Abstract
Bats are special in their ability to host emerging viruses. As the only flying mammal, bats endure high metabolic rates yet exhibit elongated lifespans. It is currently unclear whether these unique features are interlinked. The important inflammasome sensor, NLR family pyrin domain containing 3 (NLRP3), has been linked to both viral-induced and age-related inflammation. Here, we report significantly dampened activation of the NLRP3 inflammasome in bat primary immune cells compared to human or mouse counterparts. Lower induction of apoptosis-associated speck-like protein containing a CARD (ASC) speck formation and secretion of interleukin-1β in response to both ‘sterile’ stimuli and infection with multiple zoonotic viruses including influenza A virus (−single-stranded (ss) RNA), Melaka virus (PRV3M, double-stranded RNA) and Middle East respiratory syndrome coronavirus (+ssRNA) was observed. Importantly, this reduction of inflammation had no impact on the overall viral loads. We identified dampened transcriptional priming, a novel splice variant and an altered leucine-rich repeat domain of bat NLRP3 as the cause. Our results elucidate an important mechanism through which bats dampen inflammation with implications for longevity and unique viral reservoir status.
Subject terms: Viral infection, Inflammasome, Viral reservoirs
Dampened activation of the NLR family pyrin domain containing 3 (NLRP3) inflammasome in bat primary immune cells in response to infection with multiple zoonotic viruses is caused by decreased transcriptional priming, the presence of a unique splice variant and an altered leucine-rich repeat domain of bat NLRP3.
Main
Bats, as the only flying mammals, have ‘emerged’ in both the scientific and general public arenas due to their ability to asymptomatically host a large number of high-profile viruses. Such examples include Ebola virus, Nipah virus, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) coronaviruses, which are capable of causing severe, and often deadly, disease in humans and animals1–4. These viruses, predominantly RNA viruses, are highly pathogenic in humans and are often linked to an aberrant innate immune activation5–8. Importantly, bats, when challenged by these viruses, exhibit no or minimal signs of disease, even when high viral loads are detected in the sera or tissues9–12. In addition, they are special in that they harbour a higher proportion of zoonotic viruses than all other mammalian orders13. Bats also have an extraordinarily long lifespan relative to their body size, despite their elevated metabolic rates14,15.
These observations have triggered increasing interest and efforts to characterize the bat immune system, primarily through genomic16,17 and transcriptomic analysis18–20, and especially with regard to type I interferon and antiviral activities21–23. However, how bats limit excessive inflammation while asymptomatically hosting a greater variety of viruses is unknown.
NLR family pyrin domain containing 3 (NLRP3) is an important sensor that recognizes both cellular stresses (such as extracellular adenosine triphosphate (ATP), mitochondrial damage and oxidized DNA)24–26 and viral or bacterial infections27,28. NLRP3-mediated inflammation has been causally linked to aging and multiple age-related chronic diseases29,30. NLRP3 is increasingly recognized for its role in response to multiple viruses31, including those associated with bats, such as influenza A virus (IAV)32,33 and rabies virus34,35. Over-activation of the NLRP3 inflammasome has been linked to a hyper-inflammatory state and immunopathology in viral infection with minimal effect on the viral load36–39. However, nothing is known about NLRP3-mediated inflammation in bats. On the basis of the observations of increased longevity and viral asymptomaticity in bats, we hypothesize that the NLRP3 inflammasome, as a central player in viral infection and aging, differs between bats and other mammals.
Here, we report a mechanism by which bats dampen host inflammation in response to both ‘sterile’ danger signals and infections with three different zoonotic RNA viruses, without affecting the viral loads in primary immune cells. This involves dampened transcriptional priming and a lower functional capacity of bat NLRP3. We also discover that bat-borne Pteropine orthoreovirus 3 (PRV3M, previously known as Melaka virus) and MERS coronavirus (MERS-CoV) induce NLRP3-mediated inflammation in mouse or human. Bats have naturally dampened stress-related and virus-induced host inflammatory responses, with implications for longevity and asymptomatic viral reservoir status.
Results
Activation of NLRP3 inflammasome is dampened in bat primary immune cells
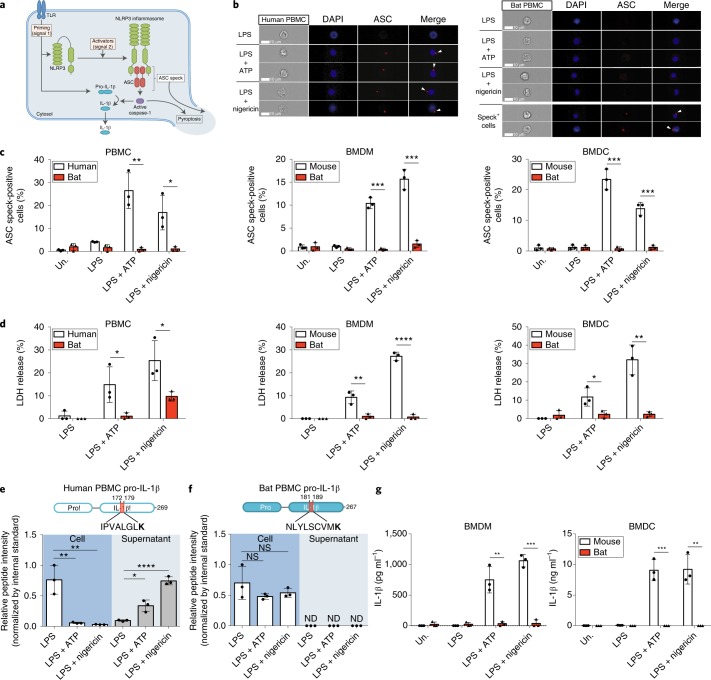
Following priming (signal 1) and activation (signal 2), NLRP3 triggers assembly of the diffuse cytosolic apoptosis-associated speck-like protein containing a CARD (ASC) protein to form ASC specks40. These then recruit and activate caspase-1 to promote inflammatory cell death via pyroptosis and cleavage/secretion of the potent pro-inflammatory cytokine interleukin-1β (IL-1β) (Fig. 1a). To assess the activation of the NLRP3-inflammasome in primary immune cells, peripheral blood mononuclear cells (PBMCs) of bat (Pteropus alecto) and human, or bone-marrow-derived macrophages (BMDMs) or dendritic cells (BMDCs) of bat and mouse, were tested. Cells were primed with lipopolysaccharide (LPS) and then stimulated by potent NLRP3 activators, ATP or nigericin, representing a host- or pathogen-derived danger signal, respectively25. ASC specks in bat cells were visualized with a monoclonal antibody specifically raised against P. alecto ASC protein (Supplementary Fig. 1a–c). Quantification of ASC specks in human and bat PBMCs was first validated by confocal microscopy (Supplementary Fig. 1d) and ImageStream imaging flow cytometry (Fig. 1b). Both approaches similarly detected ASC specks formation on treatment of human PBMCs with ATP or nigericin (Fig. 1c and Supplementary Fig. 1e). To quantify the specks in high throughput, we used ImageStream for the remainder of the experiments. We detected low-level ASC specks in LPS-primed bat cells, with negligible induction in ATP- or nigericin-treated cells, in contrast to treated human PBMCs, mouse BMDMs or BMDCs (Fig. 1c). A key feature of pyroptotic cell death is destabilization of the cell membrane and release of cytosolic contents41. We measured pyroptosis by the release of lactate dehydrogenase (LDH) and observed less LDH release in treated bat immune cells (Fig. 1d). To examine the downstream secretion of IL-1β in limited amounts of bat PBMCs, we used highly sensitive and specific parallel reaction monitoring (PRM)-based targeted mass spectrometry with heavy isotope-labelled peptide standards42. Levels of human N- or C-terminal pro-IL-1β peptides decreased in cell lysates from treated PBMCs, correlating with significantly increased levels in supernatants (Fig. 1e and Supplementary Fig. 2a,c). In contrast, levels of pro-IL-1β peptides in bat cells did not decrease with treatment, correlating with a lack of detection of endogenous peptides in supernatants, despite the detection of spiked internal standards (Fig.1f and Supplementary Fig. 2b,d). In addition, an in-house sandwich enzyme-linked immunosorbent assay (ELISA) for bat IL-1β was generated with cross-reactive antibodies (Supplementary Fig. 3). Secretion of IL-1β was also lower in bat BMDMs or BMDCs as measured by ELISA of IL-1β (Fig. 1g). Altogether, activation of the NLRP3 inflammasome is dampened in primary bat immune cells.
