Key Points
Acanthamoeba polyphaga mimivirus (APMV) and subsequently discovered giant viruses of amoebae challenge the previous definition of viruses and their classification.
The replication cycle, structure, genomic make-up and plasticity of giant viruses differ from those of traditional viruses. They extend the definition of viruses into a broader range of biological entities, some of which are very simple and others of which have a complexity that is comparable to that of other microorganisms.
Giant viruses of amoebae have virus particles as large as some microorganisms that are visible by light microscopy and that have a stunning level of complexity. Their genomes are mosaics and contain large repertoires of genes, some of which are hallmarks of cellular organisms, although the majority of which have unknown functions.
Mimiviruses are associated with a specific mobilome and are parasitized by viruses that they can defend against.
Several hypotheses on the ancient origin and evolutionary relationship between cellular organisms and giant viruses of amoebae have been proposed, and these topics continue to be debated.
The detection of giant viruses of amoebae in humans and the study of their potential pathogenicity are emerging fields.
Supplementary information
The online version of this article (doi:10.1038/nrmicro.2016.197) contains supplementary material, which is available to authorized users.
Subject terms: Viral evolution, Virus structures
The discovery of the giant amoebal virus mimivirus, in 2003, opened up a new area of virology. Extended studies, including those of mimiviruses, have since revealed that these viruses have genetic, proteomic and structural features that are more complex than those of conventional viruses.
Supplementary information
The online version of this article (doi:10.1038/nrmicro.2016.197) contains supplementary material, which is available to authorized users.
Abstract
The accidental discovery of the giant virus of amoeba — Acanthamoeba polyphaga mimivirus (APMV; more commonly known as mimivirus) — in 2003 changed the field of virology. Viruses were previously defined by their submicroscopic size, which probably prevented the search for giant viruses, which are visible by light microscopy. Extended studies of giant viruses of amoebae revealed that they have genetic, proteomic and structural complexities that were not thought to exist among viruses and that are comparable to those of bacteria, archaea and small eukaryotes. The giant virus particles contain mRNA and more than 100 proteins, they have gene repertoires that are broader than those of other viruses and, notably, some encode translation components. The infection cycles of giant viruses of amoebae involve virus entry by amoebal phagocytosis and replication in viral factories. In addition, mimiviruses are infected by virophages, defend against them through the mimivirus virophage resistance element (MIMIVIRE) system and have a unique mobilome. Overall, giant viruses of amoebae, including mimiviruses, marseilleviruses, pandoraviruses, pithoviruses, faustoviruses and molliviruses, challenge the definition and classification of viruses, and have increasingly been detected in humans.
Supplementary information
The online version of this article (doi:10.1038/nrmicro.2016.197) contains supplementary material, which is available to authorized users.
Main
The discovery of giant viruses of amoebae changed the field of virology1,2. Since the nineteenth century, viruses have been defined by their submicroscopic size, and this was a dogmatic obstacle that probably prevented (for decades) researchers from searching for, and discovering, giant protozoan viruses, which can be visualized by light microscopy using the usual dyes3 (Box 1; Fig. 1). Indeed, for years the Acanthamoeba polyphaga mimivirus (APMV) was considered to be an intracellular bacterium of amoebae1 (Box 2). In a similar manner, pandoraviruses were thought to be atypical parasites of amoebae4. However, the absence of ribosomal DNA from an isolate that was presumed to be a bacterium eventually led to the discovery of APMV1,2, and, interestingly, the absence of a ribosome essentially separates giant viruses from the three defined domains of life. The genomic and proteomic analyses of giant viruses revealed that they are more complex than other viruses. Furthermore, giant viruses have several novel biological features, which include the fact that mimiviruses can be infected by parasitic viruses (termed virophages), they can contain mobile DNA elements, (termed transpovirons), and they have a defence mechanism against virophages termed the mimivirus virophage resistance element (MIMIVIRE)5,6,7. Thus, how should giant viruses, which do not have a ribosome but have a level of complexity that is approaching that of numerous bacteria and even eukaryotes with an intracellular growth, be defined8 (Box 1; Supplementary information S1 (table))? Furthermore, there is disagreement as to whether the large genomes of giant viruses are a result of smaller viruses acquiring genes or of a genome with cellular ancestry adapting to escape the cell nucleus2,9,10,11,12,13. In this Review, we reflect on 13 years of research into giant viruses and detail the advances that have been made in characterizing their genomes, structures and mechanisms of replication, as well as the virophages and mobile elements with which they are associated. We place these findings in the context of the ongoing debate on the evolutionary origin of giant viruses and on the extent of similarity between giant viruses and bacteria and eukaryotic cells.
Figure 1. Particle and genome size of giant viruses of amoebae.
Families (namely, the Mimiviridae and Marseilleviridae; represented by dark blue circles) or putative families of giant viruses of amoebae (namely, those that include pandoraviruses, pithoviruses, faustoviruses and mollivirus; dark blue circles) are shown, along with other families in the proposed order Megavirales (namely, Poxviridae, Asfarviridae, Phycodnaviridae, Iridoviridae and Ascoviridae; light blue circles). Some families or genera of smaller viruses (grey circles) are shown in the inset, which magnifies a section of the larger graph and shows viruses that have a genome size ≤400 kb and a particle size ≤400 nm. Circle sizes are proportional to virus particle sizes. For each family or genus, the size of the largest member is shown. Viruses are referred to by their family or genus name unless there is no family or genus name.
Box 1: Defining viruses and microorganisms and their origin and evolution.
The definition of viruses and giant viruses of amoebae
During the 1950s, Lwoff introduced a definition of viruses that was based mainly on negative criteria112 (Supplementary information S1 (table)). Viruses were considered as simple entities that relied on a cellular host to replicate, were devoid of machineries for energy production and translation, and consisted of a nucleic acid enclosed in a symmetrical protein shell known as the capsid112,113. In 2003, the discovery of Acanthamoeba polyphaga mimivirus (APMV) challenged this paradigm and caused debates in regard to the definition and classification of viruses2,8,55. Although APMV has many of the phenotypic and genotypic characteristics of viruses, it also shares many features with microorganisms55. For example, APMV particles are visible using a light microscope1, have larger genomes than small bacteria and contain more diverse genes than other viruses2. Moreover, APMV is more complex than other viruses, as its capsid contains two types of nucleic acid — a DNA genome and some mRNAs2 — as well as 114 proteins19. In addition, APMV contains translation factors and aminoacyl-tRNA synthetases (which is unprecedented for a virus), and six tRNAs (tRNA is rarely detected in other double-stranded DNA (dsDNA) viruses)2. Furthermore, the mobilome of APMV is more diverse than that of other viruses and includes several self-excising introns and inteins, as well as pro-virophages and transpovirons5,6. Finally, unlike many viruses, APMV can enter host cells through phagocytosis that does not require specific viral–host interactions, owing to its large size76; this may mean that APMV has a broad host range.
The origin of giant viruses of amoebae
The presence of genes that are homologous to bacterial, archaeal and eukaryotic genes in APMV, marseillevirus and other nucleocytoplasmic large DNA viruses (NCLDVs)2 has enabled phylogenetic reconstructions and comparative analyses of gene repertoires, the results of which have been interpreted as evidence for a 'fourth domain' in the tree of life2,50,54 alongside Bacteria, Archaea and Eukarya, which are domains that were proposed by Woese in 1977. Such a phylogeny would suggest an archaic origin that is contemporary to that of proto-eukaryotes13,52. As these analyses did not involve ribosomal genes, it was proposed that APMV and the other members of the proposed order Megavirales comprise a fourth 'TRUC' (things resisting uncompleted classification) of microorganisms rather than a fourth domain3. Phylogenetic studies that were based on entire repertoires of the structural domains of proteins51 also suggested that a fourth major branch of the tree of life may exist. However, these claims are controversial and have been contested by arguments that the phylogenies were biased by lateral gene transfer from cellular organisms (mostly eukaryotes) to giant viruses and inappropriate methodologies10,12,13,114,115. Therefore, the evolutionary origins of giant viruses remain under debate.
Box 2: The discovery of mimivirus.
In the early 1980s, Rowbotham first described the interaction between Legionella pneumophila and amoebae in the genus Acanthamoeba119. Amoebae are host cells for Legionella spp., and, during the 1980s and 1990s, Rowbotham assembled a large collection of amoeba-associated bacteria from environmental water samples that were gathered as part of investigations into outbreaks of Legionnaires' disease. These bacteria were mostly strict intracellular Legionella spp.104,120, but two isolates were unidentified and, by their morphology, seemed to be small Gram-positive coccoid bacteria121. In 1995, Birtles brought this collection to the laboratory of D. Raoult with the objective of characterizing the two unidentified isolates by 16S ribosomal DNA (rDNA) amplification and sequencing. All attempts to amplify the isolate that became known as the 'Bradford coccus' were unsuccessful and, as an alternative strategy, the ultrastructure of this isolate was studied by electron microscopy1. Unexpectedly, the electron micrographs showed that the coccoid 'bacterium' in infected amoebae consisted of very regular icosahedral forms that resembled a giant iridovirus. Additional observations suggested that, despite its large diameter of ∼600 nm, the Bradford coccus was in fact a virus, as the isolate exhibited eclipse-phase replication, which is typical of viruses, the production of virus particles into 'virus factories' and contained a large double-stranded DNA (dsDNA) chromosome that, according to preliminary genome analyses, belonged to the group of viruses that are known as nucleocytoplasmic large DNA viruses (NCLDVs)47. These studies revealed that the Bradford coccus was a virus, and it was subsequently renamed 'mimivirus', with an etymology that is based both on the concept of 'mimicking a microorganism' and on a childhood recollection of stories of 'Mimi the amoeba'.
