Key Points
Infections are possibly the major selective pressure acting on humans, and host–pathogen interactions contribute to shaping the genetic diversity of both organisms.
Comparisons among species provide a snapshot of selective events that have been unfolding over long timescales. These approaches use extant genetic diversity and phylogenetic relationships among species to identify positively selected sites.
Positive selection often acts on a limited number of sites in a protein that is otherwise selectively constrained; one example is the localized signal of selection at Niemann–Pick C1 protein (NPC1), the receptor for the Ebola virus.
As epitomized by the evolutionary history of tripartite motif-containing 5 (TRIM5), past infection events may leave a signature that affects the ability of extant species to fight emerging pathogens.
Protein regions at the host–pathogen interface are expected to be targeted by the strongest selective pressure (this is the case for dipeptidyl peptidase 4 (DPP4) and angiotensin-converting enzyme 2 (ACE2), which act as receptors for coronaviruses).
Other mammals host a wide range of viruses that are highly pathogenic for humans. Sequencing the genomes of these pathogens will be instrumental in refining our understanding of the process of host–pathogen interaction.
Pathogen-driven natural selection is not limited to the immune system: genes that encode incidental pathogen receptors and components of the contact system and coagulation cascade can also be targeted.
Supplementary information
The online version of this article (doi:10.1038/nrg3905) contains supplementary material, which is available to authorized users.
Subject terms: Pathogens, Evolution, Comparative genomics
Host–pathogen interactions influence genetic diversity, and comparative genomic analyses are beginning to dissect genetic determinants involved in this process. This Review describes examples of such host–pathogen interactions and outline evolutionary approaches that are useful for identifying genomic regions associated with susceptibility to infection in mammals.
Supplementary information
The online version of this article (doi:10.1038/nrg3905) contains supplementary material, which is available to authorized users.
Abstract
Infections are one of the major selective pressures acting on humans, and host-pathogen interactions contribute to shaping the genetic diversity of both organisms. Evolutionary genomic studies take advantage of experiments that natural selection has been performing over millennia. In particular, inter-species comparative genomic analyses can highlight the genetic determinants of infection susceptibility or severity. Recent examples show how evolution-guided approaches can provide new insights into host–pathogen interactions, ultimately clarifying the basis of host range and explaining the emergence of different diseases. We describe the latest developments in comparative immunology and evolutionary genetics, showing their relevance for understanding the molecular determinants of infection susceptibility in mammals.
Supplementary information
The online version of this article (doi:10.1038/nrg3905) contains supplementary material, which is available to authorized users.
Main
Infections are thought to represent the major selective pressure for humans1 and, possibly, for all living organisms. Encounters of hosts and pathogens result in so-called 'arms races', whereby hosts are under pressure to evolve resistance to pathogens while pathogens strive to develop countermeasures to evade host surveillance and to achieve a successful infection. Thus, when resistance and counter-resistance are at least partially genetically determined, cyclical adaptation and counter-adaptation occur, and a genetic conflict is fuelled (Box 1). This is generally referred to as a 'Red Queen' scenario, a definition proposed by Leigh Van Valen2 after the character in Lewis Carroll's novel Through the Looking-Glass who says: “It takes all the running you can do, to keep in the same place”. At its core, the Red Queen hypothesis highlights the relevance of biotic versus abiotic interactions as drivers of perpetual evolutionary change (see Ref. 3 for a recent review). Although the hypothesis is perfectly conjured up by the Red Queen imagery proposed in 1973, some of its principles can be traced back to the work of J. B. S. Haldane at the beginning of the twentieth century. In fact, Haldane was the first to propose that infectious diseases should be considered as a major selective pressure in our species4.
In this Review we present some of the most recent advances in the field of evolutionary biology applied to the study of infectious diseases. In particular, we focus on inter-species comparisons among mammals and on the way in which these analyses have helped to clarify the genetic determinants of species-specific infection and disease, as well as the reasons behind pathogen emergence. Although arms races involve both the host and the pathogen, in this Review we only focus on genetic diversity in mammalian hosts. Host–pathogen genetic conflicts are not confined to mammals (and their pathogens): they drive molecular evolution in most realms of life, including bacterial–bacteriophage systems5, plants and their infectious agents6, as well as invertebrates and their pests7,8.
Although we review studies and methods (Boxes 1,2,3) that analyse genetic diversity at the inter-species level, the investigation of intra-species and intra-population signatures of pathogen-driven selection has also provided extremely valuable insight into infectious disease susceptibility, especially in our species. The interested reader is directed towards several recent reviews for more information9,10,11,12,13.
Box 1: Detecting natural selection.
Comparisons among species take a snapshot of selective events that have been unfolding over long timescales. Most of these approaches use extant genetic diversity and phylogenetic relationships among species to infer underlying evolutionary patterns. Briefly, inter-species approaches rely on the alignment of orthologous coding sequences, analyse these alignments site-by-site, and at each site determine which, among all possible substitutions, would be non-synonymous (amino acid replacing) or synonymous (non-amino acid replacing) (see the figure). The observed number of non-synonymous differences per non-synonymous site (dN) and the observed number of synonymous differences per synonymous site (dS) are then estimated. Under neutral evolution, the rate at which amino acid replacements accumulate is expected to be comparable to the rate for silent changes and, therefore, dN/dS should be equal to 1 (green codons in the figure). Nonetheless, most amino acid replacements are deleterious and, as a consequence, are eliminated by selection; this results in a large preponderance of sites with dN/dS <1, a situation referred to as purifying (or negative) selection (shown in blue in the figure). Conversely, the selective pressure exerted, for instance, by a pathogen, may favour amino acid replacements (for example, changes that modify the sequence and structure of a cellular receptor): in this case, dN/dS may reach values greater than 1, a hallmark of positive (or diversifying) selection (red in the figure).
The figure shows a hypothetical example whereby a virus uses a cellular receptor to infect the host. To prevent viral binding and infection, selection favours variants that modify the sequence and structure of the host receptors; on the other side, the virus adapts to such changes by gaining mutations that keep re-establishing receptor binding. This process fuels a genetic conflict, which is evident at the interaction surfaces. Some lineages may be under stronger selective pressure than others and may display lineage-specific selected sites (episodic selection; cyan). In this case the branch of the phylogeny leading to these species may show significant evidence of positive selection (Box 3).
Box 2: Detection of positively selected genes and sites.
The 'site models' implemented in the phylogenetic analysis by maximum likelihood (PAML) package91 are widely used to infer positive selection and to identify positively selected sites. These models allow dN/dS to vary from site to site, assuming a constant rate at synonymous sites. Data (alignment and phylogenetic tree) are fitted to models that allow (selection models) or do not allow (neutral model) a class of codons to evolve with dN/dS >1. Likelihood ratio tests are then applied to determine whether the neutral model can be rejected in favour of the positive selection model. If so, the gene is declared to be positively selected. Also, if (and only if) the null hypothesis of neutral selection is rejected, a Bayes empirical Bayes (BEB) approach can be used to detect specific sites targeted by selection (BEB calculates the posterior probability that each site belongs to the class with dN/dS >1)92,93.
The PAML approach implicitly assumes that the strength and direction of natural selection is uniform across all lineages. Because this is often not the case, Murrell and co-workers recently developed the mixed effects model of evolution (MEME, HyPhy package)94. MEME allows the distribution of dN/dS to vary from site to site and from branch to branch; thus, the method has greater power to detect episodic selection, especially if it is confined to a small subset of branches in the phylogeny. A major issue related to these approaches is their extreme sensitivity to errors in sequence (coverage), annotation and alignment. Misalignments and incorrect sequence information may result in apparently fast evolutionary rates and thus inflate the false-positive rate95,96,97. The use of specific alignment algorithms (for example, PRANK) and filtering procedures (for example, GUIDANCE) may partially overcome this problem98. Likewise, genetic variability that is generated by recombination can be mistaken for positive selection99. Thus, to limit false positives, alignments should be screened for recombination before running positive selection tests (and, if necessary, split on the basis of recombination breakpoints) or recombination should be incorporated into the model.
Box 3: Detection of lineages under positive selection and lineage-specific sites.
Signatures of selection along specific branches can be detected through the so called 'branch-site' models implemented in the phylogenetic analysis by maximum likelihood (PAML) package100. In analogy to the site models described in Box 2, alignment errors result in high false-positive rates when branch-site models are applied101; this issue can be partially mitigated by the use of specific aligners101. Branch-site models require the phylogeny to be divided into 'foreground' and 'background' branches. A likelihood ratio test is then applied to compare a model that allows positive selection on a class of codons for the foreground branches with a model that does not allow such selection100. Designation of the foreground branches needs a priori information, possibly based on biological evidence. If no clues are available as to which branches are more likely to have undergone selection, it is still possible to run the analysis by designating each branch of the tree as 'foreground'; this generates a multiple-hypothesis testing problem that must be appropriately corrected102.