Fig. 1. Activation of the NLRP3 inflammasome is dampened in bat PBMCs, BMDMs and BMDCs.
a, Schematic model for NLRP3 inflammasome activation. b, Representative single-cell images of gated monocytes of human (left) and bat (right) PBMCs, primed with LPS for 3 h, with or without stimulation by ATP or nigericin for 30 min. Cells were acquired using ImageStream. DAPI, blue; ASC, red; arrowheads, ASC specks. Scale bars, 10 μm. c, Quantification of ASC specks in gated monocytes of PBMCs (left), BMDMs (middle) or BMDCs (right) by ImageStream, unprimed (Un.) or primed with LPS for 3 h, with or without stimulation with ATP or nigericin for 30 min. d, LDH release in cell-free supernatants of PBMCs (left), BMDMs (middle) or BMDCs (right), primed with LPS for 3 h, with or without stimulation by ATP or nigericin for 1 h. e,f, Detection of human (e) and bat (f) C-terminal pro-IL-1β peptides in cell lysates (left) or supernatants (right) of PBMCs as in d, using PRM-based targeted mass spectrometry with heavy isotope-labelled internal standards. K indicates heavy labelled lysine (13C6, 15N2); ND, not detected; NS, not significant. g, Secretion of IL-1β by ELISA in the supernatant of BMDMs (left) or BMDCs (right) as stimulated in d. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; two-tailed unpaired t-test. Exact P values are provided in Supplementary Table 5. Data are representative of three biological replicates (n = 3) in b, or mean + s.d. of three biological replicates (n = 3) in c–g.
Transcriptional priming of bat NLRP3 is impaired, independent of TLRs
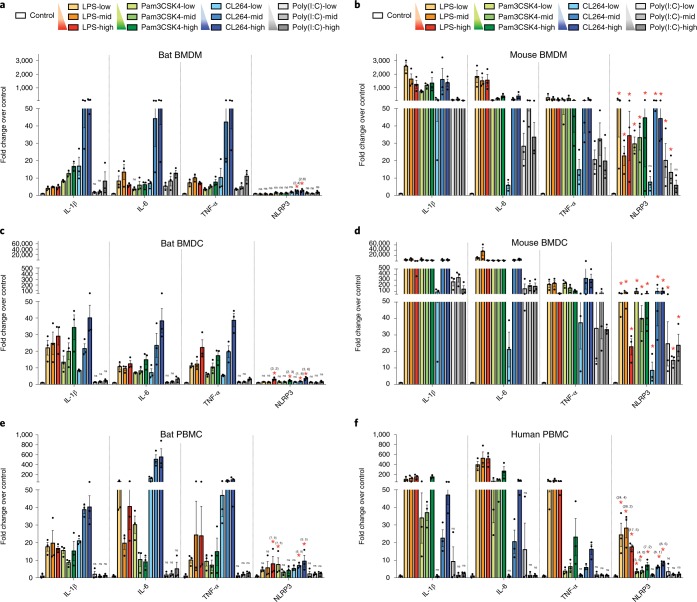
Lack of ASC speck formation and downstream activation can be attributed to either the adaptor ASC or the sensor NLRP3, or both. NLRP3 expression is rate-limiting and transcriptional upregulation by NF-κB through Toll-like receptors (TLRs) is an essential first step required for the inflammasome activation43,44. We thus examined the transcriptional priming of both NLRP3 and IL-1β, along with classical NF-κB-induced genes IL-6 and TNF-α. BMDMs, BMDCs or PBMCs of bat and mouse or human were treated with various doses of four different NF-κB-activating TLR agonists: LPS (bacterial lipopolysaccharide) for TLR4, Pam3CSK4 (bacterial triacylated lipopeptide) for TLR1/2, CL264 (adenine analogue) for TLR7 and poly(I:C) (viral dsRNA analogue) for TLR3. The majority of the TLR stimulations induced IL-1β, IL-6 and TNF-α, up to 115-, 67- and 57-fold, respectively, in bat BMDMs (Fig. 2a), 40-, 38- and 39-fold in bat BMDCs (Fig. 2c) and 40-, 552- and 95-fold in bat PBMCs (Fig. 2e), suggesting robust NF-κB activation in these bat immune cells. However, NLRP3 was not induced with most of the TLR stimuli, except at higher doses when it was up to only threefold in bat BMDMs, fourfold in bat BMDCs and ninefold in bat PBMC (Fig. 2a,c,e). In contrast, NLRP3 was significantly induced with higher amplitude for most stimuli in mouse BMDMs or BMDCs and human PBMCs (Fig. 2b,d,f). Despite the differences in the amplitude of IL-1β, IL-6 or TNF-α responses to various stimulations between species, a more robust induction of NLRP3 in mouse or human cells compared to bat cells was observed with the TLR stimuli of similar potency for IL-1β, IL-6 or TNF-α between species, such as CL264 in bat and mouse BMDMs (Fig. 2a,b), CL264 and poly(I:C) for bat and mouse BMDCs, respectively (Fig. 2c,d), and CL264 and LPS for bat and human PBMCs (Fig. 2e,f). We excluded the possibility of a difference in NLRP3 basal expression level by comparing mRNA levels across bat and mouse tissues (Supplementary Fig. 4). Comparable levels of NLRP3 transcripts relative to housekeeping genes between bat and mouse tissues were observed. Basal NLRP3 expression across bat tissues was 100 to 1,000 times lower than constitutively expressed inflammasome genes such as ASC and caspase-1, which is also consistent with the pattern seen in mouse tissues (Supplementary Fig. 4). Collectively, these data indicate that bat NLRP3 has dampened transcriptional priming independent of TLR signalling.
Fig. 2. Transcriptional priming of bat NLRP3 is dampened independent of TLRs.
a–f, Fold changes of IL-1β, IL-6, TNF-α and NLRP3 measured by qPCR in BMDMs (a,b), BMDCs (c,d) or PBMCs (e,f) of bat (a,c,e) and mouse (b,d) or human (f) after stimulating with different concentrations of TLR ligands for 3 h. LPS = 10, 100, 1,000 ng ml−1; Pam3CSK4 = 10, 100, 1,000 ng ml−1; CL264 = 0.1, 1, 10 μg ml−1; poly(I:C) = 0.1, 1, 10 μg ml−1. Fold induction of cytokines is relative to mock-treated samples (control) after normalizing with GAPDH. *P < 0.05 (only labelled for NLRP3), NS, not significant (labelled for IL-1β, IL-6, TNF-α and NLRP3), by one-way analysis of variance (ANOVA) with Bonferroni’s multiple comparisons test for log fold changes. Exact P values are provided in Supplementary Table 5. Data are presented as mean + s.e.m. of three biological replicates (n = 3) in a–f.
The function of all four bat NLRP3 isoforms, but not ASC, is dampened
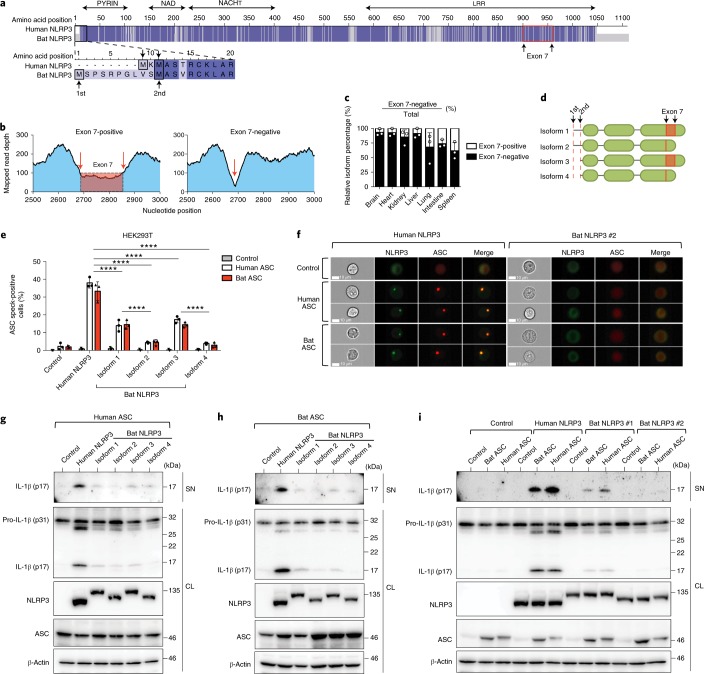
To characterize NLRP3 and ASC functions, we examined their major isoforms in bat immune cells. Transcriptome analysis revealed only one ASC isoform, yet a NLRP3 splice-variant was identified. The alignment of full-length human and P. alecto NLRP3 proteins revealed 79% identity with two potential start codons in bat NLRP3 (Fig. 3a). De novo assembly of mapped RNA–seq reads from P. alecto splenocyte subsets revealed an exon 7-skipping splice variant in bat NLRP3, not previously described in other mammalian orders (Fig. 3b). The exon 7 region was located within the leucine-rich repeat (LRR) domain of NLRP3. Both exon 7-positive and exon 7-negative variants were validated by PCR and sequencing from spleen and thymus cDNA. This exon 7-negative isoform constituted more than 60% of NRLP3 transcripts in different P. alecto tissues (Fig. 3c). The four isoforms with different start codons and/or alternative splicing are designated 1 to 4 (Fig. 3d). Human embryonic kidney (HEK293T) reporter cells stably expressing human/bat ASC–mPlum fusions and transiently expressing human/bat NLRP3–mCitrine fusions were used for ImageStream and immunoblot analysis. For ImageStream analysis, double-positive cells of similar fluorescent intensity for ASC–mPlum and NLRP3–mCitrine were gated. This revealed that formation of bat ASC specks and their downstream cleavage of IL-1β was not different from human ASC, in response to all NLRP3 variants (Fig. 3e,f,i). In addition, the microscopic structures of specks including size, density and shape were comparable between human and bat ASC (Supplementary Fig. 5). In contrast, ASC speck and IL-1β cleavage by bat NLRP3 isoforms was lower than that of human NLRP3 (Fig. 3e–h). Notably, exon 7-negative isoforms 2 and 4 were less functional than exon 7-positive isoforms 1 and 3. The alternative translational start resulted in no difference in activity. Together, these data show that the function of all four NLRP3 isoforms but not ASC is dampened compared to the human NLRP3 and that alternative splicing of exon 7 contributes to further dampening of bat NLRP3 activity.
