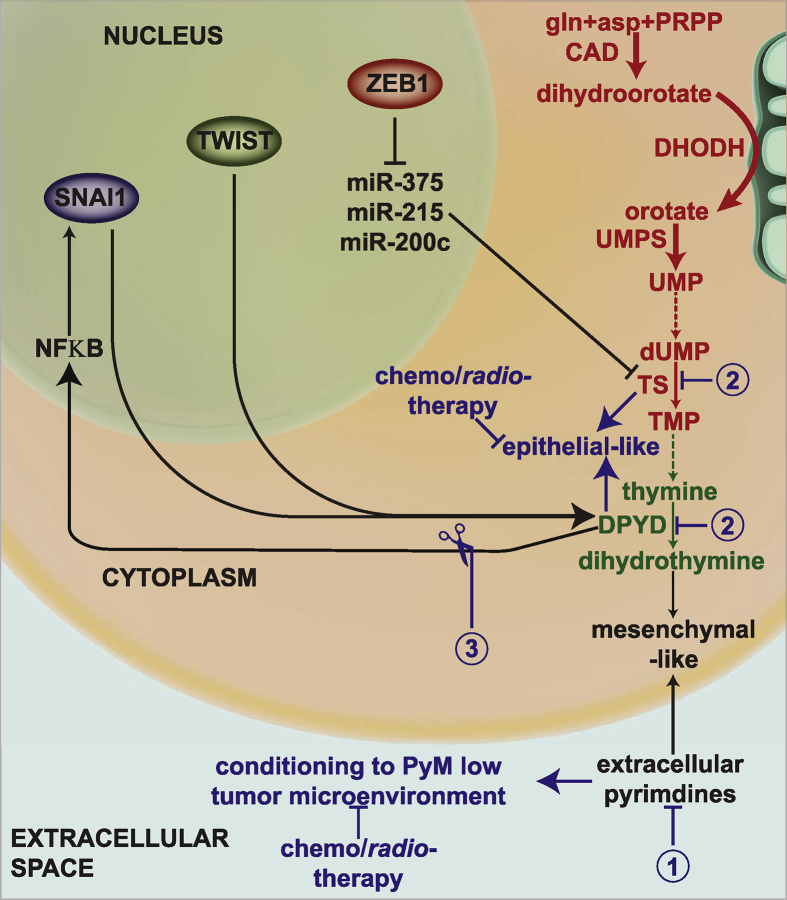
Based on the themes considered in this review, different strategies could be adopted to render solid tumors sensitive to chemotherapy (outlined in Figure 4). For instance, sources that contribute to the extracellular pyrimidines can be identified and eradicated to debilitate the protumorigenic microenvironment. In addition, a reduced dose of drugs (or novel improved agents) that could target PyM without stimulating expression of the target genes could be determined and used in the clinic to sensitize cells to combination chemotherapy. Finally, the network between PyM enzymes and EMT-TFs that maintains the cells in the stem-cell-like state could be disconnected to render cells more susceptible to existing chemotherapies and radiotherapy. Thus, overall, in-depth investigation of the non-proliferative functions of PyM can provide therapeutic options for difficult-to-treat cancers.
Figure 4.
Schematic representation of the proposed therapeutic strategies to tackle aggressive tumors by harnessing PyM-mediated EMT. The first strategy aims at identifying and eliminating the entities that enrich the tumor microenvironment with pyrimidine metabolites. The loss of pyrimidine-rich microenvironment, which might support EMT, could sensitize the cancer cells to chemo-/radiotherapy. Similarly, the second strategy aims at the cytosolic depletion of PyM and, thus, forcing the cells into an epithelial-like state. Mesenchymal-like cells could be converted to epithelial-like cells by administering either lower doses of anti-PyM drugs or novel and more effective (and less toxic) drugs. The cells can then be targeted by chemo-/radiotherapy. The third strategy aims at recognizing the interactions between PyM enzymes and EMT-TFs, such as NFκB, and targeting them to break the connection between PyM and EMT-TFs. De novo reactions have been depicted in red, salvage reactions in green, proposed therapeutic strategies in blue and interactions in black. The broken lines represent multiple-step conversions.
Acknowledgements
Work supported by the Interdisciplinary Center for Clinical Research (IZKF) of the University of Erlangen-Nuremberg (Junior Group 1), and by the IALSC Young Investigator Award (to P. Ceppi). The authors would like to thank Annemarie Schwab (University of Erlangen-Nuremberg) for the help with the manuscript.
Conflict of interest
None declared.