Structure and genomes of giant viruses
APMV and other giant viruses of amoebae have several structural and genomic features that had not been described in viruses before their discovery.
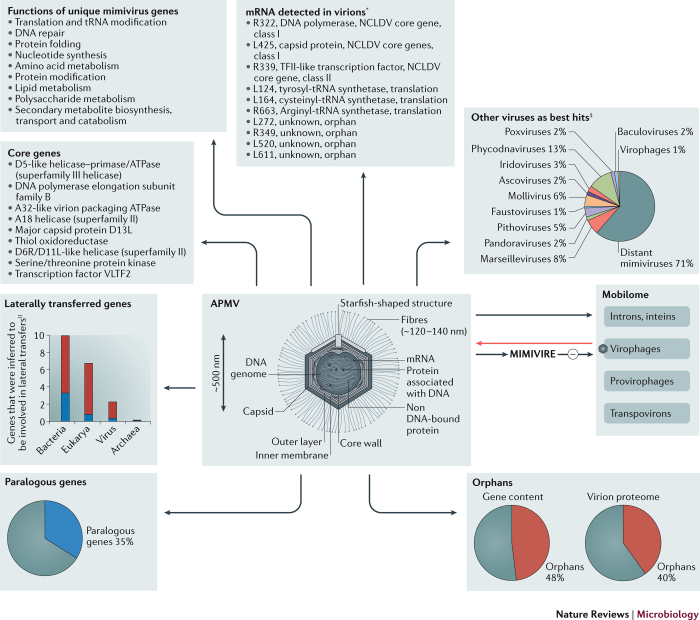
Understanding APMV. APMV has remarkable features compared with other viruses. The APMV capsid is ∼500 nm in size and is covered by fibrils that are ∼120–140 nm long and 1.4 nm thick1. These fibrils are morphologically unique among viruses and, although their structures have not been fully elucidated1,14 (Supplementary information S2 (figure)), they form a dense layer, are extensively glycosylated, and enable the attachment of APMV to amoebae, bacteria, arthropods and fungi through glycans15. The capsid comprises proteins that have a double jelly-roll fold and is icosahedral except at one vertex, which is covered by a unique five-branch starfish-shaped structure (termed 'stargate') that is devoid of fibres16. Beneath the capsid, and surrounded by an inner lipid membrane and fibres, is a spherical lipid bilayer compartment that is ∼340 nm in size and contains the genome (with an estimated packing density of ∼0.06 nm3 per bp) and proteins17,18. This nucleocapsid has a large depression that faces the 'stargate', which creates a cavity and indicates that the nucleocapsid has a fixed position relative to the external capsid. APMV particles contain 114 proteins, which is only 12% of the number of predicted genes (see below), among which 12 are involved in transcription, 5 are involved in DNA topology and repair, 2 are involved in RNA modification, 5 are involved in particle structure and 7 are involved in protein or lipid modifications19.
APMV has a double-stranded DNA (dsDNA) genome that is 1.2 Mb in length and contains 979 genes that putatively encode proteins with a coding density of 89%2,20 (Fig. 2; Supplementary information S3 (table)). The genome is also AT rich, comprising 72% AT nucleobases. Several APMV genes are not found in viruses other than giant viruses of amoebae, including those that encode translation factors and aminoacyl tRNA synthetases; some of the genes encoding aminoacyl tRNA synthetases are expressed21,22 and encode functional proteins23. In addition, the APMV genome encodes four different tRNAs2. Other genes that are unique to APMV and other giant viruses of amoebae encode proteins that are involved in nucleotide synthesis, amino acid metabolism, protein modification, lipid or polysaccharide metabolism, DNA repair or protein folding2. In addition, the APMV genome contains four major groups of genes, including core genes that are shared with poxviruses, ascoviruses, iridoviruses, asfarviruses, phycodnaviruses and other giant viruses of amoebae, as well as large sets of horizontally transferred genes, paralogous genes (in addition to genes that were involved in large genome duplication events) and orphan genes (also known as ORFan genes)2,9,24,25,26. Orphan genes are estimated to represent ∼48% of the predicted gene content2,27, and the proportion of orphan-encoded proteins is also very high (40%) in APMV particles19. The APMV genome also contains early and late gene promoters, and mRNAs are expressed as polyadenylated transcripts that most often end with short palindromic sequences that form hairpin-like structures2,22,28. Recoding events, including a frameshift and a readthrough, were described for a gene that encodes a translation termination factor in the APMV genome29. In addition, introns and inteins were detected in a few conserved genes, including those that encode the major capsid protein and the DNA polymerase30,31, and a 'mobilome' that is unique to APMV was identified6 (see below). Finally, a few mRNAs were detected in mimivirus capsids2 (Fig. 2).
Figure 2. Major genomic and structural features of APMV.
Major structural features of the Acanthamoeba polyphaga mimivirus (APMV) particle are shown in the middle of the figure. Black arrows link APMV to the major features of the gene repertoire and nucleic acid content of APMV, including its mobilome and mRNAs, which are shown in grey boxes. The red arrow indicates that virophages can infect mimivirus factories. The minus sign indicates that mimivirus virophage resistance element (MIMIVIRE) can protect against virophages. *Gene definitions and putative functions are given, as well as their relationship to other viral genes. ‡Corresponds to mimivirus genes, the closest match to which in the NCBI GenBank protein sequence database is from a virus that does not belong in the family Mimiviridae (except distant mimiviruses). || Maximum and minimum proportions of genes that are inferred to be involved in lateral transfer are given, as assessed in three different studies9,25,26. NCLDV, nucleocytoplasmic large DNA viruses; VLTF2, viral late transcription factor 2.
Other giant viruses of amoebae. In 2005, APMV became the founding member of the family Mimiviridae32. Since then, about 100 new mimivirus strains have been isolated by culturing on amoebae from water, soil, insect and human samples that were collected worldwide (Supplementary information S3,S4 (table, figure)), most recently using high-throughput strategies33,34,35 (Box 3); the second mimivirus to be cultured was named mamavirus5. The sizes, morphologies and genomes of the other mimivirus isolates are similar to those of APMV30,33,36. They have capsids 370–600 nm in diameter and their genomes are 1.02–1.26 Mb in length, AT rich (72–75% AT nucleobases) and encode 930–1,120 putative proteins. Phylogenomics has enabled mimiviruses that infect amoebae to be divided into three lineages that were named lineage A (in which APMV is the prototype), lineage B (in which moumouvirus is the prototype)30 and lineage C (in which Megavirus chiliensis is the prototype)36. In 2010, a distant mimivirus relative named Cafeteria roenbergensis virus, the capsid and genome of which are smaller than those of the mimiviruses of Acanthamoeba, was isolated from an abundant marine dinoflagellate37. Subsequently, a few other viruses that infect marine unicellular eukaryotes, including Phaeocystis globosa virus, were linked, albeit distantly, to mimiviruses38,39,40.
In addition to mimiviruses, other giant viruses of amoebae were discovered using amoebal co-culture methods. The first of these was marseillevirus, which was described in 2009 (Ref. 41). Since 2013, the number and diversity of giant viruses of amoebae have expanded considerably, and, as of 2016, two virus families, the Mimiviridae and Marseilleviridae, have been described32,42. However, other giant viruses, including pandoraviruses, pithoviruses, faustoviruses and Mollivirus sibericum, represent putative new giant virus families5,43,44,45,46 (Supplementary information S3,S5,S6 (table, table, figure)). Since the APMV genome was described, giant viruses of amoebae have been linked, through phylogenomic analyses, to nucleocytoplasmic large DNA viruses (NCLDV), which is a group of dsDNA viruses that was described in 2001 and comprises poxviruses, ascoviruses, iridoviruses, asfarviruses and phycodnaviruses; these viruses infect a wide range of eukaryotic cells, from algae to insects and mammals2,47,48. In 2009, it was noted that giant viruses of amoebae and NCLDVs share a small subset of nine core genes, five of which are found in all of their genomes (these encode a major capsid protein, a D5 helicase, a family B DNA polymerase, an A32-like packaging ATPase and a very late transcription factor), and a larger subset of ∼200 genes are shared by at least two NCLDV families48,49. Moreover, giant viruses of amoebae and NCLDVs were described to have a common ancestor, the genome of which is thought to contain ∼50 conserved genes that are likely, based on phylogenomic analyses, to have an early origin that is possibly concomitant with eukaryogenesis48,49,50,51,52. In 2012, it was proposed that giant viruses of amoebae and NCLDV families should be classified into a new viral order, the Megavirales, on the basis of their common origin, which was suggested by the fact that they share a large set of ancestral genes that encode key viral functions, a common virus particle architecture and major biological features, including replication that occurs inside cytoplasmic factories53. Nevertheless, the architecture of pandoraviruses, Pithovirus sibericum and M. sibericum (see below) differs considerably from that of other giant viruses of amoebae and no capsid- resembling structure, or even gene for pandoraviruses, was detected in these viruses, which, together with poxviruses and ascoviruses that have brick-shaped virus particles and allantoid capsids, respectively, challenges the criteria that are used to classify viruses in the proposed order Megavirales43,44,46,53.