Fig. 3. The function of all four bat NLRP3 isoforms, but not ASC, is reduced.
a, Amino acid sequence alignment of human and bat (P. alecto) NLRP3, with identical residues highlighted in dark blue and mismatches in light blue. Black boxes, alternative translation start sites. Red box, boundaries of exon 7. b, Read-mapping analysis of two splice variants (exon 7-positive/negative) by RNA–seq read counts of the P. alecto splenocytes subsets. Arrows, exon–exon junctions. c, Percentage of exon 7-positive or exon 7-negative isoform in P. alecto tissues measured by qPCR. d, Schematic representation of four bat NRLP3 isoforms. e, Quantification of ASC specks by ImageStream in HEK293T cells stably expressing mPlum (control), human or bat ASC–mPlum, transfected with mCitrine (control), human or bat NLRP3–mCitrine. f, Representative images from e. NLRP3–mCitrine (green); ASC–mPlum (red). Scale bars, 10 μm. g–i, Immunoblot analysis of IL-1β cleavage in cell lysates and supernatant in HEK293T cells stably expressing mPlum (control), human or bat ASC–mPlum, transfected with mCitrine (control), human or bat NLRP3–mCitrine, plus human pro-caspase-1 and mouse pro-IL-1β. NLRP3, NLRP3–mCitrine stained with anti-GFP; ASC, ASC–mPlum stained with anti-mPlum; SN, supernatant; CL, cell lysate. ****P < 0.0001 by two-way ANOVA with Bonferroni’s multiple comparisons test (e). Exact P values are provided in Supplementary Table 5. Data are presented as mean + s.d. of three biological replicates (c) or three independent experiments (e), or representative of three independent experiments (f–i).
NLRP3 isoform activity in bat cells from both major bat lineages is reduced
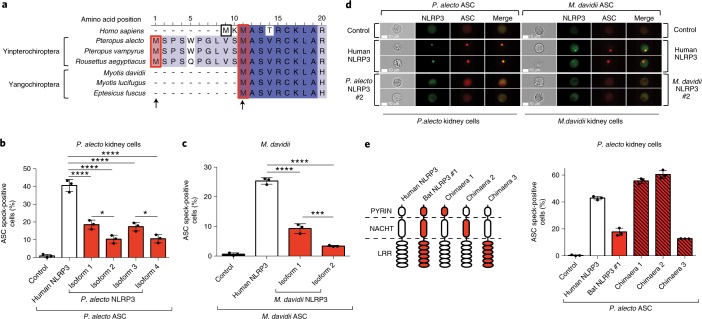
Chiroptera is a diverse order consisting of more than a thousand bat species. To further examine whether our findings for NLRP3 in P. alecto from the Yinpterochiroptera suborder are also true in evolutionally distant bats, we extended our observations to Myotis davidii in the other suborder, Yangochiroptera. Only one start codon matching the second start of the P. alecto protein was identified in M. davidii NLRP3 (Fig. 4a). Similarly, both exon 7-positive (isoform 1) and exon 7-negative (isoform 2) isoforms were identified by PCR and sequencing from spleen and kidney cDNA. To assess NLRP3 function in native bat cells of different species origin, we established P. alecto kidney (PaKi) and M. davidii kidney (MdKi) cells stably expressing the homologous ASC–mPlum reporter construct. Overexpression of human NLRP3–mCitrine in both bat cell types led to redistribution of diffuse bat ASC into specks (Fig. 4b–d). In comparison, the activity of all bat NLRP3 isoforms was significantly lower than that of the human protein (Fig. 4b–d). Importantly, both forms of M. davidii NLRP3 also displayed reduced activation. Consistent with previous findings in HEK293T cells, lack of exon 7 resulted in additional dampening of NLRP3 function. To map the region responsible for the reduced function of exon 7-positive isoforms, we generated chimaeras between human NLRP3 and P. alecto NLRP3 isoform 1 by substituting the human PYRIN, NACHT and LRR domains, respectively (Fig. 4e). While chimaeras 1 and 2 showed increased activity, only chimaera 3 exhibited lower activity than the wild-type human protein, suggesting that the LRR domain is responsible for the reduced function. Evolutionary analysis using 10 bat and 17 non-bat mammalian NLRP3 sequences confirmed NLRP3 was under higher selection pressure in the bat ancestor lineage compared to other mammals and the LRR domain was the only domain under significant positive selection (Supplementary Tables 1–3), consistent with our chimaeric functional analysis. Overall, these data show that a bat-specific splice variant and/or an altered LRR domain are responsible for the dampened activity of all NLRP3 isoforms.
Fig. 4. NLRP3 isoform activity in bat cells from both major bat lineages is dampened.
a, Amino acid sequence alignment of N-terminal NLRP3 from human and six bats species from Yinpterochiroptera or Yangochiroptera suborders, with identical residues (dark blue) and mismatches (light blue). Arrows indicate alternative translation start sites. b, Quantification of ASC specks by ImageStream in P. alecto kidney (PaKi) cells stably expressing bat ASC, transfected with mCitrine (control), human or bat NLRP3–mCitrine. c, Quantification of ASC specks by ImageStream in M. davidii kidney cells stably expressing M. davidii ASC, transfected with mCitrine (control), human or M. davidii NLRP3–mCitrine. d, Representative ImageStream images of P. alecto or M. davidii cells stably expressing P. alecto or M. davidii ASC, respectively, transfected with empty vector, human or bat NLRP3–mCitrine. NLRP3–mCitrine, green; ASC–mPlum, red. Scale bars, 10 μm. e, Left, Schematic representation for chimaeras between human NLRP3 and P. alecto NLRP3 isoform 1. Right, Quantification of ASC specks by ImageStream in PaKi cells stably expression P. alecto ASC, transfected with mCitrine (control), human or bat NLRP3 or chimaera–mCitrine. *P < 0.05, ***P < 0.001, ****P < 0.0001 by one-way ANOVA with Bonferroni’s multiple comparisons test (b,c). Exact P values are provided in Supplementary Table 5. Data are presented as mean + s.d. of three independent experiments (b,c,e) or representative of three independent experiments (d).