Two alternative methods can detect selection at specific lineages without a priori branch partition. The branch site-random effects likelihood (BS-REL) method considers three different evolutionary scenarios (purifying, neutral and diversifying selection) for all branches in a given tree, and each branch is considered independently from the others; the algorithm applies sequential likelihood ratio tests to identify branches with significant evidence of positive selection103. The second method, the covarion-like codon model (FitModel)104, allows each site to switch between selective regimes at any time on the phylogeny. Thus, switches are not necessarily associated with tree nodes. Recently, this approach was shown to be more powerful than the branch-site tests if a priori information is available105. Both FitModel and the PAML branch-site methods envisage a Bayesian approach to identify sites evolving under episodic positive selection. However, extensive simulations revealed that the branch-site approach is accurate but has limited power at detecting sites106. This problem has been referred to as the 'selection inference uncertainty principle' — that is, it is difficult to simultaneously infer both the site and the branch that are subject to positive selection94.
The dynamics of host–pathogen interactions
A central tenet of the Red Queen hypothesis is that organisms must continually adapt to survive and thrive in the face of continually evolving opposing organisms. Nonetheless, evolution is not all about biotic interactions. At a macroevolutionary level, mixed models of evolution are likely to operate; biotic factors mainly shape species diversity locally and over short time spans, whereas shifts in the physical environment (for example, climate changes and oceanographic and tectonic events) drive evolution at a large scale, across much longer time periods14. Recently, a new interpretation of the Red Queen hypothesis was proposed15; the analysis of several phylogenies from different taxa indicated that speciation mostly occurs at a constant rate through rare stochastic events that cause reproductive isolation15. This view curtails the role of biotic interactions as major determinants of species diversity15.
Despite these observations, the Red Queen hypothesis has proven to be an extremely useful framework for the study of host–pathogen interactions. In this context, Red Queen dynamics can be divided into different types (see Ref. 3 for a recent review). Frequency-dependent selection, for example, determines allele frequency fluctuations in both host and pathogen populations. In this scenario, rare alleles are favoured by selection (the pathogen, for instance, may be adapted to the most common host genotype and may fail to infect hosts carrying a rare allele), and diversity within populations is maintained. Escalatory arms races are another form of selection that usually apply to quantitative or polygenic traits and proceed through recurrent selective sweeps. Selection results in an escalation in the phenotypes of both the host (for example, resistance) and the pathogen (for example, virulence). Finally, in chase Red Queen scenarios the host is under pressure to reduce the strength of the interaction through de novo evolution of novelty, whereas the pathogen evolves to tighten the interaction by reducing phenotypic distance. Chase scenarios occur when host–pathogen interactions have a complex genetic basis (polygenic); they determine selective sweeps and tend to reduce genetic diversity within populations.
Over the years, the Red Queen hypothesis has been supported by the description of rapid rates of evolution in genes involved in genetic conflicts and, in a few instances, by the temporal reconstruction of host–pathogen co-evolution in natural settings16. More recently, the development of experimental evolution approaches has allowed its formal testing17,18. Although extremely valuable, laboratory-based studies often use an isogenic host population that is infected by one or a few pathogen strains, and such studies only partially recapitulate the complex nature of host–pathogen interactions that occur in real life. For instance, phenotypic plasticity (an environmentally based change in the phenotype) and multiway host–pathogen interactions are common in nature. A remarkable example of phenotypic plasticity is the vertebrate adaptive immune system: through rearrangement and somatic hypermutation, the same genetic arsenal is used to combat a wide array of pathogens and to develop lifelong resistance to some infections. Despite the relevance of adaptive immunity for host defence, its action does not preclude pathogen-driven selection at several genes involved in innate immunity or, more generally, in the interaction with pathogens (these represent the focus of this Review). As for multiway interactions, these represent the norm: the same host can be infected by multiple pathogens (or even by multiple strains of the same infectious agent) during its lifetime, whereas pathogens differ in their ability to infect one or more host species. Thus, multiple host–pathogen interactions might drive the evolution of the same or different molecular systems, blurring the expectations of the Red Queen hypothesis. Finally, hosts with long generation times (such as mammals, which are the focus of this Review), evolve at lower rates compared with most of their pathogens and also display smaller population sizes, resulting in an asymmetry of the arms race (although parasites with life cycles involving two or more species may be constrained in their ability to adapt (reviewed in Ref. 19)). Even in the presence of a strong selective pressure (for example, a fatal infection), several generations may be required before the molecular signatures of the genetic conflict can be detected in mammalian host genomes19. Nevertheless, natural selection signatures have been described at several mammalian genes that interact with recently emerged human infectious agents (for example, HIV-1), possibly as a result of the pressure imposed by extinct pathogens or because these agents have established long-lasting interactions with non-human hosts.
Ancient and recent infections
Since 1940, 335 emerging infectious diseases (EIDs) have been reported in humans; EID events are increasing significantly over time and are dominated by zoonoses, most of which originate from wildlife20. Recent zoonoses are exemplified by Ebola virus (EBOV) outbreaks, which have occurred episodically in Africa since 1976, and by the emergence of Middle East respiratory syndrome coronavirus (MERS-CoV) as a dangerous human pathogen. Both EBOV and MERS-CoV are thought to have originated in bats and spread to humans either directly or through an intermediate host. Because EIDs are almost inevitably caused by an existing pathogen that adapts to infect a new host, comparative analyses of different species may help to unveil the genetic and immunological determinants underlying pathogen spillover and infection susceptibility.
HIV-1, for example, originated from the cross-species transmission of the simian immunodeficiency virus SIVcpz, which naturally infects chimpanzees21. Old World monkeys are resistant to HIV-1 infection owing to a post-entry viral block operated by cellular restriction factors. This differential susceptibility to infection was exploited to isolate tripartite motif-containing protein 5 (TRIM5; also known as TRIM5α), a major retrovirus restriction factor, from a rhesus macaque cDNA library22. The protein product of TRIM5 binds directly to the incoming viral capsid and targets it for disassembly. Whereas macaque TRIM5 is highly efficient against HIV-1, the human protein is not22. Most species-specific determinants of antiviral activity were mapped to a short amino acid stretch in the so-called B30.2 (or SPRY) domain of TRIM5 (Ref. 23). In primates, this region has evolved under positive selection, and the human lineage shows some of the strongest selection signatures23. Why then is human TRIM5 so highly inefficient against HIV-1? Possibly because the human gene evolved to fight another retrovirus. In a seminal paper, Kaiser and co-workers resurrected an extinct Pan troglodytes endogenous retrovirus (PtERV1) and showed that the amino acid status of a single residue in the TRIM5 B30.2 domain modulates its activity against PtERV1 and HIV-1, with the gain of restriction for one virus resulting in decreased control of the other one24. Human TRIM5 is very active against PtERV1, suggesting that our ancestors adapted to fight this virus or some related retrovirus, and this left them (us) unprepared against the HIV-1 epidemic.
More recently, several genes identified as HIV-1 host factors were analysed in primates, and evidence emerged of positive selection at five of these (ankyrin repeat domain 30A (ANKRD30A), CD4, microtubule-associated protein 4 (MAP4), nucleoporin 153 kDa (NUP153) and RAN binding protein 2 (RANBP2))25. Importantly, most of the positive selection targets in CD4, MAP4 and NUP153 are located in protein regions or domains that are responsible for direct interaction with the virus. The authors suggested that the selective pressure on these genes was exerted by ancient lentiviruses25,26.
Overall, a number of concepts can be taken from these studies: past infection events may leave a signature that affects the ability of extant species to fight emerging pathogens. Evolution may act through trade-offs, whereby changes that are favourable in one specific environment (in this case, the presence of a specific pathogen) may be unfavourable when conditions change. Protein regions at the host–pathogen interface are expected to be targeted by the strongest selective pressure. Evolutionary studies based on inter-species comparisons allow the identification of molecular determinants of infection susceptibility at single amino acid resolution.
Susceptibility to infection in mammals
Mammals display different susceptibility to distinct pathogens, and infection with the same agent can have extremely different outcomes in diverse species (see Ref. 27 for a recent review). Thus, domestic and wild mammalian (and non-mammalian) species represent natural reservoirs of human pathogens and/or may provide the adaptive environment for pathogen spillover. Because host reservoir species and their pathogens often co-evolve for millions of years, evolutionary analyses may help to explain host adaptive events associated with low susceptibility and mild disease outcomes. The most extensive body of knowledge on host–pathogen specificity focuses on viral infections, as the example of TRIM5 mentioned above testifies, but recent work has also shed new light on bacterial diseases.