Members of the proposed order Megavirales that were isolated as a result of them being cultured on various amoebae and described during the past 13 years have a wide range of sizes, shapes, structures, genome lengths, percentage of GC nucleobases, gene repertoires and replicative sites (Supplementary information S3,S5,S6,S7 (table, table, figure, figure)). Nonetheless, they still comprise a monophyletic clade that is based on a limited set of core genes and informational genes48,49,53,54,55. In addition, all of these giant viruses enter amoebae through phagocytosis, after which fusion occurs between the vacuole membrane and the internal membrane of the virus; this leads to the release of the genome into the cytoplasm of the amoeba56,57. Finally, the virus factory has a cytoplasmic location, with the exception of M. sibericum46.
Giant viruses of amoebae were isolated from various environmental samples, ecosystems and geographical locations, and from hosts, including amoebae, invertebrates and mammals33,57,58. They were also detected in several metagenomes that were generated from environmental, animal and human samples59,60,61,62,63,64, and in plant genomes65. Finally, sequences from new putative giant viruses were detected in marine environmental metagenomes by approaches that revealed that previously overlooked sequences are related to members of the Megavirales54,66,67. These facts suggest that giant viruses of amoebae are common in our biosphere.
Giant viruses of amoebae have various morphological features (Supplementary information S3,S5,S6 (table, table, figure)). Marseilleviruses and faustoviruses have icosahedral capsids that are 250 nm and 200 nm in size, respectively, and have no 'stargate' structure41,45. The surface of the marseillevirus prototype isolate is covered by fibres that are 12 nm long, whereas the faustovirus prototype isolate has no fibres. Contrary to all other DNA viruses, the faustovirus prototype isolate has a double protein shell to encapsidate, with an outer shell that is composed of a double jelly-roll protein and is similar to that of many double-stranded DNA viruses (including giant viruses of amoebae), and an inner shell that is composed of a repeated hexameric unit, the structure of which differs from that of known capsid proteins. In addition, the major capsid protein of the outer shell is encoded by a 17,000 bp genomic fragment that contains several introns and exons68. Pandoraviruses, pithoviruses and M. sibericum exhibit different morphologies to other giant viruses of amoebae; virus particles have ovoid or spherical (in the case of M. sibericum) shapes43,44,46. In addition, pandoraviruses and pithoviruses are larger than mimiviruses and all other giant viruses of amoebae. Pandoravirus and pithovirus particles have a wall that is 60–70 nm thick and an apical pore at one of their extremities, which enables the internal virus components to be delivered into the amoebal cytoplasm and has the appearance of a honeycomb grid in pithoviruses43,44. The M. sibericum tegument is covered by two layers of fibres that are ∼10–14 nm long and has a funnel shaped aperture at its apex46.
The genomes of giant viruses of amoebae other than mimiviruses have different lengths, GC content, gene numbers, functions and origins2,40,42,43,44,45 (Supplementary information S6,S7 (figures)). They all include numerous orphan genes, similarly to APMV, which indicates that many of their structural and functional characteristics are novel and remain to be deciphered2,41,43,44,45,46. In addition, these genomes all have a substantial level of mosaicism; this is well exemplified by marseillevirus41. This suggests that many genes were transferred laterally between themselves and other viruses and cellular organisms. Among examples of particular features in the genomes of giant viruses of amoebae, it should be mentioned that the genomes of marseillevirusesen code histone-like proteins41,69; the genes in pandoraviruses frequently include spliceosomal introns43 and the genome of Pandoravirus salinus contains particular transposons that were termed miniature inverted-repeat transposable elements70; and the genomes of faustoviruses encode membrane occupation and recognition nexus (MORN)-repeat-containing proteins, similar to marseilleviruses and pandoraviruses44. In the faustovirus prototype isolate it was found that the capsid protein is encoded by a ∼17,000 bp large genomic region with several exons and introns, which was most unexpected45,68. Of note, it has been hypothesized that giant virus genomes may evolve through a complex accordion-like process, with successive steps of genome expansions through duplications and gene transfers followed by genome reduction71. Hence, the genomes of giant viruses of amoebae both exhibit features that are shared between some or all giant viruses of amoebae and others that are particular to each family or putative family.
Giant viruses of amoebae that are living fossils. P. sibericum and M. sibericum were isolated from a Siberian permafrost sample that was more than 30,000 years old, which dates back to the Pleistocene epoch, and were therefore considered to be 'living fossils' (Ref. 44). P. sibericum has an AT-rich (64%) dsDNA genome that is 610,033 bp in length, which is unexpectedly small considering that it has the largest particle size (∼1.5 μm in length and 0.5 μm in diameter) among giant viruses of amoebae44. Moreover, its coding density is only 68%, owing to a considerable number of ∼150-nm-long regularly interspersed palindromic sequences that are distributed in tandem repeats within arrays that are ∼2,000 bp long. Although P. sibericum morphologically resembles pandoraviruses, it is most closely related to marseilleviruses and iridoviruses based on phylogenetic analyses. A total of 159 proteins were identified in purified virus particles, of which two-thirds and the four most abundant have unknown functions44. The genome of M. sibericum is 651,523 bp in size, with inverted repeats approximately 10,000 bp long at its extremities, and, similar to pandoraviruses, is GC rich (60%)46. The majority of the sequences that are most similar to genes in M. sibericum correspond to pandoravirus genes, albeit with a low level of homology (about 40% on average), and this result is congruent with phylogenetic analyses. Purified virus particles contain 136 proteins, of which more than half and the three most abundant are orphans. In addition, a homologue of the major capsid protein in members of the proposed order Megavirales is translated, although it is apparently not involved in the structure of the virus particle46.
Another pithovirus, named Pithovirus massiliensis, was recently isolated from sewage in southern France and was compared with its fossil counterpart72. This analysis indicated that the genomic content of pithoviruses evolves slowly, as it was conserved after thousands of years, with selective pressure on the conserved genes. This suggests that the mechanisms of evolution are comparable in giant viruses and bacteria, and include selection, gene fixation and then selective constraints. This enabled the first estimation of a molecular clock in giant viruses, and the mutation rate that was estimated based on the dN/dS ratio was 2 × 10−6 substitutions per site per year, which is lower than that of RNA viruses and is very similar to that estimated for poxviruses (0.5–7 × 10−6 substitutions per site per year)73 and in the same order of magnitude as those found in bacteria and archaea72. Second, this showed that genes that were thought to be horizontally acquired were selected and highly conserved, which indicates that they are essential genes. The GC content and codon usage of these genes tend to adapt progressively to that of the recipient genome72. In light of these results, pithoviruses can select for genes that are acquired by horizontal gene transfer, followed by their long-term fixation and adaptation to viral codon usage. Third, orphans that are highly abundant in pithoviruses44,72 were also constrained by strong selective pressure, which indicates that their accumulation is not random and is biologically relevant. These findings do not support the hypothesis that giant viruses are 'bags' of genes and pseudogenes that have been randomly taken from cellular organisms and not used, but rather indicate that horizontally acquired genes and orphan genes are functional and biologically active. Similar findings were reported previously for two marseilleviruses that were isolated in south-eastern France and Australia74.
Box 3: Strategies for the discovery of giant viruses.
The use of amoebae as a support to isolate intra-amoebal microorganisms (known as the 'amoebal enrichment method' (Ref. 116)) led to the isolation of the bacterium Legionella pneumophila. However, as samples that were grown using this technique were often contaminated by extracellular and intracellular bacteria, antibiotics and antifungal compounds were subsequently added to the media33,43,45,94,109. The antifungal amphotericin B limited the viability of amoebae, but this drawback was overcome by the adaptation of Acanthamoeba castellanii to higher concentrations of amphotericin B43, the use of thiabendazole as an antifungal, which is less toxic to amoebae45, and then the use of voriconazole, which is not toxic to amoebae, even at high doses. In addition, procedures to pre-enrich samples for amoebae, including the addition of nutrients to water samples followed by their incubation in the dark36,94, or the use of Prescot and James medium to resuspend the sample prior to culture, were also established44.
More recently, an automated system for the high-throughput isolation of giant viruses, based on the detection of lysed protozoa in liquid media by flow cytometry (virus particles stained by SYBR Green are sorted based on their size and DNA content)35, was implemented. Furthermore, whereas the use of Acanthamoeba polyphaga as an amoebal host for culture only enabled the isolation of mimiviruses and marseilleviruses, the culture of A. castellanii led to the isolation of Pandoravirus spp.43, Pithovirus sibericum44 and Mollivirus sibericum46. This shows that A. polyphaga is less permissive to infection by several giant viruses of amoebae as a support for culture than A. castellanii. Changing the amoebal host was especially fruitful in the isolation of several faustoviruses45, which form a new putative family of giant viruses that are specific to the host Vermamoeba vermiformis117. However, each host must be adapted to a liquid medium that enables minimal survival before the multiplication of viruses. For example, the quick encystment of V. vermiformis and its inability to survive in Page's amoeba saline (PAS) prevented its use as a host for the isolation of giant viruses, although this limitation could be circumvented by using a starvation medium in which this host could survive for culture45. Varying the host species has also provided insight into host specificity; as assessed with a large set of giant viruses of amoebae, a sample could be positive for viruses when inoculated on a given Acanthamoeba species and negative when inoculated on others118.
Finally, a major challenge is to separate viruses when virus mixtures are suspected by current flow cytometric gating strategies, owing to the fact that the end-point dilution method is time consuming and favours the isolation of fast-growing viruses. Fluorescence-activated cell sorting (FACS) for the isolation of giant viruses is under development in our laboratory with encouraging preliminary results.
Infection cycle of giant viruses
The replicative cycles of giant viruses in amoebal cells show several similarities, including phagocytic entry, DNA release and replication in 'viral factories'. However, differences exist between giant viruses in the duration of their replication cycles, the involvement of the amoebal nucleus in virus replication, and the assembly and release of virus particles.