References
- 1.Villa E., Ali E.S., Sahu U., Ben-Sahra I. Cancer cells tune the signaling pathways to empower de Novo synthesis of nucleotides. Cancers. 2019;11:688. doi: 10.3390/cancers11050688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buj R., Aird K.M. Deoxyribonucleotide triphosphate metabolism in cancer and metabolic disease. Frontiers in Endocrinology. 2018;9 doi: 10.3389/fendo.2018.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiotani T., Hashimoto Y., Fujita J., Yamauchi N., Yamaji Y., Futami H. Reversal of enzymic phenotype of thymidine metabolism in induced differentiation of U-937 cells. Cancer Research. 1989;49:6758–6763. [PubMed] [Google Scholar]
- 4.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nature Reviews Molecular Cell Biology. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 5.Guarino M., Rubino B., Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 6.Aigner K., Dampier B., Descovich L., Mikula M., Sultan A., Schreiber M. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. Journal of Clinical Investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson G.W. Control of invasion by epithelial-to-mesenchymal transition programs during metastasis. Journal of Clinical Medicine. 2019;8:646. doi: 10.3390/jcm8050646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Son H., Moon A. Epithelial-mesenchymal transition and cell invasion. Toxicological Research. 2010;26:245–252. doi: 10.5487/TR.2010.26.4.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richmond A.M., Blake E.A., Torkko K., Smith E.E., Spillman M.A., Post M.D. Fascin is associated with aggressive behavior and poor outcome in uterine carcinosarcoma. International Journal of Gynecological Cancer. 2017;27:1895–1903. doi: 10.1097/IGC.0000000000001077. [DOI] [PubMed] [Google Scholar]
- 11.Guo C.C., Majewski T., Zhang L., Yao H., Bondaruk J., Wang Y. Dysregulation of EMT drives the progression to clinically aggressive sarcomatoid bladder cancer. Cell Reports. 2019;27:1781–1793. doi: 10.1016/j.celrep.2019.04.048. e1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., Nikhil K., Viccaro K., Chang L., Jacobsen M., Sandusky G. The Aurora-A-Twist1 axis promotes highly aggressive phenotypes in pancreatic carcinoma. Journal of Cell Science. 2017;130:1078–1093. doi: 10.1242/jcs.196790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mani S.A., Guo W., Liao M.-J., Eaton E.N., Ayyanan A., Zhou A.Y. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.May C.D., Sphyris N., Evans K.W., Werden S.J., Guo W., Mani S.A. Epithelial-mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Research: BCR. 2011;13:202. doi: 10.1186/bcr2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korpal M., Ell B.J., Buffa F.M., Ibrahim T., Blanco M.A., Celia-Terrassa T. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nature Medicine. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimono Y., Zabala M., Cho R.W., Lobo N., Dalerba P., Qian D. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wellner U., Schubert J., Burk U.C., Schmalhofer O., Zhu F., Sonntag A. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature Cell Biology. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 18.Chaffer C.L., Marjanovic N.D., Lee T., Bell G., Kleer C.G., Reinhardt F. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weidenfeld K., Barkan D. EMT and stemness in tumor dormancy and outgrowth: are they intertwined processes? Frontiers in Oncology. 2018;8:381. doi: 10.3389/fonc.2018.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loret N., Denys H., Tummers P., Berx G. The role of epithelial-to-mesenchymal plasticity in ovarian cancer progression and therapy resistance. Cancers. 2019;11:838. doi: 10.3390/cancers11060838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J., Li H., Ren G. Epithelial-mesenchymal transition and drug resistance in breast cancer (Review) International Journal of Oncology. 2015;47:840–848. doi: 10.3892/ijo.2015.3084. [DOI] [PubMed] [Google Scholar]
- 22.Brozovic A., Duran G.E., Wang Y.C., Francisco E.B., Sikic B.I. The miR-200 family differentially regulates sensitivity to paclitaxel and carboplatin in human ovarian carcinoma OVCAR-3 and MES-OV cells. Molecular Oncology. 2015;9:1678–1693. doi: 10.1016/j.molonc.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haslehurst A.M., Koti M., Dharsee M., Nuin P., Evans K., Geraci J. EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer. 2012;12:91. doi: 10.1186/1471-2407-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu X., Shen H., Yin X., Long L., Xie C., Liu Y. miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene. 2016;35:323–332. doi: 10.1038/onc.2015.84. [DOI] [PubMed] [Google Scholar]
- 25.Elaskalani O., Razak N.B.A., Falasca M., Metharom P. Epithelial-mesenchymal transition as a therapeutic target for overcoming chemoresistance in pancreatic cancer. World Journal of Gastrointestinal Oncology. 2017;9:37–41. doi: 10.4251/wjgo.v9.i1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren J., Chen Y., Song H., Chen L., Wang R. Inhibition of ZEB1 reverses EMT and chemoresistance in docetaxel-resistant human lung adenocarcinoma cell line. Journal of Cellular Biochemistry. 2013;114:1395–1403. doi: 10.1002/jcb.24481. [DOI] [PubMed] [Google Scholar]