Complement evasion. Leptospirosis, one of the most prevalent human bacterial zoonoses worldwide, is caused by bacteria of the Leptospira genus. Wild rodents are considered to be the main reservoirs for human leptospirosis, but a study of Malagasy small mammals indicated that several endemic species of tenrecs and bats are also infected with Leptospira species that are markedly specific to their hosts, suggesting long-term adaptation of the bacterium to different hosts28. A feature that pathogenic Leptospira species share with other bacteria is complement evasion. Indeed, these spirochetes have evolved different strategies to elude complement-mediated killing; thus, leptospiral immunoglobulin-like (Lig) proteins can bind complement factor H (CFH) and C4b-binding protein (C4BP) to mediate complement inactivation at the bacterial surface. A genome-wide analysis of positive selection in six mammalian species indicated that the complement system has been the target of extremely intense selective pressure29. Similar results were obtained by analysing positively selected genes in the bat Myotis brandtii30. Thus, selection-driven species-specific differences at complement genes might explain differential susceptibility to infections. In line with this view, human-specific pathogens such as Neisseria gonorrhoeae and Neisseria meningitidis bind CFH of human origin, but not CFH from other primates, and a single amino acid change (N1203R) in the chimpanzee molecule restores CFH binding to sialylated gonococci and bacterial killing31. Several sequenced mammalian genomes are now available; it will be important to study the detailed pattern of molecular evolution at complement genes, with the aim of gaining insight into the determinants of species-specific complement evasion.
Toll-like receptor evolution. Yersinia pestis provides another remarkable example of differential susceptibility to a bacterial infection. Again, rodents act as a natural reservoir for this human pathogen. As with other Gram-negative bacteria, lipid A, the biologically active component of Y. pestis lipopolysaccharide (LPS), is recognized by Toll-like receptor 4 (TLR4) and its co-receptor lymphocyte antigen 96 (LY96; also known as MD2) (see below). Recent data showed that, compared with mouse cells, human cells respond less efficiently to hypoacylated lipid A; this effect is almost entirely due to differences in TLR4 and LY96 sequences, as assessed by the generation of humanized mice32. Different responsiveness to variably acylated LPS from other sources (for example, Escherichia coli) had previously been described33. Starting from this premise, Ohto and co-workers34 solved the crystal structure of the mouse TLR4–LY96–LPS and TLR4–LY96–lipid IVa (a synthetic tetra-acylated lipid A precursor) complexes and compared them to the human counterparts. Structural differences were detected in the interaction of lipid IVa with the two mammalian receptors, with some amino acid replacements in LY96 and TLR4 possibly being responsible for the observed differential binding34. Analysis of TLR4 in mammals revealed that the receptor has evolved adaptively35. We mapped positively selected sites onto the structure of the human and mouse complexes and observed that some of these may indeed account for structural differences between humans and mice (Fig. 1).
Figure 1. Examples of positive selection at the host–pathogen interaction surface.
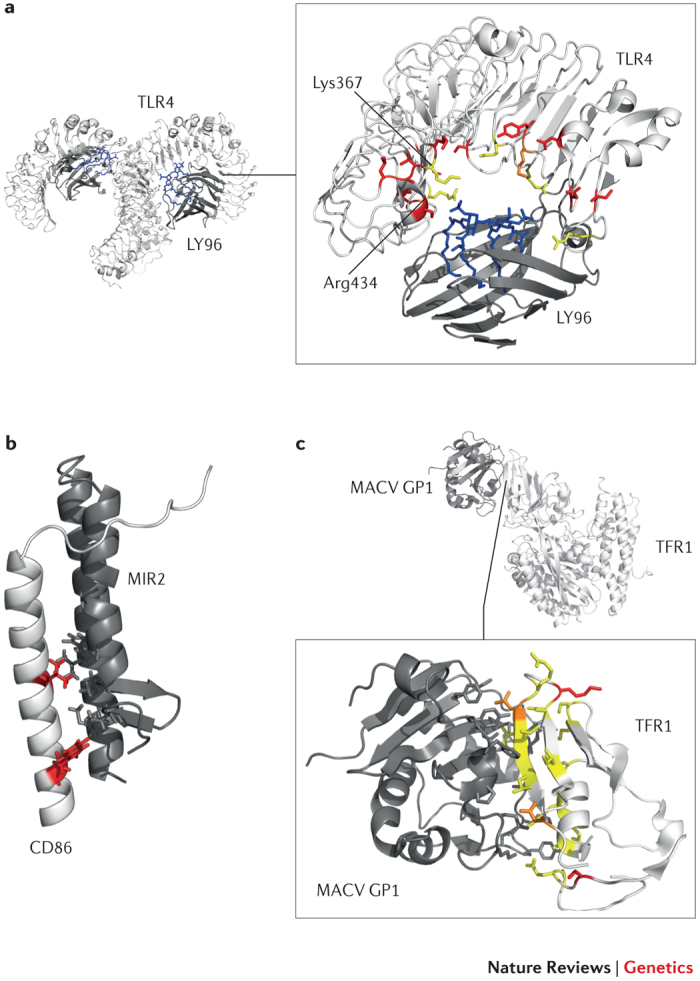
As discussed in Box 1, regions at the host–pathogen contact interface are expected to be targeted by the strongest selective pressure. Three examples are shown here. a | Detail of the Toll-like receptor 4 (TLR4)–lymphocyte antigen 96 (LY96)–lipid IVa complex. Mouse TLR4 and LY96 are in white and grey, respectively; lipid IVa is in blue. Sites that are positively selected in mammals35 are mapped onto the TLR4 structure (red): several of these flank or correspond (orange) to residues that differ between humans and mice and that surround the phosphate groups of lipid IVa (yellow)34. If Lys367 and Arg434 are replaced with the human residues (Glu369 and Gln436, respectively), the responsiveness of mouse TLR4–LY96 to lipid IVa is abolished. b | Structures of human CD86 (white; transmembrane and juxtamembrane region) and MIR2 (grey; encoded by Kaposi sarcoma-associated herpesvirus). CD86 sites that are involved in the interaction and that are positively selected in mammals are shown in red. c | Complex of transferrin receptor protein 1 (TFR1) with the surface glycoprotein (GP1) of Machupo virus (MACV), a rodent arenavirus that can also infect humans through zoonotic transmission. TFR1 residues involved in the interaction with GP1 are in yellow, positively selected sites are in red and positively selected sites that directly interact with GP1 are in orange.
Exploring natural reservoirs of infectious agents. Rodents are the most established animal model for human disease, including for susceptibility to infection. In recent years, however, technological advances have made the sequencing of whole genomes a relatively quick and inexpensive process. The genome sequences of non-model mammals that serve as natural reservoirs of human infectious agents are now available, allowing the unprecedented opportunity to exploit these data for molecular evolution studies. Bats, for example, are known to host a wide range of viruses that are highly pathogenic to humans36. The genomes of six bat species have been sequenced so far, and three of these (M. brandtii, Pteropus alecto and Myotis davidii) were analysed in detail to unveil the evolutionary history of specific traits37. Results showed that different families of immune receptors — including killer cell immunoglobulin-like receptors (KIRs), killer cell lectin-like receptors (KLRs), sialic acid-binding immunoglobulin-like lectins (SIGLECs) and leukocyte immunoglobulin-like receptors (LILRs) — have expanded or contracted in distinct bat species. Also, in these three bat species, as well as in the common ancestor of P. alecto and M. davidii, genes involved in immunity represented preferential targets of positive selection37. This is not unexpected: immune-response genes have been shown to have evolved rapidly in most mammalian species analysed to date9. Thus, although these sequenced bat genomes have not yet provided an explanation as to why bats are tolerant to EBOV, for instance, they pave the way for further analyses to test specific hypotheses and/or to address the molecular determinants of host–pathogen interactions. In a recent study, Demogines and co-workers38 showed how this can be accomplished. The authors focused on angiotensin-converting enzyme 2 (ACE2), which serves as a receptor for severe acute respiratory syndrome coronavirus (SARS-CoV) cell entry. In particular, the receptor-binding domain of the viral spike protein is responsible for ACE2 binding and is a major determinant of host range39. Although the human SARS epidemic was suggested to have originated from the zoonotic transmission of SARS-CoV from bats to humans, possibly via an intermediate host (for example, palm civets)40,41, no ACE2-binding SARS-CoV-like virus had been identified in bats when Demogines and collaborators started their work38. The authors analysed ACE2 genes in 11 bat species, and results revealed that the gene evolved adaptively and that the positively selected residues of the bat genes map at the ACE2–SARS-CoV interaction surface (Fig. 2). These data led to the conclusion that ACE2-binding coronaviruses originated in bats38. This finding was confirmed in a subsequent study that isolated an ACE2-binding SARS-like coronavirus from horseshoe bats in China42, highlighting the power of evolutionary studies in predicting host range and disease emergence.
Figure 2. Positive selection at the cellular receptors for coronaviruses (SARS-CoV and MERS-CoV).