The replicative cycle of APMV, which lasts about 12 h and occurs in the amoebal cytoplasm1,56,75 (Fig. 3), has several features that have not been observed before in viruses. APMV is internalized through phagocytosis76 before its genome-containing internal core is released through the 'stargate' into the cytoplasm, through the fusion of viral and phagosome membranes; transcription may be initiated in these cores77. Subsequently, the genome is released from the core and is replicated at high levels, which generates an early cytoplasmic replicative centre that is thought to be engulfed by a membrane layer of the endoplasmic reticulum. Replicative centres then merge into a single virus factory that contains, from the inside to the outside, zones that are involved in replication, membrane biogenesis, capsid assembly and DNA packaging, and fibre acquisition56,77. The virus internal layer seems to originate from multivesicular membrane structures that bud from the endoplasmic reticulum and become open membrane sheets; the major capsid protein is assembled around these sheets while acting as a scaffold78. Genome packaging occurs through a transient aperture that is distinct from the 'stargate' (Ref. 78). Finally, the layer of fibres is assembled. The involvement of the amoebal nucleus in the life cycle of APMV has been debated56,57,75. Indeed, the size of the amoebal nucleus decreases approximately twofold during APMV replication, even though this process occurs in a viral factory in the cytoplasm; amoebal nuclear factors may be involved.
Figure 3. The mimivirus replication cycle and key replicative features of other giant viruses of amoebae.
a | Schematic of the replication cycle of Acanthamoeba polyphaga mimivirus (APMV). Virus particles can be seen at the surface of an Acanthamoeba sp. amoeba at the first stage of the cycle. Then, after virus entry through phagocytosis, the eclipse phase begins and the release of the APMV genome into the amoebal cytoplasm seeds the virus factory, which appears at a different location to the cell nucleus. Starting from ∼8 h post-infection, the assembly of virus particles can be seen at the periphery of the virus factory. Amoebal lysis occurs ∼12 h post-infection. b–e | Electron microscopy images of APMV particles and Acanthamoeba sp. cells that are infected with APMV. b | A mimivirus particle is shown. c | A mimivirus factory in the cytoplasm of an Acanthamoeba sp. amoeba 8 h post-infection. d | This image shows the edge and periphery of a virus factory, in which internal membrane biogenesis and assembly, capsid assembly and DNA packaging, and fibre acquisition of the APMV particles occur 8 h post-infection. e | This image shows an Acanthamoeba sp. amoeba ∼12 h post-infection with mimivirus, with the amoebal cytoplasm filled with mimivirus particles. f | The key replicative features of giant viruses of amoebae are shown in this table. Dashes indicate where there is no noticeable feature for a virus. A. castellanii, Acanthamoeba castellanii; V. vermiformis, Vermamoeba vermiformis. Microscopy images in parts b–e courtesy of I. Pagnier, Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, Aix Marseille University, France.
In marseilleviruses, the replication cycle lasts about 6–16 h (Refs 41,69) (Fig. 3). The cell nucleus undergoes transient morphological changes during early stages of the replication cycle, and marseillevirus viral factories tend to have a more extensive distribution in the cytoplasm than APMV viral factories. Marseilleviruses can form giant vesicles, comprised of dozens of virus particles, that are wrapped by membranes derived from the amoebal endoplasmic reticulum79. The viruses can be released outside of the amoeba within these giant vesicles; this may cause the amoeba to undergo phagocytosis through an acidification-independent process79. In addition, single marseillevirus particles may enter Acanthamoeba spp. through an endosome-stimulated pathway or may group together to trigger their own phagocytic uptake79.
The replication cycle of faustoviruses occurs in Vermamoeba vermiformis (not in Acanthamoeba spp.) and lasts 18–20 h (Ref. 45). It occurs within the amoebal cytoplasm independently of the cell nucleus, and the viral factory occupies almost the entire amoebal cell, although it should be noted that some virus particles that are produced by the factory may only correspond to empty capsids.
The life cycles of pandoraviruses, P. sibericum and M. sibericum last ∼10–18 h, 10–20 h and 6 h, respectively43,44,46. The wall and interior of these virus particles seem to assemble simultaneously. The replication of P. sibericum does not seem to affect the amoebal nucleus44. By contrast, infection by pandoraviruses and M. sibericum results in a disorganized amoebal nucleus that exhibits numerous membrane invaginations43,46. The pandoravirus factory seems to replace the cell nucleus, whereas the nucleus persists during infection with M. sibericum, with new virus particles emerging at its periphery. These observations are consistent with the fact that transcription-associated proteins are absent from pandoravirus and mollivirus particles, which suggests that they require the amoebal nucleus, but are present in pithovirus particles, which suggests that they can initiate transcription independently from the nucleus57.
Overall, giant viruses of amoebae enter cells through a phagocytic-like process and release hundreds of new virus particles through amoebal lysis; the exception is M. sibericum, for which the release of virus particles seems to occur through exocytosis without lysis46. Acanthamoeba spp. or V. vermiformis can protect giant viruses against physical and chemical threats, as they are highly stable organisms that can become encysted80. In addition, APMV particles were stable for 9 months in environmental freshwater, saline water and hospital ventilator devices81, and resisted antibiotics, 48 h of desiccation, and several chemical biocides, including alcohols82. Finally, the phagocytosis-like entry process of giant viruses contrasts with the need of specific interactions for other viruses to enter their host and could enable giant viruses to infect a broader range of hosts. Although amoebae are the primary hosts of giant viruses, mimiviruses and marseilleviruses have been detected in oysters, insects, monkeys, cattle and humans83,84,85. Moreover, APMV can enter various human myeloid cells and replicate in peripheral blood mononuclear cells76,86, and marseillevirus can cause a productive infection in human T lymphocyte cells87. In addition, the virus factories in which these giant viruses replicate were described for other viruses, including members of the proposed order Megavirales that are not discussed here, other DNA viruses (including herpesviruses) and some RNA viruses (such as flaviviruses or coronaviruses)88. These factories are replication organelles that recruit viral and cellular components for virus assembly and maturation. Viral factories were considered as the nuclei of 'virocells' (that is, cells infected by a virus, the aim of which is to produce virions), which would themselves correspond to the cellular forms of viruses when not strictly assimilated into a virus particle (or virion)26. In the case of APMV and other giant viruses of amoebae, if the virocell is the infected amoeba, the giant viral factory can be considered as its nucleus. This view is connected to two evolutionary scenarios: viral eukaryogenesis13, in which the eukaryotic nucleus originated from the viral factory of an ancient giant virus26, and nuclear viriogenesis, in which some giant viruses originated from the nucleus of ancient proto-eukaryotic cells89.
Virophages and other mobile elements
The discovery and study of mimiviruses led to the identification of a 'mobilome' that is specific to these giant viruses and comprises viruses that can infect their viral factories and integrate into their genomes.
Virophage discovery and diversity. Virophages, which were discovered in 2008 (Ref. 5), are viruses that infect mimivirus factories. They have small virus particles that are 35–74 nm in diameter and have an icosahedral capsid and a dsDNA genome of 17–29 kb. Their genomes are predicted to contain 16–34 genes, most of which are orphans or genes of unknown function, and six core genes90,91. Virophages cannot replicate alone in their amoebal host but, instead, replicate only in the presence of mimiviruses5. A co-culture procedure that uses a culturable helper virus was established to explore virophage diversity, and it enabled the isolation of new virophages and the analysis of their ability to infect mimivirus lineages92. The first virophage, named Sputnik, was isolated with mamavirus and was shown to impair its replication cycle and morphogenesis5. Specifically, the ability of mamavirus to lyse amoebae was decreased and mamavirus particles with abnormal morphologies were observed. So far, three Sputnik-like virophages, namely Sputnik2, Sputnik 3 and Rio Negro virophage, have been isolated, and they all infect lineage A, lineage B and lineage C of the Mimiviridae92,93,94. A divergent virophage that is associated with mimiviruses of Acanthamoeba spp., named Zamilon, was also isolated95. Another virophage, named Mavirus, depends on Cafeteria roenbergensis virus for its replication and was the second virophage to be discovered after Sputnik96. The study of virophage diversity is still in its infancy, and several studies suggest that virophages are common and especially abundant in the oceans. For example, virophages were identified in the genome of the marine alga Bigelowiella natans97, and virophage genomes were assembled from metagenomic datasets98. Virophages that infect mimiviruses have been recently classified in the family Lavidaviridae91. This classification highlights that, as suggested by their particle and genome size, virophages are similar in complexity to genuine dsDNA viruses, and they share a set of six core genes that strongly suggests their monophyletic origin. In addition, the promoter and transcription termination signals of mimivirus virophages are similar to those of mimiviruses, which suggests that these virophages rely on their associated giant virus for mRNA synthesis99.