- 27.Oft M., Heider K.-H., Beug H. TGFβ signaling is necessary for carcinoma cell invasiveness and metastasis. Current Biology. 1998;8:1243–1252. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 28.Oft M., Peli J., Rudaz C., Schwarz H., Beug H., Reichmann E. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes & Development. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 29.Miettinen P.J., Ebner R., Lopez A.R., Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. The Journal of Cell Biology. 1994;127:2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin H., Pi J., Huang X., Huang F., Shao W., Li S. BMP2 promotes migration and invasion of breast cancer cells via cytoskeletal reorganization and adhesion decrease: an AFM investigation. Applied Microbiology and Biotechnology. 2012;93:1715–1723. doi: 10.1007/s00253-011-3865-3. [DOI] [PubMed] [Google Scholar]
- 31.Katsuno Y., Hanyu A., Kanda H., Ishikawa Y., Akiyama F., Iwase T. Bone morphogenetic protein signaling enhances invasion and bone metastasis of breast cancer cells through Smad pathway. Oncogene. 2008;27:6322–6333. doi: 10.1038/onc.2008.232. [DOI] [PubMed] [Google Scholar]
- 32.Choi B.-J., Park S.-A., Lee S.-Y., Cha Y.N., Surh Y.-J. Hypoxia induces epithelial-mesenchymal transition in colorectal cancer cells through ubiquitin-specific protease 47-mediated stabilization of Snail: a potential role of Sox9. Scientific Reports. 2017;7:15918. doi: 10.1038/s41598-017-15139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nature Cell Biology. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 35.Cano A., Pérez-Moreno M.A., Rodrigo I., Locascio A., Blanco M.J., del Barrio M.G. The transcription factor Snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nature Cell Biology. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 36.Spaderna S., Schmalhofer O., Wahlbuhl M., Dimmler A., Bauer K., Sultan A. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Research. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 37.Ceppi P., Peter M.E. MicroRNAs regulate both epithelial-to-mesenchymal transition and cancer stem cells. Oncogene. 2014;33:269–278. doi: 10.1038/onc.2013.55. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J., Zhang H., Qin Y., Chen C., Yang J., Song N. MicroRNA-200c-3p/ZEB2 loop plays a crucial role in the tumor progression of prostate carcinoma. Annals of Translational Medicine. 2019;7:141. doi: 10.21037/atm.2019.02.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sundararajan V., Gengenbacher N., Stemmler M.P., Kleemann J.A., Brabletz T., Brabletz S. The ZEB1/miR-200c feedback loop regulates invasion via actin interacting proteins MYLK and TKS5. Oncotarget. 2015;6:27083–27096. doi: 10.18632/oncotarget.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Reports. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diepenbruck M., Tiede S., Saxena M., Ivanek R., Kalathur R.K.R., Luond F. miR-1199-5p and Zeb1 function in a double-negative feedback loop potentially coordinating EMT and tumour metastasis. Nature Communications. 2017;8:1168. doi: 10.1038/s41467-017-01197-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rokavec M., Oner M.G., Li H., Jackstadt R., Jiang L., Lodygin D. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. Journal of Clinical Investigation. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Z., Nian Z., Jingjing Z., Tao L., Quan L. MicroRNA-424/E2F6 feedback loop modulates cell invasion, migration and EMT in endometrial carcinoma. Oncotarget. 2017;8:114281–114291. doi: 10.18632/oncotarget.23218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kondaveeti Y., Guttilla Reed I.K., White B.A. Epithelial-mesenchymal transition induces similar metabolic alterations in two independent breast cancer cell lines. Cancer Letters. 2015;364:44–58. doi: 10.1016/j.canlet.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 45.Carvalho-Cruz P., Alisson-Silva F., Todeschini A.R., Dias W.B. Cellular glycosylation senses metabolic changes and modulates cell plasticity during epithelial to mesenchymal transition. Developmental Dynamics. 2018;247:481–491. doi: 10.1002/dvdy.24553. [DOI] [PubMed] [Google Scholar]
- 46.Rosland G.V., Dyrstad S.E., Tusubira D., Helwa R., Tan T.Z., Lotsberg M.L. Epithelial to mesenchymal transition (EMT) is associated with attenuation of succinate dehydrogenase (SDH) in breast cancer through reduced expression of SDHC. Cancer & Metabolism. 2019;7:6. doi: 10.1186/s40170-019-0197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sciacovelli M., Frezza C. Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer. FEBS Journal. 2017;284:3132–3144. doi: 10.1111/febs.14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halldorsson S., Rohatgi N., Magnusdottir M., Choudhary K.S., Gudjonsson T., Knutsen E. Metabolic re-wiring of isogenic breast epithelial cell lines following epithelial to mesenchymal transition. Cancer Letters. 2017;396:117–129. doi: 10.1016/j.canlet.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 49.Bhowmik S.K., Ramirez-Pena E., Arnold J.M., Putluri V., Sphyris N., Michailidis G. EMT-induced metabolite signature identifies poor clinical outcome. Oncotarget. 2015;6:42651–42660. doi: 10.18632/oncotarget.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loffler M., Fairbanks L.D., Zameitat E., Marinaki A.M., Simmonds H.A. Pyrimidine pathways in health and disease. Trends in Molecular Medicine. 2005;11:430–437. doi: 10.1016/j.molmed.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Fairbanks L.D., Bofill M., Ruckemann K., Simmonds H.A. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans: disproportionate expansion OF pyrimidine pools and contrasting effects of de novo synthesis inhibitors. Journal of Biological Chemistry. 1995;270:29682–29689. [PubMed] [Google Scholar]