NLRP3-mediated inflammation is dampened in bat immune cells in response to three different types of RNA virus
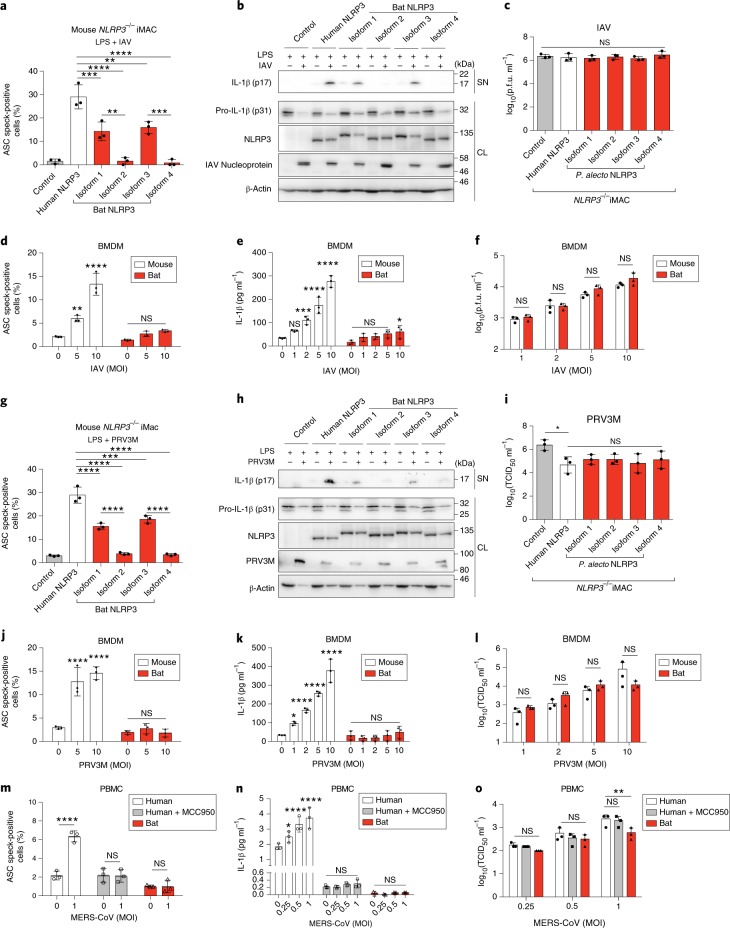
To confirm our results in a more physiological setting of immune cells, we reconstituted mouse NLRP3-knockout immortalized macrophages (iMACs) with human or P. alecto NLRP3–mCitrine. Induction of ASC specks and secretion of IL-1β in reconstituted iMACs by ATP or nigericin was significantly reduced in cells expressing bat NLRP3 isoforms compared to the human NLRP3 (Supplementary Fig. 6). To characterize the bat NLRP3 in response to virus infection, LPS-primed iMACs were infected with H1N1 influenza A virus (IAV), strain A/NWS/33. It is a negative-sense single-stranded RNA (−ssRNA) virus, known to activate the NLRP3 inflammasome27. All bat NLRP3 isoforms were significantly less responsive to IAV infection, while viral titres from supernatants were not affected (Fig. 5a–c). To validate this finding in freshly derived myeloid cells, we infected LPS-primed mouse or bat BMDMs or BMDCs with IAV at different multiplicities of infection (MOI = 1, 2, 5 or 10). MOI-dependent induction of ASC speck and IL-1β secretion was observed in infected mouse BMDMs or BMDCs (Fig. 5d,e and Supplementary Fig. 7a,b). Although the IAV titre was similar between mouse and bat cells, ASC speck formation and IL-1β secretion were minimally or not induced in infected bat BMDMs or BMDCs (Fig. 5d–f and Supplementary Figs. 7a–c and 8a).
Fig. 5. Bat NLRP3-mediated inflammation in immune cells is dampened in response to IAV, PRV3M and MERS-CoV infection.
a–c,g–i, Quantification of ASC specks by ImageStream (a,g), immunoblot analysis (b,h) and virus titration (c,i) of mouse NLRP3−/− iMACs reconstituted with mCitrine alone (control), human or bat (P. alecto) NLRP3–mCitrine, primed with LPS for 3 h, followed by infection with IAV (MOI = 0.5, a–c) or PRV3M (MOI = 2, g–i) for 24 h. NLRP3, NLRP3–mCitrine stained with anti-GFP; PRV3M, PRV3M viral protein stained with polyclonal macaque serum; SN, supernatant; CL, cell lysates. Titres in the supernatant are expressed as p.f.u. ml−1. d–f,j–l Quantification of ASC specks by ImageStream (d,j), IL-1β secretion by ELISA (e,k) and virus titration (f,l) of mouse or bat BMDMs, primed with LPS for 3 h, followed by infection with IAV (MOI = 1–10, d–f) or PRV3M (MOI = 1–10, j–l) for 24 h. Titres in the supernatant are expressed as median tissue culture infectious dose (TCID50) ml−1 m–o, Quantification of ASC specks by ImageStream (m), IL-1β secretion by ELISA (n) and virus titration (o) of human or bat PBMCs, primed with LPS for 3 h, followed by infection with MERS-CoV (MOI = 0.25, 0.5, 1) for 24 h. MCC950 inhibitor was added 1 h before infection for human PBMCs. Titres in the supernatant are expressed as TCID50 ml−1. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS, not significant, by one-way ANOVA (a,c,g,i) or two-way ANOVA (d–f and j–o) with Bonferroni’s multiple comparisons test. Exact P values are provided in Supplementary Table 5. Data are presented as mean + s.d. of three biological replicates (d–f and j–o) or three independent experiments (a,c,g,i), or representative of three independent experiments (b,h).
To confirm that the observation is not IAV-specific, PRV3M45, a bat-borne zoonotic reovirus of double-stranded RNA (dsRNA), was used to infect reconstituted iMACs. Viruses from the dsRNA group have not been demonstrated to activate the NLRP3 inflammasome. Our data show that PRV3M activated the inflammasome in an NLRP3-dependant manner as measured by ASC speck and IL-1β secretion, with decreased activation from bat NLRP3 compared to the human counterpart (Fig. 5g,h and Supplementary Fig. 8d). Additionally, there was a decrease in viral titres with NLRP3 but no obvious difference between human and bat isoforms (Fig. 5i). PRV3M infection (MOI = 1, 2, 5 or 10) in bat BMDMs or BMDCs also triggered less inflammasome activation without the MOI-dependent increase in ASC speck formation and IL-1β secretion seen in infected mouse cells, despite the similar viral titres between mouse and bat cells (Fig. 5j–l and Supplementary Figs. 7d–f and 8b).
To expand this investigation to a third type of RNA virus, we interrogated inflammasome activation in response to MERS coronavirus (MERS-CoV) infection, a positive-sense (+)ssRNA zoonotic virus from a recent epidemic with potential bat ancestry46. As mouse cells are not permissive to MERS-CoV47, human and bat PBMCs were infected. MERS-CoV infection (MOI = 0.25, 0.5 or 1) activated ASC speck and IL-1β secretion in a MOI-dependent manner in human PBMCs (Fig. 5m,n). This activation, abolished by pharmacological inhibition of NLRP3 with a small-molecule inhibitor MCC95048, suggests NLRP3-dependent activation of the inflammasome by MERS. In contrast, MERS-CoV infection failed to trigger an inflammasome response in bat PBMCs, although viral titres only differed minimally between human and bat PBMCs (Fig. 5m–o and Supplementary Fig. 8c). Taken together, our findings demonstrate dampened activity of bat NLRP3 in immune cells and reduced activation of the NLRP3 inflammasome in response to infection with three very different viruses of public health concern, without affecting viral load.
Discussion
In summary, our results demonstrate an overall dampening of NLRP3 inflammasome activation in bat primary immune cells (Supplementary Fig. 9). Dampened transcriptional priming, a bat-specific splice variant and an evolutionarily adapted LRR domain of NLRP3 are responsible for the reduction of inflammation from ‘sterile’ danger signals (ATP) and infection of different zoonotic RNA viruses. Additionally, we discovered that both PRV3M and MERS-CoV activate the NLRP3 inflammasome in mouse and human, which also expands our current knowledge of the role of the NLRP3 inflammasome in the dsRNA virus group and family Coronaviridae. A clear demonstration of dampened host inflammatory responses without affecting viral load is consistent with the unique asymptomatic viral reservoir status of bats.
We report multiple mechanisms, at both the mRNA and protein level, underlying the dampened NLRP3-mediated inflammation. Our data demonstrate dampened transcriptional priming, specific for NLRP3, following different TLR stimulations in P. alecto immune cells. A robust induction of IL-1β, IL-6 and TNF-α suggests functional NF-κB signalling in bat immune cells, consistent with the previous report using CL097 (TLR7/8 agonist) in P. alecto myeloid cells49. This NLRP3-specific effect with intact NF-κB signalling might be attributed to a change in the promoter or cis-regulatory elements. The induction of IL-1β, IL-6 or TNF-α varies substantially between different TLRs within a species and between bat and human/mouse for a particular TLR, which supports the increasing evidence for intra- and inter-species variation in TLRs50,51. Further investigation is required to determine if there is a difference in overall NF-κB signalling in bats.
The exon 7-skipping alternative splice variant identified in both P. alecto and M. davidii has not been described previously in other mammalian orders based on the NCBI protein database. At the protein level, while an altered LRR domain is responsible for reduced function for the full-length protein, loss of exon 7 located within the LRR domain results in further dampened activity. Notably, the ‘weaker’ exon 7-negative isoforms are the predominant isoforms in P. alecto tissues. Our functional study is supported by evolutionary analysis (PAML branch model), which indicates a positive selection in the LRR domain in the bat ancestral lineage. The branch-site model, however, failed to identify a positively selected residue of NLRP3 in the bat ancestral lineage. This suggests that an altered LRR domain is probably a conserved feature in bats and possibly involves accumulated changes in different residues in different lineages of bats. At the steady sate, the LRR domain plays a role in auto-inhibition by preventing oligomerization of NLRP340. Auto-inhibition of the LRR domain in bat NLRP3 could possibly be enhanced, hence making it harder for NLRP3 to trigger downstream activation. Interestingly, the same priming step for transcriptional upregulation also licenses NRLP3 activation by inducing post-translational modifications including deubiquitination in the LRR domain52. No key residues have been identified for auto-inhibition or ubiquitin modifications in the human/mouse LRR domain so far, and their roles are yet to be determined in the altered bat LRR.