Although practical for isolating new virophages, the use of a culture procedure that is based on A. polyphaga and mimiviruses as a reporter system92 was hindered by the unexpected discovery that mimiviruses of lineage A, but not of lineage B and lineage C, were resistant to the Zamilon virophage95. The search for a system that would explain this resistance was inspired by an analogy to the CRISPR–Cas system, which is widely present in bacteria and archaea and relies on the integration of short DNA sequences from invaders7,100. The resistant lineage A isolates indeed have an insertion of four 15-nucleotide-long repeated Zamilon virophage sequences within an operon, named the MIMIVIRE, the resistance mechanism of which was suggested to rely on the sequence-specific recognition of a nucleic acid sequence7. The probability of finding such a sequence in this single mimivirus lineage by chance was very low (<1 × 10−9). MIMIVIRE-associated genes encode a helicase and nuclease that are involved in the degradation of foreign nucleic acids101 and the functions of which were experimentally validated7; MIMIVIRE also includes a gene that contains the repeated insert. Silencing each of these three genes by RNA interference restored the susceptibility of mimivirus to the Zamilon virophage. However, the exact molecular mechanism of MIMIVIRE is unclear. The MIMIVIRE system is thought to differ from CRISPR–Cas systems owing to the absence of a Cas1 homologue, of protospacer-adjacent motifs and of a well-conserved organization that includes bona fide CRISPR-like repeats100,102. Furthermore, the helicase and nuclease that are described in this MIMIVIRE system are only distantly related to proteins that are classified in the same superfamilies as Cas3 and Cas4 proteins, and they do not seem to be related to them100. An alternative mechanism to explain the resistance of mimivirus to virophages is one that would rely on interactions between proteins, rather than on nucleic acid-based recognition, and would involve a restriction factor protein of the virophage replication machinery102.
Provirophages, transpovirons and mobilome. Mobile genetic elements are common features in microorganisms, and mimiviruses have a complex mobilome (Fig. 3). Indeed, the DNA of virophages can integrate into the mimiviral host genome as 'pro-virophages' — that is, proviruses of virophages6. In addition, a novel category of mobile elements, named transpovirons, was reported. Transpovirons are linear DNA elements of ∼7 kb that encompass 6–8 protein-coding genes and have terminal inverted repeats of ∼530 bp (Ref. 6). Transpovirons are randomly integrated into giant virus genomes and are strictly dependent on giant viruses for their replication and spread; they replicate in the mimivirus factory and accumulate inside mimivirus particles, virophage particles and the amoebal cytoplasm6. Distinct transpovirons associate with different mimiviruses, including, for example, mamavirus and courdo7 virus isolates, which are classified in mimivirus lineage A and lineage C, respectively.
In evolution, the giant virus mobilome represents a vehicle and route for genetic novelty and adaptation. Thus, a substantial proportion of genes in the genomes of mimiviruses and other giant viruses are predicted to have been exchanged with bacteria, archaea, eukaryotes (including their amoebal hosts) and other viruses, although the direction of putative transfers is often unclear26,103. This high level of genome mosaicism might be related to the co-infection of amoebae by giant viruses and microorganisms, such as bacteria or fungi41,103. Indeed, giant viruses can multiply within amoebae that are infected with other microorganisms, which provides opportunities for horizontal gene transfer. Acanthamoeba spp. are well-known hosts for several bacteria and some fungi, in addition to viruses104. Acanthamoeba spp. that were isolated from a contact lens cleaning liquid were found to be co-infected with a mimivirus and two bacteria93, and Acanthamoeba spp. have been observed experimentally to be co-infected with marseillevirus and two bacteria41. In addition, the fact that, in their natural environment, giant viruses are sympatric with other microorganisms that infect Acanthamoeba helps to explain the large size of their genomes, which accumulate genes that increase their fitness against other microorganisms that replicate in amoebae103,105. Interestingly, a 16% decrease in genome size was observed by co-culturing APMV in amoebae in the absence of other microorganisms105; it was determined that the APMV genes that were inactivated were primarily those with the lowest expression, which suggests the loss of obsolete genes106. Moreover, these gene losses were associated with the loss of the external fibres that cover the APMV capsid and a decreased rate of replication105.
Conclusions and future directions
The replication cycle, structure, genomic make-up and plasticity of giant viruses differ from those of traditional viruses. They have virus particles that are as large as some microorganisms and have a stunning level of complexity. Their genomes are mosaic and contain large repertoires of genes, some of which are hallmarks of cellular organisms, although the majority have unknown functions. These giant viruses enter amoebae through phagocytosis and replicate inside viral factories. In addition, as shown for mimiviruses, they are associated with a specific mobilome and are parasitized by viruses that they can defend against. Giant viruses not only challenge the classification of viruses but also raise intriguing questions about their origin. They extend the definition of viruses into a broader range of biological entities, some of which are very simple and others of which have a complexity comparable to that of other microorganisms.
More giant viruses of amoebae must be identified to gain a better knowledge of their prevalence and diversity, which is largely unexplored but has expanded over the past three years and is likely to expand further now that we have learnt not to neglect them and owing to high-throughput detection and isolation strategies (Box 3). The role that giant viruses of amoebae have in evolution also warrants further attention. Several hypotheses on their ancient origin and their evolutionary relationship with cellular organisms have been proposed, and these topics should continue to be debated. In these viruses, it will be important to search for a translation system that does not involve ribosomes, and/or that is anterior to the ribosome and of which we would find remnants. Finally, the detection of giant viruses of amoebae in humans and the study of their potential pathogenicity are emerging fields107. To date, giant viruses of amoebae that have been linked to humans were discovered before 2013 (Box 4). Mimiviruses were associated with pneumonia108,109, whereas marseillevirus was detected in the blood and the lymphoid tissue, and was associated with adenitis and lymphoma110,111. More systematic investigations of human samples for giant viruses of amoebae must be conducted; such research is more accessible to the next generation of virologists who have entered the field after the description of APMV, and will surely reveal more giant viruses and refine their definition for years to come.
Box 4: Do giant viruses of amoebae and virophages infect humans?
The detection of giant viruses of amoebae in humans and the study of their potential pathogenicity are emerging fields107. Mimiviruses have been detected in serological, molecular and culture studies of patients with pneumonia107; serological studies were almost exclusively conducted in our laboratory, although some of the sera samples were collected by other laboratories61,107. Serological evidence notably included seroconversion to Acanthamoeba polyphaga mimivirus (APMV) in patients with unexplained pneumonia, including in a laboratory technician who manipulated large quantities of APMV108. In addition, two febrile Laotian patients were seropositive for the mimivirus virophage122. In culture studies, a mimivirus was isolated from two Tunisian patients who had pneumonia; in one case it was isolated from bronchoalveolar fluid107,109. As recently reviewed (see Refs. 61,107), several studies have failed to detect APMV DNA in respiratory samples from patients with pneumonia and only three cases were positive for APMV DNA when samples were analysed by PCR123,109,124. This may indicate that APMV is only rarely associated with pneumonia. Alternatively, this may be the consequence of considerable genetic diversity in mimiviruses, which may have prevented their detection by PCR in these studies125. Finally, mice that were inoculated intracardially with APMV developed pneumonia lesions126, and APMV can enter, and replicate in, human monocytes, macrophages76 and peripheral blood mononuclear cells86, although the virus was not propagated using these human blood cells.
Marseilleviruses were first serendipitously isolated from the stool of a healthy Senegalese individual61 and were subsequently detected in human blood by metagenomics, serology, fluorescence in situ hybridization (FISH) and PCR87,110,127,128. However, the detection of marseillevirus in human blood is controversial, as two other studies failed to detect marseillevirus DNA in blood from polytransfused patients in Paris and Cameroon and in blood from blood donors in southern France, Paris, the United States and Burkina Faso129,130. By contrast, positive serologies for Lausannevirus, which is a marseillevirus, were detected in young healthy adults in Switzerland (immunoglobulin G (IgG), 1.7–2.5%)127. In 2013, a high antibody titre to marseillevirus was reported in an 11-month-old child who presented with symptoms of lymphadenitis; marseillevirus was also detected by FISH and immunohistochemistry in a lymph node of this individual, and by PCR in the serum110. Next, marseillevirus was detected in a lymph node of a 30-year-old woman who was diagnosed with Hodgkin lymphoma by PCR, FISH, immunofluorescence and immunohistochemistry, concurrently, with specific antibodies111. Finally, marseillevirus was detected twice, with a one-year interval, by PCR in the pharynx of a patient with neurological disorders and once in the blood128. Very few data are available for the infection of humans by giant viruses that have only recently been described, but sequences that might belong to these giant viruses, including pithoviruses, pandoraviruses, faustoviruses, as well as to mimiviruses, marseilleviruses and virophages, have been identified in metagenomes that were generated from human samples61,62,63,64,98.
Supplementary information
Criteria for the definition of viruses and the applicability to Mimivirus and other giant viruses of amoebas1,2,3 (PDF 89 kb)
Schematic structure of Mimivirus (PDF 731 kb)
List of major representatives of mimiviruses and other giant viruses of amoebas and virophages (PDF 120 kb)
Worldwide distribution of giant viruses of amoebae and virophages isolated by amoebal co-culture from environmental samples (PDF 143 kb)
Discovery and major features of giant viruses of amoebae (apart from mimiviruses) (PDF 106 kb)
Main features of giant viruses of amoebae that are prototype members for described viral families or represent new putative viral families (PDF 621 kb)
Comparisons of giant viruses of amoebae based on their genome size, virion size, and G+C, gene and ORFan content (PDF 128 kb)
Acknowledgements
The authors thank I. Pagnier, J. Bou Khalil and J. Andreani for providing electron microscopy images.
Glossary
- Virophages
Viruses that depend on the co-infection of their amoebal host by mimiviruses. Virophages replicate within mimivirus factories.
- Transpovirons
Transposable elements that are ∼7 kb long and contain long terminal inverted repeats.
- Mimivirus virophage resistance element
(MIMIVIRE). A viral defence system that confers a nucleic-acid-based immunity against virophage infection.
- Fourth domain
A suggested additional domain to the three domains of life (Bacteria, Archaea and Eukarya) that were proposed by Woese. The term was initially coined in 2010.