- 52.Kollareddy M., Dimitrova E., Vallabhaneni K.C., Chan A., Le T., Chauhan K.M. Regulation of nucleotide metabolism by mutant p53 contributes to its gain-of-function activities. Nature Communications. 2015;6:7389. doi: 10.1038/ncomms8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pugacheva E.N., Ivanov A.V., Kravchenko J.E., Kopnin B.P., Levine A.J., Chumakov P.M. Novel gain of function activity of p53 mutants: activation of the dUTPase gene expression leading to resistance to 5-fluorouracil. Oncogene. 2002;21:4595–4600. doi: 10.1038/sj.onc.1205704. [DOI] [PubMed] [Google Scholar]
- 54.Cunningham J.T., Moreno M.V., Lodi A., Ronen S.M., Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157:1088–1103. doi: 10.1016/j.cell.2014.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y.-C., Li F., Handler J., Huang C.R.L., Xiang Y., Neretti N. Global regulation of nucleotide biosynthetic genes by c-Myc. PloS One. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mannava S., Grachtchouk V., Wheeler L.J., Im M., Zhuang D., Slavina E.G. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–2400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Real S., Meo-Evoli N., Espada L., Tauler A. E2F1 regulates cellular growth by mTORC1 signaling. PloS One. 2011;6:e16163. doi: 10.1371/journal.pone.0016163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ben-Sahra I., Howell J.J., Asara J.M., Manning B.D. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robitaille A.M., Christen S., Shimobayashi M., Cornu M., Fava L.L., Moes S. Quantitative phosphoproteomics reveal mTORC1 activates de Novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 60.Banerjee D., Gorlick R., Liefshitz A., Danenberg K., Danenberg P.C., Danenberg P.V. Levels of E2F-1 expression are higher in lung metastasis of colon cancer as compared with hepatic metastasis and correlate with levels of thymidylate synthase. Cancer Research. 2000;60:2365–2367. [PubMed] [Google Scholar]
- 61.Floyd S., Favre C., Lasorsa F.M., Leahy M., Trigiante G., Stroebel P. The insulin-like growth factor-I-mTOR signaling pathway induces the mitochondrial pyrimidine nucleotide carrier to promote cell growth. Molecular Biology of the Cell. 2007;18:3545–3555. doi: 10.1091/mbc.E06-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graves L.M., Guy H.I., Kozlowski P., Huang M., Lazarowski E., Pope R.M. Regulation of carbamoyl phosphate synthetase by MAP kinase. Nature. 2000;403:328–332. doi: 10.1038/35002111. [DOI] [PubMed] [Google Scholar]
- 63.Makinoshima H., Takita M., Matsumoto S., Yagishita A., Owada S., Esumi H. Epidermal growth factor receptor (EGFR) signaling regulates global metabolic pathways in EGFR-mutated lung adenocarcinoma. Journal of Biological Chemistry. 2014;289:20813–20823. doi: 10.1074/jbc.M114.575464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santana-Codina N., Roeth A.A., Zhang Y., Yang A., Mashadova O., Asara J.M. Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis. Nature Communications. 2018;9:4945. doi: 10.1038/s41467-018-07472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koundinya M., Sudhalter J., Courjaud A., Lionne B., Touyer G., Bonnet L. Dependence on the pyrimidine biosynthetic enzyme DHODH is a synthetic lethal vulnerability in mutant KRAS-driven cancers. Cell Chemical Biology. 2018;25:705–717. doi: 10.1016/j.chembiol.2018.03.005. e711. [DOI] [PubMed] [Google Scholar]
- 66.Weber G., Shiotani T., Kizaki H., Tzeng D., Williams J.C., Gladstone N. Biochemical strategy of the genome as expressed in regulation of pyrimidine metabolism. Advances in Enzyme Regulation. 1978;16:3–19. doi: 10.1016/0065-2571(78)90064-x. [DOI] [PubMed] [Google Scholar]
- 67.Lowenberg B., Downing J.R., Burnett A. Acute myeloid leukemia. New England Journal of Medicine. 1999;341:1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- 68.Koeffler H.P. Induction of differentiation of human acute myelogenous leukemia cells: therapeutic implications. Blood. 1983;62:709–721. [PubMed] [Google Scholar]
- 69.Christian S., Merz C., Evans L., Gradl S., Seidel H., Friberg A. The novel dihydroorotate dehydrogenase (DHODH) inhibitor BAY 2402234 triggers differentiation and is effective in the treatment of myeloid malignancies. Leukemia. 2019;33:2403–2415. doi: 10.1038/s41375-019-0461-5. [DOI] [PubMed] [Google Scholar]
- 70.Sykes D.B., Kfoury Y.S., Mercier F.E., Wawer M.J., Law J.M., Haynes M.K. Inhibition of dihydroorotate dehydrogenase overcomes differentiation blockade in acute myeloid leukemia. Cell. 2016;167:171–186. doi: 10.1016/j.cell.2016.08.057. e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou J., Quah J.Y., Ng Y., Chooi J.Y., Toh S.H., Lin B. ASLAN003, a potent dihydroorotate dehydrogenase inhibitor for differentiation of acute myeloid leukemia. Haematologica. 2019;104 doi: 10.3324/haematol.2019.230482. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim J., Hu Z., Cai L., Li K., Choi E., Faubert B. CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature. 2017;546:168–172. doi: 10.1038/nature22359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roy B.C., Kohno T., Iwakawa R., Moriguchi T., Kiyono T., Morishita K. Involvement of LKB1 in epithelial-mesenchymal transition (EMT) of human lung cancer cells. Lung Cancer. 2010;70:136–145. doi: 10.1016/j.lungcan.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 74.Qiu B., Wei W., Zhu J., Fu G., Lu D. EMT induced by loss of LKB1 promotes migration and invasion of liver cancer cells through ZEB1-induced YAP signaling. Oncology Letters. 2018;16:6465–6471. doi: 10.3892/ol.2018.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li J., Liu J., Li P., Mao X., Li W., Yang J. Loss of LKB1 disrupts breast epithelial cell polarity and promotes breast cancer metastasis and invasion. Journal of Experimental & Clinical Cancer Research. 2014;33:70. doi: 10.1186/s13046-014-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ceppi P., Volante M., Saviozzi S., Rapa I., Novello S., Cambieri A. Squamous cell carcinoma of the lung compared with other histotypes shows higher messenger RNA and protein levels for thymidylate synthase. Cancer. 2006;107:1589–1596. doi: 10.1002/cncr.22208. [DOI] [PubMed] [Google Scholar]