The NLRP3 inflammasome is increasingly being recognized to play a critical role in the immune response to viruses. In this study, we have discovered NLRP3-dependent activation of inflammation for a bat-related dsRNA virus (PRV3M)45 and a coronavirus (MERS-CoV) with significant human spillover events. NLRP3 has no effect on viral titres for IAV and MERS-CoV infection in reconstituted iMACs and PBMCs, respectively. Interestingly, both human and bat NLRP3 mediated a similar decrease in viral titres from reconstituted iMACs infected by PRV3M, suggesting an NLRP3-dependent and inflammasome-independent mechanism. NLRP3 inflammasome activation by dsRNA virus has only been inferred from transfecting isolated dsRNA from another reovirus53. No unified mechanisms for activating the NLRP3 inflammasome have been identified for viruses. While genomic or intermediate dsRNA of PRV3M and viroporin encoded by MERS-CoV E protein54 are possible candidate NLRP3 activators, further studies are needed to determine the activation mechanisms.
Observations in both wild-caught and experimentally infected bats suggest the ability of bats to tolerate viral disease, even during a transient phase of high viral load. For instance, in vivo virus challenge with high-dose Ebola virus9 and MERS-CoV11 caused no clinical disease and limited pathology, despite high viral titres detected in tissues or sera. The viral load can reach as high as 107 TCID50 equivalents per gram of lung tissues for MERS-CoV and 107 f.f.u. (fluorescent focus-forming units) ml−1 of sera for Ebola virus. While a ‘flight as fever’ theory55 and an ‘always on’ interferon (IFN) system56 might suggest elevated antiviral immunity in bats, a dampened STING-dependent IFN response to a DNA virus21 and an inhibitory state of natural killer cells inferred from genome analysis support enhanced immune tolerance16. These hypotheses still largely lack functional confirmation, particularly with RNA virus infection. A recent study demonstrated a temperature-independent replication of Filoviruses on bat cells, as opposed to the ‘flight as fever’ hypothesis57. We demonstrate here a dampened NLRP3-mediated inflammatory response to three different types of RNA virus in bat immune cells, with no or minimal difference in viral titres. This supports an enhanced innate immune tolerance rather than an enhanced antiviral defence in bats. This may also contribute to our understanding of the role of the inflammasome in disease tolerance in bats as reservoir hosts, in contrast to severe pathogenesis in spillover hosts for many high-profile emerging zoonotic viruses. As NLRP3 is recognized to sense an increasing number of viruses, this finding may have broad application in a great variety of bat-borne viruses or viruses yet to be detected in bats. With the ongoing development of more experimental tools and reagents for bat immunology, a deeper characterization of immune responses in other cell types/tissues and in vivo will provide greater insight into the underlying mechanisms of viral disease tolerance in bats.
Methods
Reagents
Ultrapure LPS-B5, ATP, nigericin, Pam3CSK4, poly(I:C), CL264 and Hygromycin B Gold were obtained from InvivoGen. P. alecto ASC-specific monoclonal antibody (mouse IgG2b) was generated by GenScript’s monoclonal antibody service. Rabbit polyclonal anti-ASC (AL177) (human/mouse) was purchased from Adipogen. Goat polyclonal anti-dog IL-1β (ab193852) (cross-reactive to P. alecto), rabbit polyclonal anti-mouse IL-1β (ab9722) and monoclonal antibody (mAb) to IAV nucleoprotein (ab20343) were from Abcam. mAb to β-actin (A2228) was from Sigma-Aldrich and mAb to GFP and variants (including mCitrine) were from Roche (11814460001). Anti-dsRNA mAb J2 was purchased from SCICONS and anti-mouse/rabbit/goat/monkey horseradish peroxidase (HRP)-conjugated secondary antibodies were from Santa Cruz. mPlum antibody from Origene was used to stain mPlum. Human mAb to MERS-CoV spike glycoprotein (m336) was provided by D. S. Dimitrov (NIH)58. Alexa 488/568/647 Zenon labelling kits (Invitrogen) were used for direct labelling of antibodies for confocal microscopy and ImageStream. Protein G agarose beads (Millipore) were used to purify P. alecto IL-1β-Fc cloned and expressed from HEK293T cells. Polyclonal macaque serum (#5244 D42, infected by PRV3M, produced in-house) was used for immunoblot analysis.
Viruses
Human H1N1 IAV strain A/NWS/33 (ATCC # VR-219) was propagated in Vero, clone E6 cells in DMEM, 0.3% BSA, 25 mM HEPES and 1 µg ml−1 TPCK-treated trypsin. P. orthoreovirus virus, PRV3M (Melaka virus) and MERS-CoV were propagated in Vero B4 cells in DMEM, 2% FBS. At a virus-induced cytopathatic effect of 80–90%, viruses were harvested, clarified by centrifugation, and the virus-containing supernatant was stored at −80 °C. PRV3M was further purified using ultracentrifugation and resuspended in DPBS. To titrate IAV, A/NWS/33 Madin Darby canine kidney (MDCK, ATCC #CCL-34) cells were infected with IAV for 1 h at 37 °C. After 1 h incubation, the medium was replaced with plaque medium (DMEM, 0.8% Avicel, 0.3% BSA, 25 mM HEPES and 1 µg ml−1 TPCK-treated trypsin). Plaques were fixed with 4% paraformaldehyde at 48 h post-infection and stained with 0.5% crystal violet, and titres were expressed in p.f.u. ml−1. PRV3M and MERS-CoV titres were determined by limiting dilution. In brief, tenfold serial diluted virus was added into a 96-well plate containing 1 × 104 Vero B4 cells per well. Cells were observed for cytopathatic effect and the titres were expressed as TCID50 ml−1. All work with live MERS-CoV was performed in BSL3 containment.
Plasmids
Expression constructs for ASC–mPlum, NLRP3–mCitrine, IL-1β-HA and empty vectors were generated in the retroviral backbone of pQCXIH (Clontech). NLRP3 chimaeras between human NLRP3 and the P. alecto NLRP3 isoform 1 were generated by overlap extension PCR. ASC-3×Myc and 3×Myc construct were generated in pDual GC (Agilent Technologies). P. alecto IL-1β-Fc construct containing the IL-2 signal sequence was generated from pFUSE-hIgG1-Fc plasmid (Invivogen). pVSV-G envelope vector for retroviral packaging was obtained from Clontech. pCI-Caspase-1 construct encoding human pro-caspase-1 and pCMV-pro-IL-1β construct encoding mouse pro-IL-1β were from Addgene. P. alecto and M. davidii genes were cloned from Omniscript (Qiagen)-generated cDNA from bat tissues. All constructs were prepared with endotoxin-free plasmid maxi-prep kits (Omega Bio-tek).
Cells
All procedures in this study dealing with animal samples are in compliance with all relevant ethical regulations. Specifically, capturing and processing of bats (P. alecto) in Australia was approved by the Queensland Animal Science Precinct & University of Queensland Animal Ethics Committee (AEC#SVS/073/16/USGMS) and the Australian Animal Health Laboratory (AAHL) Animal Ethics Committee (AEC#1389 and AEC#1557). Where possible, wild bats with irreparable physical damage (torn wings) already scheduled for euthanasia were utilized. Processing of bats has been described previously59. Human blood was obtained from healthy donors with consent, approved by the National University of Singapore Institutional Review Board (NUS-IRB reference code H-18–029). PBMCs were isolated by Ficoll-Paque Plus (GE Healthcare) density gradient centrifugation from P. alecto or human blood samples and stored in liquid nitrogen. Wild-type C57BL/6 mice were obtained with permission from the Singhealth IACUC committee. P. alecto and mouse splenocytes were isolated by grinding the spleen through a 100 μm cell strainer followed by red blood cell lysis. P. alecto bone marrow cells were processed and differentiated with recombinant P. alecto macrophage colony-stimulating factor (M-CSF) over 6 days into BMDMs or over 8 days with recombinant P. alecto granulocyte-macrophage colony-stimulating factor (GM-CSF) into BMDCs, as described previously49. Mouse bone marrow cells were harvested and differentiated into BMDMs or BMDCs over 6 days or 8 days of cultures using 10 ng ml−1 mouse M-CSF or 20 ng ml−1 mouse GM-CSF recombinant proteins (R&D System)60. Immortalized P. alecto and M. davidii kidney cells have been described previously59,61. GP2–293 retroviral packaging cells were obtained from Clontech. The NLRP3-knockout iMACs have been described previously62 and were provided by E. Latz (University of Bonn). Frozen PBMCs, splenocytes and bone marrow cells were thawed in 37 °C for 2 min, washed twice with warmed medium and recovered at 37 °C for 2 h in RPMI 1640 medium containing 10% (vol/vol) FBS before experiments. PBMCs, splenocytes, BMDMs and reconstituted iMACs were cultured in RPMI 1640 medium containing 10% FBS (Biological Industries). MDCK, Vero, GP2–293, HEK293T, PaKi and MdKi cells were cultured in DMEM (Gibco) medium supplemented with 10% FBS. Sodium pyruvate and NEAA (Life Technologies) were supplemented into the culture media of GP2–293 cells during retroviral packaging.