- TRUC
(Things resisting uncompleted classifications). A term that was coined in 2013 and proposes an alternative classification of microorganisms to the one that is based on ribosomal genes, which groups giant viruses in a fourth TRUC of microorganisms.
- Double jelly-roll fold
A protein fold that is found exclusively in double-stranded DNA viruses and is comprised of two connected single jelly-roll folds, which are composed of eight β-strands arranged in two four-stranded sheets.
- Orphan genes
Genes that lack a homologue in any sequence database.
- Mobilome
This term represents the mobile genetic elements in the mimivirus genome and corresponds to provirophages and transpovirons, in addition to inteins and introns.
- Monophyletic clade
A group of organisms, or taxon, that consists of all of the descendants of an ancestral species.
- Phagocytosis
The engulfment of a solid particle by a cell, which forms an internal compartment named a phagosome.
- Mosaicism
Applied to microbial genomes, this term is used to describe the coexistence in a genome of genes with different origins (viral, bacteria, archaeal or eukaryotic) as the result of lateral gene transfers.
- Permafrost
Rock or soil at a temperature equal to, or below, 0 °C for two or more years. Permafrost is mainly located in and around the Arctic and Antarctic regions.
- dN/dS ratio
A ratio of the number, over a given period of time, of non-synonymous substitutions per non-synonymous site (dN) to the number of synonymous substitutions per synonymous site (dS), which quantifies selection pressures.
- Viral factories
Structural and functional elements that are associated with the replication of viral nucleic acids and the substantial production of virus particles.
- Exocytosis
Active transport of molecules, such as proteins, out of a cell through a process that uses energy.
- Sympatric
Inhabiting the same ecological niche.
- Seroconversion
The development of antibodies in the blood that are specifically directed against an infectious agent and become detectable by serological tests.
Biographies
Philippe Colson is Professor of Virology at Aix-Marseille University. He works at the University Hospital Institute Méditerranée Infection on hepatitis viruses and HIV, and on the genomics, classification, and association with humans of giant viruses of amoebae in the Research Unit on Emerging Infectious and Tropical Diseases (URMITE) in Marseille, France. He has described the genomes of several new giant viruses of amoebae and the family Marseilleviridae.
Bernard La Scola is Professor of Microbiology in the School of Medicine, Aix-Marseille University, France. He heads the 'Crisis in Infectious Diseases' Department at University Hospital Institute Méditerranée Infection, Marseille, France, and the research group 'viruses of the environment and the immunocompromised' at the Research Unit on Emerging Infectious and Tropical Diseases (URMITE), Marseille, France. He works on the isolation and characterization of fastidious microorganisms, especially amoeba-associated and biosafety level 3 microorganisms. He was involved in the discovery of mimivirus, marseillevirus, faustovirus, kaumoebavirus, cedratvirus, Pithovirus massiliensis, virophages and the mimivirus virophage resistance element (MIMIVIRE) defence system. He currently develops new virus isolation methods and studies the bacterial and viral microbiota in humans.
Anthony Levasseur is Professor of Bioinformatics and Evolutionary Biology at Aix-Marseille University, France. He received his Ph.D in molecular microbiology and biotechnology in 2005 from Aix-Marseille University. During his postdoctoral research, he focused on evolutionary biology and computer science. During his early career, he was a research scientist in microbiology and genomics. Since 2014, he has been Professor in the School of Medicine at Aix-Marseille University, and heads the team evolutionary biology at the Research Unit on Emerging Infectious and Tropical Diseases (URMITE), Marseille, France. By combining his multidisciplinary education, his main research interests include microbial evolution, microbiology, molecular biology, evolutionary genomics, medical informatics, microbial pathogenesis, the human microbiota and giant viruses.
Gustavo Caetano-Anollés is Professor of Bioinformatics at the Department of Crop Sciences, Affiliate of the Carl R. Woese Institute for Genomic Biology, and University Scholar of the University of Illinois, Urbana-Champaign, USA. He holds a Ph.D. in biochemistry from the National University of La Plata, Argentina, and has an interdisciplinary research background that includes microbiology, plant physiology, molecular genetics, genomics and computational biology. He is interested in creative ways to mine, visualize and integrate data from structural and functional genomic research. His current focus is on the evolution of molecular structure and networks in biology.
Didier Raoult is Professor of Microbiology at the School of Medicine, Aix-Marseille University, France. In 1984, he created ex nihilo his research unit. Since 2011, he has been head of the University Hospital Institute Méditerranée Infection, Marseille, France. He has published more than 2,000 referenced papers. For the past 30 years, he has cultured in his laboratory ∼30% of bacteria isolated for the first time in humans including Tropheryma whipplei and Rickettsia felis, and, since 2011, has invented microbial culturomics. He was involved in the discovery of mimivirus, other giant viruses of amoebae, virophages and the mimivirus virophage resistance element (MIMIVIRE). Didier Raoult's homepage. http://www.mediterranee-infection.com
PowerPoint slides
Competing interests
The authors declare no competing financial interests.
References
- 1.La Scola B, et al. A giant virus in amoebae. Science. 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 2.Raoult D, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 3.Raoult D. TRUC or the need for a new microbial classification. Intervirology. 2013;56:349–353. doi: 10.1159/000354269. [DOI] [PubMed] [Google Scholar]
- 4.Scheid P, Balczun C, Schaub GA. Some secrets are revealed: parasitic keratitis amoebae as vectors of the scarcely described pandoraviruses to humans. Parasitol. Res. 2014;113:3759–3764. doi: 10.1007/s00436-014-4041-3. [DOI] [PubMed] [Google Scholar]
- 5.La Scola B, et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 6.Desnues C, et al. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc. Natl Acad. Sci. USA. 2012;109:18078–18083. doi: 10.1073/pnas.1208835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levasseur A, et al. MIMIVIRE is a defence system in mimivirus that confers resistance to virophage. Nature. 2016;531:249–252. doi: 10.1038/nature17146. [DOI] [PubMed] [Google Scholar]
- 8.Raoult D, Forterre P. Redefining viruses: lessons from Mimivirus. Nat. Rev. Microbiol. 2008;6:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- 9.Filee J, Siguier P, Chandler M. I am what I eat and I eat what I am: acquisition of bacterial genes by giant viruses. Trends Genet. 2007;23:10–15. doi: 10.1016/j.tig.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Moreira D, Lopez-Garcia P. Ten reasons to exclude viruses from the tree of life. Nat. Rev. Microbiol. 2009;7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- 11.Raoult D. There is no such thing as a tree of life (and of course viruses are out!) Nat. Rev. Microbiol. 2009;7:615. doi: 10.1038/nrmicro2108-c6. [DOI] [PubMed] [Google Scholar]
- 12.Moreira D, Lopez-Garcia P. Evolution of viruses and cells: do we need a fourth domain of life to explain the origin of eukaryotes? Phil. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20140327. doi: 10.1098/rstb.2014.0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forterre P, Gaia M. Giant viruses and the origin of modern eukaryotes. Curr. Opin. Microbiol. 2016;31:44–49. doi: 10.1016/j.mib.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Klose T, et al. A Mimivirus enzyme that participates in viral entry. Structure. 2015;23:1058–1065. doi: 10.1016/j.str.2015.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues RA, et al. Mimivirus fibrils are important for viral attachment to microbial world by a diverse glycoside interaction repertoire. J. Virol. 2015;89:11812–11819. doi: 10.1128/JVI.01976-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zauberman N, et al. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008;6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuznetsov YG, et al. Atomic force microscopy investigation of the giant mimivirus. Virology. 2010;404:127–137. doi: 10.1016/j.virol.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Xiao C, et al. Structural studies of the giant mimivirus. PLoS Biol. 2009;7:e92. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renesto P, et al. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J. Virol. 2006;80:11678–11685. doi: 10.1128/JVI.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legendre M, Santini S, Rico A, Abergel C, Claverie JM. Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing. Virol. J. 2011;8:99. doi: 10.1186/1743-422X-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva LC, et al. Modulation of the expression of mimivirus-encoded translation-related genes in response to nutrient availability during Acanthamoeba castellanii infection. Front. Microbiol. 2015;6:539. doi: 10.3389/fmicb.2015.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legendre M, et al. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 2010;20:664–674. doi: 10.1101/gr.102582.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abergel C, Rudinger-Thirion J, Giege R, Claverie JM. Virus-encoded aminoacyl-tRNA synthetases: structural and functional characterization of mimivirus TyrRS and MetRS. J. Virol. 2007;81:12406–12417. doi: 10.1128/JVI.01107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suhre K. Gene and genome duplication in Acanthamoeba polyphaga Mimivirus. J. Virol. 2005;79:14095–14101. doi: 10.1128/JVI.79.22.14095-14101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreira D, Brochier-Armanet C. Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol. Biol. 2008;8:12. doi: 10.1186/1471-2148-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forterre P. Giant viruses: conflicts in revisiting the virus concept. Intervirology. 2010;53:362–378. doi: 10.1159/000312921. [DOI] [PubMed] [Google Scholar]