- 77.Yu Z., Sun J., Zhen J., Zhang Q., Yang Q. Thymidylate synthase predicts for clinical outcome in invasive breast cancer. Histology & Histopathology. 2005;20:871–878. doi: 10.14670/HH-20.871. [DOI] [PubMed] [Google Scholar]
- 78.Burdelski C., Strauss C., Tsourlakis M.C., Kluth M., Hube-Magg C., Melling N. Overexpression of thymidylate synthase (TYMS) is associated with aggressive tumor features and early PSA recurrence in prostate cancer. Oncotarget. 2015;6:8377–8387. doi: 10.18632/oncotarget.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Siddiqui A., Vazakidou M.E., Schwab A., Napoli F., Fernandez-Molina C., Rapa I. Thymidylate synthase is functionally associated with ZEB1 and contributes to the epithelial-to-mesenchymal transition of cancer cells. The Journal of Pathology. 2017;242:221–233. doi: 10.1002/path.4897. [DOI] [PubMed] [Google Scholar]
- 80.Siddiqui A., Gollavilli P.N., Schwab A., Vazakidou M.E., Ersan P.G., Ramakrishnan M. Thymidylate synthase maintains the de-differentiated state of triple negative breast cancers. Cell Death & Differentiation. 2019;26:2223–2236. doi: 10.1038/s41418-019-0289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shaul Yoav D., Freinkman E., Comb William C., Cantor Jason R., Tam Wai L., Thiru P. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 2014;158:1094–1109. doi: 10.1016/j.cell.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu W.-P., Liu Z.-Y., Zhao Y.-M., He X.-G., Pan Q., Zhang N. Dihydropyrimidine dehydrogenase predicts survival and response to interferon-α in hepatocellular carcinoma. Cell Death & Disease. 2018;9:69. doi: 10.1038/s41419-017-0098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kahlert U.D., Joseph J.V., Kruyt F.A.E. EMT- and MET-related processes in nonepithelial tumors: importance for disease progression, prognosis, and therapeutic opportunities. Molecular Oncology. 2017;11:860–877. doi: 10.1002/1878-0261.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alonso S.R., Tracey L., Ortiz P., Pérez-Gómez B., Palacios J., Pollán M. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Research. 2007;67:3450–3460. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- 85.Caramel J., Papadogeorgakis E., Hill L., Browne Gareth J., Richard G., Wierinckx A. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24:466–480. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 86.Wang X., Yang K., Wu Q., Kim L.J.Y., Morton A.R., Gimple R.C. Targeting pyrimidine synthesis accentuates molecular therapy response in glioblastoma stem cells. Science Translational Medicine. 2019;11:eaau4972. doi: 10.1126/scitranslmed.aau4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bageritz J., Puccio L., Piro R.M., Hovestadt V., Phillips E., Pankert T. Stem cell characteristics in glioblastoma are maintained by the ecto-nucleotidase E-NPP1. Cell Death & Differentiation. 2014;21:929–940. doi: 10.1038/cdd.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eyden B., Pandit D., Banerjee S.S. Malignant melanoma with neuroendocrine differentiation: clinical, histological, immunohistochemical and ultrastructural features of three cases. Histopathology. 2005;47:402–409. doi: 10.1111/j.1365-2559.2005.02240.x. [DOI] [PubMed] [Google Scholar]
- 89.Quintana E., Shackleton M., Sabel M.S., Fullen D.R., Johnson T.M., Morrison S.J. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.White R.M., Cech J., Ratanasirintrawoot S., Lin C.Y., Rahl P.B., Burke C.J. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morgenroth A., Deisenhofer S., Glatting G., Kunkel F.H., Dinger C., Zlatopolskiy B. Preferential tumor targeting and selective tumor cell cytotoxicity of 5-[131/125I]iodo-4'-thio-2'-deoxyuridine. Clinical Cancer Research. 2008;14:7311–7319. doi: 10.1158/1078-0432.CCR-08-0907. [DOI] [PubMed] [Google Scholar]
- 92.Reske S.N., Deisenhofer S., Glatting G., Zlatopolskiy B.D., Morgenroth A., Vogg A.T. 123I-ITdU-mediated nanoirradiation of DNA efficiently induces cell kill in HL60 leukemia cells and in doxorubicin-, beta-, or gamma-radiation-resistant cell lines. Journal of Nuclear Medicine. 2007;48:1000–1007. doi: 10.2967/jnumed.107.040337. [DOI] [PubMed] [Google Scholar]
- 93.Sciacovelli M., Goncalves E., Johnson T.I., Zecchini V.R., da Costa A.S., Gaude E. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature. 2016;537:544–547. doi: 10.1038/nature19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schwab A., Siddiqui A., Vazakidou M.E., Napoli F., Bottcher M., Menchicchi B. Polyol pathway links glucose metabolism to the aggressiveness of cancer cells. Cancer Research. 2018;78:1604–1618. doi: 10.1158/0008-5472.CAN-17-2834. [DOI] [PubMed] [Google Scholar]