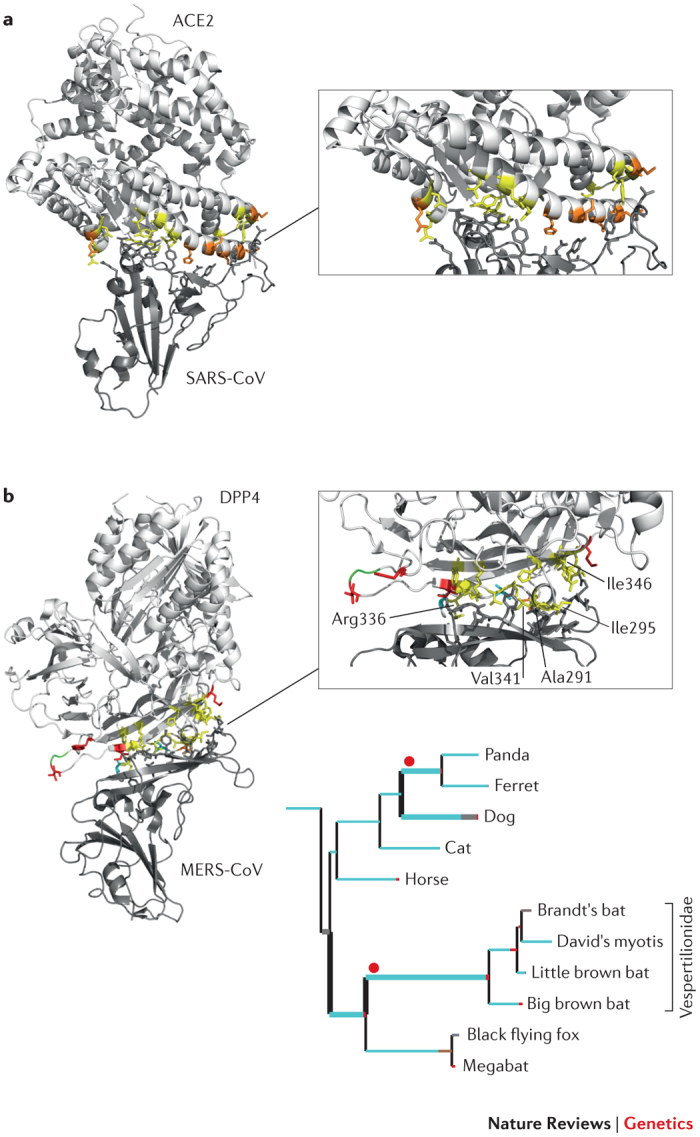
The receptor-binding domains (RBDs) are structurally similar in the spike proteins of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV)47, but these proteins bind distinct cellular receptors. The structure of the SARS-CoV and MERS-CoV RBDs in complex with angiotensin-converting enzyme 2 (ACE2) and dipeptidyl peptidase 4 (DPP4), respectively, are shown with the binding interfaces enlarged. In both panels, sites that directly interact with the RBD are shown in yellow. a | ACE2 residues that are responsible for RBD binding and that are also positively selected in bats are shown in orange38. b | DPP4 residues that are positively selected at the RBD binding interface are shown in red (positively selected) and orange (positively selected and interacting); sites in cyan were found to be positively selected along specific branches (Supplementary information S1,S2 (box, table)), as shown in the tree panel. The tree includes a subset of relevant branches, with those showing evidence of episodic positive selection represented with thick lines and red dots. Branch colours indicate the strength of selection (dN/dS): red indicates positive selection (dN/dS>5); blue indicates purifying selection (dN/dS=0); and grey indicates neutral evolution (dN/dS=1). Human residues that modify the binding energy if they are replaced with their hamster counterparts are labelled45. One of these (Val341) is positively selected (orange). A three amino acid deletion in bats is shown in green (see Supplementary information S1,S2 (box, table)).
Similarly to SARS-CoV, MERS-CoV is thought to have originated in bats and to have spread to humans via an intermediate host, possibly dromedary camels43. Infection is initiated by binding of the MERS-CoV spike protein to human dipeptidyl peptidase 4 (DPP4; also known as CD26)44. Recent data indicate that five amino acids in DPP4 that differ between humans (MERS-CoV susceptible) and hamsters (non-susceptible) are key determinants for host specificity45 (Fig. 2). We extended a previous evolutionary analysis of mammalian DPP4 (Ref. 46): strong evidence of positive selection was found with episodic selection in the Vespertilionidae bat family and the panda and ferret branches, as well as in the dog lineage (Fig. 2; see Supplementary information S1,S2 (box, table)). As shown in Fig. 2, most positively selected sites are located at the DPP4–spike protein interaction surface47, and one of these is among those described as binding determinants45. Thus, as observed for ACE2, MERS-CoV and related viruses (for example, coronavirus HKU4) are likely to act as drivers of molecular evolution on mammalian DPP4 genes; it will be especially interesting to evaluate the contribution of positively selected sites in ferrets because these animals are resistant to MERS-CoV infection.
Detecting and fighting infections
Immune responses in mammals are highly coordinated processes involving multiple systems that sense infection, activate antiviral and antimicrobial responses, and trigger adaptive immunity. The evolutionary history of several such systems has been analysed in detail, and below we describe the most recent findings.
Innate immune receptors. The mammalian immune system is endowed with a repertoire of molecular sensors called pattern-recognition receptors (PRRs). These molecules detect pathogen-associated molecular patterns (PAMPs) and initiate a downstream signalling cascade that culminates in the production of cytokines and antimicrobial factors. The main families of PRRs include TLRs, NOD-like receptors (NLRs), RIG-like receptors (RLRs) and AIM2-like receptors (ALRs). In the host–pathogen arms race, these molecules represent one of the foremost detection–defence systems; consistently, several studies have reported adaptive evolution at genes encoding mammalian PRRs.
Analyses in primates, rodents and representative mammalian species indicate that positive selection shaped nucleotide diversity at most TLRs, with the strongest pressure acting on TLR4 (Refs 35,48,49). Similarly to TLR4 (Fig. 1), several positively selected sites in other TLRs are located in PAMP-binding regions, raising questions as to whether species-specific host–pathogen co-evolution is occurring, and how these sequence changes translate into differential PAMP recognition. In fact, as mentioned above for LPS, species-specific differences in ligand binding by TLRs seem to be common and potentially affect the overall immune response to specific pathogens50. Integration of evolutionary, immunological and genetic studies will be instrumental in the future for medical applications, especially in light of the proposed use of TLR ligands as vaccine adjuvants, a step that may require tailoring to distinct species50.
Compared with TLRs, mammalian ALRs are much less conserved and more dynamic, with distinct species carrying different sets of functional genes (ranging from 13 in mice to none in some bats)37,51. As a consequence, analysis of several mammals indicated that, with the exception of absent in melanoma 2 (AIM2), which is non-functional in several species, no unequivocal orthologues can be inferred for the remaining ALR genes. This prevents the application of standard codon-based tests across the entire mammalian phylogeny, although closely related species can be analysed. Thus, interferon-γ-inducible protein 16 (IFI16) and AIM2 were shown to have evolved under positive selection in primates. Positively selected sites were observed to mainly localize near to regions or domains involved in DNA binding and protein–protein interaction, suggesting modulation of substrate specificity or genetic conflicts with viral inhibitors52. Positive selection was also described for the three mammalian RLRs (retinoic acid-inducible gene I (RIGI; also known as DDX58), melanoma differentiation-associated 5 (MDA5; also known as IFIH1) and LGP2 (also known as DHX58)), the primate NLR family apoptosis inhibitory protein (NAIP) and rodent Naip2 genes53,54. Indeed, as is the case for ALRs, rodents have multiple NAIP paralogues that show widespread evidence of inter-locus recombination. This led to the application of a dN/dS sliding window approach: the Naip2 sites evolving with dN/dS >1 were found to be located in the bacterial ligand domain54.
Antiviral effectors and restriction factors. Studies on antiviral restriction factors have been extensive because these molecules represent obvious targets in host–pathogen arms races. Specifically, genetic conflicts between host restriction factors and viral components often play out in terms of binding-seeking dynamics (the host factor adapts to bind the viral component) and binding-avoidance dynamics (the virus counter-adapts to avoid binding and restriction by the host factors). The evolutionary history of antiviral restriction factors has been comprehensively reviewed elsewhere55,56,57, and we only highlight a few recent developments here.
The first restriction factor to be identified was the product of the mouse gene Friend virus susceptibility 1 (Fv1), a protein that protects against murine leukaemia virus (MLV) infection58. The origin and evolution of FV1 is extremely interesting: early sequence analysis revealed that it derives from the gag gene of an ancient endogenous retrovirus that is not directly related to MLV58. Thus, FV1 exemplifies a paradoxical twist of the arms race scenario whereby a viral gene is co-opted by the host to serve an antiviral function (this is not the only instance, see Ref. 59). Recent results showed that the Fv1 gene was inserted into the mouse genome between 4 million and 7 million years ago, long before the appearance of MLV. Thus, the selective pressure exerted by other viruses must have maintained FV1 function and driven its evolution60. Indeed, analysis of FV1 from wild-type mice indicates that different Fv1 products can recognize and block multiple genera of retroviruses (for example, equine infectious anaemia virus and feline foamy virus), and a number of positively selected sites in the carboxy-terminal region of FV1 are directly involved in restriction specificity60. Thus, in a similar way to TRIM5, FV1 was identified for its ability to restrict an extant virus, but its evolution was driven by different waves of retroviral species, some of which are likely to be extinct.