Generation of HEK293T, PaKi and MdKi reporter cells
GP2–293 cells, stably expressing gag and pol proteins, were co-transfected with pVSV-G envelope vector and human or bat ASC–mPlum, or mPlum-only empty vector for 48 h. Retroviral supernatants were collected, centrifuged at 500g for 10 min and filtered through a 0.45 μm hydrophilic polyethersulfone filter (Millipore). HEK293T, PaKi or MdKi cells were transduced with a low MOI of retrovirus for 24 h to obtain a single viral copy per transduced cell, followed by Hygromycin B antibiotic selection 72 h post transduction for at least 1 month to ensure stable expression. HEK293T stable cells were further (fluorescence-activated cell sorting (FACS)-sorted for low mPlum-positive cells of similar fluorescent intensity to ensure no spontaneous activation of ASC–mPlum.
Reconstitution of NLPR3 in macrophages
Retroviral supernatants for human or bat NLRP3–mCitrine, or mCitrine-only, were similarly generated to transduce mouse NLRP3-knockout iMACs by first centrifuging cells with the retroviral supernatants at 300g for 1 h at 37 °C, then incubating for 24 h. At 72 h after the transduction, cells were FACS-sorted for low mCitrine-positive cells of similar fluorescent intensity.
Inflammasome activation assays
PBMCs, BMDMs, BMDCs, splenocytes or reconstituted iMACs were seeded at 1 × 106 ml−1 into 96-well, 24-well or 12-well plates. Cells were primed with 10 ng ml−1 LPS for 3 h. The medium was removed and replaced with serum-free RPMI 1640 medium or medium containing 5 mM ATP or 6.7 μM nigericin for 30 min for detection of ASC specks or 60 min for LDH release assay and IL-1β detection by immunoblots, ELISA or targeted mass spectrometry. For ImageStream detection of ASC speck formation, HEK293T, PaKi and MdKi reporter cells were seeded overnight into 24-well plates and transfected with 100 ng (HEK293T) or 300 ng (PaKi or MdKi) per 24-well plate of the indicated NLRP3–mCitrine or mCitrine-only empty vectors using Fugene 6 (Promega) for HEK293T and Lipofectamine 3000 (Thermo) for Paki or Mdki, according to the manufacturer’s instructions. At 24 h (HEK293T) or 48 h (PaKi or MdKi) after transfection, cells were harvested by trypsinization and resuspended in FACS buffer for analysis. For immunoblotting, inflammasome system was reconstituted in HEK293T reporter cells by transfecting 50 ng indicated NLRP3–mCitrine, 10 ng human pro-caspase-1 and 190 ng mouse pro-IL-1β constructs per 24-well plate. Both supernatants and cells were collected for immunoblot analysis.
Viral infection of cells
For IAV and PRV3M infections, BMDMs, BMDCs or reconstituted iMACs were first primed with 10 ng ml−1 LPS for 3 h, washed and then infected with IAV (MOI = 0.5, 1, 2, 5 or 10) for 2 h or PRV3M (MOI = 1, 2, 5 or 10) for 4 h. Subsequently, the medium was replaced with AIM V serum-free medium (Invitrogen). At 24 h post infection, cells were collected for detection of ASC specks and immunoblots, and the cell-free supernatant for IL-1β immunoblots or ELISA and virus titrations. For MERS-CoV virus infections, PBMCs were first primed with 10 ng ml−1 LPS for 3 h, washed and then infected with a series of MOIs (0.25, 0.5 or 1) for 1 h. Lower MOIs of MERS-CoV were able to elicit a robust activation of the inflammasome in human PBMCs and hence were used in this study. The medium was replaced with AIM V serum-free medium (Invitrogen) for an additional 23 h before harvesting the cells for detection of ASC specks post-fixation and the cell-free supernatant for IL-1β ELISA and virus titration. Inhibitor MCC950 (50 μM; Selleckchem) was added 1 h before the virus infection in human PBMCs.
Confocal microscopy
Human or bat PBMCs were seeded into 24-well plates with coverslips (#1.5 thickness) inside the wells. After treatment, the cells were fixed with 4% paraformaldehyde with 0.37% gluteraldehyde in Tris-buffered saline (TBS) at room temperature for 20 min. Cells were then washed twice with TBS, permeabilized with 0.1% Triton X-100 for 10 min, blocked with 5% BSA in TBS for 30 min, stained with primary antibody (anti-ASC) pre-labelled with Zenon labelling kits in TBS for 1 h and washed three times. Nuclei were stained with DAPI. Coverslips were subsequently mounted onto glass slides with Mowiol 4.88. Z-stack confocal images were acquired on a Leica TCS SP8 machine with a ×100 objective, followed by quantification of ASC specks with Imaris 9.2.0 software for at least 10 fields per sample. Representative images were processed by maximum intensity projection of Z-stack with Image J 2.0.0.
ImageStream imaging flow cytometry
Cell supernatants were transferred to a 96-well V-bottom plate and centrifuged at 500g for 5 min at 4 °C. Cells were incubated with ice-cold 5 mM EDTA in PBS for 10 min, followed by pipetting to detach cells. Detached cells were combined with cell pellets from the supernatants, centrifuged again, and resuspended with 4% paraformaldehyde in TBS. Fixed cells were permeabilized with 0.3% Triton X-100 + 2% FBS + 1% NGS (normal goat serum) for 15 min at 4 °C, washed twice in wash buffer (TBS + 2% FBS), stained with pre-labelled primary antibodies for 1 h in TBS + 2% FBS + 1% NGS, washed twice, then resuspended in FACS buffer (PBS + 2 mM EDTA + 1% FBS). Nuclei were stained with DAPI. Cells were acquired using INSPIRE software on an Amnis ImageStreamX Mk II imaging flow cytometer using ×40 magnification. At least 10,000 single cells were acquired per sample and analysed with the inbuilt IDEAS software. First, cells in focus were gated using the bright-field gradient r.m.s. values. Single cells with an intermediate bright-field area and a high aspect ratio were selected. For PBMCs, the monocyte population was gated and separated from the lymphocyte population using bright-field area and side scatter. Based on the DAPI intensity, sub-G0/G1 cells were excluded. Double-positive cells of similar fluorescent intensity for ASC–mPlum and NLRP3–mCitrine or virus-infected cells with positive staining for IAV nucleoprotein, dsRNA or MERS-CoV spike protein were also gated. ASC speck formation was then analysed by gating on a small area and a high maximum pixel value of ASC signal. Mean pixel intensity, size and circularity score (average distance of the object boundary from its centre divided by the variation of this distance) were measured for detected ASC specks.
LDH release assay
Cytosolic LDH is rapidly released into the supernatant medium following disruption of the plasma membrane. Cell culture media were collected and centrifuged at 500g for 5 min at 4 °C. The release of LDH was measured in the cell-free supernatants using a Cytotoxicity Detection Kit PLUS (LDH) from Roche. The kit was used according to the manufacturer’s instructions. The percentage of LDH release was calculated as LDH release (%) = (experiment value – low control)/(high control – low control) × 100. Low control is the baseline LDH release from untreated cells, while high control is the maximum LDH release from cells treated with lysis solution.
Immunoblot analysis
Cells were lysed in lysis buffer63 containing cOmplete ULTRA protease inhibitor cocktail and PhosSTOP phosphatase inhibitors (Roche). Cell-free supernatants were directly used or concentrated by methanol–chloroform precipitation and dissolved in SDS–sample buffers. Proteins were separated by 6–15% SDS–PAGE gels, transferred onto 0.45 μm polyvinylidene difluoride membrane with a Trans-Blot Turbo transfer system (Biorad). Membranes were blocked with 5% BSA in TBS-T (0.1% Tween-20) for 1 h and incubated with specific primary antibodies overnight in a cold room in 5% BSA in TBS-T. Membranes were washed and incubated with HRP-conjugated secondary antibody for 1 h. Membranes were developed using Amersham ECL Prime Western blotting detection reagent (GE Healthcare) and signals were detected with a myECL Imager (Thermo Scientific). Anti-GFP antibody was used to detect NLRP3–mCitrine by immunoblots.
ELISAs
ELISAs for human/mouse IL-1β in cell-free supernatants were performed according to the manufacturer’s instructions (R&D Systems; DY201 and DY401). Sandwich ELISA for bat IL-1β was generated using the goat anti-dog IL-1β (ab193852) as the capturing antibody and rabbit anti-mouse IL-1β (ab9722) as the detection antibody. Bicarbonate/carbonate coating buffer (50 mM), OptEIA assay diluent (BD bioscience), donkey anti-rabbit HRP-conjugated secondary antibody (Santa Cruz), 3,3′,5,5′-tetramethylbenzidine (TMB) chromogen solution (Invitrogen) and TMB stop solution (VWR) were used in the assay. Purified P. alecto IL-1β-Fc recombinant proteins were used as standards.