- 95.Masin M., Vazquez J., Rossi S., Groeneveld S., Samson N., Schwalie P.C. GLUT3 is induced during epithelial-mesenchymal transition and promotes tumor cell proliferation in non-small cell lung cancer. Cancer & Metabolism. 2014;2:11. doi: 10.1186/2049-3002-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kang H., Kim H., Lee S., Youn H., Youn B. Role of metabolic reprogramming in Epithelial-Mesenchymal transition (EMT) International Journal of Molecular Sciences. 2019;20 doi: 10.3390/ijms20082042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Takahashi R.U., Miyazaki H., Takeshita F., Yamamoto Y., Minoura K., Ono M. Loss of microRNA-27b contributes to breast cancer stem cell generation by activating ENPP1. Nature Communications. 2015;6:7318. doi: 10.1038/ncomms8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamaguchi N., Weinberg E.M., Nguyen A., Liberti M.V., Goodarzi H., Janjigian Y.Y. PCK1 and DHODH drive colorectal cancer liver metastatic colonization and hypoxic growth by promoting nucleotide synthesis. eLife. 2019;8:e52135. doi: 10.7554/eLife.52135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kang M., Zheng W., Chen Q., Qin W., Li P., Huang S. Thymidylate synthase prompts metastatic progression through the dTMP associated EMT process in pancreatic ductal adenocarcinoma. Cancer Letters. 2018;419:40–52. doi: 10.1016/j.canlet.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 100.Pastushenko I., Blanpain C. EMT transition states during tumor progression and metastasis. Trends in Cell Biology. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 101.Varga J., Greten F.R. Cell plasticity in epithelial homeostasis and tumorigenesis. Nature Cell Biology. 2017;19:1133. doi: 10.1038/ncb3611. [DOI] [PubMed] [Google Scholar]
- 102.Ye X., Weinberg R.A. Epithelial–mesenchymal plasticity: a central regulator of cancer progression. Trends in Cell Biology. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kröger C., Afeyan A., Mraz J., Eaton E.N., Reinhardt F., Khodor Y.L. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proceedings of the National Academy of Sciences. 2019;116:7353–7362. doi: 10.1073/pnas.1812876116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 105.Aiello N.M., Maddipati R., Norgard R.J., Balli D., Li J., Yuan S. EMT subtype influences epithelial plasticity and mode of cell migration. Developmental Cell. 2018;45:681–695. doi: 10.1016/j.devcel.2018.05.027. e684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mizukoshi K., Okazawa Y., Haeno H., Koyama Y., Sulidan K., Komiyama H. Metastatic seeding of human colon cancer cell clusters expressing the hybrid epithelial/mesenchymal state. International Journal of Cancer. 2019;146:2547–2562. doi: 10.1002/ijc.32672. [DOI] [PubMed] [Google Scholar]
- 107.Le M.T., Hamar P., Guo C., Basar E., Perdigao-Henriques R., Balaj L. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. Journal of Clinical Investigation. 2014;124:5109–5128. doi: 10.1172/JCI75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Blay J., White T.D., Hoskin D.W. The extracellular fluid of solid carcinomas contains immunosuppressive concentrations of adenosine. Cancer Research. 1997;57:2602–2605. [PubMed] [Google Scholar]
- 110.Di Virgilio F. Purines, purinergic receptors, and cancer. Cancer Research. 2012;72:5441–5447. doi: 10.1158/0008-5472.CAN-12-1600. [DOI] [PubMed] [Google Scholar]
- 111.Burnstock G., Di Virgilio F. Purinergic signalling and cancer. Purinergic Signalling. 2013;9:491–540. doi: 10.1007/s11302-013-9372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schneider G., Glaser T., Lameu C., Abdelbaset-Ismail A., Sellers Z.P., Moniuszko M. Extracellular nucleotides as novel, underappreciated pro-metastatic factors that stimulate purinergic signaling in human lung cancer cells. Molecular Cancer. 2015;14:201. doi: 10.1186/s12943-015-0469-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang W.C., Shyh-Chang N., Yang H., Rai A., Umashankar S., Ma S. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 114.Dalin S., Sullivan M.R., Lau A.N., Grauman-Boss B., Mueller H.S., Kreidl E. Deoxycytidine release from pancreatic stellate cells promotes gemcitabine resistance. Cancer Research. 2019;79:5723–5733. doi: 10.1158/0008-5472.CAN-19-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Halbrook C.J., Pontious C., Kovalenko I., Lapienyte L., Dreyer S., Lee H.J. Macrophage-released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell Metabolism. 2019;29:1390–1399. doi: 10.1016/j.cmet.2019.02.001. e1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Diasio R.B., Harris B.E. Clinical pharmacology of 5-fluorouracil. Clinical Pharmacokinetics. 1989;16:215–237. doi: 10.2165/00003088-198916040-00002. [DOI] [PubMed] [Google Scholar]
- 117.Schellens J.H. Capecitabine. Oncologist. 2007;12:152–155. doi: 10.1634/theoncologist.12-2-152. [DOI] [PubMed] [Google Scholar]
- 118.Adjei A.A. Pharmacology and mechanism of action of pemetrexed. Clinical Lung Cancer. 2004;5(Suppl 2):S51–S55. doi: 10.3816/clc.2004.s.003. [DOI] [PubMed] [Google Scholar]
- 119.Scagliotti G.V., Ceppi P., Capelletto E., Novello S. Updated clinical information on multitargeted antifolates in lung cancer. Clinical Lung Cancer. 2009;10(Suppl 1):S35–S40. doi: 10.3816/CLC.2009.s.006. [DOI] [PubMed] [Google Scholar]
- 120.Wilson P.M., Danenberg P.V., Johnston P.G., Lenz H.J., Ladner R.D. Standing the test of time: targeting thymidylate biosynthesis in cancer therapy. Nature Reviews Clinical Oncology. 2014;11:282–298. doi: 10.1038/nrclinonc.2014.51. [DOI] [PubMed] [Google Scholar]