- 27.Boyer M, Gimenez G, Suzan-Monti M, Raoult D. Classification and determination of possible origins of ORFans through analysis of nucleocytoplasmic large DNA viruses. Intervirology. 2010;53:310–320. doi: 10.1159/000312916. [DOI] [PubMed] [Google Scholar]
- 28.Byrne D, et al. The polyadenylation site of Mimivirus transcripts obeys a stringent 'hairpin rule'. Genome Res. 2009;19:1233–1242. doi: 10.1101/gr.091561.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeudy S, Abergel C, Claverie JM, Legendre M. Translation in giant viruses: a unique mixture of bacterial and eukaryotic termination schemes. PLoS Genet. 2012;8:e1003122. doi: 10.1371/journal.pgen.1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoosuf N, et al. Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage. Genome Biol. Evol. 2012;4:1324–1330. doi: 10.1093/gbe/evs109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azza S, Cambillau C, Raoult D, Suzan-Monti M. Revised Mimivirus major capsid protein sequence reveals intron-containing gene structure and extra domain. BMC Mol. Biol. 2009;10:39. doi: 10.1186/1471-2199-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.La Scola B, de Lamballerie XN, Claverie JM, Drancourt M, Raoult D. Genus Mimivirus in Virus Taxonomy. 2005. pp. 275–276. [Google Scholar]
- 33.Pagnier I, et al. A decade of improvements in Mimiviridae and Marseilleviridae isolation from amoeba. Intervirology. 2013;56:354–363. doi: 10.1159/000354556. [DOI] [PubMed] [Google Scholar]
- 34.Khalil JY, Andreani J, La Scola B. Updating strategies for isolating and discovering giant viruses. Curr. Opin. Microbiol. 2016;31:80–87. doi: 10.1016/j.mib.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Khalil JY, et al. High-throughput isolation of giant viruses in liquid medium using automated flow cytometry and fluorescence staining. Front. Microbiol. 2016;7:26. doi: 10.3389/fmicb.2016.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl Acad. Sci. USA. 2011;108:17486–17491. doi: 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl Acad. Sci. USA. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yau S, et al. Virophage control of antarctic algal host–virus dynamics. Proc. Natl Acad. Sci. USA. 2011;108:6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santini S, et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc. Natl Acad. Sci. USA. 2013;110:10800–10805. doi: 10.1073/pnas.1303251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yutin N, Colson P, Raoult D, Koonin EV. Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol. J. 2013;10:106–110. doi: 10.1186/1743-422X-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyer M, et al. Giant marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl Acad. Sci. USA. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colson P, et al. “Marseilleviridae”, a new family of giant viruses infecting amoebae. Arch. Virol. 2013;158:915–920. doi: 10.1007/s00705-012-1537-y. [DOI] [PubMed] [Google Scholar]
- 43.Philippe N, et al. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 44.Legendre M, et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl Acad. Sci. USA. 2014;111:4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reteno DG, et al. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J. Virol. 2015;89:6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Legendre M, et al. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl Acad. Sci. USA. 2015;112:E5327–E5335. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yutin N, Wolf YI, Raoult D, Koonin EV. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 2009;17:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yutin N, Koonin EV. Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol. J. 2012;9:161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boyer M, Madoui MA, Gimenez G, La Scola B, Raoult D. Phylogenetic and phyletic studies of informational genes in genomes highlight existence of a 4 domain of life including giant viruses. PLoS ONE. 2010;5:e15530. doi: 10.1371/journal.pone.0015530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nasir A, Caetano-Anolles G. A phylogenomic data-driven exploration of viral origins and evolution. Sci. Adv. 2015;1:e1500527. doi: 10.1126/sciadv.1500527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koonin EV, Yutin N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology. 2010;53:284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colson P, et al. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. 2013;158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma V, Colson P, Giorgi R, Pontarotti P, Raoult D. DNA-dependent RNA polymerase detects hidden giant viruses in published databanks. Genome Biol. Evol. 2014;6:1603–1610. doi: 10.1093/gbe/evu128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma V, Colson P, Pontarotti P, Raoult D. Mimivirus inaugurated in the 21st century the beginning of a reclassification of viruses. Curr. Opin. Microbiol. 2016;31:16–24. doi: 10.1016/j.mib.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 56.Suzan-Monti M, La Scola B, Barrassi L, Espinosa L, Raoult D. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS ONE. 2007;2:e328. doi: 10.1371/journal.pone.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abergel C, Legendre M, Claverie JM. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol. Rev. 2015;39:779–796. doi: 10.1093/femsre/fuv037. [DOI] [PubMed] [Google Scholar]
- 58.Aherfi S, Colson P, La Scola B, Raoult D. Giant viruses of amoebas: an update. Front. Microbiol. 2016;7:349. doi: 10.3389/fmicb.2016.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghedin E, Claverie JM. Mimivirus relatives in the Sargasso sea. Virol. J. 2005;2:62. doi: 10.1186/1743-422X-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colson P, et al. Evidence of the megavirome in humans. J. Clin. Virol. 2013;57:191–200. doi: 10.1016/j.jcv.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 62.Law J, et al. Identification of hepatotropic viruses from plasma using deep sequencing: a next generation diagnostic tool. PLoS ONE. 2013;8:e60595. doi: 10.1371/journal.pone.0060595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verneau J, Levasseur A, Raoult D, La Scola B, Colson P. MG-Digger: an automated pipeline to search for giant virus-related sequences in metagenomes. Front. Microbiol. 2016;7:428. doi: 10.3389/fmicb.2016.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rampelli S, et al. ViromeScan: a new tool for metagenomic viral community profiling. BMC Genomics. 2016;17:165–2446. doi: 10.1186/s12864-016-2446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maumus F, Epert A, Nogue F, Blanc G. Plant genomes enclose footprints of past infections by giant virus relatives. Nat. Commun. 2014;5:4268. doi: 10.1038/ncomms5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu D, et al. Stalking the fourth domain in metagenomic data: searching for, discovering, and interpreting novel, deep branches in marker gene phylogenetic trees. PLoS ONE. 2011;6:e18011. doi: 10.1371/journal.pone.0018011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mozar, M. & Claverie, J. M. Expanding the Mimiviridae family using asparagine synthase as a sequence bait. Virology 466–467, 112–122 (2014). [DOI] [PubMed]
- 68.Klose T, et al. Structure of faustovirus, a large dsDNA virus. Proc. Natl Acad. Sci. USA. 2016;113:6206–6211. doi: 10.1073/pnas.1523999113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas V, et al. Lausannevirus, a giant amoebal virus encoding histone doublets. Environ. Microbiol. 2011;13:1454–1466. doi: 10.1111/j.1462-2920.2011.02446.x. [DOI] [PubMed] [Google Scholar]
- 70.Sun C, Feschotte C, Wu Z, Mueller RL. DNA transposons have colonized the genome of the giant virus Pandoravirus salinus. BMC Biol. 2015;13:38–0145. doi: 10.1186/s12915-015-0145-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Filee J. Genomic comparison of closely related giant viruses supports an accordion-like model of evolution. Front. Microbiol. 2015;6:593. doi: 10.3389/fmicb.2015.00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levasseur A, et al. Comparison of a modern and fossil pithovirus reveals its genetic conservation and evolution. Genome Biol. Evol. 2016;8:2333–2339. doi: 10.1093/gbe/evw153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Babkin IV, Babkina IN. Molecular dating in the evolution of vertebrate poxviruses. Intervirology. 2011;54:253–260. doi: 10.1159/000320964. [DOI] [PubMed] [Google Scholar]
- 74.Doutre G, Philippe N, Abergel C, Claverie JM. Genome analysis of the first Marseilleviridae representative from Australia indicates that most of its genes contribute to the virus fitness. J. Virol. 2014;88:14340–14349. doi: 10.1128/JVI.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mutsafi Y, Zauberman N, Sabanay I, Minsky A. Vaccinia-like cytoplasmic replication of the giant Mimivirus. Proc. Natl Acad. Sci. USA. 2010;107:5978–5982. doi: 10.1073/pnas.0912737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghigo E, et al. Ameobal pathogen mimivirus infects macrophages through phagocytosis. PLoS Pathog. 2008;4:e1000087. doi: 10.1371/journal.ppat.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mutsafi, Y., Fridmann-Sirkis, Y., Milrot, E., Hevroni, L. & Minsky, A. Infection cycles of large DNA viruses: emerging themes and underlying questions. Virology 466–467, 3–14 (2014). [DOI] [PubMed]
- 78.Mutsafi Y, Shimoni E, Shimon A, Minsky A. Membrane assembly during the infection cycle of the giant Mimivirus. PLoS Pathog. 2013;9:e1003367. doi: 10.1371/journal.ppat.1003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arantes TS, et al. The large marseillevirus explores different entry pathways by forming giant infectious vesicles. J. Virol. 2016;90:5246–5255. doi: 10.1128/JVI.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boratto PV, et al. Amoebas as mimivirus bunkers: increased resistance to UV light, heat and chemical biocides when viruses are carried by amoeba hosts. Arch. Virol. 2013;159:1039–1043. doi: 10.1007/s00705-013-1924-z. [DOI] [PubMed] [Google Scholar]