TLR stimulations and qPCR
BMDMs, BMDCs or PBMC of bat and mouse/human were seeded at 1 × 106 ml−1 into a 96-well plate. Cells were stimulated by various concentrations of TLR ligands for 3 h: LPS-B5 (10, 100 and 1,000 ng ml−1), Pam3CSK4 (10, 100 and 1,000 ng ml−1), CL264 (0.1, 1 and 10 μg ml−1) and poly(I:C) (0.1, 1 and 10 μg ml−1). Harvested tissues of mice or bats were homogenized using silicon-carbide sharp particles (BioSpec Products) in the FastPrep-24 5G Homogenizer (MP Biomedicals). Tissue and cellular RNA was extracted using an RNeasy Mini kit and an RNeasy Plus Micro Kit (Qiagen), respectively. RNA was converted into cDNA using a QuantiTect reverse transcription kit (Qiagen). qPCR reactions were prepared using the SensiFAST SYBR No-ROX Kit (Bioline) and were run on the CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using the following cycling condition: 95 °C for 5 min, 40 cycles of 95 °C for 10 s and 55 °C for 30 s, with a melt curve cycle. Targeted genes (NLRP3, ASC, caspase-1 and IL-1β) were normalized to the geometric mean of three housekeeping genes (GAPDH, β-actin and SNRDP3) in P. alecto or mouse tissues after correction for PCR efficiency by standard curves from sample serial dilution. Fold induction of cytokines (NLRP3, IL-1β, IL-6 and TNF-α) was normalized to housekeeping (GAPDH) and compared to mock treated. The relative abundance of exon 7-positive and exon 7-negative NLRP3 isoforms was determined by quantifying ‘total’ NLRP3 and exon 7-positive isoforms using specific primers targeting a common region and exon 7, respectively. Serially diluted plasmids of both isoforms were used to perform standard curve analysis. qPCR primers for target and reference genes are listed in Supplementary Table 4.
Isoform analysis
Illumina HiSeq RNA–seq sequencing was performed on both CD11b+MHC-II− (putative monocytes/macrophages) and CD11b+MHC-II+ (putative DCs) of P. alecto splenocytes (n = 5) as described previously49. We used TopHat to map the pooled reads from all data sets to the genomic locus of P. alecto NLRP3 and used Cufflinks64 and Velvet65 to perform de novo assembly. TopHat was then used to map the pooled reads to the two splice variants identified. For PCR validation of isoforms, RNA extraction from the spleen and thymus of P. alecto or the spleen and kidney from M. davidii was performed using the RNeasy mini kit (Qiagen) and cDNA was produced using an Omniscript (Qiagen) reverse transcription kit. PCR amplification was performed using Q5 polymerase (NEB).
Targeted mass spectrometry analysis
Cells and concentrated cell-free supernatants (methanol–chloroform precipitation) were lysed and denatured in 8 M urea/50 mM Tris-HCl buffer pH 8.0. Proteins were reduced with 25 mM TCEP for 20 min at 25 °C and alkylated with 55 mM 2-chloroacetamide (CAA) for 30 min, in the dark, at room temperature. Before digestion, samples were diluted with 100 mM triethylamonium bicarbonate buffer. Protease digestion was carried out with LysC enzyme (Wako) for 4 h, followed by trypsin (Promega) treatment for 18 h at 25 °C (1:100, enzyme:protein ratio). On the following day, samples were acidified with 1% trifluoroacetic acid and peptides were purified by Sep-Pak C18 cartridges (Waters). Elution of peptides was performed with 0.5% acetic acid, 80% acetonitrile followed by peptide concentration using a vacuum concentrator system. Vacuum dried peptides were subsequently analysed on an EASY-nLC 1000 (Thermo) chromatography system coupled with Orbitrap Fusion mass spectrometer (Thermo). Each sample was separated on 70 min gradient (0.1% formic acid in water and 99.9% acetonitrile with 0.1% formic acid) using a 50 cm × 75 μm inner diameter EASY-Spray Reverse Phase Column (C-18, 2 μm particles, Thermo). Data were acquired in targeted PRM mode together with IL-1β peptide internal standards and peptide mass inclusion list. For acquisition, an Orbitrap analyser with ion targets and resolution (OT-MS 4e5 ions 120k; PRM tMS/MS 3e5 ions 15k) was used. The total area MS1 of each peptide was analysed and exported using Skyline software (version 3.7.0)66. Values for endogenous peptides were normalized by those for the heavy isotope-labelled internal standards.
Evolutionary analysis of mammalian NLRP3
The coding sequences for NLRP3 were collected form GenBank and Ensembl for 27 species representing the major groups of mammals (Supplementary Tables 2 and 3). After manual correction, the multiple sequence alignments for the full-length gene and its three domains (PYD, NACHT and LRR) were separately subjected to evolutionary analysis. The branch model from PAML was used to test whether the bat ancestor lineage is under positive selection and statistically different to the rest of the phylogeny, using a likelihood ratio test67.
Statistical analysis
Data are presented as mean and s.d. or s.e.m. of multiple biological replicates or independent experiments (as indicated). Statistical analysis was performed using GraphPad Prism software. Results were tested for significance using unpaired two-tailed Student’s t-tests when two conditions were compared. One-way or two-way ANOVA involving one or two independent variables and Bonferroni’s multiple comparisons test were performed if multiple samples or conditions were compared. Data were considered significant if *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Exact P values are provided in Supplementary Table 5.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Supplementary Figures 1–10, Supplementary Tables 1–5 and Supplementary Notes.
Acknowledgements
This work was funded by the Singapore National Research Foundation (grants NRF2012NRF-CRP001–056 to F.G. and L.-F.W. and NRF2016NRF-NSFC002–013 to L.-F.W.), a New Investigator’s Grant (to A.T.I.) from the National Medical Research Council of Singapore (NMRC/BNIG/2040/2015) and the National Natural Science Foundation of China (31621061). R.M.S. is supported by a Young Investigator Grant YIG 2015 (BMRC, A*STAR) and NMRC MS-CETSA platform grant (MOHIAFCAT2/004/2015). The authors thank the following for help with bat sampling: Crameri Research Consulting, J. Meers, H. Field and Duke-NUS team members (for a detailed listing see Supplementary Information). The authors thank A. Bertoletti and A. T. Tan for use of the Amnis ImageStream. The authors give special thanks to E. Latz for providing the immortalized NLRP3-knockout macrophages. The authors also acknowledge the facilities and technical assistance of the Advanced Bioimaging Core and Flow Cytometry Core at SingHealth Duke-NUS Academic Medical Centre, and X. F. Lim and S. Velraj for their valuable assistance in the Duke-NUS ABSL3 Facility.
Author contributions
M.A., A.T.I. and L.-F.W. conceived the study. J.H.J.N., Z.-L.S. and L.-F.W. provided resources and materials. M.A., D.E.A., Q.Z., C.W.T., B.L.L., W.N.C., S.M., R.M.S., K.L. and A.T.I. performed experiments. M.A., D.E.A., Q.Z., C.W.T., R.M.S., W.M., L.C.W. and A.T.I. performed analysis. C.A.D and F.G. provided access to splenocyte subset RNA–seq data. M.A., A.T.I. and L.-F.W. wrote the manuscript with input from all authors. Correspondence and requests for materials should be addressed to A.T.I. and L.-F.W.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. RNA–seq data used in this study have been deposited in the NCBI Sequence Read Archive (SRR8382151). The bat NLRP3 sequences generated in this study have been deposited in GenBank under accession numbers MK355440–MK355443. Supplementary figures and tables are provided in the Supplementary Information.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Aaron T. Irving, Email: aaron.irving@duke-nus.edu.sg
Lin-Fa Wang, Email: linfa.wang@duke-nus.edu.sg.
Electronic supplementary material
Supplementary information is available for this paper at 10.1038/s41564-019-0371-3.