- 121.Elander N.O., Aughton K., Ghaneh P., Neoptolemos J.P., Palmer D.H., Cox T.F. Expression of dihydropyrimidine dehydrogenase (DPD) and hENT1 predicts survival in pancreatic cancer. British Journal of Cancer. 2018;118:947–954. doi: 10.1038/s41416-018-0004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kobayashi H., Koike T., Nakatsuka A., Kurita H., Sagara J., Taniguchi S. Dihydropyrimidine dehydrogenase expression predicts survival outcome and chemosensitivity to 5-fluorouracil in patients with oral squamous cell carcinoma. Oral Oncology. 2005;41:38–47. doi: 10.1016/j.oraloncology.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 123.Taddia L., D'Arca D., Ferrari S., Marraccini C., Severi L., Ponterini G. Inside the biochemical pathways of thymidylate synthase perturbed by anticancer drugs: novel strategies to overcome cancer chemoresistance. Drug Resistance Updates. 2015;23:20–54. doi: 10.1016/j.drup.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 124.Mitra T., Prasad P., Mukherjee P., Chaudhuri S.R., Chatterji U., Roy S.S. Stemness and chemoresistance are imparted to the OC cells through TGFbeta1 driven EMT. Journal of Cellular Biochemistry. 2018;119:5775–5787. doi: 10.1002/jcb.26753. [DOI] [PubMed] [Google Scholar]
- 125.Chu E., Grem J.L., Johnston P.G., Allegra C.J. New concepts for the development and use of antifolates. Stem Cells. 1996;14:41–46. doi: 10.1002/stem.140041. [DOI] [PubMed] [Google Scholar]
- 126.Kitchens M.E., Forsthoefel A.M., Barbour K.W., Spencer H.T., Berger F.G. Mechanisms of acquired resistance to thymidylate synthase inhibitors: the role of enzyme stability. Molecular Pharmacology. 1999;56:1063–1070. doi: 10.1124/mol.56.5.1063. [DOI] [PubMed] [Google Scholar]
- 127.Marverti G., Ligabue A., Paglietti G., Corona P., Piras S., Vitale G. Collateral sensitivity to novel thymidylate synthase inhibitors correlates with folate cycle enzymes impairment in cisplatin-resistant human ovarian cancer cells. European Journal of Pharmacology. 2009;615:17–26. doi: 10.1016/j.ejphar.2009.04.062. [DOI] [PubMed] [Google Scholar]
- 128.Longley D.B., Harkin D.P., Johnston P.G. 5-fluorouracil: mechanisms of action and clinical strategies. Nature Reviews Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 129.Brown K.K., Spinelli J.B., Asara J.M., Toker A. Adaptive reprogramming of De novo pyrimidine synthesis is a metabolic vulnerability in triple-negative breast cancer. Cancer Discovery. 2017;7:391–399. doi: 10.1158/2159-8290.CD-16-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chai J., Dong W., Xie C., Wang L., Han D.L., Wang S. MicroRNA-494 sensitizes colon cancer cells to fluorouracil through regulation of DPYD. IUBMB Life. 2015;67:191–201. doi: 10.1002/iub.1361. [DOI] [PubMed] [Google Scholar]
- 131.Zheng X., Carstens J.L., Kim J., Scheible M., Kaye J., Sugimoto H. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lu W., Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Developmental Cell. 2019;49:361–374. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lu S., Yu L., Mu Y., Ma J., Tian J., Xu W. Role and mechanism of Twist1 in modulating the chemosensitivity of FaDu cells. Molecular Medicine Reports. 2014;10:53–60. doi: 10.3892/mmr.2014.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang X., Ling M.T., Guan X.Y., Tsao S.W., Cheung H.W., Lee D.T. Identification of a novel function of TWIST, a bHLH protein, in the development of acquired taxol resistance in human cancer cells. Oncogene. 2004;23:474–482. doi: 10.1038/sj.onc.1207128. [DOI] [PubMed] [Google Scholar]
- 136.Harada K., Ferdous T., Ueyama Y. Establishment of 5-fluorouracil-resistant oral squamous cell carcinoma cell lines with epithelial to mesenchymal transition changes. International Journal of Oncology. 2014;44:1302–1308. doi: 10.3892/ijo.2014.2270. [DOI] [PubMed] [Google Scholar]
- 137.Uchibori K., Kasamatsu A., Sunaga M., Yokota S., Sakurada T., Kobayashi E. Establishment and characterization of two 5-fluorouracil-resistant hepatocellular carcinoma cell lines. International Journal of Oncology. 2012;40:1005–1010. doi: 10.3892/ijo.2011.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim A.Y., Kwak J.H., Je N.K., Lee Y.H., Jung Y.S. Epithelial-mesenchymal transition is associated with acquired resistance to 5-fluorocuracil in HT-29 colon cancer cells. Toxicological Research. 2015;31:151–156. doi: 10.5487/TR.2015.31.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nishijima N., Seike M., Soeno C., Chiba M., Miyanaga A., Noro R. miR-200/ZEB axis regulates sensitivity to nintedanib in non-small cell lung cancer cells. International Journal of Oncology. 2016;48:937–944. doi: 10.3892/ijo.2016.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yoshida T., Song L., Bai Y., Kinose F., Li J., Ohaegbulam K.C. ZEB1 mediates acquired resistance to the epidermal growth factor receptor-tyrosine kinase inhibitors in non-small cell lung cancer. PloS One. 2016;11 doi: 10.1371/journal.pone.0147344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhou G., Zhang F., Guo Y., Huang J., Xie Y., Yue S. miR-200c enhances sensitivity of drug-resistant non-small cell lung cancer to gefitinib by suppression of PI3K/Akt signaling pathway and inhibites cell migration via targeting ZEB1. Biomedicine & Pharmacotherapy. 2017;85:113–119. doi: 10.1016/j.biopha.2016.11.100. [DOI] [PubMed] [Google Scholar]