Other restriction factors that have been the topic of recent investigation are encoded by two paralogous genes, myxovirus resistance 1 (MX1; also known as MxA) and MX2 (also known as MxB). The protein products of the two genes display high sequence identity but different antiviral specificity. MX1 has broad activity against RNA and DNA viruses. Recently, Mitchell and collaborators61 showed the potential of evolutionary analyses to generate experimentally testable hypotheses on the nature of genetic changes that affect species-specific susceptibility to infection. The authors applied an evolution-guided approach and identified a cluster of positively selected residues in an unstructured surface-exposed MX1 loop (loop 4), which confers antiviral specificity; genetic variation in loop 4 is a major determinant of MX1 antiviral activity against Thogoto and avian influenza A viruses, and replacements at a single positively selected site alter the ability of MX1 to restrict these pathogens61.
More recently, the selection pattern at the MX2 gene, which encodes an antiretroviral effector62, was shown to parallel that of MX1, with most selected sites located in loop 4 (Ref. 63). In MX2, sites selected in the primate lineage were detected outside loop 4, and MX1 also showed evidence of selection in other domains61,63; these sites are promising candidates for being additional determinants of antiviral activity.
Antigen presentation, T cell activation and immunoglobulin G receptors. Antigen presentation and the ensuing T cell activation are central processes in mammalian cell-mediated immune response (Fig. 3). Therefore, a convenient strategy for pathogens to elude immune surveillance is to hijack the molecular pathways responsible for these processes64,65. In line with the arms race scenario, there is evidence of positive selection at several mammalian genes involved in antigen presentation and the regulation of T cell activation66,67 (Fig. 3). The pathogen-driven mechanisms underlying evolution at these genes are likely to be manifold. One mechanism is genetic conflict with a pathogen-encoded component, evidence of which can be seen in the protein CD86. Positively selected sites in the transmembrane and juxtamembrane region of CD86 interact with MIR2 (Fig. 3), a Kaposi sarcoma-associated herpesvirus (KSHV) protein that downmodulates CD86 expression67,68. A second mechanism is the use of cell-surface molecules as viral receptors: some adenovirus strains, for example, have been reported to exploit CD80 and CD86 for cellular attachment69,70. A third mechanism is the broadening or tuning of the host's ability to process and present pathogen-derived components. For example, a positively selected site in the carbohydrate-recognition domain of CD207 (also known as langerin; a Birbeck granule molecule) affects an amino acid position that is directly involved in the binding of pathogen-derived glycoconjugates71.
Figure 3. Genes involved in antigen processing and presentation and T cell regulation are common targets of positive selection in mammals.
All pathway components are designated using official gene names (excluding the major histocompatibility complex (MHC) and T cell receptor (TCR)) and are highlighted in red if they are targets of positive selection in mammals or primates25,66,67. The molecular components of different antigen processing and presentation pathways are shown (details from Refs 107,108) to provide a general overview of the extent of positive selection and to highlight the function of positively selected genes, as most of their protein products directly interact with the antigen. Thus, the figure is not meant to show all molecules involved in the process or to convey mechanistic insights. Also, some genes may show tissue-specific expression or may be induced under specific circumstances: their products are nonetheless included for the sake of completeness. As for T cell regulatory molecules, the representation does not reflect the stoichiometry of binding (for example, CD28 functions as a dimer). Notably, the same molecule may be expressed by different populations of T cells, although here each molecule is shown on one T cell type only (to avoid redundancy). The dashed arrows and '?' indicate steps that lack clear molecular definition or are only inferred. The orange circles, and red and blue shapes at the bottom of the figure represent proteolytic fragments. B2M, β2-microglobulin; BLMH, bleomycin hydrolase; CALR, calreticulin; CD40LG, CD40 ligand; CTLA4, cytotoxic T lymphocyte protein 4; CTS, cathepsin; CYB, cytochrome b; ERAP, endoplasmic reticulum aminopeptidase; HAVCR2, hepatitis A virus cellular receptor 2; HLA-DM, major histocompatibility complex, class II, DM; ICOS, inducible T cell co-stimulator; ICOSLG, ICOS ligand; IFI30, interferon-γ-inducible protein 30; iNKT, invariant natural killer T; iTCR, invariant TCR; LGMN, legumain; LNPEP, leucyl-cystinyl aminopeptidase; NCF, neutrophil cytosol factor; NPEPPS, puromycin-sensitive aminopeptidase (also known as PSA); NRD1, nardilysin; PDCD1, programmed cell death 1; PDCD1LG2, programmed cell death 1 ligand 2; PDIA3, protein disulfide-isomerase A3; ROS, reactive oxygen species; TAP, antigen peptide transporter; TAPBP, TAP-binding protein (also known as tapasin); THOP1, thimet oligopeptidase 1; TPP2, tripeptidyl-peptidase 2.
These mechanisms are not mutually exclusive. For example, a plethora of viral pathogens (such as herpes simplex virus 1, human papillomavirus, HIV-1 and KSHV) interfere with CD1D trafficking and recycling72,73. As a consequence, the cytoplasmic and transmembrane regions of CD1D display positively selected sites, one of which is within a primate-specific trafficking signal. Additional positively selected sites are located in the CD1D extracellular region and flank the T cell receptor interaction surface and the lipid-binding pocket, which suggests that they exert an effect on antigen-binding specificity67.
Finally, we draw attention to one of the few attempts at assessing the part that helminth infections have played as selective pressures for mammals and at integrating epidemiological information into molecular evolutionary approaches. Machado and co-workers74 found evidence of positive selection at the mammalian gene Fc fragment of IgG, low affinity IIIb, receptor (FCGR3B), which is expressed by eosinophils and is involved in the binding of immunoglobulin G (IgG)-coated parasites. Notably, the authors also tested a specific hypothesis whereby mammalian lineages hosting a wider range of helminth species should show stronger evidence of selection compared with other species (this was accomplished by running the phylogenetic analysis by maximum likelihood (PAML) branch-site models with helminth-rich lineages as foreground branches74; Box 3). Their hypothesis was verified, providing a plausible explanation for the evolutionary pattern at FCGR3B and suggesting that similar approaches may be used to detect other mammalian genes involved in genetic conflicts with helminth parasites.
Examples other than immune effectors
As exemplified by ACE2, host–pathogen interactions are not limited to immune system components. The reasons why genes with no specific defence function may be targeted by the selective pressure imposed by infectious agents are manifold. The best known instances probably refer to gene products that act as incidental receptors for pathogens, as is the case with ACE2. Other host gene products that engage in genetic conflicts include those that participate in the coagulation cascade and the contact system, which are commonly hijacked by bacterial pathogens to promote tissue invasion or to elude detection by immune cells (see Ref. 75 for a review). An alternative possibility is that the host builds a line of defence based on the sequestration of essential micronutrients from the pathogen, a phenomenon known as 'nutritional immunity'.
Housekeeping genes. Incidental receptors are often represented by the products of housekeeping genes, which are typically expressed at high levels by different cell types. Among these, the transferrin receptor (TFRC) gene encodes a cell-surface molecule (transferrin receptor protein 1 (TFR1)) that is essential for iron uptake. TFR1 is used as a receptor by mouse mammary tumour virus, canine parvovirus and rodent New World arenaviruses. In line with the arms race scenario, TFRC evolved adaptively in rodents and caniforms, and positively selected sites are mainly located in the extracellular domain regions that interact with rodent-infecting arenaviruses (Fig. 1) and carnivore-infecting parvoviruses, respectively76,77. Interestingly, positive selection at the primate transferrin (TF) gene, which encodes the TFR1 ligand, was also recently described78; in this case, selection is driven by bacteria, not viruses78. Transferrin is the major circulating iron transporter in mammals and is also thought to participate in nutritional immunity by sequestering iron from bacteria. Consistently, most positively selected sites were found to have evolved to counteract binding by bacterial transferrin surface receptors that scavenge host iron78. Thus, different selective pressures exerted by distinct molecular mechanisms contributed to shaping the evolution of a central homeostatic process — in this case, iron transport in mammals.