References
- 1.Leroy EM, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 2.Clayton BA, Wang LF, Marsh GA. Henipaviruses: an updated review focusing on the pteropid reservoir and features of transmission. Zoonoses Public Health. 2013;60:69–83. doi: 10.1111/j.1863-2378.2012.01501.x. [DOI] [PubMed] [Google Scholar]
- 3.Li W, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 4.Mohd HA, Al-Tawfiq JA, Memish ZA. Middle East respiratory syndrome coronavirus (MERS-CoV) origin and animal reservoir. Virol. J. 2016;13:87. doi: 10.1186/s12985-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron MJ, Bermejo-Martin JF, Danesh A, Muller MP, Kelvin DJ. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, et al. Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol. 2017;18:4. doi: 10.1186/s13059-016-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Totura AL, Baric RS. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zampieri CA, Sullivan NJ, Nabel GJ. Immunopathology of highly virulent pathogens: insights from Ebola virus. Nat. Immunol. 2007;8:1159–1164. doi: 10.1038/ni1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swanepoel R, et al. Experimental inoculation of plants and animals with Ebola virus. Emerg. Infect. Dis. 1996;2:321–325. doi: 10.3201/eid0204.960407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe S, et al. Bat coronaviruses and experimental infection of bats, the Philippines. Emerg. Infect. Dis. 2010;16:1217–1223. doi: 10.3201/eid1608.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munster VJ, et al. Replication and shedding of MERS-CoV in Jamaican fruit bats (Artibeus jamaicensis) Sci. Rep. 2016;6:21878. doi: 10.1038/srep21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Middleton DJ, et al. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus) J. Comp. Pathol. 2007;136:266–272. doi: 10.1016/j.jcpa.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Olival KJ, et al. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson GS, South JM. Life history, ecology and longevity in bats. Aging Cell. 2002;1:124–131. doi: 10.1046/j.1474-9728.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- 15.Thomas SP, Suthers RA. The physiology and energetics of bat flight. J. Exp. Biol. 1972;57:317–335. [Google Scholar]
- 16.Pavlovich SS, et al. The Egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell. 2018;173:1098–1110. doi: 10.1016/j.cell.2018.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang G, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glennon NB, Jabado O, Lo MK, Shaw ML. Transcriptome profiling of the virus-induced innate immune response in Pteropus vampyrus and its attenuation by Nipah virus interferon antagonist functions. J. Virol. 2015;89:7550–7566. doi: 10.1128/JVI.00302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wynne JW, et al. Proteomics informed by transcriptomics reveals Hendra virus sensitizes bat cells to TRAIL-mediated apoptosis. Genome Biol. 2014;15:532. doi: 10.1186/s13059-014-0532-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papenfuss AT, et al. The immune gene repertoire of an important viral reservoir, the Australian black flying fox. BMC Genomics. 2012;13:261. doi: 10.1186/1471-2164-13-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie J, et al. Dampened STING-dependent interferon activation in bats. Cell Host Microbe. 2018;23:297–301. doi: 10.1016/j.chom.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De La Cruz-Rivera PC, et al. The IFN response in bats displays distinctive IFN-stimulated gene expression kinetics with atypical RNASEL induction. J. Immunol. 2018;200:209–217. doi: 10.4049/jimmunol.1701214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P, et al. Contraction of the type I IFN locus and unusual constitutive expression of IFN-alpha in bats. Proc. Natl Acad. Sci. USA. 2016;113:2696–2701. doi: 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimada K, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 26.Iyer SS, et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuriakose T, Kanneganti TD. Regulation and functions of NLRP3 inflammasome during influenza virus infection. Mol. Immunol. 2017;86:56–64. doi: 10.1016/j.molimm.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sha W, et al. Human NLRP3 inflammasome senses multiple types of bacterial RNAs. Proc. Natl Acad. Sci. USA. 2014;111:16059–16064. doi: 10.1073/pnas.1412487111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Youm YH, et al. Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab. 2013;18:519–532. doi: 10.1016/j.cmet.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lupfer C, Malik A, Kanneganti TD. Inflammasome control of viral infection. Curr. Opin. Virol. 2015;12:38–46. doi: 10.1016/j.coviro.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakrabarti A, et al. RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe. 2015;17:466–477. doi: 10.1016/j.chom.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong S, et al. A distinct lineage of influenza A virus from bats. Proc. Natl Acad. Sci. USA. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence TM, Hudacek AW, de Zoete MR, Flavell RA, Schnell MJ. Rabies virus is recognized by the NLRP3 inflammasome and activates interleukin-1β release in murine dendritic cells. J. Virol. 2013;87:5848–5857. doi: 10.1128/JVI.00203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson N, et al. Human rabies due to lyssavirus infection of bat origin. Vet. Microbiol. 2010;142:151–159. doi: 10.1016/j.vetmic.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Ren R, et al. The H7N9 influenza A virus infection results in lethal inflammation in the mammalian host via the NLRP3-caspase-1 inflammasome. Sci. Rep. 2017;7:7625. doi: 10.1038/s41598-017-07384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, et al. Zika virus infection induces host inflammatory responses by facilitating NLRP3 inflammasome assembly and interleukin-1β secretion. Nat. Commun. 2018;9:106. doi: 10.1038/s41467-017-02645-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coates BM, et al. Inhibition of the NOD-like receptor protein 3 inflammasome is protective in juvenile influenza A virus iInfection. Front. Immunol. 2017;8:782. doi: 10.3389/fimmu.2017.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tate MD, et al. Reassessing the role of the NLRP3 inflammasome during pathogenic influenza A virus infection via temporal inhibition. Sci. Rep. 2016;6:27912. doi: 10.1038/srep27912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaRock CN, Cookson BT. Burning down the house: cellular actions during pyroptosis. PLoS Pathog. 2013;9:e1003793. doi: 10.1371/journal.ppat.1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauernfeind FG, et al. Cutting edge: NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Connor W, Jr, Harton JA, Zhu X, Linhoff MW, Ting JP. Cutting edge: CIAS1/cryopyrin/PYPAF1/NALP3/CATERPILLER 1.1 is an inducible inflammatory mediator with NF-κB suppressive properties. J. Immunol. 2003;171:6329–6333. doi: 10.4049/jimmunol.171.12.6329. [DOI] [PubMed] [Google Scholar]
- 45.Chua KB, et al. A previously unknown reovirus of bat origin is associated with an acute respiratory disease in humans. Proc. Natl Acad. Sci. USA. 2007;104:11424–11429. doi: 10.1073/pnas.0701372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu B, Ge X, Wang LF, Shi Z. Bat origin of human coronaviruses. Virol. J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peck KM, et al. Permissivity of dipeptidyl peptidase 4 orthologs to Middle East respiratory syndrome coronavirus is governed by glycosylation and other complex determinants. J. Virol. 2017;91:e00534-17. doi: 10.1128/JVI.00534-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coll RC, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou P, et al. Unlocking bat immunology: establishment of Pteropus alecto bone marrow-derived dendritic cells and macrophages. Sci. Rep. 2016;6:38597. doi: 10.1038/srep38597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Netea MG, Wijmenga C, O’Neill LA. Genetic variation in Toll-like receptors and disease susceptibility. Nat. Immunol. 2012;13:535–542. doi: 10.1038/ni.2284. [DOI] [PubMed] [Google Scholar]
- 51.Werling D, Jann OC, Offord V, Glass EJ, Coffey TJ. Variation matters: TLR structure and species-specific pathogen recognition. Trends Immunol. 2009;30:124–130. doi: 10.1016/j.it.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell. 2013;49:331–338. doi: 10.1016/j.molcel.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 53.Mitoma H, et al. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39:123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Surya W, Li Y, Verdia-Baguena C, Aguilella VM, Torres J. MERS coronavirus envelope protein has a single transmembrane domain that forms pentameric ion channels. Virus Res. 2015;201:61–66. doi: 10.1016/j.virusres.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Shea TJ, et al. Bat flight and zoonotic viruses. Emerg. Infect. Dis. 2014;20:741–745. doi: 10.3201/eid2005.130539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schountz T, Baker ML, Butler J, Munster V. Immunological control of viral infections in bats and the emergence of viruses highly pathogenic to humans. Front. Immunol. 2017;8:1098. doi: 10.3389/fimmu.2017.01098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller MR, et al. Broad and temperature independent replication potential of filoviruses on cells derived from old and new world bat species. J. Infect. Dis. 2016;214:S297–S302. doi: 10.1093/infdis/jiw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ying T, et al. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J. Virol. 2014;88:7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crameri G, et al. Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS One. 2009;4:e8266. doi: 10.1371/journal.pone.0008266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc. Natl Acad. Sci. USA. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, et al. Host range, prevalence, and genetic diversity of adenoviruses in bats. J. Virol. 2010;84:3889–3897. doi: 10.1128/JVI.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sadler AJ, Latchoumanin O, Hawkes D, Mak J, Williams BR. An antiviral response directed by PKR phosphorylation of the RNA helicase A. PLoS Pathog. 2009;5:e1000311. doi: 10.1371/journal.ppat.1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trapnell C, et al. Differential gene and transcript expression analysis of RNA–seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–10, Supplementary Tables 1–5 and Supplementary Notes.
Data Availability Statement
The data supporting the findings of this study are available from the corresponding authors upon request. RNA–seq data used in this study have been deposited in the NCBI Sequence Read Archive (SRR8382151). The bat NLRP3 sequences generated in this study have been deposited in GenBank under accession numbers MK355440–MK355443. Supplementary figures and tables are provided in the Supplementary Information.