- 142.Hsu D.S., Lan H.Y., Huang C.H., Tai S.K., Chang S.Y., Tsai T.L. Regulation of excision repair cross-complementation group 1 by Snail contributes to cisplatin resistance in head and neck cancer. Clinical Cancer Research. 2010;16:4561–4571. doi: 10.1158/1078-0432.CCR-10-0593. [DOI] [PubMed] [Google Scholar]
- 143.Maseki S., Ijichi K., Tanaka H., Fujii M., Hasegawa Y., Ogawa T. Acquisition of EMT phenotype in the gefitinib-resistant cells of a head and neck squamous cell carcinoma cell line through Akt/GSK-3β/snail signalling pathway. British Journal of Cancer. 2012;106:1196–1204. doi: 10.1038/bjc.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dennis M., Wang G., Luo J., Lin Y., Dohadwala M., Abemayor E. Snail controls the mesenchymal phenotype and drives erlotinib resistance in oral epithelial and head and neck squamous cell carcinoma cells. Otolaryngology - Head and Neck Surgery. 2012;147:726–732. doi: 10.1177/0194599812446407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yin T., Wang C., Liu T., Zhao G., Zha Y., Yang M. Expression of snail in pancreatic cancer promotes metastasis and chemoresistance. Journal of Surgical Research. 2007;141:196–203. doi: 10.1016/j.jss.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 146.Chang T.H., Tsai M.F., Su K.Y., Wu S.G., Huang C.P., Yu S.L. Slug confers resistance to the epidermal growth factor receptor tyrosine kinase inhibitor. American Journal of Respiratory and Critical Care Medicine. 2011;183:1071–1079. doi: 10.1164/rccm.201009-1440OC. [DOI] [PubMed] [Google Scholar]
- 147.Li Y., Wu Y., Abbatiello T.C., Wu W.L., Kim J.R., Sarkissyan M. Slug contributes to cancer progression by direct regulation of ERalpha signaling pathway. International Journal of Oncology. 2015;46:1461–1472. doi: 10.3892/ijo.2015.2878. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 148.Tsukasa K., Ding Q., Yoshimitsu M., Miyazaki Y., Matsubara S., Takao S. Slug contributes to gemcitabine resistance through epithelial-mesenchymal transition in CD133+ pancreatic cancer cells. Human Cell. 2015;28:167–174. doi: 10.1007/s13577-015-0117-3. [DOI] [PubMed] [Google Scholar]
- 149.Kikuno N., Shiina H., Urakami S., Kawamoto K., Hirata H., Tanaka Y. Knockdown of astrocyte-elevated gene-1 inhibits prostate cancer progression through upregulation of FOXO3a activity. Oncogene. 2007;26:7647–7655. doi: 10.1038/sj.onc.1210572. [DOI] [PubMed] [Google Scholar]
- 150.Yoo B.K., Emdad L., Su Z.Z., Villanueva A., Chiang D.Y., Mukhopadhyay N.D. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. Journal of Clinical Investigation. 2009;119:465–477. doi: 10.1172/JCI36460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yoo B.K., Gredler R., Vozhilla N., Su Z.Z., Chen D., Forcier T. Identification of genes conferring resistance to 5-fluorouracil. Proceedings of the National Academy of Sciences of the U S A. 2009;106:12938–12943. doi: 10.1073/pnas.0901451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Taube J.H., Malouf G.G., Lu E., Sphyris N., Vijay V., Ramachandran P.P. Epigenetic silencing of microRNA-203 is required for EMT and cancer stem cell properties. Scientific Reports. 2013;3:2687. doi: 10.1038/srep02687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li T., Gao F., Zhang X.P. miR-203 enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Oncology Reports. 2015;33:607–614. doi: 10.3892/or.2014.3646. [DOI] [PubMed] [Google Scholar]
- 154.El Amrani M., Corfiotti F., Corvaisier M., Vasseur R., Fulbert M., Skrzypczyk C. Gemcitabine-induced epithelial-mesenchymal transition-like changes sustain chemoresistance of pancreatic cancer cells of mesenchymal-like phenotype. Molecular Carcinogenesis. 2019;58:1985–1997. doi: 10.1002/mc.23090. [DOI] [PubMed] [Google Scholar]
- 155.Todaro M., Alea M.P., Di Stefano A.B., Cammareri P., Vermeulen L., Iovino F. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 156.Zhang H.-L., Wang P., Lu M.-Z., Zhang S.-D., Zheng L. c-Myc maintains the self-renewal and chemoresistance properties of colon cancer stem cells. Oncology letters. 2019;17:4487–4493. doi: 10.3892/ol.2019.10081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tièche C.C., Peng R.-W., Dorn P., Froment L., Schmid R.A., Marti T.M. Prolonged pemetrexed pretreatment augments persistence of cisplatin-induced DNA damage and eliminates resistant lung cancer stem-like cells associated with EMT. BMC Cancer. 2016;16:125. doi: 10.1186/s12885-016-2117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Haraguchi N., Ishii H., Mimori K., Tanaka F., Ohkuma M., Kim H.M. CD13 is a therapeutic target in human liver cancer stem cells. Journal of Clinical Investigation. 2010;120:3326–3339. doi: 10.1172/JCI42550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang J., Fang C., Qu M., Wu H., Wang X., Zhang H. CD13 inhibition enhances cytotoxic effect of chemotherapy agents. Frontiers in Pharmacology. 2018;9 doi: 10.3389/fphar.2018.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang W., Feng M., Zheng G., Chen Y., Wang X., Pen B. Chemoresistance to 5-fluorouracil induces epithelial–mesenchymal transition via up-regulation of Snail in MCF7 human breast cancer cells. Biochemical and Biophysical Research Communications. 2012;417:679–685. doi: 10.1016/j.bbrc.2011.11.142. [DOI] [PubMed] [Google Scholar]
- 161.Kurtova A.V., Xiao J., Mo Q., Pazhanisamy S., Krasnow R., Lerner S.P. Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance. Nature. 2015;517:209–213. doi: 10.1038/nature14034. [DOI] [PMC free article] [PubMed] [Google Scholar]