Another housekeeping gene product that acts as a viral receptor is Niemann–Pick C1 protein (NPC1), a sterol transporter located in the membrane of late endosomes and lysosomes. NPC1 is expressed by most cell types and is used by filoviruses (such as EBOV and Marburg virus). Evolutionary analysis of mammalian NPC1 genes indicated that three positively selected residues are located in the amino-terminal portion of the second NPC1 luminal loop; binding of this loop by the EBOV glycoprotein (GP) is necessary and sufficient for the viral receptor activity of the sterol transporter79,80 (Fig. 4). The second luminal loop of NPC1 is also bound with high affinity by the GP encoded by Lloviu virus, a bat-derived, EBOV-like filovirus81. Thus, NPC1 may represent a universal receptor for filoviruses, and the constant selective pressure exerted by such viruses might have greatly contributed to shaping mammalian genetic diversity at loop 2. These data may have great and immediate practical values. In fact, small molecules that directly target NPC1 and disrupt GP binding are regarded as possible therapeutic compounds against EBOV82,83,84 (Fig. 4). Because mammalian NPC1 diversity at the interaction surface is driven by selection, future efforts in this direction are likely to benefit from the incorporation of evolutionary analysis; this would be especially important when testing therapeutic molecules on model organisms and non-human mammals. In humans, mutations in NPC1 cause Niemann–Pick disease type C1, a progressive neurodegenerative condition. This is in line with the central role of this transporter in housekeeping functions; thus purifying selection. is expected to constrain variation in the gene. Indeed, the human–mouse dN/dS calculated for the NPC1 whole-gene region is definitely lower than 1, as is the case for most genes (Fig. 4). In fact, mammalian NPC1 genes show a large preponderance of codons evolving with dN/dS <1, and positive selection is extremely localized in loop 2 (Fig. 4). This specific example illustrates a general concept, whereby molecules involved in central homeostatic processes may be engaged in genetic conflicts with pathogens, although in several instances the sequence space accessible for adaptive mutation without a high fitness cost is expected to be limited.
Figure 4. Positive and purifying selection.
a | Distribution of dN/dS values for human–mouse one-to-one orthologues. The values for some of the genes discussed in this Review are indicated. Data were derived from the Ensembl BioMart database (see Further information). b | Natural selection acting on mammalian Niemann–Pick C1 (NPC1) genes. NPC1 is shown with its predicted membrane topology and protein regions coloured in hues of blue that represent the percentage of negatively selected sites (as detected by the single-likelihood ancestor counting method using Datamonkey); the darker the blue, the higher the percentage. The location of three positively selected residues (red) is indicated on the left, and an alignment of the corresponding region is shown on the protein to the right (with red and blue representing positively and negatively selected sites, respectively). The interaction with the glycoprotein (GP; green) of filoviruses (such as Ebola virus, Marburg virus or Lloviu virus) is shown. GP binds NPC1 after processing by cellular proteases. ACE2, angiotensin-converting enzyme 2; DARC, Duffy blood group, atypical chemokine receptor; MX1, myxovirus resistance 1; SSD, sterol-sensing domain; TFRC, transferrin receptor; TLR4, Toll-like receptor 4.
The coagulation cascade and contact system. As anticipated above, several components of the coagulation cascade and contact system evolved adaptively in mammals, most likely as a result of genetic conflicts with bacterial pathogens85,86. For instance, Staphylococcus aureus is endowed with an arsenal of proteins that target such systems, including two cysteine proteinases (ScpA and SspB) that cleave plasma kininogen at each terminal side of the bradykinin domain to generate kinins, with a consequent increase of vascular leakage87. These events are central for bacterial virulence and are linked to the pathogenesis of sepsis. In kininogen 1 (KNG1), positively selected sites are located in all domains, with the exception of the highly conserved bradykinin region85. One of the positively selected sites defines the N-terminal cleavage site of ScpA and SspB, suggesting that sites flanking the bradykinin sequence are evolving to avoid recognition and cleavage by bacterial-encoded proteases. In analogy to the strong purifying selection acting on the bradykinin region, analysis of calculation cascade genes indicated that disease-causing mutations are more likely to occur at sites targeted by purifying selection and are rarer at positively selected sites86. Again, these observations highlight the coexistence of distinct selective regimes at the same gene regions and exemplify the concept of evolutionary trade-offs.
Conclusions
The advent of high-throughput sequencing technologies has allowed for the generation of an unprecedented wealth of genetic data, including the whole-genome sequences of host reservoir species for human pathogens, as well as genetic information for multiple microbial and viral species and strains. Moreover, large-scale approaches such as RNA interference and mass spectrometry are providing detailed pictures of host–pathogen interactomes88,89. Finally, an increasing number of crystal structures of interacting host and pathogen proteins solved in complex are available, allowing the opportunity to determine the structural basis of these interactions to identify regions or amino acids that lie at the host–pathogen contact surface. Integration of these data with evolutionary analysis will allow the testing of specific hypotheses, including which species have responded to the pressure exerted by one or more pathogens (see the SARS-CoV example), which molecules and residues have participated in the arms race and which host–pathogen interacting partners are expected to have co-evolved. These advances are also expected to progressively change evolutionary genetics from a hypothesis-driven to a hypothesis-generating discipline. In this respect, we note that although the arms race scenarios we have described in this Review imply some form of host–pathogen co-evolution over time, the nature of the interaction and its dynamics have often been inferred from the observed pattern of variation. Indeed, the fact that the same residues that affect specific host–pathogen interactions are targeted by positive selection does not necessarily imply a causal link, and in many instances the specific selective agents underlying molecular adaptations remain to be determined. As shown above, these may well be accounted for by extinct pathogens or by agents that had a major co-evolutionary role in the past but that are now fading away from the landscape of common infections. With a few exceptions16,24, evolutionary studies only investigate extant genetic variation and modern pathogens, with little reconstruction of past events. Nevertheless, we do not necessarily need to go back in time: evolutionary analyses can be used as predictive tools to pinpoint which genes and residues are more likely to contribute to present-day host–pathogen interaction and help explain species-specific susceptibility to infection. Several studies mentioned above, including those investigating selection at MX1 (Ref. 61), TFRC (Refs 76,77), TF78 and other protein-coding genes23,24,54,60, used experimental analyses to show that evolutionary information can indeed be exploited to gain high-resolution insight into the molecular determinants of binding affinities at host–pathogen interfaces.
The studies of iron transporters78 hold particular value because the authors analysed the genetic variability of both the host and the pathogen and showed that both parties evolved in response to mutually exerted pressures, in line with the Red Queen principles. So far, few attempts have been made at integrating evolutionary analyses of host and pathogen interacting partners into a common framework. However, efforts in this direction hold the promise of improving our understanding of the strategies used by both hosts and pathogens to adapt and counter-adapt. In turn, this knowledge has possible biomedical and therapeutic implications, given the ability of different pathogens or distinct strains of the same infectious agent to elude not only natural host defences but also drugs and vaccination strategies.
As a final note, we mention that we have exclusively focused on adaptive events involving coding gene regions. Nevertheless, several recent studies (see Ref. 10 for a review) have highlighted the role of non-coding variants as important determinants of susceptibility to infection within species. Thus, host–pathogen conflicts are more than likely to have contributed to adaptive evolution at regulatory elements during speciation. Detection of these adaptive events will benefit from the availability of high-throughput techniques (for example, RNA sequencing and chromatin immunoprecipitation followed by sequencing) and the development of methodological approaches for dissecting molecular evolution in non-coding regions; notably, recent data have shown the usefulness of a framework similar to dN/dS to analyse the evolutionary history of mammalian transcriptional enhancers90. Application of this methodology (or extensions thereof) to the study of host–pathogen interactions will provide valuable information on which non-coding sequence changes have been targeted by selection and thus modulate susceptibility to infection or related phenotypes.
Supplementary information
Evolutionary analysis of mammalian DDP4. (PDF 2101 kb)
LRT statistics for DPP4. (PDF 260 kb)
Acknowledgements
D.F. is supported by fellowships of the Doctorate School of Molecular Medicine, University of Milan, Italy.
Glossary
- Positive selection
The accumulation of favourable amino acid-replacing substitutions, which results in more non-synonymous changes than expected under neutrality (dN/dS > 1).
- Episodic selection
Positive selection localized to a subset of sites or confined to a few species in a phylogeny.
- Orthologues
Genes that evolved from a common ancestral gene through speciation.
- Paralogues
Homologous genes created by a duplication event within the same genome.
- dN
The observed number of non-synonymous substitutions per non-synonymous site.
- dS
The observed number of synonymous substitutions per synonymous site.
- Purifying selection
The elimination of deleterious amino acid-replacing substitutions, which results in fewer non-synonymous changes than expected under neutrality (dN/dS < 1).
Biographies
Manuela Sironi received her Ph.D. in molecular medicine from the University of Milan, Italy. Since 2007 she has been coordinating and supervising one of the research teams at the Bioinformatic Unit of Scientific Institute IRCCS Eugenio Medea, Bosisio Parini, Italy. In recent years her research has mainly focused on the evolutionary history of human disease alleles and the application of evolutionary approaches to study host–pathogen interactions. She and her group have also investigated the genetic determinants of HIV-1 infection susceptibility.
Rachele Cagliani received her speciality in applied genetics from the University of Milan, Italy. She worked for about 2 years in the Department of Biotechnology at the San Raffaele Scientific Institute in Milan, Italy, as a researcher. She joined IRCCS Eugenio Medea, Bosisio Parini, Italy, in 1999 as a researcher. Her research interests include the evolutionary history of human disease alleles, the molecular genetics of infectious and autoimmune diseases and the genetics of complex traits.