- 81.Dornas FP, et al. Acanthamoeba polyphaga mimivirus stability in environmental and clinical substrates: implications for virus detection and isolation. PLoS ONE. 2014;9:e87811. doi: 10.1371/journal.pone.0087811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Slimani M, Pagnier I, Boughalmi M, Raoult D, La Scola B. Alcohol disinfection procedure for isolating giant viruses from contaminated samples. Intervirology. 2013;56:434–440. doi: 10.1159/000354566. [DOI] [PubMed] [Google Scholar]
- 83.Boughalmi M, et al. First isolation of a giant virus from wild Hirudo medicinalis leech: Mimiviridae isolation in Hirudo medicinalis. Viruses. 2013;5:2920–2930. doi: 10.3390/v5122920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dornas FP, et al. Mimivirus circulation among wild and domestic mammals, Amazon Region, Brazil. Emerg. Infect. Dis. 2014;20:469–472. doi: 10.3201/eid2003.131050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andrade KR, et al. Oysters as hot spots for mimivirus isolation. Arch. Virol. 2015;160:477–482. doi: 10.1007/s00705-014-2257-2. [DOI] [PubMed] [Google Scholar]
- 86.Silva LC, et al. A resourceful giant: APMV is able to interfere with the human type I Interferon system. Microbes Infect. 2014;16:187–195. doi: 10.1016/j.micinf.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 87.Popgeorgiev N, et al. Marseillevirus-like virus recovered from blood donated by asymptomatic humans. J. Infect. Dis. 2013;208:1042–1050. doi: 10.1093/infdis/jit292. [DOI] [PubMed] [Google Scholar]
- 88.Netherton CL, Wileman T. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr. Opin. Virol. 2011;1:381–387. doi: 10.1016/j.coviro.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Claverie JM. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yutin N, Raoult D, Koonin EV. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol. J. 2013;10:158. doi: 10.1186/1743-422X-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krupovic M, Kuhn JH, Fischer MG. A classification system for virophages and satellite viruses. Arch. Virol. 2016;161:233–247. doi: 10.1007/s00705-015-2622-9. [DOI] [PubMed] [Google Scholar]
- 92.Gaia M, et al. Broad spectrum of Mimiviridae virophage allows its isolation using a mimivirus reporter. PLoS ONE. 2013;8:e61912. doi: 10.1371/journal.pone.0061912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cohen G, Hoffart L, La Scola B, Raoult D, Drancourt M. Ameba-associated Keratitis, France. Emerg. Infect. Dis. 2011;17:1306–1308. doi: 10.3201/eid1707.100826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Campos RK, et al. Samba virus: a novel mimivirus from a giant rain forest, the Brazilian Amazon. Virol. J. 2014;11:95. doi: 10.1186/1743-422X-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gaia M, et al. Zamilon, a novel virophage with Mimiviridae host specificity. PLoS ONE. 2014;9:e94923. doi: 10.1371/journal.pone.0094923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fischer MG, Suttle CA. A virophage at the origin of large DNA transposons. Science. 2011;332:231–234. doi: 10.1126/science.1199412. [DOI] [PubMed] [Google Scholar]
- 97.Blanc G, Gallot-Lavallee L, Maumus F. Provirophages in the Bigelowiella genome bear testimony to past encounters with giant viruses. Proc. Natl Acad. Sci. USA. 2015;112:E5318–E5326. doi: 10.1073/pnas.1506469112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou J, et al. Diversity of virophages in metagenomic data sets. J. Virol. 2013;87:4225–4236. doi: 10.1128/JVI.03398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Claverie J, Abergel C. Mimivirus and its virophage. Annu. Rev. Genet. 2009;43:49–66. doi: 10.1146/annurev-genet-102108-134255. [DOI] [PubMed] [Google Scholar]
- 100.Mohanraju P, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR–Cas systems. Science. 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 101.Marraffini LA. CRISPR–Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 102.Claverie JM, Abergel C. CRISPR–Cas-like system in giant viruses: why MIMIVIRE is not likely to be an adaptive immune system. Virol. Sin. 2016;31:193–196. doi: 10.1007/s12250-016-3801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Raoult D, Boyer M. Amoebae as genitors and reservoirs of giant viruses. Intervirology. 2010;53:321–329. doi: 10.1159/000312917. [DOI] [PubMed] [Google Scholar]
- 104.Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 2004;17:413–433. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boyer M, et al. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc. Natl Acad. Sci. USA. 2011;108:10296–10301. doi: 10.1073/pnas.1101118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Colson P, Raoult D. Lamarckian evolution of the giant Mimivirus in allopatric laboratory culture on amoebae. Front. Cell. Infect. Microbiol. 2012;2:91. doi: 10.3389/fcimb.2012.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Colson P, Aherfi S, La Scola B, Raoult D. The role of giant viruses of amoebas in humans. Curr. Opin. Microbiol. 2016;31:199–208. doi: 10.1016/j.mib.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 108.Raoult D, Renesto P, Brouqui P. Laboratory infection of a technician by mimivirus. Ann. Intern. Med. 2006;144:702–703. doi: 10.7326/0003-4819-144-9-200605020-00025. [DOI] [PubMed] [Google Scholar]
- 109.Saadi H, et al. First isolation of Mimivirus in a patient with pneumonia. Clin. Infect. Dis. 2013;57:e127–e134. doi: 10.1093/cid/cit354. [DOI] [PubMed] [Google Scholar]
- 110.Popgeorgiev N, Michel G, Lepidi H, Raoult D, Desnues C. Marseillevirus adenitis in an 11-month-old child. J. Clin. Microbiol. 2013;51:4102–4105. doi: 10.1128/JCM.01918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aherfi S, et al. Marseillevirus in lymphoma: a giant in the lymph node. Lancet Infect. Dis. 2016;16:e225–e234. doi: 10.1016/S1473-3099(16)30051-2. [DOI] [PubMed] [Google Scholar]
- 112.Lwoff A. The concept of virus. J. Gen. Microbiol. 1957;17:239–253. doi: 10.1099/00221287-17-2-239. [DOI] [PubMed] [Google Scholar]
- 113.Lwoff A, Tournier P. The classification of viruses. Annu. Rev. Microbiol. 1966;20:45–74. doi: 10.1146/annurev.mi.20.100166.000401. [DOI] [PubMed] [Google Scholar]
- 114.Williams TA, Embley TM, Heinz E. Informational gene phylogenies do not support a fourth domain of life for nucleocytoplasmic large DNA viruses. PLoS ONE. 2011;6:e21080. doi: 10.1371/journal.pone.0021080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yutin, N., Wolf, Y. I. & Koonin, E. V. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 466–467, 38–52 (2014). [DOI] [PMC free article] [PubMed]
- 116.Rowbotham TJ. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J. Clin. Pathol. 1983;36:978–986. doi: 10.1136/jcp.36.9.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pagnier I, Valles C, Raoult D, La Scola B. Isolation of Vermamoeba vermiformis and associated bacteria in hospital water. Microb. Pathog. 2015;80:14–20. doi: 10.1016/j.micpath.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 118.Dornas FP, et al. Isolation of new Brazilian giant viruses from environmental samples using a panel of protozoa. Front. Microbiol. 2015;6:1086. doi: 10.3389/fmicb.2015.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 1980;33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Birtles RJ, Rowbotham TJ, Raoult D, Harrison TG. Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology. 1996;142:3525–3530. doi: 10.1099/13500872-142-12-3525. [DOI] [PubMed] [Google Scholar]
- 121.Birtles RJ, Rowbotham TJ, Storey C, Marrie TJ, Raoult D. Chlamydia-like obligate parasite of free-living amoebae. Lancet. 1997;349:925–926. doi: 10.1016/S0140-6736(05)62701-8. [DOI] [PubMed] [Google Scholar]
- 122.Parola P, et al. Acanthamoeba polyphaga mimivirus virophage seroconversion in travelers returning from Laos. Emerg. Infect. Dis. 2012;18:1500–1502. doi: 10.3201/eid1809.120099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.La Scola B, Marrie TJ, Auffray JP, Raoult D. Mimivirus in pneumonia patients. Emerg. Infect. Dis. 2005;11:449–452. doi: 10.3201/eid1103.040538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang XA, et al. Lack of Mimivirus detection in patients with respiratory disease, China. Emerg. Infect. Dis. 2016;22:10. doi: 10.3201/eid2211.160687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ngounga T, et al. Real-time PCR systems targeting giant viruses of amoebae and their virophages. Intervirology. 2013;56:413–423. doi: 10.1159/000354563. [DOI] [PubMed] [Google Scholar]
- 126.Khan M, La Scola B, Lepidi H, Raoult D. Pneumonia in mice inoculated experimentally with Acanthamoeba polyphaga mimivirus. Microb. Pathog. 2007;42:56–61. doi: 10.1016/j.micpath.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 127.Mueller L, Baud D, Bertelli C, Greub G. Lausannevirus seroprevalence among asymptomatic young adults. Intervirology. 2013;56:430–433. doi: 10.1159/000354565. [DOI] [PubMed] [Google Scholar]
- 128.Aherfi S, Colson P, Raoult D. Marseillevirus in the pharynx of a patient with neurologic disorders. Emerg. Infect. Dis. 2016;22:2008–2010. doi: 10.3201/eid2211.160189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sauvage V, et al. No evidence of marseillevirus-like virus presence in blood donors and recipients of multiple blood transfusions. J. Infect. Dis. 2014;210:2017–2018. doi: 10.1093/infdis/jiu443. [DOI] [PubMed] [Google Scholar]
- 130.Phan TG, et al. Absence of giant blood Marseille-like virus DNA detection by polymerase chain reaction in plasma from healthy US blood donors and serum from multiply transfused patients from Cameroon. Transfusion. 2015;55:1256–1262. doi: 10.1111/trf.12997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Criteria for the definition of viruses and the applicability to Mimivirus and other giant viruses of amoebas1,2,3 (PDF 89 kb)
Schematic structure of Mimivirus (PDF 731 kb)
List of major representatives of mimiviruses and other giant viruses of amoebas and virophages (PDF 120 kb)
Worldwide distribution of giant viruses of amoebae and virophages isolated by amoebal co-culture from environmental samples (PDF 143 kb)
Discovery and major features of giant viruses of amoebae (apart from mimiviruses) (PDF 106 kb)
Main features of giant viruses of amoebae that are prototype members for described viral families or represent new putative viral families (PDF 621 kb)
Comparisons of giant viruses of amoebae based on their genome size, virion size, and G+C, gene and ORFan content (PDF 128 kb)