Diego Forni is a third-year Ph.D. student at the Doctoral School of Molecular and Translational Medicine, University of Milan, Italy. He carries out his research at IRCCS Eugenio Medea in Bosisio Parini, Italy, in the Bioinformatic Unit. His research focuses on the evolutionary dynamics that shape genetic diversity in human populations and mammalian species.
Mario (Mago) Clerici, M.D., is a professor of immunology at the University of Milan, Italy, and the Scientific Director of the Don Gnocchi Foundation, a network of Italian research hospitals. He graduated from the University of Milan and trained in the United States at the National Institutes of Health in Bethesda, Maryland (1987–1994). He has been awarded national and international prizes, and he is a highly cited researcher (Thomson Reuters, 2001). His research focuses on the immunopathogenesis of human diseases with an emphasis on HIV infection and neurodegenerative pathologies.
Related links
FURTHER INFORMATION
PowerPoint slides
Competing interests
The authors declare no competing financial interests.
References
- 1.Fumagalli M, et al. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7:e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Valen L. A new evolutionary law. Evol. Theory. 1973;1:1–30. [Google Scholar]
- 3.Brockhurst, M. A. et al. Running with the Red Queen: the role of biotic conflicts in evolution. Proc. Biol. Sci.10.1098/rspb.2014.1382 (2014). [DOI] [PMC free article] [PubMed]
- 4.Haldane JBS. The Causes of Evolution. 1932. [Google Scholar]
- 5.Samson JE, Magadan AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nature Rev. Microbiol. 2013;11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 6.Karasov TL, Horton MW, Bergelson J. Genomic variability as a driver of plant-pathogen coevolution? Curr. Opin. Plant Biol. 2014;18:24–30. doi: 10.1016/j.pbi.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray JC, Cutter AD. Mainstreaming Caenorhabditis elegans in experimental evolution. Proc. Biol. Sci. 2014;281:20133055. doi: 10.1098/rspb.2013.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keebaugh ES, Schlenke TA. Insights from natural host-parasite interactions: the Drosophila model. Dev. Comp. Immunol. 2014;42:111–123. doi: 10.1016/j.dci.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nature Rev. Genet. 2010;11:17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- 10.Quintana-Murci L, Clark AG. Population genetic tools for dissecting innate immunity in humans. Nature Rev. Immunol. 2013;13:280–293. doi: 10.1038/nri3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siddle KJ, Quintana-Murci L. The Red Queen's long race: human adaptation to pathogen pressure. Curr. Opin. Genet. Dev. 2014;29:31–38. doi: 10.1016/j.gde.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Fumagalli M, Sironi M. Human genome variability, natural selection and infectious diseases. Curr. Opin. Immunol. 2014;30:9–16. doi: 10.1016/j.coi.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and infectious disease in human populations. Nature Rev. Genet. 2014;15:379–393. doi: 10.1038/nrg3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benton MJ. The Red Queen and the Court Jester: species diversity and the role of biotic and abiotic factors through time. Science. 2009;323:728–732. doi: 10.1126/science.1157719. [DOI] [PubMed] [Google Scholar]
- 15.Venditti C, Meade A, Pagel M. Phylogenies reveal new interpretation of speciation and the Red Queen. Nature. 2010;463:349–352. doi: 10.1038/nature08630. [DOI] [PubMed] [Google Scholar]
- 16.Decaestecker E, et al. Host–parasite 'Red Queen' dynamics archived in pond sediment. Nature. 2007;450:870–873. doi: 10.1038/nature06291. [DOI] [PubMed] [Google Scholar]
- 17.Luijckx P, Fienberg H, Duneau D, Ebert D. A matching-allele model explains host resistance to parasites. Curr. Biol. 2013;23:1085–1088. doi: 10.1016/j.cub.2013.04.064. [DOI] [PubMed] [Google Scholar]
- 18.Paterson S, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–278. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nature Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 20.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stremlau M, et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 23.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5α antiviral protein. Science. 2007;316:1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- 25.Meyerson NR, et al. Positive selection of primate genes that promote HIV-1 replication. Virology. 2014;454–455:291–298. doi: 10.1016/j.virol.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaller T, et al. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bean AG, et al. Studying immunity to zoonotic diseases in the natural host — keeping it real. Nature Rev. Immunol. 2013;13:851–861. doi: 10.1038/nri3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietrich M, et al. Diversification of an emerging pathogen in a biodiversity hotspot: Leptospira in endemic small mammals of Madagascar. Mol. Ecol. 2014;23:2783–2796. doi: 10.1111/mec.12777. [DOI] [PubMed] [Google Scholar]
- 29.Kosiol C, et al. Patterns of positive selection in six mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seim I, et al. Genome analysis reveals insights into physiology and longevity of the Brandt's bat Myotis brandtii. Nature Commun. 2013;4:2212. doi: 10.1038/ncomms3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaughnessy J, et al. Molecular characterization of the interaction between sialylated Neisseria gonorrhoeae and factor H. J. Biol. Chem. 2011;286:22235–22242. doi: 10.1074/jbc.M111.225516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hajjar AM, et al. Humanized TLR4/MD-2 mice reveal LPS recognition differentially impacts susceptibility to Yersinia pestis and Salmonella enterica. PLoS Pathog. 2012;8:e1002963. doi: 10.1371/journal.ppat.1002963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohto U, Fukase K, Miyake K, Shimizu T. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proc. Natl Acad. Sci. USA. 2012;109:7421–7426. doi: 10.1073/pnas.1201193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Areal H, Abrantes J, Esteves PJ. Signatures of positive selection in Toll-like receptor (TLR) genes in mammals. BMC Evol. Biol. 2011;11:368. doi: 10.1186/1471-2148-11-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang LF, Walker PJ, Poon LL. Mass extinctions, biodiversity and mitochondrial function: are bats 'special' as reservoirs for emerging viruses? Curr. Opin. Virol. 2011;1:649–657. doi: 10.1016/j.coviro.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang G, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demogines A, Farzan M, Sawyer SL. Evidence for ACE2-utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J. Virol. 2012;86:6350–6353. doi: 10.1128/JVI.00311-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo L, Godeke GJ, Raamsman MJ, Masters PS, Rottier PJ. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan Y, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 41.Lau SK, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ge XY, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cotten M, et al. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg. Infect. Dis. 2013;19:736–742B. doi: 10.3201/eid1905.130057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raj VS, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Doremalen N, et al. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 2014;88:9220–9232. doi: 10.1128/JVI.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui J, Eden JS, Holmes EC, Wang LF. Adaptive evolution of bat dipeptidyl peptidase 4 (dpp4): implications for the origin and emergence of Middle East respiratory syndrome coronavirus. Virol. J. 2013;10:304. doi: 10.1186/1743-422X-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu G, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wlasiuk G, Nachman MW. Adaptation and constraint at Toll-like receptors in primates. Mol. Biol. Evol. 2010;27:2172–2186. doi: 10.1093/molbev/msq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fornuskova A, et al. Contrasted evolutionary histories of two Toll-like receptors (Tlr4 and Tlr7) in wild rodents (MURINAE) BMC Evol. Biol. 2013;13:194. doi: 10.1186/1471-2148-13-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Werling D, Jann OC, Offord V, Glass EJ, Coffey TJ. Variation matters: TLR structure and species-specific pathogen recognition. Trends Immunol. 2009;30:124–130. doi: 10.1016/j.it.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 51.Brunette RL, et al. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J. Exp. Med. 2012;209:1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cagliani R, et al. Ancient and recent selective pressures shaped genetic diversity at AIM2-like nucleic acid sensors. Genome Biol. Evol. 2014;6:830–845. doi: 10.1093/gbe/evu066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cagliani R, et al. RIG-I-like receptors evolved adaptively in mammals, with parallel evolution at LGP2 and RIG-I. J. Mol. Biol. 2014;426:1351–1365. doi: 10.1016/j.jmb.2013.10.040. [DOI] [PubMed] [Google Scholar]
- 54.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol. Cell. 2014;54:17–29. doi: 10.1016/j.molcel.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu. Rev. Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 56.Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nature Rev. Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sawyer SL, Elde NC. A cross-species view on viruses. Curr. Opin. Virol. 2012;2:561–568. doi: 10.1016/j.coviro.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 59.Aswad A, Katzourakis A. Paleovirology and virally derived immunity. Trends Ecol. Evol. 2012;27:627–636. doi: 10.1016/j.tree.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 60.Yap MW, Colbeck E, Ellis SA, Stoye JP. Evolution of the retroviral restriction gene Fv1: inhibition of non-MLV retroviruses. PLoS Pathog. 2014;10:e1003968. doi: 10.1371/journal.ppat.1003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitchell PS, et al. Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell Host Microbe. 2012;12:598–604. doi: 10.1016/j.chom.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goujon C, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sironi M, et al. Evolutionary analysis identifies an MX2 haplotype associated with natural resistance to HIV-1 infection. Mol. Biol. Evol. 2014;31:2402–2414. doi: 10.1093/molbev/msu193. [DOI] [PubMed] [Google Scholar]
- 64.Khan N, Gowthaman U, Pahari S, Agrewala JN. Manipulation of costimulatory molecules by intracellular pathogens: veni, vidi, vici!! PLoS Pathog. 2012;8:e1002676. doi: 10.1371/journal.ppat.1002676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hansen TH, Bouvier M. MHC class I antigen presentation: learning from viral evasion strategies. Nature Rev. Immunol. 2009;9:503–513. doi: 10.1038/nri2575. [DOI] [PubMed] [Google Scholar]
- 66.Forni D, et al. An evolutionary analysis of antigen processing and presentation across different timescales reveals pervasive selection. PLoS Genet. 2014;10:e1004189. doi: 10.1371/journal.pgen.1004189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Forni D, et al. A 175 million year history of T cell regulatory molecules reveals widespread selection, with adaptive evolution of disease alleles. Immunity. 2013;38:1129–1141. doi: 10.1016/j.immuni.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 68.Kajikawa M, et al. The intertransmembrane region of Kaposi's sarcoma-associated herpesvirus modulator of immune recognition 2 contributes to B7-2 downregulation. J. Virol. 2012;86:5288–5296. doi: 10.1128/JVI.00219-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaudhry A, et al. The Nef protein of HIV-1 induces loss of cell surface costimulatory molecules CD80 and CD86 in APCs. J. Immunol. 2005;175:4566–4574. doi: 10.4049/jimmunol.175.7.4566. [DOI] [PubMed] [Google Scholar]
- 70.Short JJ, Vasu C, Holterman MJ, Curiel DT, Pereboev A. Members of adenovirus species B utilize CD80 and CD86 as cellular attachment receptors. Virus Res. 2006;122:144–153. doi: 10.1016/j.virusres.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feinberg H, et al. Structural basis for langerin recognition of diverse pathogen and mammalian glycans through a single binding site. J. Mol. Biol. 2011;405:1027–1039. doi: 10.1016/j.jmb.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Horst D, Geerdink RJ, Gram AM, Stoppelenburg AJ, Ressing ME. Hiding lipid presentation: viral interference with CD1d-restricted invariant natural killer T (iNKT) cell activation. Viruses. 2012;4:2379–2399. doi: 10.3390/v4102379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu J, et al. A threonine-based targeting signal in the human CD1d cytoplasmic tail controls its functional expression. J. Immunol. 2010;184:4973–4981. doi: 10.4049/jimmunol.0901448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Machado LR, et al. Evolutionary history of copy-number-variable locus for the low-affinity Fcγ receptor: mutation rate, autoimmune disease, and the legacy of helminth infection. Am. J. Hum. Genet. 2012;90:973–985. doi: 10.1016/j.ajhg.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Engelmann B, Massberg S. Thrombosis as an intravascular effector of innate immunity. Nature Rev. Immunol. 2013;13:34–45. doi: 10.1038/nri3345. [DOI] [PubMed] [Google Scholar]
- 76.Demogines A, Abraham J, Choe H, Farzan M, Sawyer SL. Dual host-virus arms races shape an essential housekeeping protein. PLoS Biol. 2013;11:e1001571. doi: 10.1371/journal.pbio.1001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaelber JT, et al. Evolutionary reconstructions of the transferrin receptor of caniforms supports canine parvovirus being a re-emerged and not a novel pathogen in dogs. PLoS Pathog. 2012;8:e1002666. doi: 10.1371/journal.ppat.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barber MF, Elde NC. Nutritional immunity. Escape from bacterial iron piracy through rapid evolution of transferrin. Science. 2014;346:1362–1366. doi: 10.1126/science.1259329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Al-Daghri NM, et al. Mammalian NPC1 genes may undergo positive selection and human polymorphisms associate with type 2 diabetes. BMC Med. 2012;10:140. doi: 10.1186/1741-7015-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krishnan A, et al. Niemann–Pick C1 (NPC1)/NPC1-like1 chimeras define sequences critical for NPC1's function as a flovirus entry receptor. Viruses. 2012;4:2471–2484. doi: 10.3390/v4112471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ng, M. et al. Cell entry by a novel European filovirus requires host endosomal cysteine proteases and Niemann–Pick C1. Virology 468–470, 637–646 (2014). [DOI] [PMC free article] [PubMed]
- 82.Shoemaker CJ, et al. Multiple cationic amphiphiles induce a Niemann–Pick C phenotype and inhibit Ebola virus entry and infection. PLoS ONE. 2013;8:e56265. doi: 10.1371/journal.pone.0056265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee K, et al. Inhibition of ebola virus infection: identification of Niemann–Pick C1 as the target by optimization of a chemical probe. ACS Med. Chem. Lett. 2013;4:239–243. doi: 10.1021/ml300370k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cote M, et al. Small molecule inhibitors reveal Niemann–Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cagliani R, et al. Evolutionary analysis of the contact system indicates that kininogen evolved adaptively in mammals and in human populations. Mol. Biol. Evol. 2013;30:1397–1408. doi: 10.1093/molbev/mst054. [DOI] [PubMed] [Google Scholar]
- 86.Rallapalli PM, Orengo CA, Studer RA, Perkins SJ. Positive selection during the evolution of the blood coagulation factors in the context of their disease-causing mutations. Mol. Biol. Evol. 2014;31:3040–3056. doi: 10.1093/molbev/msu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Imamura T, et al. Induction of vascular leakage through release of bradykinin and a novel kinin by cysteine proteinases from Staphylococcus aureus. J. Exp. Med. 2005;201:1669–1676. doi: 10.1084/jem.20042041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pichlmair A, et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- 89.Karlas A, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 90.Smith JD, McManus KF, Fraser HB. A novel test for selection on cis-regulatory elements reveals positive and negative selection acting on mammalian transcriptional enhancers. Mol. Biol. Evol. 2013;30:2509–2518. doi: 10.1093/molbev/mst134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 92.Anisimova M, Bielawski JP, Yang Z. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol. Biol. Evol. 2002;19:950–958. doi: 10.1093/oxfordjournals.molbev.a004152. [DOI] [PubMed] [Google Scholar]
- 93.Yang Z, Wong WS, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 94.Murrell B, et al. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Markova-Raina P, Petrov D. High sensitivity to aligner and high rate of false positives in the estimates of positive selection in the 12 Drosophila genomes. Genome Res. 2011;21:863–874. doi: 10.1101/gr.115949.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blackburne BP, Whelan S. Class of multiple sequence alignment algorithm affects genomic analysis. Mol. Biol. Evol. 2013;30:642–653. doi: 10.1093/molbev/mss256. [DOI] [PubMed] [Google Scholar]
- 97.Schneider A, et al. Estimates of positive Darwinian selection are inflated by errors in sequencing, annotation, and alignment. Genome Biol. Evol. 2009;1:114–118. doi: 10.1093/gbe/evp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jordan G, Goldman N. The effects of alignment error and alignment filtering on the sitewise detection of positive selection. Mol. Biol. Evol. 2012;29:1125–1139. doi: 10.1093/molbev/msr272. [DOI] [PubMed] [Google Scholar]
- 99.Anisimova M, Nielsen R, Yang Z. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics. 2003;164:1229–1236. doi: 10.1093/genetics/164.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 101.Fletcher W, Yang Z. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol. Biol. Evol. 2010;27:2257–2267. doi: 10.1093/molbev/msq115. [DOI] [PubMed] [Google Scholar]
- 102.Anisimova M, Yang Z. Multiple hypothesis testing to detect lineages under positive selection that affects only a few sites. Mol. Biol. Evol. 2007;24:1219–1228. doi: 10.1093/molbev/msm042. [DOI] [PubMed] [Google Scholar]
- 103.Kosakovsky Pond SL, et al. A random effects branch-site model for detecting episodic diversifying selection. Mol. Biol. Evol. 2011;28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guindon S, Rodrigo AG, Dyer KA, Huelsenbeck JP. Modeling the site-specific variation of selection patterns along lineages. Proc. Natl Acad. Sci. USA. 2004;101:12957–12962. doi: 10.1073/pnas.0402177101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu A, Guindon S. Performance of standard and stochastic branch-site models for detecting positive selection among coding sequences. Mol. Biol. Evol. 2014;31:484–495. doi: 10.1093/molbev/mst198. [DOI] [PubMed] [Google Scholar]
- 106.Yang Z, dos Reis M. Statistical properties of the branch-site test of positive selection. Mol. Biol. Evol. 2011;28:1217–1228. doi: 10.1093/molbev/msq303. [DOI] [PubMed] [Google Scholar]
- 107.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nature Rev. Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 108.Barral DC, Brenner MB. CD1 antigen presentation: how it works. Nature Rev. Immunol. 2007;7:929–941. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Evolutionary analysis of mammalian DDP4. (PDF 2101 kb)
LRT statistics for DPP4. (PDF 260 kb)




